Abstract
Nuclear pore complexes (NPCs), are large multiprotein channels that penetrate the nuclear envelope connecting the nucleus to the cytoplasm. Accumulating evidence shows that besides their main role in regulating the exchange of molecules between these two compartments, NPCs and their components also play important transport-independent roles, including gene expression regulation, chromatin organization, DNA repair, RNA processing and quality control, and cell cycle control. Here we will describe the recent findings about the role of these structures in the regulation of gene expression.
Introduction
In 1950, Callan and Tomlin used Xenopus laevis oocytes to perform the first electron microcopy studies of the nuclear envelope (NE) and observed that it was perforated by many large pores [1]. This was the first description of nuclear pore complexes (NPCs). Our understanding of NPCs has come a long way since that initial observation. Thanks to a momentous amount of work performed by many different groups over the last six decades we now know that NPCs are giant multiprotein channels of about 110MDa in mammals that represent the sole gateway into the nucleus. At the structural level, NPCs have an eight-fold rotational symmetry, and consist of a core ring embedded in the NE, two outer rings, one cytoplasmic and one nuclear, and eight filaments attached to these rings [2]. The nuclear filaments are also joined in a distal ring assembling a structure known as the nuclear basket. Despite being one of the largest protein complexes of eukaryotic cells these structures have a rather simple composition and are built by the repetition of roughly 30 different proteins known as nucleoporins [3,4]. Recent studies combining structural information from the NPC itself and from individual nucleoporins has resulted in an unprecedented resolution of this structure [5–7].
In addition to their main function as mediators of nucleocytoplasmic molecule exchange, increasing evidence shows that NPCs regulate multiple cellular processes in a transport-independent manner [2]. Probably the most studied so far, and the focus of this review, is their role in genome organization and gene expression regulation.
Gene expression regulation by NPCs in yeast
The first evidence for a role of nuclear pore complexes in the regulation of gene expression came from studies in Saccharomyces cerevisiae. Though the initial observations of NPCs association with repressed telomeric and subtelomeric chromatin fueled the idea that the nuclear periphery was always associated with gene silencing, [8,9], Menon et al were the first ones to show that the Nup84 scaffold subcomplex of the NPC could act as transcriptional activator by itself [10]. Two other later studies showed that many more nucleoporins were associated with transcriptionally active genes [11,12]. These findings supported a potential role for NPCs in active gene expression. In the decade following this discovery, several groups have confirmed the role of NPCs in the positive regulation of gene expression in yeast. Many different genes, including CTT1 [13], FIG2 [14], GAL (GAL1, GAL2, GAL7 and GAL10) [15–20], HIS4 [21], HSP104 [15], HXK1 [17,22], INO1 [18,19,23,24], PRM1 [21], STL1 [13,25], SUC2 [26], and TSA2 [24] have been shown to relocalize from the nuclear interior to NPCs upon activation (Figure 1). In most cases NPC association is not required for activation, but results in a more efficient gene expression and enhances transcriptional memory of some of these genes [20,22,23], although the relocation of the GAL locus (GAL7, GAL10, and GAL1) to the nuclear periphery has also been associated with repression post-induction under specific conditions [16].
Figure 1. NPC-gene association in yeast.
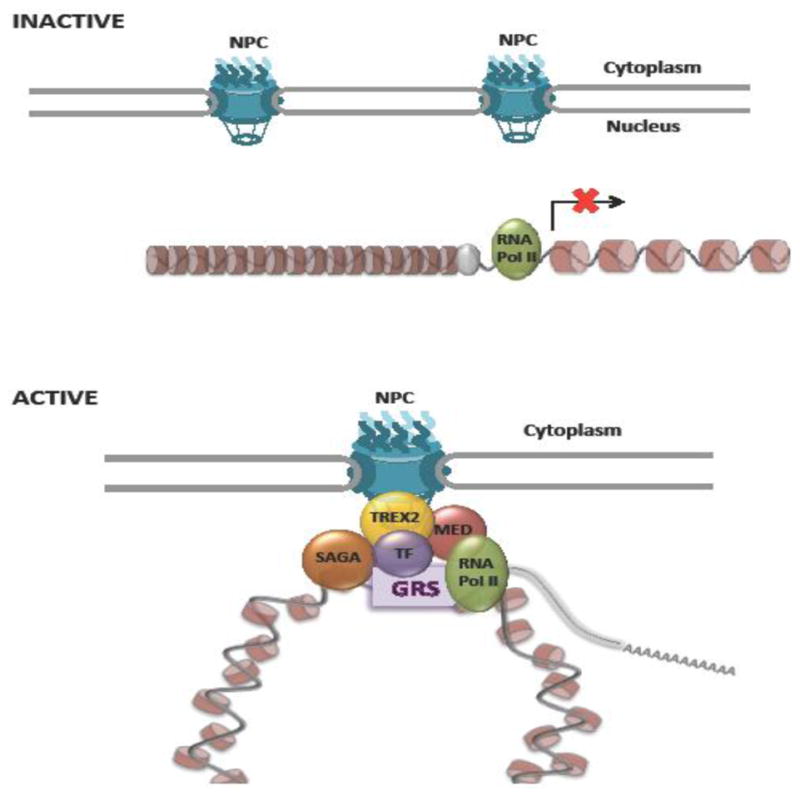
Upon activation several inducible yeast genes re-localize to nuclear pore complexes. Factors involved in NPC tethering include nucleoporins (Mlp1, Mlp2, Nup1, Nup2, Nup42, Nup60, Nup100, Nup116, Nup157, Gle2), cis-DNA elements or Gene Recruitment Sequences (GRS), transcription factors (TF) including Put3, Cbf1, Gcn4, Ste12, Gal4, Sfl1, chromatin modulators (SAGA, HDACs), transport export complexes TREX-2/THO-TREX (TREX) and Mediator.
In the last few years there have been major advances in our understanding of the molecular mechanisms that mediate gene-pore association and gene regulation at the nuclear periphery. Targeting to the nuclear periphery for several yeast genes has been shown to rely on cis-DNA sequences in their promoters known as Gene Recruitment Sequences (GRSs) (recently reviewed in [27]) (Figure 1). Interestingly, several GRSs have been shown to represent binding sites for transcription factors which ultimately mediate gene-relocation [21]. For INO1 it was identified that NPC-tethering is mediated by the transcription factors Put3 [28] and Cbf1 [27], which do not directly regulate the activity of this gene. Conversely, the Ste12 and Gcn4 transcription factors required for repositioning of PRM1 and HIS4 also regulate the activity of these genes [21]. Yet, the activation domain of Gcn4 is dispensable for gene repositioning [27]. These findings indicate that the transcription factors involved in gene positioning not necessarily need to regulate the expression of the gene, separating the processes of gene positioning and transcriptional regulation. Notably, genes that contain the same GRS sequences cluster together at the nuclear periphery [28]. Interestingly, Brickner et al showed that the molecular mechanisms that control NPC targeting are different from the ones controlling interchromosomal clustering and that both processes are regulated through the cell cycle [21,28,29].
GRS sequences are sufficient to artificially induce the relocation of an unrelated gene to the nuclear periphery. For example insertion of GRS I and GRS II sequences that mediate INO1-NPC association [24] into the URA3 promoter results in the repositioning of this intranuclear gene to NPCs. But differently from the repositioning of endogenous INO1, the GRS-mediated association of URA3 with NPCs is constitutive (note that INO1 gene localizes to the periphery only when activated by inositol starvation). This indicates that GRS activity is modulated by additional factors that can control repositioning in a stimuli-dependent manner. For INO1, relocation is negatively modulated by the histone deacetylase complex Rpd3(L) which is recruited to the INO1 promoter by transcriptional repressors and blocks binding of Put3. [21]. On the other hand, NPC-relocation mediated by Ste12 and Gcn4 transcription factor binding sites is regulated independently of DNA binding. Mitogen-activated protein kinase phosphorylation of the Dig2 inhibitor induces its dissociation from Ste12 and allows the relocation of PRM1, while Gcn4-dependent targeting is up-regulated by increasing Gcn4 protein levels [21]. The different speeds at which, cell signaling (Ste12), chromatin remodeling (INO1) and transcription factor abundance (Gcn4) occur, might explain the different kinetics observed for the NPC-association of different genes [21].
Most of the studies investigating gene repositioning to NPCs have centered their analysis on specific genes without determining if the association with NPCs involved larger chromatin domains. Recent, work from the Weis lab has shown that during activation, the association of the GAL locus with the NPC involves large genome rearrangements that position several chromosomal domains in close proximity to the nuclear envelope [30]. The authors propose that this association likely results from multiple independent anchoring sites. Moreover, using a genome-wide screen, they identified a novel role for histone deacetylases in chromatin association with the nuclear periphery and provided further evidence for the previously described role of the transcriptional coactivator SAGA complex in peripheral gene repositioning in response to galactose activation [30].
SAGA and TREX complexes in NPC-gene tethering and gene expression regulation
The SAGA complex, and the TREX-2 and THO-TREX complexes, which couple transcription to mRNA export (reviewed in [31]), were the first factors shown to mediate the interaction of chromatin with NPCs [32–37] (Figure 1). In yeast, the SAGA and TREX-2 complexes have been shown to interact with the nuclear basket of the NPC, [32,34,38,39]. Through a common partner, Sus1, these complexes tether genes that are being actively transcribed to NPCs and coordinate transcription with nuclear export [33,36,37,40,41]. Work by Schneider et al recently showed that the Mediator complex plays a critical role in connecting RNA Polymerase II to TREX-2 at NPCs [42]. By analyzing the re-positioning of GAL1 and HXK1 in Mediator mutant yeast strains or in mutants that affect the interaction between TREX-2 and Mediator, this group showed that both complexes are required for the NPC-targeting of these inducible genes. Moreover, they revealed that the TREX-2/Mediator-interacting surface is important for mRNA export [42]. Considering that Mediator is an important regulator of RNA Pol II during transcription initiation, this complex would provide the NPC-associated TREX-2 complex with access to the core transcription machinery (Figure 1). The NPC-associated SUMO protease Ulp1 is another factor required for the repositioning of GAL1 to the nuclear periphery and for its efficient activation in response to galactose [20]. Texari et al showed that Ulp1 facilitates the derepression of GAL1 at the nuclear periphery by desumoylating the transcriptional repressors Tup1 and Ssn6 and proposed that this modification could induce conformational changes that allow the interaction with coactivators such as the SAGA and Mediator complexes [20].
NPC-gene tethering and transcriptional memory
The association of some genes, including INO1, GAL and HXK1, with NPCs has been linked to their transcriptional memory (reviewed in [43]). Upon activation these genes move from the nuclear interior to NPCs as described above. Once repressed, they are maintained at NPCs for several generations where they are poised for transcription and will reactivate faster in response to a second induction (transcriptional memory). HXK1 and GAL1 transcriptional memory depends on the formation of NPC-associated gene loops that require the nuclear basket nucleoporin Mlp1 [17]. Transcriptional memory of INO1, on the other hand, requires association with Nup100 [18,44]. Interestingly, even though INO1 is recruited to NPCs through GRSI and II during activation [24], its maintenance at NPCs in the repressed state depends on a different cis-DNA element named Memory Recruitment Sequence (MRS) [44]. Binding of the transcription factor Sfl1 to MRS during memory is required for the maintenance of INO1 at the nuclear periphery and binding of Mediator is needed for efficient reactivation [45]. Transcriptional memory for INO1 also requires chromatin modifications mediated by the Set1/COMPASS and SET3C complexes [45].
Gene expression regulation by NPCs in metazoans
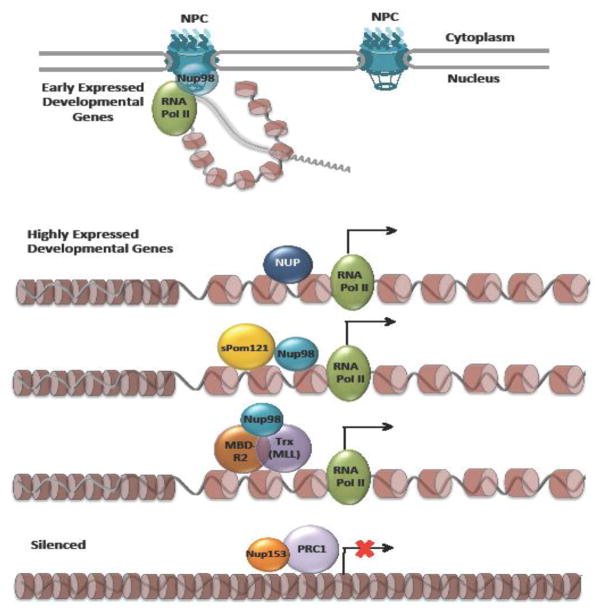
Our knowledge of NPC-mediated regulation of gene expression in metazoans is considerably more limited than in yeast. The first connection of NPC components with chromatin modulation/gene expression regulation was described in Drosophila melanogaster with the identification of two nucleoporins, mTor/TPR and Nup153, as partners of the MSL dosage compensation complex [46]. This work showed that both nucleoporins were required for the localization of the MSL complex to the X chromosome and for the transcription of dosage compensation genes [46]. A year later, the SAGA/Eny-2(Sus-1) complex was shown to mediate the association of the HSP70 loci with NPCs [47]. Although NPC-anchoring of HSP70 was activation-independent, the association was necessary for efficient gene transcription and mRNA export [47]. More recently, CHIP-on-chip experiments and DamID assays have shown that the nucleoporin Nup98 binds to many actively transcribed sequences, mostly developmental and cell cycle genes [48,49] (Figure 2). Interestingly, the binding of these active genes takes place inside the nucleus and Nup98 was shown to be required for their proper expression [48,49]. NPC-associated Nup98, on the other hand, was described to bind a small subset of predominantly non-active genes [49]. Recently, Nup98 was also found to bind and regulate the expression of genes critical for the anti-viral response in Drosophila, but whether this association takes place inside the nucleus or at the nuclear periphery has not been investigated [50]. Other nucleoporins that were also found to bind active genes inside the nucleus include Nup50, Nup62, Sec13 and Nup153 [48,49,51] (Figure 2). Interestingly, Nup153 was identified to bind ~25% of the genome in Nucleoporin-Associated Regions (NARs) of 10–500 kb in length that are enriched in markers for active transcription [51].
Figure 2. Gene regulation by nucleoporins and NPCs in metazoans.
In Drosophila and mammalian cells, several nucleoporins regulate gene expression inside the nucleus and independent of NPCs. Nup98, Nup62, Nup50, and Sec13 (NUP) have been shown to bind and regulate developmental and cell cycle genes. A soluble form of the transmembrane nucleoporin Pom121 (sPom121) binds many target genes together with Nup98. Nup98 also interacts with the histone-modifying complexes MBD-R2/NSL and Trx/MLL. MBD-R2/NSL is required for Nup98 recruitment to its target genes. Nup153 on the other hand recruits the polycomb complex PRC1 to gene promoters repressing the activity of developmental genes.
In most cases, the molecular mechanisms through which nucleoporins regulate gene expression in Drosophila are still unknown. Recently, it was reported that Nup98 physically associates with the histone-modifying complexes MBD-R2/NSL and Trx/MLL, and that MBD-R2 mediates the recruitment of Nup98 to several active genes, including Hox genes [52] (Figure 2). These findings link Nup98 to epigenetic modulation. Notably, the chromatin domains associated with NPC-associated Nup98 at the nuclear periphery are enriched in the insulator protein Su(Hw), further suggesting a potential role for this nucleoporin in chromatin organization [53]. The function of Nup98 in controlling chromatin organization might vary depending on its location/environment being different at the pore, where Nup98 binds inactive genes, and at the nuclear interior, were it associates with highly transcribed genes.
Mammalian Nup98 has also been shown to bind genes both in the nuclear interior and at NPCs. A study of Nup98 dynamics during embryonic stem cell differentiation showed that this nucleoporin is associated with developmentally regulated genes, which is consistent with the findings in Drosophila. Interestingly, this study identified that genes induced during the early stages of differentiation associate with Nup98 at NPCs, while genes that are highly activated bind Nup98 in the nucleoplasm [54]. Nup98 has also been shown to associate with INF-γ regulated genes inside the nucleus [55]. Consistent with the role of its yeast homologue Nup100 in transcriptional memory, Nup98 is critical for the transcriptional memory to INF-γ induction [55]. However, in mammalian cells, the transcriptional memory function of Nup98 is independent of NPCs. Interestingly, a soluble variant of the transmembrane nucleoporin Pom121 (sPom121) that arises from alternate transcription initiation was recently discovered to interact with nucleoplasmic Nup98 at the promoters of many target genes to co-regulate their activity [56] (Figure 2), though its role in the regulation of developmental or INF-γ genes has not yet been studied. Like Nup98, Nup153 has also been shown to bind several developmental genes in embryonic stem cells. But differently from Nup98, Nup153 mediates the recruitment of the polycomb-repressive complex 1 (PRC1) silencing these genes to maintain stem cell pluripotency [57] (Figure 2).
In Caenorhabditis elegans, the HSP-16 gene was recently shown to associate with NPCs upon heat shock induction [58]. Similar to yeast genes, the association with NPCs requires a transcription factor, HSF-1, and the Sus1 homologue Eny-2 suggesting that repositioning might be mediated by the SAGA-THO/TREX complexes. C. elegans nuclear pore complex components have also been described to associate with RNA Pol III transcribed genes [59]. In this case, nucleoporin association is not required for transcriptional regulation but plays a critical role in the processing of small nucleolar RNAs (snoRNAs) and tRNAs. Whether RNA processing takes places inside the nucleus or at NPCs remains to be investigated, but in yeast, tRNA genes have been shown to associate with NPCs during M-phase [60].
Conclusions
Since the discovery of the first Nucleoporin-gene interactions just 10 years ago, it has become clear that the NPC and its components play a pivotal role in the regulation of gene expression in multiple organisms. In this review, we have mostly focused in the positive regulation of gene expression by these proteins, but NPCs and nucleoporins have also been shown to associate and regulate silent/inactive chromatin [8,26,57,61–65]. Understanding how NPCs contribute to modulating these two chromatin environments (active/silent) and further dissecting the molecular mechanism of transcriptional regulation are two foreseeable challenges for the next decade.
Highlights.
Nuclear pore complexes and nucleoporins have many transport-independent functions
Nuclear pore complexes play key roles in gene expression regulation
In yeast, the activity of many inducible genes is regulated at nuclear pore complexes
In metazoans, many nucleoporins regulate gene expression inside the nucleus and away from nuclear pore complexes
Acknowledgments
We apologize to all colleagues whose work could not be cited directly owing to space limitation. M.A.D. is a Pew Scholar in the Biomedical Sciences. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R01AR065083. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Callan HG, Tomlin SG. Experimental studies on amphibian oocyte nuclei. I. Investigation of the structure of the nuclear membrane by means of the electron microscope. Proc R Soc Lond B Biol Sci. 1950;137:367–378. doi: 10.1098/rspb.1950.0047. [DOI] [PubMed] [Google Scholar]
- 2.Raices M, D’Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nature reviews Molecular cell biology. 2012;13:687–699. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- 3.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, et al. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science. 2016;352:363–365. doi: 10.1126/science.aaf0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN, et al. Architecture of the symmetric core of the nuclear pore. Science. 2016;352:aaf1015. doi: 10.1126/science.aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoelz A, Glavy JS, Beck M. Toward the atomic structure of the nuclear pore complex: when top down meets bottom up. Nat Struct Mol Biol. 2016;23:624–630. doi: 10.1038/nsmb.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 2000;403:108–112. doi: 10.1038/47528. [DOI] [PubMed] [Google Scholar]
- 9.Hediger F, Dubrana K, Gasser SM. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J Struct Biol. 2002;140:79–91. doi: 10.1016/s1047-8477(02)00533-6. [DOI] [PubMed] [Google Scholar]
- 10.Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci U S A. 2005;102:5749–5754. doi: 10.1073/pnas.0501768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 12.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 13.Guet D, Burns LT, Maji S, Boulanger J, Hersen P, Wente SR, Salamero J, Dargemont C. Combining Spinach-tagged RNA and gene localization to image gene expression in live yeast. Nat Commun. 2015;6:8882. doi: 10.1038/ncomms9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Molecular and cellular biology. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green EM, Jiang Y, Joyner R, Weis K. A negative feedback loop at the nuclear periphery regulates GAL gene expression. Molecular biology of the cell. 2012;23:1367–1375. doi: 10.1091/mbc.E11-06-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A. Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brickner DG, Brickner JH. Cdk phosphorylation of a nucleoporin controls localization of active genes through the cell cycle. Mol Biol Cell. 2010;21:3421–3432. doi: 10.1091/mbc.E10-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Texari L, Dieppois G, Vinciguerra P, Contreras MP, Groner A, Letourneau A, Stutz F. The nuclear pore regulates GAL1 gene transcription by controlling the localization of the SUMO protease Ulp1. Mol Cell. 2013;51:807–818. doi: 10.1016/j.molcel.2013.08.047. [DOI] [PubMed] [Google Scholar]
- 21.Randise-Hinchliff C, Coukos R, Sood V, Sumner MC, Zdraljevic S, Meldi Sholl L, Garvey Brickner D, Ahmed S, Watchmaker L, Brickner JH. Strategies to regulate transcription factor-mediated gene positioning and interchromosomal clustering at the nuclear periphery. J Cell Biol. 2016;212:633–646. doi: 10.1083/jcb.201508068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 23.Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol. 2010;12:111–118. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regot S, de Nadal E, Rodriguez-Navarro S, Gonzalez-Novo A, Perez-Fernandez J, Gadal O, Seisenbacher G, Ammerer G, Posas F. The Hog1 stress-activated protein kinase targets nucleoporins to control mRNA export upon stress. J Biol Chem. 2013;288:17384–17398. doi: 10.1074/jbc.M112.444042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarma NJ, Haley TM, Barbara KE, Buford TD, Willis KA, Santangelo GM. Glucose-responsive regulators of gene expression in Saccharomyces cerevisiae function at the nuclear periphery via a reverse recruitment mechanism. Genetics. 2007;175:1127–1135. doi: 10.1534/genetics.106.068932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randise-Hinchliff C, Brickner JH. Transcription factors dynamically control the spatial organization of the yeast genome. Nucleus. 2016;7:369–374. doi: 10.1080/19491034.2016.1212797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brickner DG, Ahmed S, Meldi L, Thompson A, Light W, Young M, Hickman TL, Chu F, Fabre E, Brickner JH. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Developmental Cell. 2012;22:1234–1246. doi: 10.1016/j.devcel.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brickner DG, Sood V, Tutucci E, Coukos R, Viets K, Singer RH, Brickner JH. Subnuclear positioning and interchromosomal clustering of the GAL1-10 locus are controlled by separable, interdependent mechanisms. Mol Biol Cell. 2016;27:2980–2993. doi: 10.1091/mbc.E16-03-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dultz E, Tjong H, Weider E, Herzog M, Young B, Brune C, Mullner D, Loewen C, Alber F, Weis K. Global reorganization of budding yeast chromosome conformation in different physiological conditions. J Cell Biol. 2016;212:321–334. doi: 10.1083/jcb.201507069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickramasinghe VO, Laskey RA. Control of mammalian gene expression by selective mRNA export. Nat Rev Mol Cell Biol. 2015;16:431–442. doi: 10.1038/nrm4010. [DOI] [PubMed] [Google Scholar]
- 32.Fischer T, Strasser K, Racz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, Lechner J, Hurt E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 34.Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 35.Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, Chaurasia S, Valentini SR, Corbett AH. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem. 2007;282:3042–3049. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- 36.Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- 37.Pascual-Garcia P, Govind CK, Queralt E, Cuenca-Bono B, Llopis A, Chavez S, Hinnebusch AG, Rodriguez-Navarro S. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev. 2008;22:2811–2822. doi: 10.1101/gad.483308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lei EP, Stern CA, Fahrenkrog B, Krebber H, Moy TI, Aebi U, Silver PA. Sac3 is an mRNA export factor that localizes to cytoplasmic fibrils of nuclear pore complex. Mol Biol Cell. 2003;14:836–847. doi: 10.1091/mbc.E02-08-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jani D, Valkov E, Stewart M. Structural basis for binding the TREX2 complex to nuclear pores, GAL1 localisation and mRNA export. Nucleic Acids Res. 2014;42:6686–6697. doi: 10.1093/nar/gku252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodriguez-Navarro S. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klockner C, Schneider M, Lutz S, Jani D, Kressler D, Stewart M, Hurt E, Kohler A. Mutational uncoupling of the role of Sus1 in nuclear pore complex targeting of an mRNA export complex and histone H2B deubiquitination. J Biol Chem. 2009;284:12049–12056. doi: 10.1074/jbc.M900502200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider M, Hellerschmied D, Schubert T, Amlacher S, Vinayachandran V, Reja R, Pugh BF, Clausen T, Kohler A. The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression. Cell. 2015;162:1016–1028. doi: 10.1016/j.cell.2015.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Urso A, Brickner JH. Mechanisms of epigenetic memory. Trends Genet. 2014;30:230–236. doi: 10.1016/j.tig.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Light WH, Brickner DG, Brand VR, Brickner JH. Interaction of a DNA zip code with the nuclear pore complex promotes H2A. Z incorporation and INO1 transcriptional memory. Molecular cell. 2010;40:112–125. doi: 10.1016/j.molcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D’Urso A, Takahashi YH, Xiong B, Marone J, Coukos R, Randise-Hinchliff C, Wang JP, Shilatifard A, Brickner JH. Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory. Elife. 2016:5. doi: 10.7554/eLife.16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 47.Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, Spehner D, Schultz P, Tora L, Georgieva SG. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 2007;26:4956–4965. doi: 10.1038/sj.emboj.7601901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 50.Panda D, Pascual-Garcia P, Dunagin M, Tudor M, Hopkins KC, Xu J, Gold B, Raj A, Capelson M, Cherry S. Nup98 promotes antiviral gene expression to restrict RNA viral infection in Drosophila. Proc Natl Acad Sci U S A. 2014;111:E3890–3899. doi: 10.1073/pnas.1410087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS genetics. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pascual-Garcia P, Jeong J, Capelson M. Nucleoporin Nup98 associates with Trx/MLL and NSL histone-modifying complexes and regulates Hox gene expression. Cell Rep. 2014;9:433–442. doi: 10.1016/j.celrep.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Kalverda B, Fornerod M. Characterization of genome-nucleoporin interactions in Drosophila links chromatin insulators to the nuclear pore complex. Cell Cycle. 2010;9:4812–4817. doi: 10.4161/cc.9.24.14328. [DOI] [PubMed] [Google Scholar]
- 54.Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. Dynamic association of NUP98 with the human genome. PLoS Genet. 2013;9:e1003308. doi: 10.1371/journal.pgen.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Light WH, Freaney J, Sood V, Thompson A, D’Urso A, Horvath CM, Brickner JH. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 2013;11:e1001524. doi: 10.1371/journal.pbio.1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franks TM, Benner C, Narvaiza I, Marchetto MC, Young JM, Malik HS, Gage FH, Hetzer MW. Evolution of a transcriptional regulator from a transmembrane nucleoporin. Genes Dev. 2016;30:1155–1171. doi: 10.1101/gad.280941.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacinto FV, Benner C, Hetzer MW. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 2015;29:1224–1238. doi: 10.1101/gad.260919.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rohner S, Kalck V, Wang X, Ikegami K, Lieb JD, Gasser SM, Meister P. Promoter- and RNA polymerase II-dependent hsp-16 gene association with nuclear pores in Caenorhabditis elegans. J Cell Biol. 2013;200:589–604. doi: 10.1083/jcb.201207024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ikegami K, Lieb JD. Integral nuclear pore proteins bind to Pol III-transcribed genes and are required for Pol III transcript processing in C. elegans. Mol Cell. 2013;51:840–849. doi: 10.1016/j.molcel.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen M, Gartenberg MR. Coordination of tRNA transcription with export at nuclear pore complexes in budding yeast. Genes Dev. 2014;28:959–970. doi: 10.1101/gad.236729.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 62.Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, Aitchison JD. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol. 2001;153:1465–1478. doi: 10.1083/jcb.153.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Therizols P, Fairhead C, Cabal GG, Genovesio A, Olivo-Marin JC, Dujon B, Fabre E. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. The Journal of cell biology. 2006;172:189–199. doi: 10.1083/jcb.200505159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van de Vosse DW, Wan Y, Lapetina DL, Chen WM, Chiang JH, Aitchison JD, Wozniak RW. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell. 2013;152:969–983. doi: 10.1016/j.cell.2013.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]