Abstract
The appropriate orchestration of different arms of the immune response is critical during viral infection to promote efficient viral clearance while limiting immunopathology. However, the signals and mechanisms that guide this coordination are not fully understood. Interferons are produced at high levels during viral infection and have convergent signaling through signal transducer and activator of transcription 1 (STAT1). We hypothesized that STAT1 signaling during viral infection would regulate the balance of innate lymphoid cells (ILC), a diverse class of lymphocytes that are poised to respond to environmental insults including viral infections with the potential for both anti-viral or immunopathologic functions. During infection with respiratory syncytial virus (RSV), STAT1-deficient mice had reduced numbers of anti-viral IFNγ+ ILC1 and increased numbers of immunopathologic IL-5+ and IL-13+ ILC2 and IL-17A+ ILC3 compared to RSV-infected WT mice. Using bone marrow chimeric mice, we found that both ILC-intrinsic and ILC-extrinsic factors were responsible for this ILC dysregulation during viral infection in STAT1-deficient mice. Regarding ILC-extrinsic mechanisms, we found that STAT1-deficient mice had significantly increased expression of IL-33 and IL-23, cytokines that promote ILC2 and ILC3 respectively, compared to WT mice during RSV infection. Moreover, disruption of IL-33 or IL-23 signaling attenuated cytokine-producing ILC2 and ILC3 responses in STAT1-deficient mice during RSV-infection. Collectively, these data demonstrate that STAT1 is a key orchestrator of cytokine-producing ILC responses during viral infection via ILC-extrinsic regulation of IL-33 and IL-23.
INTRODUCTION
A carefully orchestrated immune response is vital for effective host defense and tissue homeostasis. Both innate and adaptive immune responses are precisely regulated to respond productively to a variety of different environmental insults while simultaneously restricting detrimental host immunopathology. Lymphocytes, both innate and adaptive, exhibit the expected functional diversity to accomplish this arduous task.
Innate lymphoid cells (ILC) are an expansive class of cells that derive from the common lymphoid progenitor and respond rapidly to cytokine stimuli in an antigen-independent manner (1). ILC have been classified into three major subsets—ILC1, ILC2, and ILC3, which largely mirror adaptive CD4+ Th1, Th2, and Th17 cells, respectively, in both transcriptional regulation and cytokine production. ILC1 include classical natural killer (cNK) cells as well as non-NK cell helper-like ILC1 and produce predominantly interferon (IFN) γ. ILC2 express high levels of IL-5 and IL-13. ILC3 include NKp46− ILC3, NKp46+ ILC3, and lymphoid tissue inducer (LTi) cells, with marked production of IL-17 and/or IL-22. ILC are activated by the local cytokine milieu including IL-12, IL-15, and IL-18 (ILC1); IL-33, TSLP, and IL-25 (ILC2); and IL-23 and IL-1β (ILC3). Moreover, several endogenous signals have been identified as inhibitors of ILC including prostacyclin (PGI2), 1,25-dihydroxyvitamin D3, and the natural Ras-ERK inhibitor Spred1 (2–4). In particular, the activation of ILC2 induces detrimental immunopathology in the context of respiratory syncytial virus (RSV), influenza, and rhinovirus infection (5–9). Similarly, the activation of CD4+ Th2 and Th17 cells has previously been associated with maladaptive immunopathology and enhanced disease severity in the context of viral infection (10–14). Conversely, ILC1 have been shown to produce anti-viral mediators in the context of viral infection and/or enhance viral clearance (15–17).
Coordinated responses to viral infection, namely the enhancement of anti-viral Th1 cells and the restriction of Th2 and Th17 cells, are accomplished by several mechanisms. Multiple, unique signals are integrated to either promote or inhibit Th1, Th2, or Th17 cells. Notably, type I, II, and/or III interferons are produced at high levels during viral infection and have convergent signaling through signal transducer and activator of transcription 1 (STAT1) (18). The presence of interferons in the polarizing cytokine milieu strongly promotes the generation of Th1 cells and IFNγ while restricting Th2 and Th17 polarization and their associated cytokine production (19–25). These data implicate interferons and STAT1 signaling as a principle coordinator of CD4+ T cell subset balance during viral infection.
While many discrete pathways for activation and inhibition of ILC have been defined, it remains unclear how ILC responses are orchestrated collectively to provide an effective anti-viral response. Given the central role of STAT1 signaling during viral infection and in the regulation of CD4+ T cell subset balance, we hypothesized that STAT1 signaling promotes ILC1 responses and restrains ILC2 and ILC3 responses to viral infection. To test this hypothesis, we evaluated cytokine-producing ILC responses in Stat1−/− mice infected with RSV, a major pathogen of infants and the elderly with robust clinical morbidity in both developed and undeveloped countries and a particularly high mortality rate worldwide. ILC were significantly dysregulated in Stat1−/− mice, with diminished IFNγ+ ILC1 and enhanced IL-5+ ILC2, IL-13+ ILC2, and IL-17A+ ILC3 populations. This dysregulation was associated with increased production of ILC-associated cytokines in the lungs, pathophysiologic changes in the airways, and poor viral clearance. Concurrently, Stat1−/− mice had significantly increased expression of IL-33 and IL-23, and abrogating IL-33 or IL-23 attenuated the cytokine-producing ILC2 and ILC3 responses in these mice, respectively. Together, these data demonstrate the critical role of STAT1 signaling in the proper regulation of ILC responses to viral infection.
MATERIALS AND METHODS
Mice
Female 8–12 week old mice were used for all experiments. BALB/cJ and CByJ.SJL(B6)-Ptprca/J CD45.1+ congenic mice were obtained from the Jackson Laboratory. Stat1−/− and Il33−/− mice on a BALB/c genetic background were generated as previously described (26–28). Stat1−/− Il33−/− double-deficient mice were bred from these strains. Mice were housed in microisolator cages under specific pathogen free conditions. All animal experiments were approved by the Vanderbilt University Medical Center Institutional Animal Care and Use Committee and were conducted in compliance with the 1996 “Guide for the Care and Use of Laboratory Animals” prepared by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council.
RSV Infection and Viral Titer
Respiratory syncytial virus strain 01/2–20 was isolated from a patient in the Vanderbilt Vaccine Clinic and propagated in HEp-2 cells as previously described (29, 30). Mice were anesthetized with a ketamine/xylazine solution and inoculated with 3.0 × 106 PFU of RSV 01/2–20 by intranasal instillation with 100 μl of viral preparation or mock preparation (lysed uninfected HEp-2 cells). Mice were sacrificed and harvested for tissue on days 0–9 post-infection. Viral titer was assessed by immunodetection plaque assay (31). Briefly, lungs were collected and homogenized in 1 mL of MEM media using a BeadBeater (29). Debris was removed by centrifugation and serial dilutions of the homogenate were plated on 80% confluent HEp-2 cells for 1 hour while shaking. The cells were then overlaid with MEM supplemented with 10% FBS, penicillin G, streptomycin, gentamicin, amphotericin B, and 0.75% methylcellulose and incubated at 37°C for 6 days. Cells were subsequently fixed with methanol at −80°C for 2 hour, blocked with 5% non-fat milk/PBS buffer, incubated at 37°C for 1 hour each with primary goat anti-RSV (EMD Millipore Cat. # AB1128, 1:250 dilution) and secondary HRP-conjugated donkey anti-goat (Jackson Immuno Cat. # 705-035-147, 1:500 dilution) antibodies, and visualized with 4-chloronaphthol.
Flow Cytometry
Flow cytometric analysis was performed as previously described (5). Briefly, lungs were minced and enzymatically disrupted with 1 mg/mL collagenase IV and 50 U/mL of DNase I in RPMI with 5% FBS for 1 hour at 37°C. Enzymes were inactivated with EDTA and the disrupted lungs were passed through 70 micron strainers to generate single cell suspensions. RBCs were lysed and cells were restimulated with 10 ng/mL phorbol 12-myristate 13-acetate and 1 μmol/L ionomycin in the presence of 0.07% monensin in Iscove modified Dulbecco medium media supplemented with 10% FBS, 0.01 mmol/L nonessential amino acids, penicillin/streptomycin, and 1 mmol/L sodium pyruvate for 5 hours at 37°C. In some experiments, cells were treated during the restimulation one hour prior to collection with 0.1% BSA in PBS (vehicle) or 10 ng/mL of recombinant murine IFNα (R&D Cat. #12100-1), IFNβ (R&D Cat. # 12400-1), IFNγ (PeproTech Cat. # 50-813-664), IFNλ2 (R&D Cat. # 4635-ML-025), or IL-27 (R&D Cat. # 2799-ML-010). Restimulated cells were stained with a fixable viability dye (Tonbo Cat. # 13-0868) and combinations of cell surface markers. Cells were fixed and permeabilized (Tonbo Cat. # TNB-0607-KIT) per manufacturer instructions and stained for intracellular markers. All antibodies used for flow cytometric analysis are listed in Supplemental Table I. Cells were subsequently evaluated on a BD LSR II Flow Cytometer and analyzed using FlowJo Software Version 10. Classical NK cells (cNK) were defined as CD3− CD19− NKp46+ CD49b+ T-bet+ EOMES+ cells that produced IFNγ. Non-NK cells ILC1 were defined as Lin− CD127+ T-bet+ EOMES− NKp46+ RORγt- cells that produced IFNγ. ILC2 were defined as Lin− CD45+ CD25+ CD127+ cells that expressed either IL-5 or IL-13. ILC3 were defined as Lin− CD90+ CD127+ RORγt+ cells that expressed either IL-17A or IL-22. Lineage markers (Lin) include CD3, CD5, B220, CD11b, Gr-1, 7-4, and Ter-119. Gating strategies are shown in Supplemental Figure 1 (cNK and ILC1) and Supplemental Figure 2 (ILC2 and ILC3).
ELISA
Lungs were isolated and homogenized in 1 mL of MEM media using a BeadBeater. Cell debris was removed by centrifugation and supernatants were plated for ELISA. ELISAs for IL-5 (Cat. # M5000), IL-13 (Cat. # M1300CB), IL-17A (Cat. # M1700), IL-22 (Cat. # M2200), IL-12 p70 (Cat. # M1270), IL-15 (Cat. # DY447), IL-18 (Cat. # 7625), IL-33 (Cat. # DY3626), TSLP (Cat. # MTLP00), IL-25 (Cat. # DY1399), IL-1beta (Cat. # MLB00C), and IL-12p40 (Cat. # DY2398) were obtained from R&D and performed as per manufacturer instructions.
Quantitative PCR
Lungs were collected and homogenized in 1 mL of TRIzol reagent (Ambion Cat. # 15596018) using a BeadBeater. RNA was isolated per TRIzol manufacturer instructions. Samples were DNase treated (Invitrogen Cat. # 18068-015) and cDNA was generated using SuperScript III (Invitrogen Cat. # 18080-051) per manufacturer instructions. cDNA abundance was assessed using FAM-MGB TaqMan primers for Il23p19 (Cat. # Mm01160011_g1), Il33 (Cat. # Mm00505403_m1) and Gapdh (Cat. # Mm99999915_g1) from Applied Biosystems. Amplifications were carried out on an Applied Biosystems QuantStudio 12k Flex Real-Time PCR machine and data were analyzed using Applied Biosystems QuantStudio 12k Flex Software v1.2.2. Il23p19 expression was normalized to Gapdh and expressed as a fold change calculated by comparing Stat1−/− to WT samples by the ΔΔCt method at each time point.
Bronchoalveolar Lavage (BAL)
Endotracheal tubes were placed into euthanized mice. Saline (0.8 mL) was instilled into the lungs and recovered with a syringe placed in the endotracheal tube. 0.1 mL of the recovered solution was spun onto slides and these cells stained (Richard-Allan Scientific Cat. # 22-050-272) to visualize macrophages, lymphocytes, neutrophils, and eosinophils. 200 cells on each slide were counted to assess cell type frequencies. Total numbers were determined by multiplying the cell frequencies by the total number of cells recovered from the BAL. In some experiments, BAL fluid was centrifuged to remove cells and the supernatants were plated for ELISA.
Periodic acid-Schiff (PAS) Staining
To evaluate mucus, lungs were instilled with 0.8 mL of 10% neutral buffered formalin and immersed in excess 10% neutral buffered formalin for 24 hours at room temperature. Lungs were subsequently paraffin embedded and sectioned (5μ) for histologic analysis. Slides were stained with PAS. Small- and medium-sized airways were quantified for airway mucus by a trained pathologist blinded to the experimental conditions using the following scoring matrix: 0, no PAS+ cells observed in cross-sections of medium to small airways; 1, less than 10 PAS+ cells observed in cross-sections of medium to small airways; 2, greater than 10 PAS+ cells observed in cross-sections of medium to small airways; 3, greater than 10 PAS+ cells observed in cross-sections of medium to small airways with mucous strands observed in air spaces; or 4, greater than 10 PAS+ cells observed in cross-sections of medium to small airways with mucous plugging of airways.
Bone Marrow Chimeras
Female 6 week-old BALB/cJ or Stat1−/− mice were lethally irradiated with 10 Gy of Cs-137. A 1:1 mixture of whole bone marrow was prepared from 6 week-old CByJ.SJL(B6)-Ptprca/J CD45.1+ congenic and Stat1−/− CD45.2+ mice. This bone marrow mixture was transferred via retroorbital injection into irradiated mice and allowed to reconstitute for 6 weeks prior to RSV infection and analysis. Mice were maintained on antibiotic water (0.025% trimethoprim/0.125% sulfamethoxazole) for 2 weeks following transplant.
IL-23p19 Neutralization
Stat1−/− mice were treated on day 0 and day 3 post-infection intraperitoneally with 250 μg of an anti-IL-23p19 subunit neutralizing monoclonal antibody (Amgen) or an isotype control (BioXCell clone MOPC-21). Lungs were isolated at day 6 post-infection for flow cytometric analysis.
Statistics
All data were collated and analyzed in GraphPad Prism Version 5. Whenever possible, data were pooled from multiple experiments for analysis. Statistical analyses including Student’s t-test, one-way ANOVA with Bonferroni post-test, or two-way ANOVA with Bonferroni post-tests were performed using GraphPad Prism Version 5 as appropriate and as documented throughout the text.
RESULTS
Innate lymphocytes and their cytokine products are dysregulated in Stat1−/− mice during viral infection
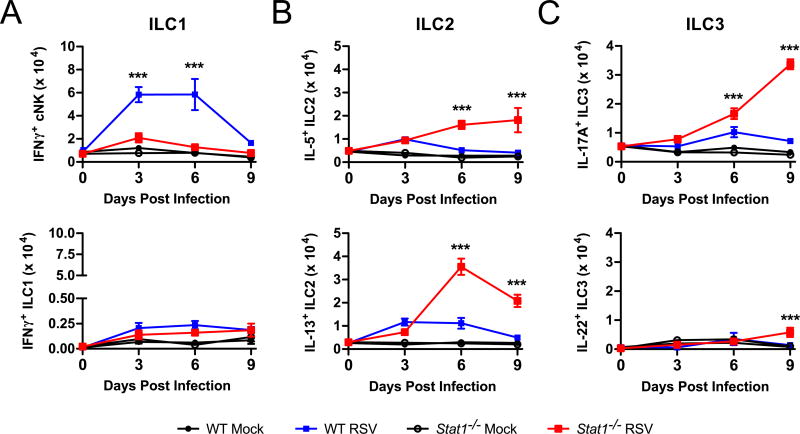
To test the role of STAT1 signaling on cytokine-producing ILC populations during viral infection, we infected Stat1−/− mice intranasally with RSV or mock inoculum (uninfected HEp-2 cell lysate) and measured ILC1, ILC2, and ILC3 cell numbers throughout the course of infection. Gating for ILC1, ILC2, and ILC3 are described in the Methods section and representative gating is shown in Supplemental Figure 1 (cNK and ILC1) and Supplemental Figure 2 (ILC2 and ILC3). We defined ILC subsets by their cytokine production, as our primary interest in these studies was in understanding the balance of type 1, 2, and 17 cytokine production by ILC. Relative to WT mice, RSV-infected Stat1−/− mice exhibited a significant decrease in the total number of IFNγ+ cNK cells at days 3 and 6 post-infection and a significant increase in the total number of IL-5+ ILC2, IL-13+ ILC2, and IL-17A+ ILC3 at days 6 and 9 post-infection (Fig. 1 A–C). There were no significant increases throughout the course of infection in IFNγ+ non-NK ILC1 or IL-22+ ILC3 with RSV infection compared to mock infection in either WT or Stat1−/− mice, except a very modest but statistically significant increase in IL-22+ ILC3 at day 9 post-infection in Stat1−/− mice (Fig. 1 A and C). The IL-17A+ ILC3 were exclusively NKp46−, consistent with previous reports of ILC3 in the lungs (data not shown) (32).
Figure 1. ILC are dysregulated in Stat1−/− mice.
WT and Stat1−/− mice were infected with RSV or mock infected with vehicle and lungs were harvested on days 0, 3, 6, and 9 post-infection for flow cytometry. The total number of functional cytokine-producing (A) ILC1, (B) ILC2, and (C) ILC3 as determined by flow cytometric analysis are shown. Data are pooled from 2 independent experiments comprising 5–7 mice per time point per group and evaluated by two-way ANOVA. **p<0.01 and ***p<0.001 between WT RSV and Stat1−/− RSV.
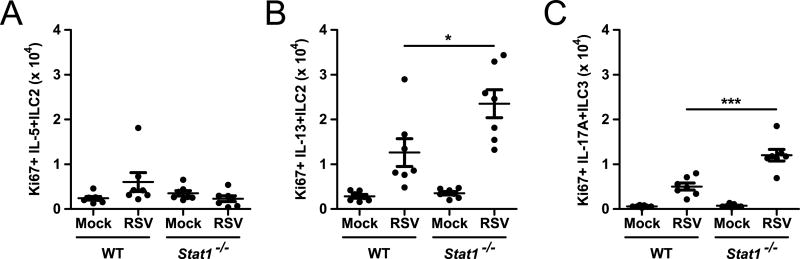
Local proliferation of ILC populations is thought to be an important driver of inflammatory responses, including during RSV infection (5, 33, 34). To assess whether ILC were replicating in situ, we measured the expression of the cell cycle associated protein Ki67 in IL-5+ or IL-13+ ILC2 and IL-17A+ ILC3. RSV-infected Stat1−/− mice had increased expression of Ki67 in the IL-13+ ILC2 and IL-17A+ ILC3 compartments, but not the IL-5+ ILC2 compartment, compared to RSV-infected WT mice (Fig. 2). These data suggest that local proliferation of IL-13+ ILC2 and IL-17A+ ILC3 contribute to the increased numbers of these cells in Stat1−/− mice during RSV infection.
Figure 2. Stat1−/− ILC2 and ILC3 proliferate robustly during RSV infection.
WT and Stat1−/− mice were infected with RSV or mock infected with vehicle and lungs were harvested on day 6 post-infection for flow cytometry. The total number of Ki67+ (A) IL-5+ ILC2, (B) IL-13+ ILC2, and (C) IL-17A+ ILC3 is shown. Data are pooled from 2 independent experiments and evaluated by one-way ANOVA. *p<0.05 and ***p<0.001.
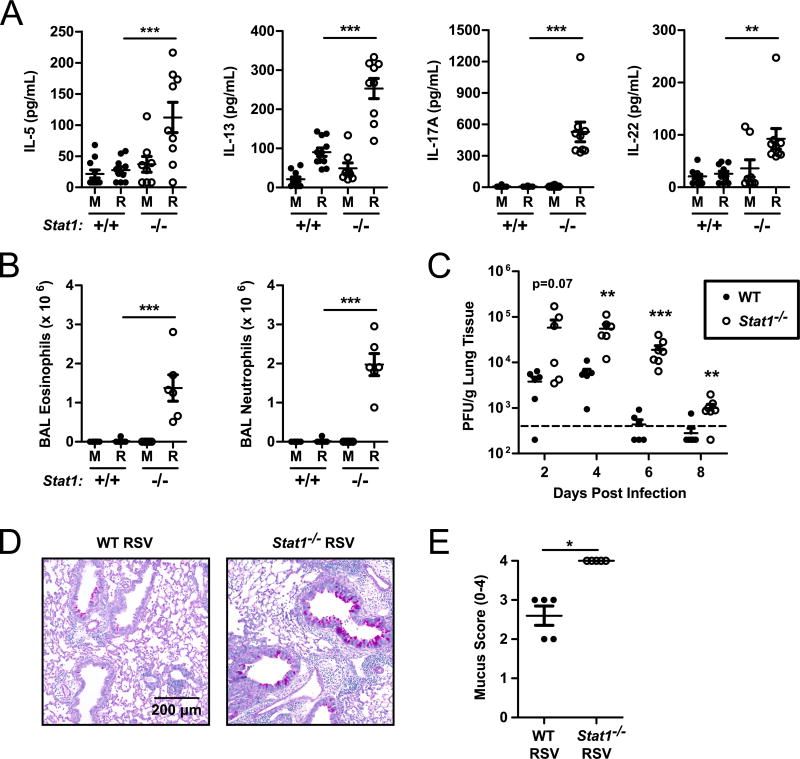
Concurrent with the increases observed in cytokine-producing ILC2 and ILC3, we identified significantly increased expression of the cytokines IL-5, IL-13, and IL-17A at day 6 post-infection in RSV-infected Stat1−/− mice compared to WT mice (Fig. 3 A). While unlikely to be the exclusive source of these cytokines, the increased expression of IL-5, IL-13, and IL-17A correlates with an increase in the number of cytokine-expressing ILC. We also identified robust BAL eosinophilia and neutrophilia and airway mucus accumulation at day 6 post-infection in RSV-infected Stat1−/− mice (Fig. 3 B, D, and E). Importantly, Stat1−/− mice exhibited poor viral control, as evidenced by both increased viral titers and delayed viral clearance compared to WT mice (Fig. 3 C). These cytokine, BAL, and viral load data are broadly consistent with prior reports of a dysregulated Th2/Th17-skewed immune response in Stat1−/− mice following RSV infection (35–42). Collectively, these data demonstrate a critical role for STAT1 signaling in promoting IFNγ+ ILC1 and restraining IL-5+ ILC2, IL-13+ ILC2, and IL-17A+ ILC3 during RSV infection that largely parallels the ineffective, pathologic immune response occurring in the lungs.
Figure 3. Stat1−/− mice displayed enhanced type 2 and type 17 immunity and poor viral clearance during RSV infection.
WT and Stat1−/− mice were infected with RSV or mock infected with vehicle and lungs were harvested on day 6 post-infection. (A) IL-5, IL-13, IL-17A, and IL-22 protein measured in the whole lung homogenate. (B) Total number of eosinophils and neutrophils in the bronchoalveolar lavage. (C) Viral load measured by plaque assay from lungs harvested on days 2, 4, 6, and 8 post-infection. (D) Representative airway sections stained with PAS and (E) quantification of PAS staining for multiple mice. Data are pooled from 2 independent experiments (A–C). Data were evaluated by one-way ANOVA (A, B) with the resultant comparisons shown or Student’s t-test (C and E) comparing WT and Stat1−/− mice on each day post-infection. **p<0.01 and ***p<0.001. M = mock-infected. R = RSV-infected.
Cell-intrinsic and cell-extrinsic factors contributed to ILC dysregulation in Stat1−/− mice
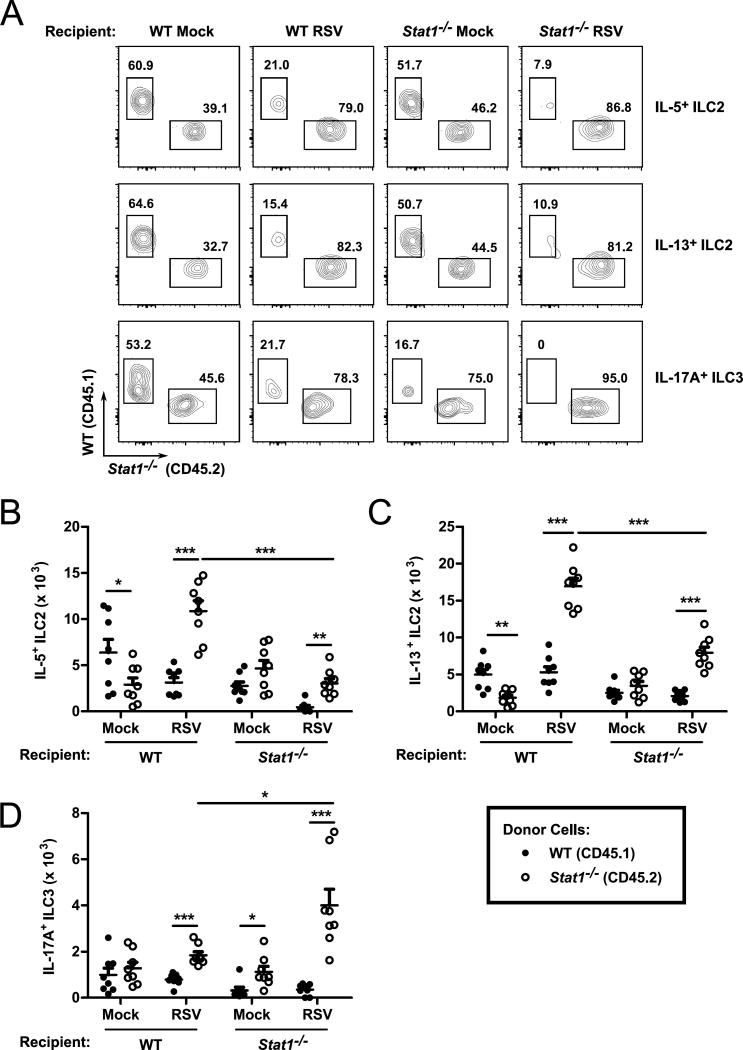
To interrogate whether cell-extrinsic factors contribute to the skewed cytokine-producing ILC response in Stat1−/− mice, we measured the number of WT and Stat1−/− ILC following RSV infection in a mixed bone marrow chimera model. WT (BALB/cJ) or Stat1−/− mice were lethally irradiated and reconstituted via retroorbital injection with a 1:1 mixture of WT CD45.1+ (CByJ.SJL[B6]-Ptprca/J) or Stat1−/− CD45.2+ whole bone marrow. Six weeks after reconstitution, mice were inoculated with RSV and the total numbers of cytokine-producing WT and Stat1−/− ILC were enumerated at day 6 post-infection (Fig. 4). Mock-infected WT recipients had significantly more WT IL-5+ and IL-13+ ILC2 than Stat1−/− IL-5+ and IL-13+ ILC2, suggesting a potential basal advantage of WT ILC2 in the WT recipient background (Fig. 4 A–C). Upon infection, however, Stat1−/− IL-5+ and IL-13+ ILC2 were significantly increased compared to WT IL-5+ and IL-13+ ILC2, demonstrating a cell-intrinsic advantage of IL-5+ and IL-13+ Stat1−/− ILC2 during viral infection (Fig. 4 A–C).
Figure 4. ILC cell-intrinsic and cell-extrinsic factors contribute to the dysregulation of Stat1−/− ILC.
Six week old BALB/c mice were lethally irradiated and transplanted with a 1:1 mixture of WT Congenic (CD45.1) and Stat1−/− (CD45.2) bone marrow. Mice were reconstituted for 6 weeks, infected with RSV or mock infected with vehicle, and harvested six days post-infection. (A) Representative flow plots with the percentages of the parent gates (IL-5+ ILC2, IL-13+ ILC2, or IL-17A+ ILC3) as indicated. Aggregated data for the total number of (B) IL-5+ ILC2, (C) IL-13+ ILC2, and (D) IL-17A+ ILC3. Data are pooled from 2 independent experiments and analyzed by two-way ANOVA. *p<0.05, **p<0.01, and ***p<0.001.
In Stat1−/− recipient mice, both WT and Stat1−/− IL-5+ and IL-13+ ILC2 were found in similar numbers with mock infection (Fig. 4 A–C). Similar to WT recipient mice, Stat1−/− IL-5+ and IL-13+ ILC2 were disproportionately activated upon RSV infection compared to WT IL-5+ and IL-13+ ILC2, though notably to a lesser degree than in the WT recipient, suggesting additional factors that may be present in the WT mouse that further potentiate cytokine-producing ILC2 responses (Fig. 4 A–C). Collectively, these data indicate that cell-intrinsic differences are at least partially responsible for the enhanced responsiveness of Stat1−/− ILC2 during RSV infection.
We similarly evaluated the number of IL-17A+ ILC3 in our mixed bone marrow chimera model (Fig. 4 A and D). In WT recipients, we found equal numbers of WT and Stat1−/− IL-17A+ ILC3 suggesting comparable fitness of these cells in the absence of a viral insult. Upon RSV infection, we identified significantly more Stat1−/− IL-17A+ ILC3 than WT IL-17A+ ILC3, though the number of Stat1−/− IL-17A+ ILC3 was comparable between mock and RSV infection in the WT recipient. In Stat1−/− recipients, Stat1−/− IL-17A+ ILC3 were found in higher frequencies in both mock and RSV-infected mice, with particularly robust expansion following RSV infection. In contrast, WT IL-17A+ ILC3 did not expand following RSV infection in the Stat1−/− recipients (Fig. 4 A and D). These data suggest that Stat1−/− IL-17A+ ILC3 have a cell-intrinsic advantage over WT cells and that this at least partially contributes to the dysregulation of IL-17A+ ILC3 during RSV infection. Moreover, these data suggest that an additional cell-extrinsic factor(s) present in the Stat1−/− recipient may be required for dysregulation of the ILC3 population.
Expression of IL-33 and IL-23 is increased during viral infection in Stat1−/− mice
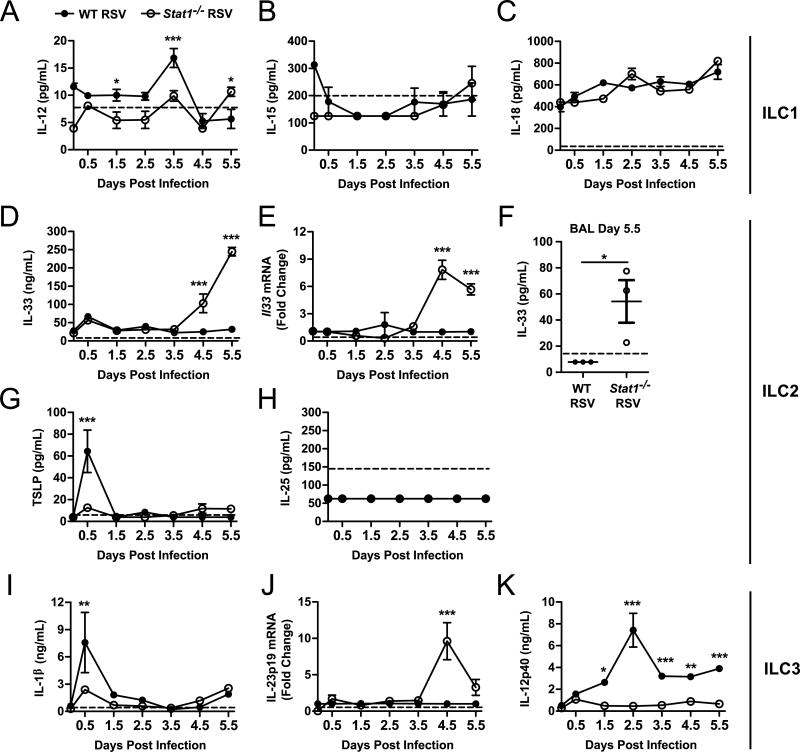
Our bone marrow chimera experiments suggest that both cell-intrinsic and cell-extrinsic factors may play a role in the dysregulation of cytokine-producing ILC during viral infection. Prior reports suggest that ILC2 are intrinsically susceptible to inhibition to type I and II interferons, providing a likely explanation for the cell-intrinsic effect seen in our bone marrow chimeric mice (34, 43–46) . However, STAT1-dependent cell-extrinsic factors that regulate ILC frequencies have not been described. The cytokine milieu is critical for the activation of ILC. We hypothesized that pro-ILC stimulatory cytokines were differentially regulated in Stat1−/− mice compared to WT mice. We infected WT and Stat1−/− mice with RSV and measured the expression of IL-12, IL-15, IL-18, IL-33, TSLP, IL-25, IL-1β, IL-23p19, and IL-12/IL-23p40 in the whole lung homogenate daily for 5 days beginning 12 hours post-infection. All cytokines were measured by ELISA except IL-23, which was measured by quantitative PCR due to lack of sensitive and specific protein-based reagents. Strikingly, there was a delayed but substantial increase in IL-33 protein and mRNA in the whole lung homogenates, beginning at day 4.5 post-infection and continuing through day 5.5 post-infection with a statistically significant nearly tenfold increase in Stat1−/− mice compared to WT mice (Fig. 5 D and E). IL-33 is stored as preformed cytokine in the nucleus of cells. To assess whether this increased expression of IL-33 in the whole lung homogenate led to an increase in the extracellularly-available IL-33, we measured IL-33 concentrations in the BAL. We identified a significantly increased concentration of IL-33 in the BAL of RSV-infected Stat1−/− mice compared to RSV-infected WT mice at day 5.5 post-infection, when peak levels of IL-33 were observed in the whole lung homogenate, indicating that IL-33 was being released into the airways (Fig. 5 F).
Figure 5. Pro-ILC2 and ILC3 stimulatory cytokines are increased in Stat1−/− mice during viral infection.
WT and Stat1−/− mice were infected with RSV or mock-infected and lungs were harvested for cytokine measurements by ELISA or quantitative PCR. (A–C) Pro-ILC1 cytokines, (D–H) pro-ILC2 cytokines, and (I–K) pro-ILC3 cytokines. Measurements were collected from whole lung homogenates for all samples except (F) which was taken from the BAL. Data in (A–E) and (G–K) are representative of 2 similar experiments with 3 mice per group per time point and analyzed by two-way ANOVA. (F) Data analyzed by Student’s t test. *p<0.05, **p<0.01, and ***p<0.001.
Similar to IL-33, we observed a statistically significant tenfold increase in Il23p19 expression in Stat1−/− mice compared to WT mice, peaking at day 4.5 post-infection (Fig. 5 J). IL-23 is a heterodimer formed between the IL-23p19 subunit and the IL-12/IL-23p40 subunit. IL-12/IL-23p40 was found in significantly lower levels throughout infection in Stat1−/− mice compared to WT mice, though levels post-infection in Stat1−/− mice were universally higher than those at the time of infection suggesting an increased availability of the IL-12/IL-23p40 subunit for complexing with IL-23p19 and forming an active cytokine (Fig. 5 K). IL-12, TSLP, and IL-1β were marginally, but statistically significantly, higher in WT mice at days 3.5, 0.5, and 0.5, respectively (Fig. 5 A, G, and I). Expression of IL-15, IL-18, and IL-25 was absent or only detected at very low levels (Fig. 5 B, C, and H). Given the central role of IL-33 and IL-23 in the activation of ILC2 and ILC3, respectively, these data suggest an ILC cell-extrinsic mechanism for their dysregulation during viral infection in Stat1−/− mice. Importantly, the expression of IL-33 and IL-23 coincided with the onset of ILC dysregulation in Stat1−/− mice, which occurred between days 3 and 6 post-infection (Fig. 1 A–C).
Disruption of IL-33 and IL-23 signaling inhibits cytokine-producing ILC2 and ILC3 activation in Stat1−/− mice
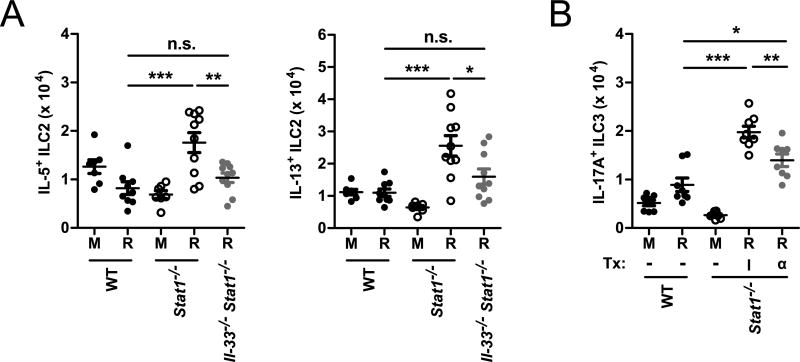
To determine the role of increased IL-33 expression during RSV infection in Stat1−/− mice, we generated Il-33−/− Stat1−/− double knockout mice. We infected WT, Stat1−/−, and Il-33−/− Stat1−/− mice with RSV or vehicle and measured the number of ILC2. RSV-infected Stat1−/− mice had significantly increased numbers of IL-5+ and IL-13+ ILC2 compared to RSV-infected WT mice (Fig. 6 A). However, compared to RSV-infected Stat1−/− mice, Il-33−/− Stat1−/− mice had significantly decreased total numbers of IL-5+ and IL-13+ ILC2 (Fig. 6 A). These data suggest that increased production of IL-33 contributes to the enhanced activity of IL-5+ and IL-13+ ILC2 in the Stat1−/− mice compared to WT mice during viral infection.
Figure 6. IL-33 and IL-23 are cell-extrinsic regulators of ILC that are dysregulated in Stat1−/− mice and promote enhanced ILC2 and ILC3 responsiveness.
(A) WT, Stat1−/−, and Il-33−/− Stat1−/− mice were infected with RSV or mock-infected and the total number of IL-5+ ILC2 and IL-13+ ILC2 were enumerated by flow cytometry at day 6 post-infection. (B) WT and Stat1−/− mice were infected with RSV or mock-infected and treated with 250 μg of an anti-IL-23p19 neutralizing antibody (α) or isotype control (I) on day 0 and day 3 post-infection. The total number of IL-17A+ ILC3 was enumerated by flow cytometry at day 6 post-infection. Data are pooled from 2 independent experiments and evaluated by one-way ANOVA. *p<0.05, **p<0.01, and ***p<0.001. n.s. = not significant. M = mock-infected. R = RSV-infected.
Similarly, we sought to determine the role of differential IL-23 expression during RSV infection in Stat1−/− mice. To assess this, we measured the number of IL-17A+ ILC3 in RSV-infected Stat1−/− mice treated with an anti-IL-23p19 neutralizing antibody or an isotype control. RSV-infected Stat1−/− mice had significantly increased numbers of IL-17A+ ILC3 compared to RSV-infected WT mice (Fig. 6 B). However, compared to isotype-treated Stat1−/− mice, Stat1−/− mice treated with an anti-IL-23p19 neutralizing antibody had significantly decreased total numbers of IL-17A+ ILC3 (Fig. 6 B). RSV-infected anti-IL-23p19-treated Stat1−/− mice still had a significantly increased number of IL-17A+ ILC3 compared to WT RSV-infected mice, consistent with our bone marrow chimera experiments that suggested both cell-intrinsic and cell-extrinsic factors play a role in the dysregulation of IL-17A+ ILC3 during RSV infection (Fig. 4 A, D and Fig. 6 B). Collectively, these data suggest that increased production of IL-23 contributes to the enhanced activity of IL-17A+ ILC3 in Stat1−/− mice compared to WT mice during viral infection.
IL-17A+ ILC3 express receptors for type I and II interferons and IL-27
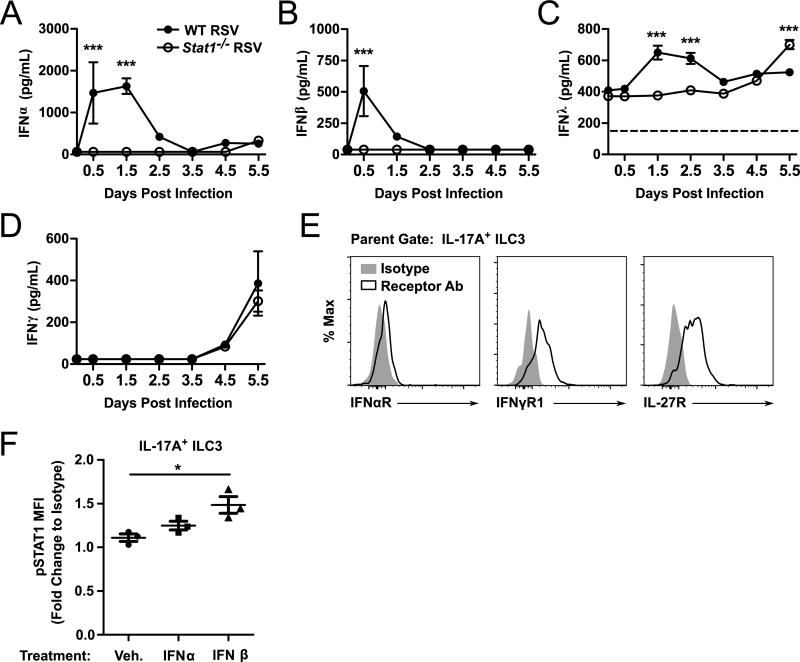
ILC2 express interferon receptors and respond directly to interferons(34, 45). We sought to determine whether IL-17A+ ILC3 also express interferon receptors. Initially, we evaluated for the expression of type I, II, and III IFNs in the first 5.5 days of RSV infection (Fig. 7 A–D). We found that WT mice produced significantly more IFNα, IFNβ, and IFNλ during the first several days of RSV infection compared to Stat1−/− mice. However, by day 5.5 post-infection, IFNλ was detected in significantly higher levels in the lungs of Stat1−/− mice compared to WT mice. No significant differences in IFNγ expression were observed throughout the first 5.5 days of infection between WT and Stat1−/− mice. We next assayed whether IL-17A+ ILC3 expressed receptors for these interferon species. Compared to isotype control staining, IL-17A+ ILC3 had detectable expression of IFNαR, IFNγR1, and the IL-27R (Fig. 7 E). IL-27, like interferons, signals through STAT1. Finally, we determined whether IFN treatment of IL-17A+ ILC3 ex vivo could induce phosphorylation of STAT1 (pSTAT1) to an activated state. Compared to vehicle treatment, culturing bulk lung cells ex vivo with IFNβ for 1 hour significantly induced pSTAT1 in IL-17A+ ILC3 (Fig. 7 F). Similar phosphorylation was not observed with IFNα treatment, and data were equivocal with high degrees of experiment to experiment variability for IFNγ and IFNλ2 treatments (Fig. 7 F and data not shown).
Figure 7. ILC3 express receptors for type I and type II IFNs and IL-27.
WT and Stat1−/− mice were infected with RSV or mock-infected and lungs were harvested for cytokine measurements by ELISA. Shown are the concentrations of (A) IFNα, (B) IFNβ, (C) IFNγ, and (D) IFNλ in the whole lung homogenate. (E) Expression of IFNαR, IFNγR1, and IL-27R on IL-17A+ ILC3. (F) Phosphorylated STAT1 expression by flow cytometry in IL-17A+ ILC3 treated for 1 hour ex vivo with IFNα or IFNβ. Data are representative to 2–3 independent experiments (A–E) or combined from 3 independent experiments (F) and evaluated by two-way ANOVA (A–D) or one-way ANOVA (F). *p<0.05 and ***p<0.001.
DISCUSSION
The appropriate orchestration of immune cells is vital for promoting productive immune responses and restricting detrimental pathophysiology. Herein, we show that STAT1 is a broad regulator of cytokine-producing ILC responses in the context of viral infection, promoting anti-viral IFNγ+ ILC1 and restricting IL-5+ and IL-13+ ILC2 and IL-17A+ ILC3. To this end, STAT1 represses cytokine-producing ILC2 and ILC3 via both cell-intrinsic and cell-extrinsic mechanisms. Extrinsically, STAT1-deficiency led to a significant increase in the expression of IL-33 and IL-23 that correlated strongly with increases in IL-5+ and IL-13+ ILC2 and IL-17A+ ILC3 in these mice. Furthermore, the genetic deletion of IL-33 or the neutralization of IL-23p19 in the context of STAT1-deficiency partially attenuated the dysregulated activation of cytokine-producing ILC2 and ILC3, respectively. Collectively, these data demonstrate that cytokine-producing ILC responses are coordinated by STAT1-dependent cell-intrinsic and cell-extrinsic mechanisms in the context of viral infection.
Previous studies have started to elucidate the direct role of interferon signaling on ILC2 responses. Specifically, these studies have shown that type I and type II interferons directly repress ILC2 responses in vitro and in vivo in the context of viral infection or airway inflammation (34, 43–46). Data from our bone marrow chimeric mouse experiments suggest cell-intrinsic factors, such as the previously described direct inhibition of ILC2 by interferons (34, 44, 45), play a role in regulating ILC frequencies during viral infection. Consistent with this literature, we identified that type I, II, and III IFNs are expressed in the lungs throughout RSV infection and that IL-17A+ ILC3 express receptors for type I and II IFNs as well as IL-27, another activator of STAT1. These data suggest that ILC3 may also be modulated directly by IFNs. Our chimeric experiments further suggest that cell-extrinsic factors regulate ILC frequencies during viral infection via a previously undefined STAT1-dependent mechanism. These data were particularly strong for IL-17A+ ILC3, and consistent with these data we observed an increase in Il23p19 during RSV infection in Stat1−/− mice. While our bone marrow chimera experiments did not specifically identify a cell-extrinsic effect for STAT1-deficiency in regulating ILC2, we observed significantly enhanced production of IL-33 in Stat1−/− compared to WT mice during RSV infection similar to Il23p19. Genetic deletion of Il33 in STAT1-deficient mice significantly reduced the number of IL-5+ and IL-13+ ILC2 during RSV infection. It is not entirely clear why a cell-extrinsic effect for cytokine-producing ILC2 was not observed in our bone marrow chimeric mice despite a clear effect of IL-33 in our other experiments. It is possible that changes in the cell phenotype such as a downregulation of the IL-33 receptor post-transplant altered their capacity to respond, or that factors present in the WT recipient background in combination with donor Stat1−/− ILC2 selectively potentiated these cells by a non-IL-33-dependent mechanism, masking our ability to detect a cell-extrinsic difference by this method. Regardless, our results using genetic deletion of Il33 in the context of STAT1-deficiency strongly suggest an important role for STAT1 repression of IL-33 in regulating cytokine-producing ILC2 responses.
Some other unexpected results were observed in our bone marrow chimera experiments. Specifically, WT donor IL-5+ ILC2 were observed at a high level in mock-infected WT recipient mice. This may be due to an advantage compared to other ILC subsets in reseeding the tissue following transplant. Additionally, Stat1−/− IL-5+ and IL-13+ ILC2 were more frequent in WT recipient mice compared to Stat1−/− recipient mice during RSV infection, suggesting that additional environmental factors present in WT mice may add to STAT1-deficiency in promoting cytokine-producing ILC2. Finally, RSV-infection did not expand the number of WT IL-13+ ILC2 as had been observed in our intact animals at day 6 post-infection shown in Figure 1B. The comprehensive network of activators and inhibitors of ILC in these mixed bone marrow chimeric mice will need to be further defined to better understand all of these data. However, broadly, these data in conjunction with previously published studies extend our understanding of the role of STAT1 signaling on the ILC response during viral infection.
A role for STAT1 in regulating IL-33 had been suggested previously, but these experiments were performed using ectopic expression of IL-33 and IFNγ in lung fibroblasts in the context of intact or disrupted STAT1 signaling.(47) Using an in vivo model of RSV infection, we are the first to show that endogenous STAT1 signaling is sufficient to negatively repress IL-33 expression and regulate immune cell numbers. Moreover, we found that STAT1 negatively regulated IL-23, consistent with our previous work.(41) Herein, we report for the first time that this repression of IL-23 by endogenous STAT1 attenuates the IL-17A+ ILC3 response to viral infection.
We identified an increase in the proliferation marker Ki67 in IL-13+ ILC2 and IL-17A+ ILC3, but not IL-5+ ILC2. It is possible that proliferative burst occurs earlier in the IL-5+ ILC2 compartment. Alternatively, cytokine-producing ILC2 and ILC3 may also arise from the activation of existing quiescent ILC in the tissue, differentiation of new ILC in situ, recruitment of ILC from other tissues, or the shifting of one ILC class to another. Additional investigative approaches will be necessary to better understand how these cytokine-producing ILC2 and ILC3 are accumulating in the lungs of Stat1−/− mice during RSV infection.
ILC subsets have been shown to participate in a variety of physiologic and pathophysiologic processes. Accordingly, significant attention has been paid to the identification of activators and inhibitors of ILC subsets that could be exploited therapeutically. For ILC2, such targets include lipid mediators PGI2 (inhibitory), PGD2 (activating), and LTD4 (activating) as well as cytokines including IL-33 and TSLP (3, 48–52). Retinoic acid has been shown to promote ILC3 in the gastrointestinal tract, and accordingly dietary modification of retinoic acid derivatives including vitamin A may present a viable therapeutic target (53). However, an effective and non-pathologic immune response often requires the coordinated activation and inhibition of multiple cell types. Targeting pathways that orchestrate this broader response may present a high-yield, efficacious approach to treating disease and promoting health. Our data suggest that STAT1 signaling represents such a pathway, promoting beneficial anti-viral ILC1 and repressing pathologic ILC2 and ILC3 responses by multiple mechanisms in the context of viral infection.
Endogenous mutations in STAT1 signaling alter disease susceptibility in humans. Chronic mucocutaneous candidiasis is a clinical syndrome characterized by recurrent infections of mucus membranes and skin with Candida species. A majority of these patients have been shown to have gain-of-function mutations in STAT1, which promote type 1 immune responses and repress type 17 responses, predisposing individuals to infection with fungal diseases (54–58). Additionally, loss-of-function mutations in STAT1 have also been identified that confer significant susceptibility to viral and mycobacterial infections (59). Analysis of CD4+ T helper subsets in the context of either gain-of function STAT1 mutations show a strong concordance between the STAT1 signaling and CD4+ T helper activity, with increases in STAT1 signaling promoting Th1 cells and restricting Th17 cells (56, 57). Our data suggest that ILC subsets are similarly regulated by STAT1 in mice, and it is intriguing to consider the role of ILC in human patients with mutations at the STAT1 locus, especially considering the prominence of ILC at mucosal sites.
Throughout, we focused our efforts on understanding the role of STAT1 signaling on cytokine-producing ILC, defining ILC1, ILC2, and ILC3 subsets stringently based on their capacity to produce canonical cytokines. While this allowed us to key in on the effects of STAT1 signaling on the major functional outputs of ILC, we were limited using this approach in our ability to understand the entirety of the ILC pool including both cytokine-producing and non-cytokine producing ILC. We chose to focus on cytokine production, as we found that during viral infection the expression of hallmark transcription factors, specifically GATA3, was not always present in IL-5 and IL-13 producing ILC. We reason that these cells may be ILC2 that have downregulated GATA3 as a negative feedback mechanism or possibly ex-ILC1/ILC3 that have shifted their cytokine production to a type 2 program but do not yet express detectable levels of GATA3 (data not shown). Similarly, it is unclear if the changes in cytokine-producing ILC are due to changing total numbers of ILC subsets, increases in the percentage of those subsets expressing cytokines, or other changes in the phenotypes of these cells. Such analyses are beyond the scope of this work, as our primary focus was on the production of type 1, 2, and 17 cytokines from ILC during viral infection.
Our data demonstrate that STAT1 is a chief regulator of cytokine-producing ILC1, ILC2, and ILC3 during viral infection. STAT1 promoted IFNγ+ ILC1 and acted to repress IL-5+ and IL-13+ ILC2 and IL-17A+ ILC3 via multiple mechanisms, both cell-intrinsic and cell-extrinsic. These data delineate a critical role of STAT1 in broadly orchestrating a productive cytokine-producing ILC response to viral infection.
Supplementary Material
Acknowledgments
Funding:
R01 AI 111820 – R. S. P.
R01 AI 124456 – R. S. P.
2I01 BX 000624 – R. S. P.
U19 AI 095227 – R. S. P.
R01 HL122554 – D. C. N.
R21 AI 121420 – D. C. N.
T32 GM 007347 – Vanderbilt MSTP
F30 A I114262 – M. T. S.
We greatly appreciate the support and assistance of the Vanderbilt Technologies for Advanced Genomics Core and the VMC Flow Cytometry Shared Resource.
References
- 1.Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat. Med. 2015;21:698–708. doi: 10.1038/nm.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J, Waddell A, Lin Y-D, Cantorna MT. Dysbiosis caused by vitamin D receptor deficiency confers colonization resistance to Citrobacter rodentium through modulation of innate lymphoid cells. Mucosal Immunol. 2015;8:618–626. doi: 10.1038/mi.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou W, Toki S, Zhang J, Goleniewksa K, Newcomb DC, Cephus JY, Dulek DE, Bloodworth MH, Stier MT, Polosuhkin V, Gangula RD, Mallal SA, Broide DH, Peebles RS. Prostaglandin I2 Signaling and Inhibition of Group 2 Innate Lymphoid Cell Responses. Am. J. Respir. Crit. Care Med. 2016;193:31–42. doi: 10.1164/rccm.201410-1793OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki M, Morita R, Hirata Y, Shichita T, Yoshimura A. Spred1, a Suppressor of the Ras-ERK Pathway, Negatively Regulates Expansion and Function of Group 2 Innate Lymphoid Cells. J. Immunol. 2015;195:1273–1281. doi: 10.4049/jimmunol.1500531. [DOI] [PubMed] [Google Scholar]
- 5.Stier MT, Bloodworth MH, Toki S, Newcomb DC, Goleniewska K, Boyd KL, Quitalig M, Hotard AL, Moore ML, Hartert TV, Zhou B, McKenzie AN, Peebles RS. Respiratory syncytial virus infection activates IL-13-producing group 2 innate lymphoid cells through thymic stromal lymphopoietin. J. Allergy Clin. Immunol. 2016;138:814–824.e11. doi: 10.1016/j.jaci.2016.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Y-J, Kim HY, Albacker LA, Baumgarth N, McKenzie ANJ, Smith DE, Dekruyff RH, Umetsu DT. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat. Immunol. 2011;12:631–638. doi: 10.1038/ni.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong JY, Bentley JK, Chung Y, Lei J, Steenrod JM, Chen Q, Sajjan US, Hershenson MB. Neonatal rhinovirus induces mucous metaplasia and airways hyperresponsiveness through IL-25 and type 2 innate lymphoid cells. J. Allergy Clin. Immunol. 2014;134:429–439. doi: 10.1016/j.jaci.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson DJ, Makrinioti H, Rana BMJ, Shamji BWH, Trujillo-Torralbo M-B, Footitt J, Jerico Del-Rosario, Telcian AG, Nikonova A, Zhu J, Aniscenko J, Gogsadze L, Bakhsoliani E, Traub S, Dhariwal J, Porter J, Hunt D, Hunt T, Hunt T, Stanciu LA, Khaitov M, Bartlett NW, Edwards MR, Kon OM, Mallia P, Papadopoulos NG, Akdis CA, Westwick J, Edwards MJ, Cousins DJ, Walton RP, Johnston SL. IL-33-Dependent Type 2 Inflammation during Rhinovirus-induced Asthma Exacerbations In Vivo. Am. J. Respir. Crit. Care Med. 2014;190:1373–1382. doi: 10.1164/rccm.201406-1039OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorski SA, Hahn YS, Braciale TJ. Group 2 innate lymphoid cell production of IL-5 is regulated by NKT cells during influenza virus infection. PLoS Pathog. 2013;9:e1003615. doi: 10.1371/journal.ppat.1003615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alwan WH, Kozlowska WJ, Openshaw PJ. Distinct types of lung disease caused by functional subsets of antiviral T cells. J. Exp. Med. 1994;179:81–89. doi: 10.1084/jem.179.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham MB, Braciale VL, Braciale TJ. Influenza virus-specific CD4+ T helper type 2 T lymphocytes do not promote recovery from experimental virus infection. J. Exp. Med. 1994;180:1273–1282. doi: 10.1084/jem.180.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maloy KJ, Burkhart C, Junt TM, Odermatt B, Oxenius A, Piali L, Zinkernagel RM, Hengartner H. CD4(+) T cell subsets during virus infection. Protective capacity depends on effector cytokine secretion and on migratory capability. J. Exp. Med. 2000;191:2159–2170. doi: 10.1084/jem.191.12.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu X, McCoy KS, Xu J, Hu W, Chen H, Jiang K, Han F, Chen P, Wang Y. Galectin-9 ameliorates respiratory syncytial virus-induced pulmonary immunopathology through regulating the balance between Th17 and regulatory T cells. Virus Res. 2015;195:162–171. doi: 10.1016/j.virusres.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Kulcsar KA, Baxter VK, Greene IP, Griffin DE. Interleukin 10 modulation of pathogenic Th17 cells during fatal alphavirus encephalomyelitis. Proc. Natl. Acad. Sci. U. S. A. 2014;111:16053–16058. doi: 10.1073/pnas.1418966111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gil-Cruz C, Perez-Shibayama C, Onder L, Chai Q, Cupovic J, Cheng H-W, Novkovic M, Lang PA, Geuking MB, McCoy KD, Abe S, Cui G, Ikuta K, Scandella E, Ludewig B. Fibroblastic reticular cells regulate intestinal inflammation via IL-15-mediated control of group 1 ILCs. Nat. Immunol. 2016;17:1388–1396. doi: 10.1038/ni.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silver JS, Kearley J, Copenhaver AM, Sanden C, Mori M, Yu L, Pritchard GH, Berlin AA, Hunter CA, Bowler R, Erjefalt JS, Kolbeck R, Humbles AA. Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat. Immunol. 2016;17:626–635. doi: 10.1038/ni.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. NATURAL KILLER CELLS IN ANTIVIRAL DEFENSE: Function and Regulation by Innate Cytokines. Annu. Rev. Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 18.De Weerd NA, Nguyen T. The interferons and their receptors--distribution and regulation. Immunol. Cell Biol. 2012;90:483–491. doi: 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parronchi P, De Carli M, Manetti R, Simonelli C, Sampognaro S, Piccinni MP, Macchia D, Maggi E, Del Prete G, Romagnani S. IL-4 and IFN (alpha and gamma) exert opposite regulatory effects on the development of cytolytic potential by Th1 or Th2 human T cell clones. J. Immunol. 1992;149:2977–2983. [PubMed] [Google Scholar]
- 20.Brinkmann V, Geiger T, Alkan S, Heusser CH. Interferon alpha increases the frequency of interferon gamma-producing human CD4+ T cells. J. Exp. Med. 1993;178:1655–1663. doi: 10.1084/jem.178.5.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogge L, Barberis-Maino L, Biffi M, Passini N, Presky DH, Gubler U, Sinigaglia F. Selective expression of an interleukin-12 receptor component by human T helper 1 cells. J. Exp. Med. 1997;185:825–831. doi: 10.1084/jem.185.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smeltz RB, Chen J, Ehrhardt R, Shevach EM. Role of IFN-gamma in Th1 differentiation: IFN-gamma regulates IL-18R alpha expression by preventing the negative effects of IL-4 and by inducing/maintaining IL-12 receptor beta 2 expression. J. Immunol. 2002;168:6165–6172. doi: 10.4049/jimmunol.168.12.6165. [DOI] [PubMed] [Google Scholar]
- 23.Scott P. IFN-gamma modulates the early development of Th1 and Th2 responses in a murine model of cutaneous leishmaniasis. J. Immunol. 1991;147:3149–3155. [PubMed] [Google Scholar]
- 24.Koltsida O, Hausding M, Stavropoulos A, Koch S, Tzelepis G, Ubel C, Kotenko SV, Sideras P, Lehr HA, Tepe M, Klucher KM, Doyle SE, Neurath MF, Finotto S, Andreakos E. IL-28A (IFN-λ2) modulates lung DC function to promote Th1 immune skewing and suppress allergic airway disease. EMBO Mol. Med. 2011;3:348–361. doi: 10.1002/emmm.201100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 26.Hardman CS, Panova V, McKenzie ANJ. IL-33 citrine reporter mice reveal the temporal and spatial expression of IL-33 during allergic lung inflammation. Eur. J. Immunol. 2013;43:488–498. doi: 10.1002/eji.201242863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durbin JE, Johnson TR, Durbin RK, Mertz SE, Morotti RA, Peebles RS, Graham BS. The role of IFN in respiratory syncytial virus pathogenesis. J. Immunol. 2002;168:2944–2952. doi: 10.4049/jimmunol.168.6.2944. [DOI] [PubMed] [Google Scholar]
- 28.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 29.Stokes KL, Chi MH, Sakamoto K, Newcomb DC, Currier MG, Huckabee MM, Lee S, Goleniewska K, Pretto C, Williams JV, Hotard A, Sherrill TP, Peebles RS, Moore ML. Differential pathogenesis of respiratory syncytial virus clinical isolates in BALB/c mice. J. Virol. 2011;85:5782–5793. doi: 10.1128/JVI.01693-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graham BS, Perkins MD, Wright PF, Karzon DT. Primary respiratory syncytial virus infection in mice. J. Med. Virol. 1988;26:153–162. doi: 10.1002/jmv.1890260207. [DOI] [PubMed] [Google Scholar]
- 31.Miller AL, Bowlin TL, Lukacs NW. Respiratory syncytial virus-induced chemokine production: linking viral replication to chemokine production in vitro and in vivo. J. Infect. Dis. 2004;189:1419–1430. doi: 10.1086/382958. [DOI] [PubMed] [Google Scholar]
- 32.Kim HY, Lee HJ, Chang Y-J, Pichavant M, Shore SA, Fitzgerald KA, Iwakura Y, Israel E, Bolger K, Faul J, DeKruyff RH, Umetsu DT. Interleukin-17-producing innate lymphoid cells and the NLRP3 inflammasome facilitate obesity-associated airway hyperreactivity. Nat. Med. 2013;20:54–61. doi: 10.1038/nm.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science. 2015;350:981–985. doi: 10.1126/science.aac9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moro K, Kabata H, Tanabe M, Koga S, Takeno N, Mochizuki M, Fukunaga K, Asano K, Betsuyaku T, Koyasu S. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat. Immunol. 2016;17:76–86. doi: 10.1038/ni.3309. [DOI] [PubMed] [Google Scholar]
- 35.Durbin JE, Johnson TR, Durbin RK, Mertz SE, Morotti RA, Peebles RS, Graham BS. The role of IFN in respiratory syncytial virus pathogenesis. J. Immunol. 2002;168:2944–2952. doi: 10.4049/jimmunol.168.6.2944. [DOI] [PubMed] [Google Scholar]
- 36.Moore ML, Newcomb DC, Parekh VV, Van Kaer L, Collins RD, Zhou W, Goleniewska K, Chi MH, Mitchell D, Boyce JA, Durbin JE, Sturkie C, Peebles RS. STAT1 negatively regulates lung basophil IL-4 expression induced by respiratory syncytial virus infection. J. Immunol. 2009;183:2016–2026. doi: 10.4049/jimmunol.0803167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peebles RS, Hashimoto K, Collins RD, Jarzecka K, Furlong J, Mitchell DB, Sheller JR, Graham BS. Immune interaction between respiratory syncytial virus infection and allergen sensitization critically depends on timing of challenges. J. Infect. Dis. 2001;184:1374–1379. doi: 10.1086/324429. [DOI] [PubMed] [Google Scholar]
- 38.Newcomb DC, Boswell MG, Sherrill TP, Polosukhin VV, Boyd KL, Goleniewska K, Brody SL, Kolls JK, Adler KB, Peebles RS. IL-17A induces signal transducers and activators of transcription-6-independent airway mucous cell metaplasia. Am. J. Respir. Cell Mol. Biol. 2013;48:711–716. doi: 10.1165/rcmb.2013-0017OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newcomb DC, Boswell MG, Huckabee MM, Goleniewska K, Dulek DE, Reiss S, Lukacs NW, Kolls JK, Peebles RS. IL-13 regulates Th17 secretion of IL-17A in an IL-10-dependent manner. J. Immunol. 2012;188:1027–1035. doi: 10.4049/jimmunol.1102216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore ML, Chi MH, Goleniewska K, Durbin JE, Peebles RS. Differential regulation of GM1 and asialo-GM1 expression by T cells and natural killer (NK) cells in respiratory syncytial virus infection. Viral Immunol. 2008;21:327–339. doi: 10.1089/vim.2008.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashimoto K, Durbin JE, Zhou W, Collins RD, Ho SB, Kolls JK, Dubin PJ, Sheller JR, Goleniewska K, O’Neal JF, Olson SJ, Mitchell D, Graham BS, Peebles RS. Respiratory syncytial virus infection in the absence of STAT 1 results in airway dysfunction, airway mucus, and augmented IL-17 levels. J. Allergy Clin. Immunol. 2005;116:550–557. doi: 10.1016/j.jaci.2005.03.051. [DOI] [PubMed] [Google Scholar]
- 42.Johnson TR, Mertz SE, Gitiban N, Hammond S, Legallo R, Durbin RK, Durbin JE. Role for innate IFNs in determining respiratory syncytial virus immunopathology. J. Immunol. 2005;174:7234–7241. doi: 10.4049/jimmunol.174.11.7234. [DOI] [PubMed] [Google Scholar]
- 43.Han M, Hong JY, Jaipalli S, Rajput C, Lei J, Hinde JL, Chen Q, Hershenson NM, Bentley JK, Hershenson MB. IFN-γ Blocks Development of an Asthma Phenotype in Rhinovirus-infected Baby Mice by Inhibiting ILC2s. Am. J. Respir. Cell Mol. Biol. 2016;56:242–251. doi: 10.1165/rcmb.2016-0056OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molofsky AB, Van Gool F, Liang H-E, Van Dyken SJ, Nussbaum JC, Lee J, Bluestone JA, Locksley RM. Interleukin-33 and Interferon-γ Counter-Regulate Group 2 Innate Lymphoid Cell Activation during Immune Perturbation. Immunity. 2015;43:161–174. doi: 10.1016/j.immuni.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duerr CU, McCarthy CDA, Mindt BC, Rubio M, Meli AP, Pothlichet J, Eva MM, Gauchat J-F, Qureshi ST, Mazer BD, Mossman KL, Malo D, Gamero AM, Vidal SM, King IL, Sarfati M, Fritz JH. Type I interferon restricts type 2 immunopathology through the regulation of group 2 innate lymphoid cells. Nat. Immunol. 2015;17:65–75. doi: 10.1038/ni.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bi J, Cui L, Yu G, Yang X, Chen Y, Wan X. NK Cells Alleviate Lung Inflammation by Negatively Regulating Group 2 Innate Lymphoid Cells. J. Immunol. 2017;198:3336–3344. doi: 10.4049/jimmunol.1601830. [DOI] [PubMed] [Google Scholar]
- 47.Kopach P, Lockatell V, Pickering EM, Haskell RE, Anderson RD, Hasday JD, Todd NW, Luzina IG, Atamas SP. IFN- Directly Controls IL-33 Protein Level through a STAT1- and LMP2-dependent Mechanism. J. Biol. Chem. 2014;289:11829–11843. doi: 10.1074/jbc.M113.534396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doherty TA, Khorram N, Lund S, Mehta AK, Croft M, Broide DH. Lung type 2 innate lymphoid cells express cysteinyl leukotriene receptor 1, which regulates TH2 cytokine production. J. Allergy Clin. Immunol. 2013;132:205–213. doi: 10.1016/j.jaci.2013.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue L, Salimi M, Panse I, Mjösberg JM, McKenzie ANJ, Spits H, Klenerman P, Ogg G. Prostaglandin D2 activates group 2 innate lymphoid cells through chemoattractant receptor-homologous molecule expressed on TH2 cells. J. Allergy Clin. Immunol. 2014;133:1184–1194.e7. doi: 10.1016/j.jaci.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wojno DET, Monticelli LAV, Tran S, Alenghat T, Osborne LC, Thome JJ, Willis C, Budelsky A, Farber DL, Artis D. The prostaglandin D2 receptor CRTH2 regulates accumulation of group 2 innate lymphoid cells in the inflamed lung. Mucosal Immunol. 2015;8:1313–1323. doi: 10.1038/mi.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TKA, Bucks C, Kane CM, Fallon PG, Pannell R, Jolin HE, McKenzie ANJ. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halim YTF, Krauss RH, Sun AC, Takei F. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity. 2012;36:451–463. doi: 10.1016/j.immuni.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 53.Goverse G, Labao-Almeida C, Ferreira M, Molenaar R, Wahlen S, Konijn T, Koning J, Veiga-Fernandes H, Mebius RE. Vitamin A Controls the Presence of RORγ+ Innate Lymphoid Cells and Lymphoid Tissue in the Small Intestine. J. Immunol. 2016;196:5148–5155. doi: 10.4049/jimmunol.1501106. [DOI] [PubMed] [Google Scholar]
- 54.Depner M, Fuchs S, Raabe J, Frede N, Glocker C, Doffinger R, Gkrania-Klotsas E, Kumararatne D, Atkinson TP, Schroeder HW, Niehues T, Dückers G, Stray-Pedersen A, Baumann U, Schmidt R, Franco JL, Orrego J, Ben-Shoshan M, McCusker C, Jacob CMA, Carneiro-Sampaio M, Devlin LA, Edgar JDM, Henderson P, Russell RK, Skytte A-B, Seneviratne SL, Wanders J, Stauss H, Meyts I, Moens L, Jesenak M, Kobbe R, Borte S, Borte M, Wright DA, Hagin D, Torgerson TR, Grimbacher B. The Extended Clinical Phenotype of 26 Patients with Chronic Mucocutaneous Candidiasis due to Gain-of-Function Mutations in STAT1. J. Clin. Immunol. 2016;36:73–84. doi: 10.1007/s10875-015-0214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sampaio EP, Hsu AP, Pechacek J, Bax HI, Dias DL, Paulson ML, Chandrasekaran P, Rosen LB, Carvalho DS, Ding L, Vinh DC, Browne SK, Datta S, Milner JD, Kuhns DB, Long Priel DA, Sadat MA, Shiloh M, De Marco B, Alvares M, Gillman JW, Ramarathnam V, de la Morena M, Bezrodnik L, Moreira I, Uzel G, Johnson D, Spalding C, Zerbe CS, Wiley H, Greenberg DE, Hoover SE, Rosenzweig SD, Galgiani JN, Holland SM. Signal transducer and activator of transcription 1 (STAT1) gain-of-function mutations and disseminated coccidioidomycosis and histoplasmosis. J. Allergy Clin. Immunol. 2013;131:1624–1634. doi: 10.1016/j.jaci.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uzel G, Sampaio EP, Lawrence MG, Hsu AP, Hackett M, Dorsey MJ, Noel RJ, Verbsky JW, Freeman AF, Janssen E, Bonilla FA, Pechacek J, Chandrasekaran P, Browne SK, Agharahimi A, Gharib AM, Mannurita SC, Yim JJ, Gambineri E, Torgerson T, Tran DQ, Milner JD, Holland SM. Dominant gain-of-function STAT1 mutations in FOXP3 wild-type immune dysregulation-polyendocrinopathy-enteropathy-X-linked-like syndrome. J. Allergy Clin. Immunol. 2013;131:1611–1623. doi: 10.1016/j.jaci.2012.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu L, Okada S, Kong X-F, Kreins AY, Cypowyj S, Abhyankar A, Toubiana J, Itan Y, Audry M, Nitschke P, Masson C, Toth B, Flatot J, Migaud M, Chrabieh M, Kochetkov T, Bolze A, Borghesi A, Toulon A, Hiller J, Eyerich S, Eyerich K, Gulácsy V, Chernyshova L, Chernyshov V, Bondarenko A, Grimaldo RMC, Blancas-Galicia L, Beas IMM, Roesler J, Magdorf K, Engelhard D, Thumerelle C, Burgel P-R, Hoernes M, Drexel B, Seger R, Kusuma T, Jansson AF, Sawalle-Belohradsky J, Belohradsky B, Jouanguy E, Bustamante J, Bué M, Karin N, Wildbaum G, Bodemer C, Lortholary O, Fischer A, Blanche S, Al-Muhsen S, Reichenbach J, Kobayashi M, Rosales FE, Lozano CT, Kilic SS, Oleastro M, Etzioni A, Traidl-Hoffmann C, Renner ED, Abel L, Picard C, Maródi L, Boisson-Dupuis S, Puel A, Casanova J-L. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J. Exp. Med. 2011;208:1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toubiana J, Okada S, Hiller J, Oleastro M, Lagos Gomez M, Aldave Becerra JC, Ouachée-Chardin M, Fouyssac F, Girisha KM, Etzioni A, Van Montfrans J, Camcioglu Y, Kerns LA, Belohradsky B, Blanche S, Bousfiha A, Rodriguez-Gallego C, Meyts I, Kisand K, Reichenbach J, Renner ED, Rosenzweig S, Grimbacher B, van de Veerdonk FL, Traidl-Hoffmann C, Picard C, Marodi L, Morio T, Kobayashi M, Lilic D, Milner JD, Holland S, Casanova J-L, Puel A International STAT1 Gain-of-Function Study Group. Heterozygous STAT1 gain-of-function mutations underlie an unexpectedly broad clinical phenotype. Blood. 2016;127:3154–3164. doi: 10.1182/blood-2015-11-679902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boisson-Dupuis S, Kong X-F, Okada S, Cypowyj S, Puel A, Abel L, Casanova J-L. Inborn errors of human STAT1: allelic heterogeneity governs the diversity of immunological and infectious phenotypes. Curr. Opin. Immunol. 2012;24:364–378. doi: 10.1016/j.coi.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.