Abstract
Liver-resident CD8+ T cells are highly motile cells that patrol the vasculature and provide protection against liver pathogens. A key question is: how can these liver CD8+ T cells be simultaneously present in the circulation and tissue-resident? Because liver-resident T cells do not express CD103 - a key integrin for T cell residence in epithelial tissues - we investigated other candidate adhesion molecules. Using intra-vital imaging we found that CD8+ T cell patrolling in the hepatic sinusoids is dependent upon LFA-1-ICAM-1 interactions. Interestingly, liver-resident CD8+ T cells up-regulate LFA-1 compared to effector-memory cells, presumably to facilitate this behavior. Finally, we found that LFA-1 deficient CD8+ T cells failed to form substantial liver-resident memory populations following Plasmodium or LCMV immunization. Collectively, our results demonstrate that it is adhesion through LFA-1 that allows liver-resident memory CD8+ T cells to patrol and remain in the hepatic sinusoids.
Introduction
CD8+ T cells play critical roles in protection against infectious diseases and cancers. One problem that T cells must overcome is that the number of infected cells can represent a tiny fraction of cells in the body. To achieve efficient tissue surveillance, specialized populations of CD8+ T cells that patrol different niches develop after priming. The first populations to be defined, based on the expression of CD62L and CCR7, were central and effector memory CD8+ T cells (1). More recently populations of tissue resident memory (TRM) cells have been identified in numerous tissues - especially barrier tissues - such as the skin, lung, gut and female reproductive tract (2-6). Such cells are strictly defined by their inability to recirculate from their tissue of residence, though they are frequently identified by the expression of CD69 and the integrin CD103 (2, 3, 6). Intra-vital imaging studies have revealed that CD8+ TRM cells in the skin are largely sessile cells that may act as sentinels against invading pathogens (7). This is consistent with the finding that these cells function as the first line of defense in peripheral tissues, able to recruit other cells to the immune response (8).
Given that the liver is the target organ of important pathogens including Hepatitis B Virus, Hepatitis C Virus and Plasmodium, several recent studies have begun to characterize TRM cells in this organ (5, 9, 10). Although previously the liver was thought of as a “graveyard” for CD8+ T cells, it is now clear that it harbors large numbers of memory CD8+ T cells capable of protecting against pathogen challenge (11, 12). We have recently shown that the formation of robust CD69+ CD8+ T cell populations is essential for effective protection against Plasmodium (malaria) liver stages (9). Based on parabiosis studies these CD69+ cells in the liver have been defined as a resident population that does not recirculate within 2-3 months (5, 9). Liver TRM cells also share with epithelial TRM cells a common gene expression signature dependent on the expression of the transcription factors Hobit and Blimp1 (9, 10). However, liver CD8+ T cells have some distinct features compared to epithelial TRM cells: notably most liver TRM cells are present in the circulation as revealed by in vivo antibody labeling experiments (5, 9). Intra-vital imaging has further shown that liver TRM display motile patrolling behavior in the hepatic sinusoids (9).
A critical question therefore is: what are the molecular interactions that retain CD8+ T cells in the liver and facilitate their patrolling behavior? Interestingly liver TRM cells do not express high levels of CD103 (9, 10), an integrin that is required for TRMs to be retained in many epithelial tissues (13). One the other hand, a variety of other adhesion molecules have been implicated in the migration of CD8+ T cells to the liver. The initial trapping of CD8+ T cells in the liver appears to be mediated by interactions between CD8+ T cells and platelets bound to the endothelium via CD44, rather than selectin-mediated rolling interactions (14). Some studies have suggested that ICAM-1 is required for the retention of naive and activated CD8+ T cells in the liver but only in the presence of antigen (15, 16). Intriguingly, while the ICAM-1 ligand LFA-1 has been found to be critical for NKT cell retention in the liver (17, 18), this canonical adhesion molecule has been regarded as dispensable for the intrahepatic retention of activated CD8+ T cells (14, 17).
Here we investigated the roles of a range of adhesion molecules in the intrahepatic migration of effector and memory CD8+ T cells using intra-vital imaging. We found that, surprisingly, ICAM-1-LFA-1 interactions are indeed important for the movement of activated CD8+ T cells in the liver. Further analysis revealed that LFA-1 is highly expressed specifically on liver TRM cells and that its absence results in their inability to establish residence in the liver. Our data thus reveal an unexpected role for the adhesion molecule LFA-1, rather than CD103, in the retention of liver TRM cells and highlight the distinct adhesion molecule requirements for memory T cells that patrol vascular, rather than barrier, tissues.
Results
Activated CD8+ T cells use LFA-1:ICAM-1 interactions to patrol the liver
Previous studies have shown that CD8+ T cell migrate in the hepatic sinusoids with a characteristic patrolling behavior that facilitates their ability to scan the liver and find pathogens such as Plasmodium and Hepatitis B virus (14, 19). We used antibody blockade to investigate the roles of ICAM-1, VCAM-1, and CD44 on the patrolling behavior of transferred in vitro activated CD8+ T cells. These molecules have been proposed to have roles in the intrahepatic accumulation of CD8+ T cells, though with the exception of CD44, their roles in migratory behavior within the liver have not been studied (14, 16). We also transferred cells to β2 microglobulin-deficient recipients to examine the role of MHC Class I interactions which have also been suggested to be important for CD8+ T cells adhesion in the liver (20). We subsequently examined the migration of the activated CD8+ T cells in the liver by time-lapse multi-photon microscopy (Movie S1). Surprisingly only ICAM-1 blockade had any effect on CD8+ T cell movement, with cells in treated mice moving more slowly and spending more time arrested than cells in control mice (Figure 1A-B) suggesting that ICAM-1 and its ligands might be important for CD8+ T cell patrolling of the liver. In contrast antibodies to other adhesion molecules as well as rat IgG2b isotype control antibodies had no effect on migration of CD8+ T cells (Figure S1). The reduction in crawling behavior seen in anti-ICAM-1 treated mice is consistent with previous in vitro studies which show that ICAM-1 is required for the crawling motility of lymphocytes as well as their adhesion (21).
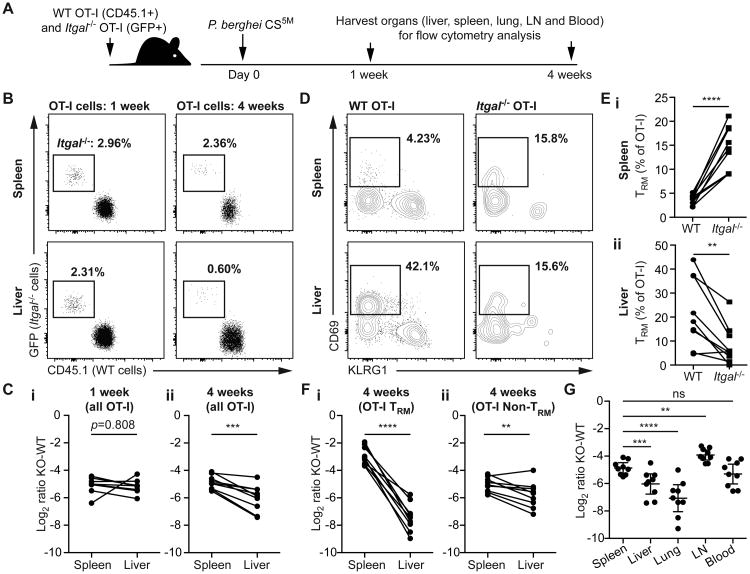
Fig 1. LFA-1:ICAM-1 interactions are required for CD8+ T cell motility in the liver.
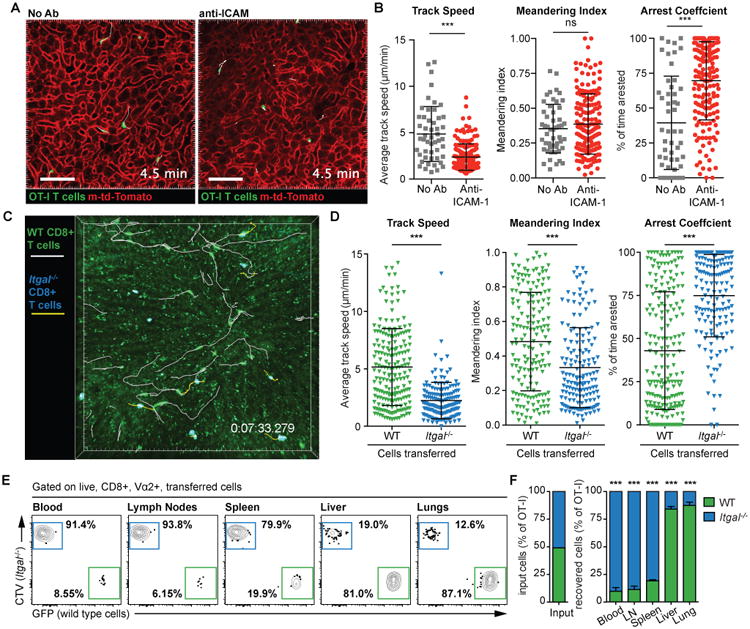
(A) 2. hours prior to the transfer of 7 × 106 in vitro activated OT-I T cells, mice (WT or mT/mG) were treated with blocking Abs to ICAM-1. 4 hours after cell transfer mice were prepared for intravital imaging and imaged by 2-photon microscopy using a standard galvanometer-scanner to acquire a 50-micron deep Z-stack approximately every 30 seconds. Representative images from time lapse imaging of mT/mG mice either with or without anti-ICAM-1 are shown. Scale bar is 30 μm. (B) Movement parameters of OT-I cells following anti-ICAM-1 treatment; data pooled from 4 experiments and analyzed using linear mixed models with experiment and mouse as random effects and speed, meandering index or arrest as the fixed effects. Means and standard deviations (SD) are shown. (C) 7 × 106 in vitro activated Itgal-/- OT-I T cells (labeled with cell trace violet) and 7 × 106 in vitro activated GFP+ WT OT-I+ T cells were co-transferred to WT recipient mice. Mice were imaged as in (A); image shows a representative frame from a time-lapse movie showing tracks of the Itgal -/- (yellow) and WT T cells (white); scale bar is 50 μm. (D) Movement parameters of Itgal -/- and WT cells in the livers of naive recipient mice as described in C; data are pooled from 3 mice in 2 independent experiments and analyzed as in (B). (E) 2 × 106 in vitro activated Itgal -/- OT-I CD8+ T cells (labeled with cell trace violet) and 2 × 106 in vitro activated GFP+ WT OT-I CD8+ T cells were co-transferred to WT recipient mice. 24 hours later the blood, lymph nodes, spleen, liver and lungs were harvested and the proportion of WT and Itgal-/- cells in each organ determined by flow cytometry (representative plots all from the same mouse shown). (F) Summary data for the proportions of WT and Itgal -/- cells in organs harvested from 5 mice in one of two similar independent experiments, analyzed by one sample t-test (compared to the input proportions of WT and Itgal -/- cells). Means and SD are presented.
Given that ICAM-1 is highly expressed on the surface of liver sinusoidal endothelial cells and hepatocytes (14, 20), we hypothesized that T cells were crawling on these surfaces using the integrin LFA-1 that is highly expressed on CD8+ T cells. LFA-1 is composed of integrin alpha-L (ITGAL; CD11a) combined with integrin beta-2 (CD18). To further investigate a possible role of LFA-1 in T cell migration in the liver we used a mouse line carrying a Cys77Phe mutation caused by a G>T change in Exon 3 of the Itgal gene (Chromosome 7, position 127299608). This mutation causes a complete lack of ITGAL on the cell surface (Figure S2A), accordingly we designate them Itgal-/- for simplicity. It is likely that Cys77 forms disulphide bonds and that the mutation destabilizes the integrin structure. These mice were identified from an ENU mutagenesis screen for immune phenotypes in the blood (22) as they have an elevated proportion of NKT cells in the blood (Figure S2B). This may be explained by the fact that LFA-1 has been shown to be important for the intrahepatic retention of NKT cells (18). Closer analysis of our Itgal-/- mice revealed that they also have elevated CD8+ T cells in the blood, particularly CD44hi activated CD8+ T cells (Figure S2B).
To investigate the effect of loss of ITGAL on CD8+ T cell migration in the liver we activated Itgal-/- OT-I cells in vitro and co-transferred them to mice with similarly activated but differentially labeled WT OT-I cells. Migration in the liver was then assessed by multi-photon microscopy (Movie S2; Figure 1C). In agreement with our ICAM-1 blockade data, Itgal-/- cells did not display patrolling behavior, rather they spent large amounts of time arrested and moved with slower average speeds than WT cells (Figure 1D). Importantly the Itgal-/- and WT cells showed similar expression of activation markers and other β2 integrins, suggesting that the reduced migration was not due to inadequate priming of Itgal-/- cells resulting in the expression of different adhesion molecules (Figure S3). We further used flow cytometry to quantify the accumulation of cells in the liver and other organs in mice that received equal numbers of WT and Itgal-/- cells. In the liver and lungs Itgal-/- cells constituted <20% of the cells recovered from these mice; in contrast Itgal-/- cells formed the major proportion of cells recovered from the spleen, blood and lymph nodes (Figure 1E and F). One hypothesis is that the spleen and lymph nodes act as a sink for the Itgal-/- cells, however our intra-vital imaging data (Figure 1B and C), and our finding that there are also elevated numbers of Itgal-/- cells circulating in the blood suggest this is not the case. Rather our data suggest that LFA-1 is important for the retention of activated CD8+ T cells in the liver and lungs.
LFA-1 is required for efficient CD8+ T cell mediated protection against malaria
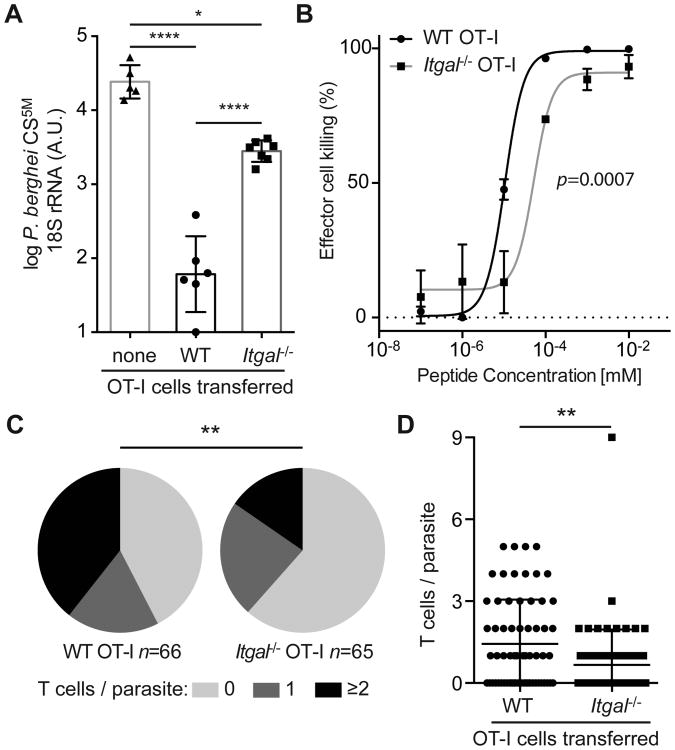
We next wanted to determine if the lack of patrolling cells in the liver would affect protection against the Plasmodium parasite. While CD8+ T cells are capable of killing parasites in the liver (19), it is not known whether the cells conferring protection are those circulating in the blood or those migrating in the sinusoids. Given the results of the previous experiment, in which the Itgal-/- cells are enriched in the blood and WT cells are enriched in the liver, we were able to test this by transferring activated Itgal-/- or WT OT-I cells to mice prior to challenge with P. berghei CS5M parasites that express the SIINFEKL epitope within the immunodominant circumsporozoite (CS) protein (23). Strikingly we found that the parasite burden was significantly greater in mice that received the Itgal-/- cells rather than WT cells (Figure 2A). While a component of this impaired protection by Itgal-/- cells may be attributed to a defect in cytotoxicity (Figure 2B) we also observed by intra-vital microscopy that WT OT-I cells were better able to associate with and cluster around P. berghei CS5M parasite than Itgal-/- OT-I cells (Figure 2 C and D). Thus our data are consistent with the hypothesis that LFA-1-mediated patrolling of the liver is required for efficient immune-surveillance, though we cannot exclude the possibility that LFA-1 is also important for CD8+ T cell killing of infected cells.
Fig 2. Itgal -/- cells do not efficiently protect against sporozoite challenge.
(A) 2 × 106 Itgal -/- or littermate WT OT-I T cells were transferred to C57BL/6 mice 1 day before mice were challenged with 5 × 103 P. berghei CS5M sporozoites. 24 hours post-challenge livers were harvested from the recipient mice and controls and the parasite load assessed by RT-PCR. Data are from one of 2 similar experiments with 5-7 mice/group, assessed by one-way ANOVA with Tukey's post-test for multiple comparisons. Means and SD of log transformed data are presented. (B) EL4 target cell killing following incubation with in vitro activated Itgal -/- or littermate Itgal +/+ OT-I T cells. Data are expressed as the number of live-pulsed target cells recovered compared to the number of live-unpulsed target cells after 6 hours. Means and SD are based on 3 technical replicates, from one of two experiments, p value is the probability the IC50 values are different (extra sum-of-squares F test). (C) Mice were infected with 1.5 × 105 P. berghei CS5M-GFP sporozoites, 15 hours later the mice received either 7 × 106 Itgal -/- or littermate WT OT-I T cells labeled with CTV; 20 hours post-infection mice were prepared for imaging and a 40-micron Z-slice was taken of each parasite. Pie charts show the proportion of parasites with 0, 1 and ≥2 T cells in contact analyzed by χ 2 test while (D) shows the number of T cells per parasite for each condition analyzed by Mann-Whitey U test. Data are from 3 mice receiving Itgal -/- cells and 4 mice receiving WT OT-I cells Bars show means and SD.
Memory T cells in the liver adopt a characteristic patrolling behavior in the hepatic sinusoids
We next wanted to determine if the migratory behavior we observed in in vitro activated cells reflects that of in vivo primed cells. Accordingly, we immunized mice that had received naïve GFP+ OT-I cells with P. berghei CS5M sporozoites. The livers of mice were then imaged at effector (1 week) and memory (4 weeks) time-points post-immunization (Figure 3A and B and Movies S3 and S4). At effector time-points the behavior of donor T cells in the liver was different to that observed with in vitro activated cells. Notably, there were many rounded up cells and cells moving with the blood flow. To capture these rapidly-moving cells we modified our imaging protocol and made high frame-rate movies (3 frames/second) using the resonant scanner on our microscope. Mathematically we were then able to distinguish three migratory phenotypes among the GFP+ cells using the parameters of cell speed and polarity (Figure 3C). The first population we designated “flowing” cells, these cells moved rapidly (>25 μm/min; median = 208 μm/min) in the blood though sometimes they briefly arrested on the walls of the sinusoids. Among the non-flowing cells moving at <25 μm/s we observed a population of “rounded” cells which we formally defined as having polarity <1.5; these cells were often arrested but sometimes detached (median speed = 5.8 μm/min). Finally we were able to observe some “patrolling” cells that we defined by their higher polarity (>1.5). Overall these cells moved similarly to in vitro activated cells, though they had a higher median speed (9.5 μm/s). At the memory time-point a strikingly different picture was seen, with a substantial increase in the number of patrolling cells which increased from <5% of cells 1 week post-immunization to >50% of the total cells at 4 weeks post-immunization (Figure 3D). We further found that ICAM-1 blockade reduced the speed and increased the arrest coefficient of memory CD8+ T cells in the liver (Figure 3E), suggesting that these cells, like in vitro activated effectors, use LFA-1:ICAM-1 interactions for their patrolling behavior.
Fig 3. Memory CD8+ T cells display patrolling behavior in the liver.
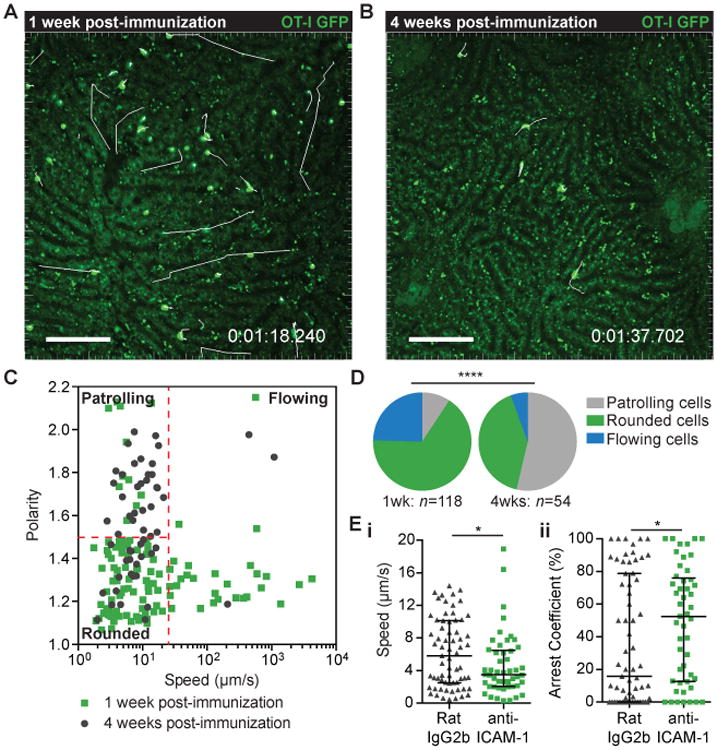
2 × 104 GFP+ OT-I cells were transferred to C57BL/6 mice prior to immunization with 5 × 104 P.berghei CS5M sporozoites. 1 week (A) and 4 weeks (B) post-immunization mice were prepared for intra-vital imaging and the livers imaged by 2-photon microscopy using a resonant scanner to collect time-lapse moves of a single Z-slice at ∼3 frames/second; images are representative time-points with T cell tracks shown in white; scale bar is 50 μm. (C) Mean speed vs. polarity of T cells in the liver, 1 week (green points) and 4 weeks (grey points) post-immunization. (D) Proportion of cells exhibiting different T cell migration behaviors 1 week and 4 weeks post immunization, analysis was performed by a χ2 test. (E) (i) mean speed and (ii) arrest coefficients of OT-I GFP T cells in the liver 4 weeks post immunization (analysis based on 50um Z stacks at 1 frame/30secs). Mice received 50 μg anti-ICAM or isotype control antibodies 3 hours before imaging. Analysis was performed by one-tailed Mann-Whitney U test as the direction of the expected effect was already known from previous experiments. Data are pooled from 6 mice in each experimental group; medians and interquartile ranges are presented.
Liver resident memory CD8+ T cells express exceptionally high levels of LFA-1
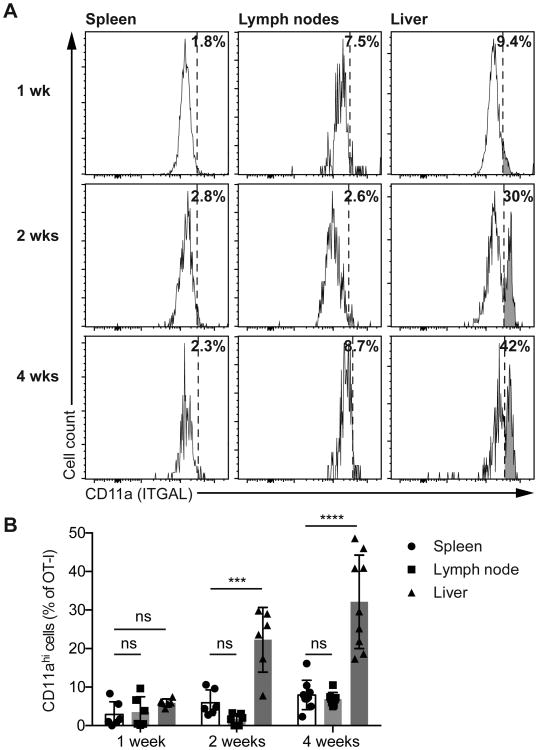
Given that LFA-1 was found to be important for migration of in vitro activated CD8+ T cells in the liver we speculated that the formation of a patrolling memory population in the liver might be associated with an increase in the expression of LFA-1 (ITGAL; CD11a) on CD8+ T cells. We examined the phenotypes of OT-I cells in the spleen, lymph node and livers of mice at 1, 2 and 4 weeks post-immunization with P. berghei CS5M sporozoites (Figure 4A). Naïve cells expressed low levels of LFA-1 that increased upon priming (Figure S4). CD11a was expressed at intermediate levels on activated CD8+ T cells in the spleen at all time-points, however, at 2 weeks and 4 weeks post-immunization we observed a distinct population of CD11ahi cells in the liver (Figure 4A and 4B; Figure S4). Importantly this population did not appear to be present in the lymph nodes and spleen- which is consistent with our previous data (Figure 1E-F) – suggesting that LFA-1 is dispensable for homing to these organs.
Fig 4. LFA-1 is highly expressed on a subset of liver memory CD8+ T cells.
2 × 104 CD45.1+ OT-I cells were transferred to C57BL/6 mice prior to immunization with 5 × 104 P.berghei CS5M sporozoites. One, 2 and 4 weeks post-immunization organs were harvested and cells prepared for flow-cytometry analysis. (A) Representative flow cytometry plots from a single mouse at each time-point, showing the expression of CD11a (ITGAL) on CD45.1+ CD8+ OT-I T cells in the spleen, lymph nodes and liver at the indicated time-points, values indicate the percentage of cells that are CD11ahi. (B) Summary data pooled from two independent experiments showing the proportion of CD45.1+ CD8+ OT-I cells that are CD11ahi. Data were analyzed using a linear mixed model including the experiment and mouse as random effects and organ and time-point as fixed effects. Bars show means and SD.
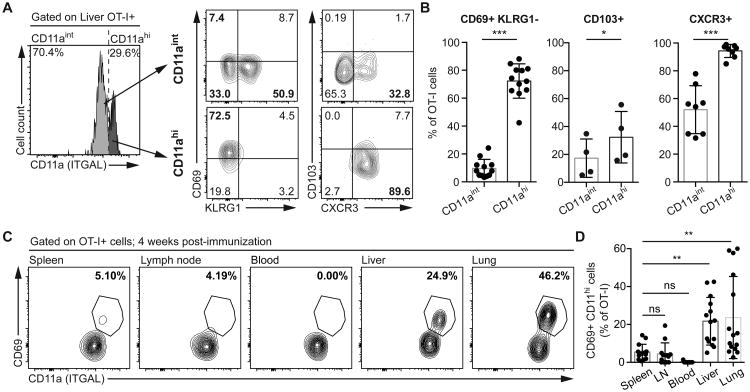
We were further interested in knowing the broader phenotype of the CD11ahi population, particularly if they might correspond to liver TRM CD8+ T cells. We found that, indeed, CD11ahi cells were almost exclusively CD69+ KLRG1-CXCR3+ (Figure 5A and B), a typical profile for liver TRM cells (9, 10). We also found that the CD11ahi cells in the liver did not express high levels of CD103 (Figure 5B), while IV injection of FITC-labeled anti-CD8a antibody revealed these cells to be in the circulation (Figure S5A) which is also in agreement with previous descriptions of liver TRM cells (5, 9, 10). To determine if this CD11ahi population is unique to the liver or may be seen in other tissues, we examined the spleen, lymph node, blood, skin and lungs of mice immunized with sporozoites. While we were unable to detect antigen specific cells in the skin of sporozoite immunized mice, we were able to detect CD11ahi CD69+ cells in the lungs of immunized mice (Figure 5C and D). However these lung CD11hi cells were unlikely to be TRM cells because lung TRM cells express CD103 and are IVAb- (6, 24) whereas the CD11hi cells we observed were CD103- and IVAb+ (Figure S5).
Fig 5. CD11ahi memory CD8+ T cells in the liver have a TRM phenotype.
2 × 104 CD45.1+ naïve OT-I cells were transferred to C57BL/6 mice prior to immunization with 5 × 104 P.berghei CS5M sporozoites. (A) Representative flow cytometry plots of the expression of CD69, KLRG1, CD103 and CXCR3 by CD45.1+ CD8+ OT-I T cells expressing intermediate and high levels of ITGAL (CD11aint and CD11ahi) in the liver 4 weeks post-immunization. (B) Summary plots showing the proportion of cells expressing the indicated phenotypes 4 weeks post-immunization in the liver. Data are pooled from 4 independent experiments for CD69 and KLRG1, a single experiment for CD103 and 2 independent experiments for CXCR3; data were analyzed by linear mixed models including mouse as a random effect and CD11a subset as a fixed effect. Bars show means and SD. (C) Representative flow cytometry plots showing the co-expression of CD69 and CD11a in the indicated organs (from a single animal) 4 weeks post-immunization. (D) Summary of the proportion of OT-I cells in the indicated organs that are CD11ahi CD69+. Data are pooled from 3 independent experiments and analyzed using a linear mixed model including mouse as a random effect and the organ as the fixed effect. Bars show means and SD.
To determine whether the elevated expression of LFA-1 was specific to Plasmodium, we also examined CD11a expression on NP396 Tetramer+ cells following intra-peritoneal infection with LCMV Armstrong. Similar to our results with sporozoites we identified a CD11ahi population in the liver that corresponded to the previously defined liver TRM populations being CD69+ CD103-KLRG1- (Figure S6 A-C). We also identified a similar population of CD11ahiCD69+KLRG1-cells in the lung after LCMV infection (Figure S6D). While in this case a modest proportion (∼20%) of Tetramer+ cells were IVAb- we did not detect any appreciable CD103 expression (Figure S6D), which is in agreement with previous observations that intra-peritoneal LCMV infection does not induce lung TRM cells (25, 26). Independently, a distinct population of CD11ahi cells in the liver has been observed previously following Vesicular Stomatitis Virus infection (27), though this observation was not followed up. Finally, we analyzed NKT cells as the other liver resident lymphocyte subset and found that NKT cells also have an identical CD11ahi CD69+ KLRG1- phenotype to liver TRM cells (Figure S7).
LFA-1 is required for the retention of TRM cells in the liver
To determine if LFA-1 is required for the intrahepatic retention of CD8+ T cells we first made mixed bone marrow chimeras containing CD45.2+ Itgal-/- cells mixed with CD45.1+ WT cells as well as control chimeras reconstituted with equal numbers of CD45.2+ and CD45.1+ WT cells. The contributions of Itgal-/- cells to the CD8+ T cell population were analysed in the spleen, lung and liver (Figure S8A). Itgal-/- CD8+ T cells migrated normally to the spleen but did not accumulate efficiently in either the lung or the liver, though the overall defect was only just significant in the liver (Figure S8B and C). Specific analysis of the contribution of Itgal-/- cells to the CD69+ and CD69- liver cell populations (Figure S9A) revealed that the intrahepatic CD69+ subset came almost entirely from the WT cells (Figure S9B and C), indicating that ITGAL is required for its residency. In contrast, when we examined CD69- CD8+ T cells in the liver we found that these were not dependent on ITGAL for retention in the liver (Figure S9B and C). A similar analysis of the lung CD69+ and CD69- cell populations revealed that, in contrast to the liver, ITGAL is critical for the accumulation of both these populations in the lung (Figure S9D-F).
To determine if LFA-1 is required for the formation of antigen-specific liver TRM cell populations after immunization we co-transferred GFP+ Itgal-/- and WT CD45.1+ OT-I cells to CD45.2+ mice prior to immunization with P. berghei CS5M sporozoites (Figure 7A). At 1 week post-infection we observed that the WT cells outnumbered the Itgal-/- cells in both the liver and spleen by around 30:1 (Figure 7B), a result consistent with the priming defect previously reported for Itgal-/- cells (28). However, while this ratio remained constant at 4 weeks post-immunization in the spleen, in the liver, the Itgal-/- cells were now outnumbered by around 60:1 suggesting these LFA-1-deficient cells have a specific defect in forming memory populations in this organ (Figure 7C).
Fig 7. Itgal-/- mice do not form liver TRMs following LCMV infection.
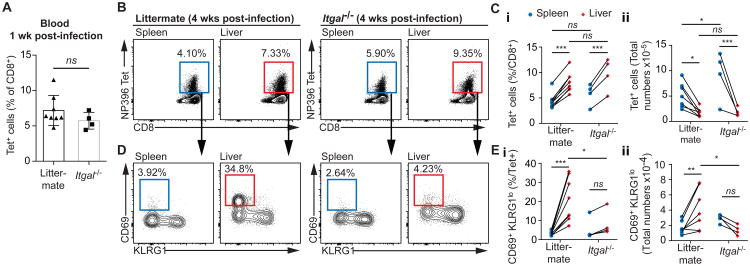
Itgal-/- and littermate mice were infected with 2 × 105 pfu/mouse LCMV Armstrong. (A) 1 week post-infection the % of CD8+ T cells in the blood that was NP396-specific was measured by flow cytometry; data are mean and SD analyzed using a 2-tailed students t-test. 4 weeks post-infection the NP396 specific immune response was measured in the spleen and liver by flow cytometry, with (B) representative flow cytometry plots from individual mice and (C) summary data presented. We further determined the proportion of antigen specific cells that had the TRM phenotype (CD69+ KLRG1lo) by flow cytometry for each organ and genotype, with (D) representative flow cytometry plots from individual mice and (E) summary data presented. Data in B-E were analyzed using linear mixed models including mouse as a random effect and organ and genotype and fixed effects; pairwise p values derived from the models are given.
Examination of the phenotypes of the cells revealed that the difference between the spleen and liver was largely driven by the relative proportions of CD69+KLRG1- (TRM) cells (Figure 7D). This population typically made up ∼30% of the WT OT-I cells in the liver, but accounted for only ∼10% of the Itgal-/- cells in the same mouse (Figure 7E). Interestingly, these cells may not be lost altogether but may enter the general circulation as in the spleen the situation was reversed with a higher proportion of TRM phenotype cells among the Itgal-/- OT-I cells compared to the WT OT-I cells (Figure 7E). As a result of this the ratio of WT to Itgal-/- OT-I TRM cells was 120:1 in the livers of immunized mice but only by 8:1 in the spleens (Figure 7F). Nonetheless the ratio of Itgal-/- to WT non-TRM cells was still significantly lower in the liver compared to the spleen, suggesting that activated non-TRM cells may still use ITGAL to accumulate in the liver.
We also investigated the ratio of Itgal-/- to WT OT-I cells in other organs in this experiment (Figure 7G). In agreement with our bone marrow chimera data, lung OT-I cells had an even stronger requirement for ITGAL than liver OT-I cells, with Itgal-/- cells being outnumbered by around 120:1 by WT cells. In contrast in the lymph nodes, Itgal-/- cells were only outnumbered around 8:1 by WT OT-I cells. This suggests that some Itgal-/- cells may accumulate in lymph nodes, though the proportion of OT-I cells in the lymph nodes was very small (Figure S5).
Finally we were concerned that our data might be biased by the poor priming of the LFA-1 deficient cells by Plasmodium, especially in a competitive environment. Accordingly we infected Itgal-/- mice and their WT littermates with LCMV-Armstrong and measured the formation of antigen specific T cell responses using NP396 tetramers. In this infection model there was no detectable defect in priming in the Itgal-/- mice as similar percentages of Tetramer+ cells were seen in the blood 1 week post immunization (Figure 7A). Moreover, 4 weeks post-immunization there were similar proportions and numbers of Tetramer+ cells between the Itgal-/- and littermate mice in both the spleen and liver (Figure 7 B and C). However while robust populations of antigen-specific CD69+ KLRG1lo cells were formed in livers of littermate control mice, few were seen in Itgal-/- mice (Figure 7 D and E). Similar to our results with Plasmodium immunization there was a ∼2-fold increase in the average numbers of CD69+ KLRG1+ cells in the spleens of Itgal-/- compared to littermates suggesting that these cells may accumulate in secondary lymphoid organs if they are unable to be retained in the liver, though this did not reach statistical significance when correction was made for multiple comparisons. Collectively these data from our bone marrow chimeras and 2 distinct models of infection suggest that LFA-1 is critically required for the retention of TRM cells in the liver.
Discussion
Several recent papers have identified populations of tissue resident CD69+ antigen specific memory cells in the liver that patrol the sinusoids (5, 9, 10). A critical question then is how can an intra-vascular population in the circulation also be tissue resident? Here we show a critical role for ITGAL - and by extension the integrin LFA-1 - in the migration of activated CD8+ T cells in the liver. We further show that LFA-1 is required for the retention of TRM cells in the liver. It is possible that these cells might occasionally exit the liver if they were to enter the hepatic venules, though it may be that these cells rapidly recirculate back to the liver due to the elevated expression of ICAM-1 in these tissues and the slow blood flow within this organ (14, 20). Overall though, our data support the hypothesis that it is the expression of high levels of LFA-1 on the surface of this cell subset that allows them to remain in the liver and patrol the hepatic sinusoids.
Our data further suggest that TRM populations in different tissues have different adhesion molecule requirements for tissue retention. In agreement with previous reports we do not detect high levels of the integrin CD103 on the surface of our CD11ahi liver TRM cells (5, 10). CD103 is highly expressed on epithelial TRM cells such as those found in the skin and the gut and has been shown to be required for T cell residence in the skin (2, 3, 13). Its expression appears to be induced, even in vitro, by TGF-β signaling (29). In vivo the level of CD103 expression increases progressively during skin TRM development as they migrate to the epidermis (30). However while CD103 may be critical for the residence of TRM cell populations in various epithelia it is presumably dispensable for TRM populations that may in fact remain exposed to the circulation. Interestingly TGF-β signaling has previously been shown to down-regulate LFA-1 expression suggesting that these integrins may be reciprocally regulated (31, 32).
Despite ICAM-1/LFA-1 interactions being considered canonical in the leukocyte adhesion cascade, their role in liver residence has been unclear. While ICAM-1 has been suggested to have a role in the retention of naïve and activated cells in the liver (16, 20, 33), antibody blockade with α-LFA-1 antibodies was not found to inhibit the accumulation of activated CD8+ T cells in the liver (14). Moreover, LFA-1 deficient mice do not possess abnormal numbers of conventional T cells in the liver (17). Our data resolve many of these contradictory observations. As with previous studies we did not see any effect of antibody blockade on retention of activated effector cells to the liver, only a defect in migration. Our data also explain why previous studies missed a role of LFA-1 in CD8+ T cell migration to the liver, as LFA-1 is only absolutely required for the retention of the TRM phenotype cells, while other CD8+ T cells which are non-resident can transiently associate with the liver in the absence of LFA-1. Thus previous studies of bulk CD8+ T cell populations were unable to detect the role of LFA-1 in this recently identified cell type.
Although our studies have focused on the role of LFA-1 in CD8+ T cell accumulation in the liver we also find that this integrin is important for CD8+ T cell retention in the lung. We identified a population of cells in the lung that was CD11ahi, which like the corresponding population in the liver were CD69+ and KLRG1-. However our CD11ahi CD69+ cells are unlikely to be lung TRM cells as lung TRM cells are typically IV Ab- and CD103+ (6, 24). The absence of lung TRM cells is not unexpected as Plasmodium does not infect the lung, while LCMV only forms significant lung TRM populations after intra-tracheal infection (25). In contrast to the liver, in the lung both CD69+ and CD69- CD8+ T cells had a strong requirement for LFA-1 to be retained. This highlights again the different adhesion molecule requirements among diverse populations of CD8+ T cells in different organs. Previous studies have shown that LFA-1 is required for the retention of early effector cells in the lung (34), while another study has shown that LFA-1 is required for memory cell entry to the lung airways (35). Our results extend these previous results in showing an important role for LFA-1 for the retention of vascular CD8+ T cell populations (as opposed to just those in the airways) at memory time-points. Interestingly, while liver sinusoids are characterized by slow blood-flow, in the lung the flow is faster and cells experience more sheer stress (36). Thus, LFA-1 may be universally required among CD8+ T cell populations in the lung to allow even transient associations with the endothelium.
In summary we have discovered a previously unappreciated role for LFA-1 in the retention of CD8+ T cells in the liver and for the movement of these T cells within hepatic sinusoids. This motility is what appears to be important for the efficient surveillance of the liver and the identification of infected cells (9). Crucially, because of the potential ability of the liver TRM cells to enter the circulation directly, the mechanism of retention in the liver appears different to that described for epithelial or mucosal TRM cells. Overall, our data suggest that the nature of tissue resident CD8+ T cell populations may be even more diverse and complex than has previously been suggested.
Materials and Methods
Details of standard immunological methods used in this study are given in the supplemental materials and methods.
Study Design
The initial aim of the study was to determine the molecules involved in CD8+ T cell migration in the liver. Accordingly experiments were performed in which we blocked candidate adhesion molecules and measured motility by intra-vital imaging (Figure 1). As the n for these experiments was determined by the number of cells each mouse was considered an experimental repeat. No randomization or blinding was performed in these experiments, however the hypothesis that blocking these molecules would affect migration was specified in advance, though the direction of the effect was not predicted. We therefore analysed these experiments using a linear mixed model approach, which accounts for variation between mice and experiments as random effects in addition to our fixed effects (movement parameters). These experiments suggested a role for LFA-1 in accumulation and migration, which we tested by co-transferring LFA-1 deficient and wild type cells to mice and measuring migration and accumulation by intra-vital microscopy and flow cytometry. In these experiments the hypothesis was specified in advance. Randomization and blinding could not be performed in these experiments as both the experimental and control groups were contained in the same mouse.
Having identified LFA-1 as a likely candidate for T cell migration and residence in the liver we performed a series of controlled laboratory experiments to investigate this further (Figures 2-7). In these experiments at least 4 mice per group were used in each individual experiment where the n was determined by the number of mice. Data presented are typically pooled from multiple experiments, and were therefore usually analysed using linear mixed models, which account for variation due to random effects (individual mice and experiments). Data from all mice studied are presented in the figures and no outliers were excluded. In these experiments the hypotheses were specified in advance. No randomization or blinding was performed, however in many experiments control and experimental groups (cells) were contained within the same animal.
Mice
C57BL/6.J mice, Rag1-/- mice, OT-I mice (37), B2m-/- mice (38) and uGFP mice (39) were bred in house at ANU. mTmG mice (40) were purchased from Jackson laboratories and maintained at the ANU. ITGAL C77F (Itgal-/-) mice were identified in our ongoing ENU mutagenesis screens (22). ITGAL-C77F mice were maintained on a C57BL/6.J background and crossed to an OT-I uGFP background as required. All animal procedures were approved by the Animal Experimentation Ethics Committee of the Australian National University (Protocol numbers: A2013/12; A2014/62 and A2015/76).
Immunizations
Mice were immunized i.v. with 5 × 104 P. berghei CS5M sporozoites expressing mCherry (23) dissected by hand from the salivary glands of Anopheles stephensi mosquitoes. Mice were then treated with 0.6mg choloroquine i.p daily for 10 days to prevent the development of blood stage infection. LCMV-Armstrong was delivered i.p. at a dose of 2 × 105 pfu/mouse.
Antibody blockade
The following antibodies were injected i.v. 2 hours prior to transfer of activated OT-I CD8+T cells: anti-ICAM-1 (clone YN1/1.7.4; BioXCell; 50 μg/mouse (18)); anti-VCAM-1 (clone 429; Biolegend; 50 μg/mouse (16)) anti-CD44 (clone KM81, blocking CD44 binding to hyaluronan; Cedarlane; 20 μg/mouse (14)), Rat IgG2b isotype control (clone LTF2; BioXCell; 50 μg/mouse).
Multi-photon microscopy
Mice were prepared for multi-photon microscopy essentially as described in the supplementary experimental procedures (41). Imaging was performed using a Fluoview FVMPE-RS multiphoton microscope system (Olympus) equipped with a XLPLN25XWMP2 objective (25×; NA1.05; water immersion; 2mm working distance). Images were acquired using FV30 software (Olympus) and exported to Imaris (Bitplane) for downstream processing as described in the supplementary experimental procedures. Movies were annotated and prepared for display using Adobe After Effects (Adobe).
Statistical analysis
Details of statistical analysis for each experiment is given in the relevant figure legend. χ2 tests, t tests, Mann-Whitney U tests and one-way ANOVAs were performed using Prism6 (GraphPad). Where data were pooled from multiple experiments, multi-level analyses were performed using a linear mixed model (LMM) in GenStat (VSNi). LMMs are a regression analysis model containing both fixed and random effects: fixed effects being the variable/treatment under examination, whilst random effects are unintended factors that may influence the variable being measured. If significance was found from running a LMM, pair-wise comparisons of the least significant differences of means (LSD) was undertaken to determine at which level interactions were occurring. Statistical significance was assumed if the p-value was < 0.05 for a tested difference. (ns = not significant, *= p < 0.5, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001).
Supplementary Material
Movie S1. Anti-ICAM-1 inhibits effector T cell migration in the hepatic sinusoids
Movie S2. LFA-1 deficient cells display impaired motility in the liver
Movie S3. Migration of OT-I cells in the livers of mice 1 week post-sporozoite immunization
Movie S4. Migration of OT-I cells in the livers of mice 4 weeks post-sporozoite immunization
Fig. S1. CD44, VCAM-1 and β2-microglobulin are not required for CD8+ T cell motility in the vliver
Fig. S2. Phenotype of ITGAL-C77F mice
Fig. S3. Similar phenotypes of WT and Itgal-/- cells following in vitro priming
Fig. S4. Expression of CD11a on different populations of OT-I T cells
Fig. S5 CD11ahi cells in the liver and lung are CD69+, KLRG1-, CD103- and IV Ab+ following sporozoite immunization
Fig. S6. LCMV infection induces populations of CD11hi cells in the liver and lung that are phenotypically similar to those seen after sporozoite immunization
Fig. S7. NKT cells have a similar CD11ahi CD69+ KLRG1- phenotype to liver TRM cells
Fig S8. LFA-1 is required for the retention of cells in the liver and lung in the steady state
Fig. S9. LFA-1 is required for the formation of TRM cells in the liver in the steady state.
Fig 6. LFA-1 is required for residence of Plasmodium specific TRM cells in the liver.
(A) 2 × 104 naïve CD45.1+ WT OT-I cells were co-transferred with 2 × 104 naïve GFP+ Itgal-/- OT-I cells to C57BL/6 1 day prior to immunization with 5 × 104 P. berghei CS5M sporozoites, at 1 week and 4 weeks post-immunization organs were harvested and the number and phenotype of transferred cells determined by flow cytometry. (B) Representative plots from a single mouse at each time-point showing the expansion of the different OT-I+ populations in the spleen and liver 1 and 4 weeks post immunization. (C) Summary data showing the overall ratio of Itgal-/- (KO) to WT OT-I cells in the spleen and liver of mice (i) 1 week and (ii) 4 weeks post-immunization. (D) Representative flow cytometry plots showing the TRM phenotype of WT and Itgal-/- OT-I cells in the spleen and liver of a single animal 4 weeks post immunization. (E) Summary data the showing percentage of WT and Itgal-/- OT-1 cells that are TRM in (i) the spleen and (ii) livers 4 weeks post immunization. (F) Summary data showing the overall ratio of Itgal-/- (KO) to WT OT-I cells that are (i) TRM and (ii) non TRM. (G) Summary data of the overall ratio of Itgal-/- (KO) to WT OT-I cells in different organs of mice analyzed 4 weeks post-immunization; bars show means and SD. All data are pooled from 9 mice in 2 independent experiments. Panels C and F were analyzed using linear mixed models with mouse as a random effect and organ as the fixed effect. Panel E was analyzed similarly to C but with genotype as the fixed effect. Panel G was analyzed similarly to C but with experiment included as a random effect.
Acknowledgments
We thank Michael Devoy, Harpreet Vohra and Catherine Gillespie of the Imaging and Cytometry Facility at the John Curtin School of Medical Research for assistance with flow cytometry and multi-photon microscopy. We also thank Theresa Neeman of the ANU statistical consulting unit for assistance with statistical analysis of the data and Professor Dale Godfrey (University of Melbourne) for reagents and advice in the initial identification of the Itgal mutant mice.
Funding: This work was supported by startup funds from the Australian National University (to IAC) and grants from the Perpetual Foundation (IAC, FR2014/1152) and Ian Potter Foundation (IAC, Grant #32616), the Ramaciotti Foundation (CG and AE), the National Institutes of Health (CGG, U19 AI100627) and the National Health and Medical Research Council (AE, GNT1035858).
Footnotes
Author Contributions: Study conception and design: HAM; PB; WRH; IAP; CGG; AE, IC and IAC. Acquisition of data: HAM; YC; MW; YS; CMR; LM; HJS; IAP and IAC. Analysis and interpretation of data: HAM, VVG, LM, AE and IAC. Drafting of manuscript: HAM and IAC. Critical revision: WRH, PB, IAP and AE.
Competing interests: The authors declare no competing interests.
References and Notes
- 1.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 2.Gebhardt T, et al. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nature immunology. 2009;10:524. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- 3.Masopust D, et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. The Journal of experimental medicine. 2010;207:553. doi: 10.1084/jem.20090858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R. Cutting edge: gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J Immunol. 2006;176:2079. doi: 10.4049/jimmunol.176.4.2079. [DOI] [PubMed] [Google Scholar]
- 5.Steinert EM, et al. Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell. 2015;161:737. doi: 10.1016/j.cell.2015.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wakim LM, Gupta N, Mintern JD, Villadangos JA. Enhanced survival of lung tissue-resident memory CD8(+) T cells during infection with influenza virus due to selective expression of IFITM3. Nature immunology. 2013;14:238. doi: 10.1038/ni.2525. [DOI] [PubMed] [Google Scholar]
- 7.Gebhardt T, et al. Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature. 2011;477:216. doi: 10.1038/nature10339. [DOI] [PubMed] [Google Scholar]
- 8.Schenkel JM, et al. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 2014;346:98. doi: 10.1126/science.1254536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Ruiz D, et al. Liver-Resident Memory CD8+ T Cells Form a Front-Line Defense against Malaria Liver-Stage Infection. Immunity. 2016;45:889. doi: 10.1016/j.immuni.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Mackay LK, et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 2016;352:459. doi: 10.1126/science.aad2035. [DOI] [PubMed] [Google Scholar]
- 11.Crispe IN, Dao T, Klugewitz K, Mehal WZ, Metz DP. The liver as a site of T-cell apoptosis: graveyard, or killing field? Immunological reviews. 2000;174:47. doi: 10.1034/j.1600-0528.2002.017412.x. [DOI] [PubMed] [Google Scholar]
- 12.Keating R, et al. Virus-specific CD8+ T cells in the liver: armed and ready to kill. J Immunol. 2007;178:2737. doi: 10.4049/jimmunol.178.5.2737. [DOI] [PubMed] [Google Scholar]
- 13.Mackay LK, et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nature immunology. 2013;14:1294. doi: 10.1038/ni.2744. [DOI] [PubMed] [Google Scholar]
- 14.Guidotti LG, et al. Immunosurveillance of the liver by intravascular effector CD8(+) T cells. Cell. 2015;161:486. doi: 10.1016/j.cell.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Bertolino P, et al. Early intrahepatic antigen-specific retention of naive CD8+ T cells is predominantly ICAM-1/LFA-1 dependent in mice. Hepatology. 2005;42:1063. doi: 10.1002/hep.20885. [DOI] [PubMed] [Google Scholar]
- 16.John B, Crispe IN. Passive and active mechanisms trap activated CD8+ T cells in the liver. J Immunol. 2004;172:5222. doi: 10.4049/jimmunol.172.9.5222. [DOI] [PubMed] [Google Scholar]
- 17.Ohteki T, Maki C, Koyasu S, Mak TW, Ohashi PS. Cutting edge: LFA-1 is required for liver NK1.1+TCR alpha beta+ cell development: evidence that liver NK1.1+TCR alpha beta+ cells originate from multiple pathways. J Immunol. 1999;162:3753. [PubMed] [Google Scholar]
- 18.Thomas SY, et al. PLZF induces an intravascular surveillance program mediated by long-lived LFA-1-ICAM-1 interactions. The Journal of experimental medicine. 2011;208:1179. doi: 10.1084/jem.20102630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cockburn IA, et al. In vivo imaging of CD8+ T cell-mediated elimination of malaria liver stages. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9090. doi: 10.1073/pnas.1303858110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warren A, et al. T lymphocytes interact with hepatocytes through fenestrations in murine liver sinusoidal endothelial cells. Hepatology. 2006;44:1182. doi: 10.1002/hep.21378. [DOI] [PubMed] [Google Scholar]
- 21.Smith A, Bracke M, Leitinger B, Porter JC, Hogg N. LFA-1-induced T cell migration on ICAM-1 involves regulation of MLCK-mediated attachment and ROCK-dependent detachment. Journal of cell science. 2003;116:3123. doi: 10.1242/jcs.00606. [DOI] [PubMed] [Google Scholar]
- 22.Cook MC, Vinuesa CG, Goodnow CC. ENU-mutagenesis: insight into immune function and pathology. Current opinion in immunology. 2006;18:627. doi: 10.1016/j.coi.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 23.Cockburn IA, Tse SW, Zavala F. CD8+ T cells eliminate liver stage Plasmodium parasites without detectable bystander effect. Infection and immunity. 2014 doi: 10.1128/IAI.01500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laidlaw BJ, et al. CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity. 2014;41:633. doi: 10.1016/j.immuni.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson KG, et al. Cutting edge: intravascular staining redefines lung CD8 T cell responses. J Immunol. 2012;189:2702. doi: 10.4049/jimmunol.1201682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slutter B, et al. Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Science Immunology. 2017;2:eaag2031. doi: 10.1126/sciimmunol.aag2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 28.Bose TO, et al. CD11a regulates effector CD8 T cell differentiation and central memory development in response to infection with Listeria monocytogenes. Infection and immunity. 2013;81:1140. doi: 10.1128/IAI.00749-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casey KA, et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J Immunol. 2012;188:4866. doi: 10.4049/jimmunol.1200402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackay LK, et al. T-box Transcription Factors Combine with the Cytokines TGF-beta and IL-15 to Control Tissue-Resident Memory T Cell Fate. Immunity. 2015;43:1101. doi: 10.1016/j.immuni.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Boutet M, et al. TGFbeta Signaling Intersects with CD103 Integrin Signaling to Promote T-Lymphocyte Accumulation and Antitumor Activity in the Lung Tumor Microenvironment. Cancer research. 2016;76:1757. doi: 10.1158/0008-5472.CAN-15-1545. [DOI] [PubMed] [Google Scholar]
- 32.Sela U, et al. The inhibition of autoreactive T cell functions by a peptide based on the CDR1 of an anti-DNA autoantibody is via TGF-beta-mediated suppression of LFA-1 and CD44 expression and function. J Immunol. 2005;175:7255. doi: 10.4049/jimmunol.175.11.7255. [DOI] [PubMed] [Google Scholar]
- 33.Mehal WZ, Juedes AE, Crispe IN. Selective retention of activated CD8+ T cells by the normal liver. J Immunol. 1999;163:3202. [PubMed] [Google Scholar]
- 34.Thatte J, Dabak V, Williams MB, Braciale TJ, Ley K. LFA-1 is required for retention of effector CD8 T cells in mouse lungs. Blood. 2003;101:4916. doi: 10.1182/blood-2002-10-3159. [DOI] [PubMed] [Google Scholar]
- 35.Ely KH, Cookenham T, Roberts AD, Woodland DL. Memory T cell populations in the lung airways are maintained by continual recruitment. J Immunol. 2006;176:537. doi: 10.4049/jimmunol.176.1.537. [DOI] [PubMed] [Google Scholar]
- 36.MacPhee PJ, Schmidt EE, Groom AC. Intermittence of blood flow in liver sinusoids, studied by high-resolution in vivo microscopy. The American journal of physiology. 1995;269:G692. doi: 10.1152/ajpgi.1995.269.5.G692. [DOI] [PubMed] [Google Scholar]
- 37.Hogquist KA, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 38.Koller BH, Marrack P, Kappler JW, Smithies O. Normal development of mice deficient in beta 2M, MHC class I proteins, and CD8+ T cells. Science. 1990;248:1227. doi: 10.1126/science.2112266. [DOI] [PubMed] [Google Scholar]
- 39.Schaefer BC, Schaefer ML, Kappler JW, Marrack P, Kedl RM. Observation of antigen-dependent CD8+ T-cell/ dendritic cell interactions in vivo. Cellular immunology. 2001;214:110. doi: 10.1006/cimm.2001.1895. [DOI] [PubMed] [Google Scholar]
- 40.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 41.van de Ven AL, Kim P, Ferrari M, Yun SH. Real-time intravital microscopy of individual nanoparticle dynamics in liver and tumors of live mice. 2013 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Anti-ICAM-1 inhibits effector T cell migration in the hepatic sinusoids
Movie S2. LFA-1 deficient cells display impaired motility in the liver
Movie S3. Migration of OT-I cells in the livers of mice 1 week post-sporozoite immunization
Movie S4. Migration of OT-I cells in the livers of mice 4 weeks post-sporozoite immunization
Fig. S1. CD44, VCAM-1 and β2-microglobulin are not required for CD8+ T cell motility in the vliver
Fig. S2. Phenotype of ITGAL-C77F mice
Fig. S3. Similar phenotypes of WT and Itgal-/- cells following in vitro priming
Fig. S4. Expression of CD11a on different populations of OT-I T cells
Fig. S5 CD11ahi cells in the liver and lung are CD69+, KLRG1-, CD103- and IV Ab+ following sporozoite immunization
Fig. S6. LCMV infection induces populations of CD11hi cells in the liver and lung that are phenotypically similar to those seen after sporozoite immunization
Fig. S7. NKT cells have a similar CD11ahi CD69+ KLRG1- phenotype to liver TRM cells
Fig S8. LFA-1 is required for the retention of cells in the liver and lung in the steady state
Fig. S9. LFA-1 is required for the formation of TRM cells in the liver in the steady state.