Abstract
The mitochondrial ATP synthase, subunit c, isoform 3 gene (Atp5g3) encodes subunit 9, the subunit of the multisubunit enzyme that catalyzes ATP synthesis during oxidative phosphorylation in mitochondria. According to the Ensembl database, Atp5g3 in mice is located on chromosome 2 between 73746504 and 73749383 bp, within the genomic regions of two sets of quantitative trait loci - alcohol preference and body weight. Both of those traits are more influenced by epigenetic factors than many other traits are. Using currently available phenotype and gene expression profiles from the GeneNetwork database, we obtained correlations between Atp5g3 and alcoholism- and obesity-relevant phenotypes. The correlation in expression levels between Atp5g3 and each of its 12 partner genes in the molecular interaction are different in various tissues and genes. Transcriptome mapping indicated that Atp5g3 is differentially regulated in the hippocampus, cerebellum, and liver. Owing to a lack of known polymorphisms of Atp5g3 among three relevant mouse strains, C57BL/6J (B6), DBA/2J (D2), and BALB/ cJ, the molecular mechanism for the connection between Atp5g3 and alcoholism and body weight requires further investigation.
Keywords: Atp5g3, Alcoholism, Epigenetic, Expression, Mouse, Transcriptome
INTRODUCTION
The mitochondrial ATP synthase, subunit c, isoform 3 gene (Atp5g3) encodes subunit 9, the subunit of the multisubunit enzyme that catalyzes ATP synthesis during oxidative phosphorylation in mitochondria. Each ATP synthase complex has multiple copies of subunit 9 in the transmembrane portion of the complex. Although the molecular mechanisms in the mitochondrial membrane ATP synthase are known, its connection to other pathways or human disorders remains unknown.
According to the Ensembl database, the Atp5g3 gene in mice is located on chromosome 2, between 73746504 and 73749383 bp. Interestingly, two sets of quantitative trait loci (QTLs) have been mapped into this chromosomal region. One is body weight (Mollah and Ishikawa, 2011) and its relevant traits fat distribution (Kobayashi et al., 2010) and obesity (Jerez-Timaure et al., 2004; Farber and Medrano, 2007). The other is alcohol preference (Ruf et al., 2004; Boyle and Gill, 2008). Both alcoholism and obesity are common, late-onset, chronic disorders that are strongly influenced by epigenetic factors (Buscemi and Turchi et al., 2011; Marti and Ordovas, 2011; Slomko et al., 2012). None of the causal genes from these two QTLs has been determined, although some candidate genes have been named.
Atp5g3 is located in the genomic regions of these QTLs but has not been considered a candidate gene. However, based on limited evidence, we hypothesized that Atp5g3 involves molecular pathways that regulate the alcohol preference QTL. Li et al. (2001) have reported that all three nuclear gene isoforms (ATP5G1, ATP5G2, and ATP5G3) were consistently upregulated, parallel with mitochondrial injury, in the pancreas of alcohol-consuming rats. No additional reports on their potential functions in regulating alcoholism, body weight, or obesity have been published to date, however. The genes that regulate the alcohol and obesity QTLs remain unknown. Rapid progress in genetic technology has yielded results that suggest an important role for epigenetics in the regulation of disorders and physiologic abnormalities; therefore, examinations of the potential role of Atp5g3 in the regulation of these 2 QTLs are important.
Rapid accumulation of genomic and bioinformatic data in recent years has provided enough resources to evaluate the potential function of the many genes regulating known phenotypes. For example, by October 5, 2011, the GeneNetwork database (http://www.GeneNetwork.org [accessed October 13, 2012]) had accumulated several hundred records for phenotyping (Williams et al., 2001; Chesler et al., 2004). Although more than 70% of those records were for drug/alcohol addiction and behavior, hundreds of records for other phenotypes have been investigated and added into the GeneNetwork database. An enormous advantage of using these data is the availability of gene expression profiles and transcriptome data from 18 tissues, including those of the brain, liver, kidney, lung, spleen, eye, cartilage, hippocampus, cerebellum, striatum, and neocortex and prefrontal cortex. This review exploits genomic and phenotypic resources from GeneNetwork and others sources to evaluate the possibility that Atp5g3 is an epigenetic factor for the QTLs of alcoholism or body weight and obesity.
MATERIAL AND METHODS
Atp5g3 expression in various tissues in BXD recombinant inbred mice
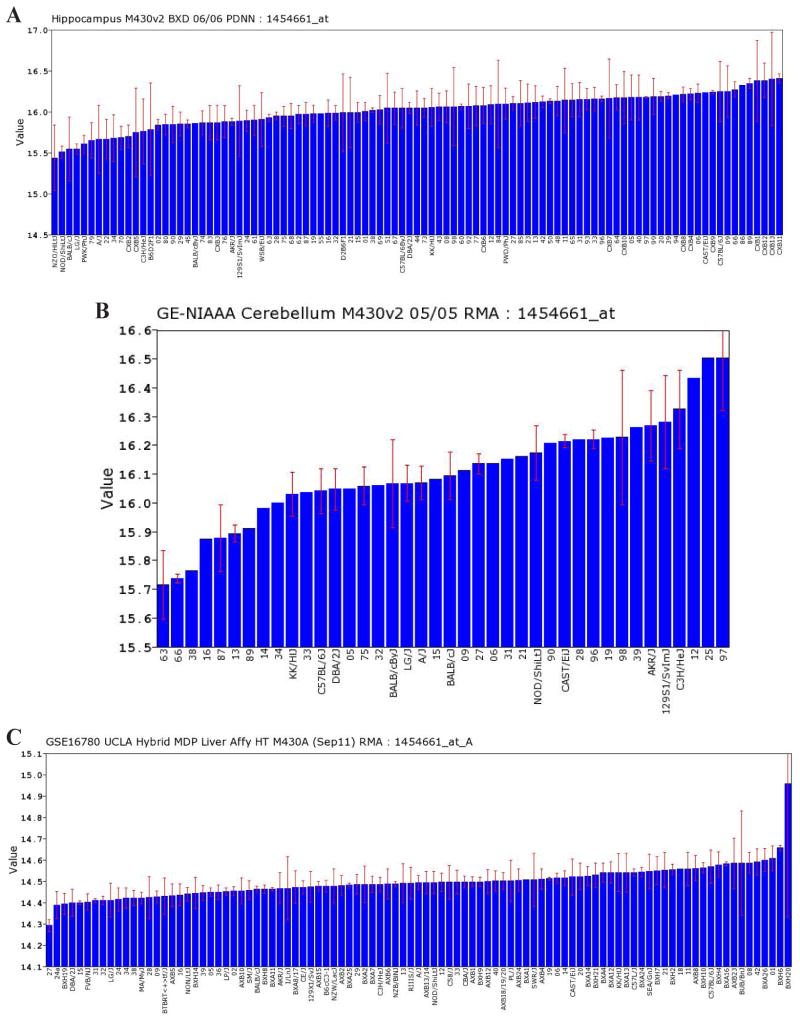
Using data from GeneNetwork, we first examined the expression level of Atp5g3 in the tissues of three organs - the hippocampus, cerebellum and liver - that are relevant to alcoholism. As shown in Figure 1, Atp5g3 is expressed in all of these tissues. The expression of Atp5g3 in the hippocampus was obtained from a data set from the Hippocampus Consortium M430v2 (Overall et al., 2009), which provided estimates of messenger RNA expression in the adult hippocampus from 67 BXD recombinant inbred strains. We used the data set GE-NIAAA Cerebellum messenger RNA M430v2 [http://www.genenetwork.org/webqtl/main.py (accessed October 13, 20120)] to examine the expression level of Atp5g3 in the cerebellum and the data set GSE16780 UCLA Hybrid MDP Liver Affy HT M430A [http://www.genenetwork.org/dbdoc/GSE16780_UCLA_ML0911.html (accessed October 13, 2012)] to examine the expression level of Atp5g3 in the liver. The relative expression level in the hippocampus and cerebellum was higher than that in the liver. Moreover, variations occurred among mouse strains in the studied tissues. The variation of expression level among mouse strains in the hippocampus and cerebellum was also larger than that in the liver.
Figure 1.
Bar graphs of expression levels of Atp5g3 in three different tissues in different strains of mice. Expression levels are relative LOG values as explained in GeneNetwork (http://www.genenetwork.org/webqtl/main.py). A. Hippocampus; B. cerebellum; C. liver.
RESULTS AND DISCUSSION
correlation between Atp5g3 expression level and phenotypes in the GeneNetwork database
We examined the correlation between expression levels of Atp5g3 and records of phenotypes in three tissues. Current data from a limited number of strains suggested potential associations between Atp5g3 expression and alcohol- and obesity-relevant phenotypes.
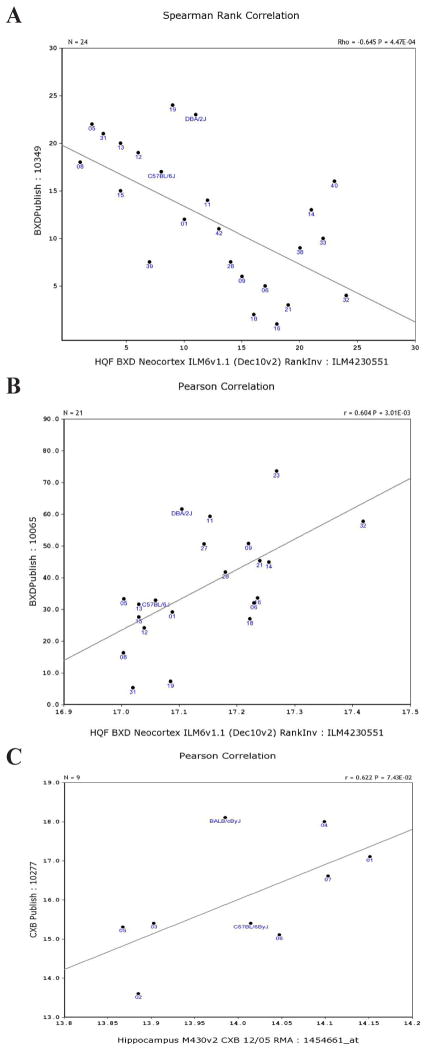
1) Phenotypes and the expression of Atp5g3 in the hippocampus. Our correlation analysis indicated that gene expression in the hippocampus was negatively correlated with ethanol response (1.75 mg/kg ip) according to the time to ataxia measured as loss of balance using a dowel test [loss corresponded to blood ethyl alcohol content time 0 (min)] (Kirstein et al., 2002). As shown in Figure 2A, the correlation was R = −0.6795. By contrast, Atp5g3 expression was positively correlated with long-term ethanol response measured by chronic withdrawal and handling-induced convulsion score (Crabbe, 1998). As shown in Figure 2B, the correlation was R = 0.6039. In addition, the Atp5g3 expression in the hippocampus was positively correlated to body weight in CXB (BALB/cBy X C57BL/6By) population strains [http://www.genenetwork.org/dbdoc/CXBPublish.html (accessed October 13, 2012); Figure 2C].
Figure 2.
Correlations between Atp5g3 expression levels in hippocampus and phenotypes. Spearman rank order correlations were obtained between the Atp5g3 expression levels and phenotypes in the GeneNetwork database. A. Response negatively correlated to ethanol (1.75 mg/kg ip), according to the time to ataxia. B. Positive correlation with long-term ethanol response measured by chronic withdrawal. C. Positively correlated to body weight.
2) Phenotypes and expression of Atp5g3 in the cerebellum. We found that the expression level of Atp5g3 was highly positively correlated to ethanol preference in males, as measured by residual mean [g/kg body weight (Fernandez et al., 1999)], with an R score as high as 0.8512 (Figure 3A). Surprisingly, Atp5g3 was also highly positively related to a set of data marked obesity (R = 0.8433; gudrun.brockmann@agrar.huberlin.de; Figure 3B). However, because detailed information on the parameters of obesity in the data set has been unavailable, it is unclear to which obesity score Atp5g3 was correlated.
Figure 3.
Correlations between Atp5g3 expression levels in cerebellum and phenotypes. Spearman rank order correlations were obtained between the Atp5g3 expression levels and phenotypes in the GeneNetwork database. A. Positively correlated to ethanol preference in males. B. Positively related to obesity.
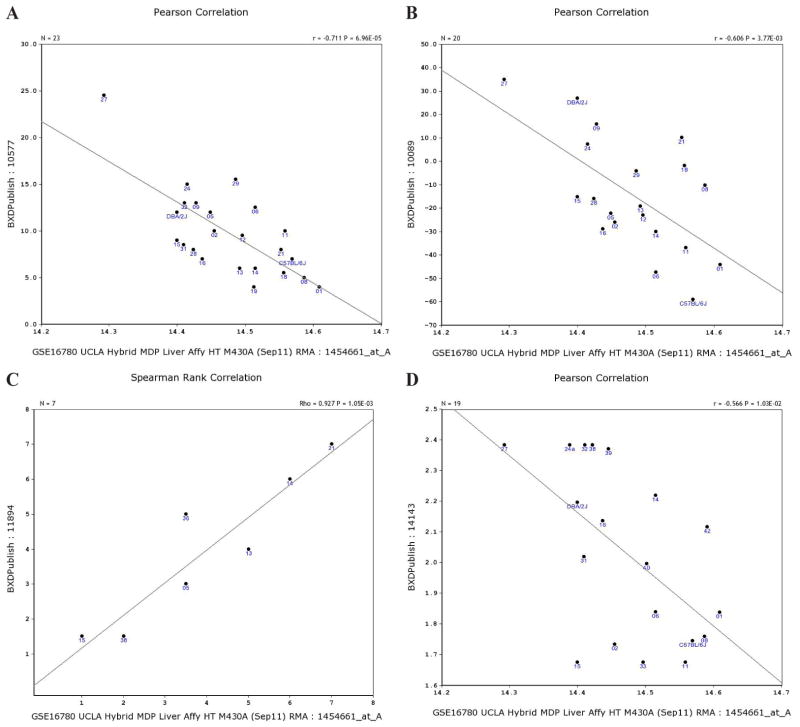
3) Phenotypes and expression of Atp5g3 in the liver. The expression of Atp5g3 in the liver was negatively correlated with corticosterone plasma levels measured 7 h after ethanol injection (μg/dL; Figure 4A) (Roberts et al., 1995). It was also negatively correlated with locomotor tolerance or sensitization in males (Figure 4B) (Cunningham, 1995). However, expression of Atp5g3 was highly positively correlated to the handling-induced convulsion score 7 h after ethanol injection in males and females (U; 4.0 g/kg ip; Figure 4C), R = 0.9553 (Philip et al., 2010). The expression of Atp5g3 was negatively correlated with the obesity-relevant phenotype measured by Brockmann et al. (http://www.genenetwork.org/webqtl/main.py. Contact information: gudrun.brockmann@agrar.huberlin.de) in males (Figure 4D).
Figure 4.
Correlations between Atp5g3 expression levels in liver and phenotypes. Spearman rank order correlations were obtained between the Atp5g3 expression levels and phenotypes in the GeneNetwork database. A. Negatively correlated to the corticosterone plasma level measured 7 h after ethanol injection. B. Negatively correlated locomotor tolerance or sensitization in males. C. Positively correlated to handling-induced convulsion (HIC) score 7 h after ethanol injection. D. Negatively correlated to obesity.
SNP information for c57bl/6j (b6) x dba/2j (d2)
To elucidate a possible molecular mechanism of differential expression, we searched the polymorphisms of Atp5g3 in relevant mouse strains using the Mouse SNP Query Form in the Mouse Genome Informatics database [http://www.informatics.jax.org/javawi2/servlet/WIFetch?page=snpQF (accessed October 12, 2012)]. Our search used the Gene Symbol/ Name Atp5g3, and the search region included SNPs located within 2 kb up- or downstream of Atp5g3. However, currently no polymorphism in Atp5g3 has been reported immediately up- or downstream among strains C57BL/6J, DBA/2J, and BALB/cJ.
Our second search was conducted using the National Center for Biotechnology Information Single Nucleotide Polymorphism Database. We searched a genomic region similar to that used in the Mouse Genome Informatics database - between 73744504 bp and 73751383 bp. Again, no SNP was identified
Patterns of expression of Atp5g3 and its partner genes
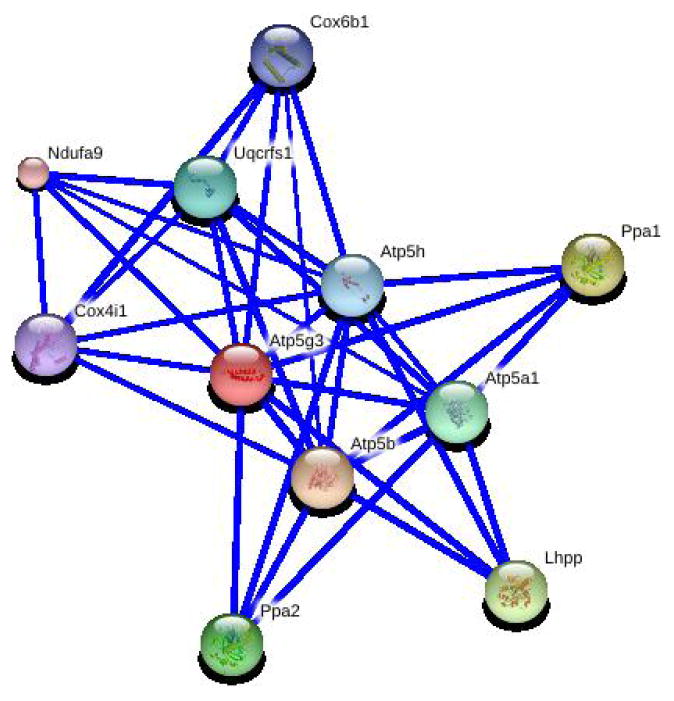
Owing to the lack of Atp5g3 polymorphisms and 2000-bp down- and upstream sequences among three relevant mouse strains, the involvement of Atp5g3 in the regulation of alcoholism appeared not to be solely attributable to Atp5g3 itself. A regulator or regulation site(s) may be located more distantly, but such a possibility is difficult to predict with current knowledge. Another possibility is that Atp5g3 interacts with other genes to influence alcoholism. We then considered the interaction of the Atp5g3 partner genes based on STRING analysis (Figure 5). As shown, Atp5g3 interacted with 12 other genes. We examined the expression levels of these genes in the hippocampus, cerebellum, and liver. Figure 6 shows the expression patterns in the hippocampus and liver, which varied greatly in the same tissue. One interesting note is that although the expression of Atp5g3 in the liver showed less variation than that in the hippocampus, its partner genes showed a variation similar to that in the hippocampus.
Figure 5.
Interaction partner genes of Atp5g3 based on STRING analysis (http://string.embl.de/newstring_cgi/show_network_section.pl?input_query_species=auto_detect&network_flavor=confidence&identifier=ENSMUSG00000018770). Associations among Atp5g3 and its partner genes: atp5a1, atp5b, atp5h, ppa1, ppa2, cox6b1, cox4il, lhpp, ndufa9, or uqcrfs1. Stronger associations are represented by thicker lines.
Figure 6.
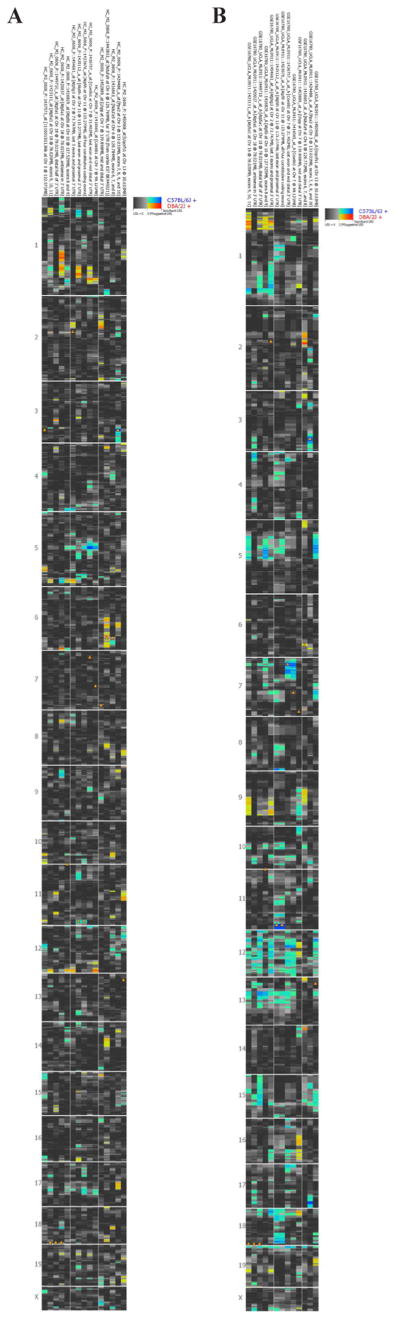
Heat maps of Atp5g3 and its partner genes in hippocampus and liver. Left = hippocampus; right = liver.
Gene interaction among Atp5g3 and its partner genes
We next examined the co-expression of Atp5g3 and its 12 partner genes in the hippocampus, cerebellum, and liver. The correlation in expression levels between Atp5g3 and each of those genes was different (Table 1). The co-expression level of the same gene with Atp5g3 was also different in different tissues. For example, the expression level of Uqcrfs1 was positively correlated with Atp5g3 in both the cerebellum and the liver, but it had no correlation or negative correlation with Atp5g3 in the hippocampus. Furthermore, different probes of the same gene detected different interactions with Atp5g3, most likely because of splicing differences. For example, probes from exons 9, 10, and 11 of Atp5a1 showed expression patterns similar to those of Atp5g3 in the three tissues, whereas probes from the distal half of the 3′-untranslated region (UTR) and antisense in the 3′-untranslated region showed expression patterns that differed from those of Atp5g3 in the hippocampus, cerebellum, and liver. According to the Ensembl database, Atp5a1 has five transcripts, three of which have protein products. The lengths of transcripts are different and also vary in the 3′ end.
Table 1.
Correlation of gene expression between Atp5g3 and its partners.
| Gene/Probes | Hippocampus/72
|
Cerebellum/30
|
Liver/32
|
|||
|---|---|---|---|---|---|---|
| Expression level | R | Expression level | R | Expression level | R | |
| 1110013G13Rik/ Ppa2 | 6.238 | −0.361 | 5.207 | −0.545 | - | - |
| Atp5a1 exons 9, 10, 11 | 15.245 | 0.717 | 15.941 | 0.557 | 13.693 | 0.772 |
| Atp5a1 distal half of 3′-UTR | 14.769 | 0.24 | 14.348 | 0.61 | 12.915 | 0.775 |
| Atp5a1 antisense in 3′-UTR | 7.895 | −0.315 | 7.858 | 0.238 | 7.234 | 0.43 |
| Atp5b exons 8 and 9 | 15.497 | 0.697 | 16.144 | 0.578 | 14.033 | 0.557 |
| Atp5g3 last 3 exons and proximal 3′-UTR | 16.013 | N | 16.106 | N | 14.496 | N |
| Atp5h last exon and proximal 3′-UTR | 15.406 | 0.143 | 14.342 | 0.554 | 13.286 | 0.41 |
| Atp5h all four constitutive coding exons | 14.978 | 0.385 | 15.123 | 0.756 | 12.973 | 0.453 |
| Cox6b1 last exon and mid distal 3′-UTR | 15.834 | 0.277 | 14.89 | 0.63 | 14.069 | 0.638 |
| Cox6b1 | 15.092 | 0.377 | 14.77 | 0.699 | 13.724 | 0.788 |
| Lhpp mid and distal 3′-UTR | 10.049 | 0.19 | 9.167 | 0.209 | 11.193 | −0.463 |
| Ndufa9 far 3′-UTR (from cortex EST BE945021) | 9.233 | −0.276 | 8.747 | −0.147 | - | - |
| Ndufa9 exons 6, 7, and 8 | 12.656 | −0.057 | 11.929 | 0.042 | 12.468 | 0.478 |
| Ppa2 exons 7, 8, 9, and 10 | 9.557 | −0.384 | 9.539 | −0.156 | 10.372 | −0.027 |
| Uqcrfs1 | 13.215 | −0.065 | 13.438 | 0.61 | 13.075 | 0.671 |
Transcriptome mapping of Atp5g3 regulation
To investigate whether Atp5g3 is regulated differently in different tissues, we analyzed the transcriptome map of Atp5g3 in the hippocampus, cerebellum, and liver. The data showed that the expression levels of Atp5g3 in the three tissues were regulated differently. Transcriptome maps with 2000 permutation tests showed significant difference among tissues (Figure 7). In the hippocampus, the permutation test indicated that transcriptome loci were significant at a log odds ratio (LOD) of 3.64 and suggestive at LOD 2.27. The expression of Atp5g3 was regulated mainly by three loci located on chromosomes 4, 5, and 12. In the cerebellum, the permutation test indicated that the significant level was at LOD 3.77 and the suggestive level at LOD 2.32. According to these criteria, two loci on chromosome 8 regulated Atp5g3 expression: one reached the significance level, whereas the other was at the suggestive level. In the liver, the permutation test indicated that the significant level was LOD 3.60 and the suggestive level was LOD 2.20. One locus on chromosome 1 reached the significance level. In addition, several loci at the suggestive level were detected on chromosomes 1, 9, 12, and 18. The differential regulation of Atp5g3 expression in various tissues seemed to agree with the fact that Atp5g3 is correlated with different phenotypes of alcoholism in those tissues.
Figure 7.
Transcriptome map of Atp5g3 in hippocampus, cerebellum, and liver. Transcriptome maps were done with 2000 permutation tests. A. Hippocampus; B. cerebellum; C. liver.
CONCLUSION
Our current information suggests that Atp5g3 involves the regulation of alcoholism. However, it is unclear whether the involvement is direct or indirect (through its partner genes). Two findings suggest the involvement of Atp5g3 in alcoholism regulation. The first finding is the correlation between Atp5g3 expression and the alcoholism-related phenotypes in three alcoholism-relevant tissues. The second finding is the chromosome location of Atp5g3, which is within the fine-mapped region of a QTL for the alcohol preference phenotype. The lack of a known polymorphism between the B6 and D2 strains negates direct involvement of Atp5g3 in alcoholism, although it cannot be completely ruled out. The fact that Atp5g3 is a relatively small gene encoding only one transcript with one protein sequence makes the possibility of direct involvement of Atp5g3 in regulating alcoholism small.
The complexity comes from the interactions between Atp5g3 and its partner genes. Those interactions are different among not only different genes but also different tissues. Two findings also allow the speculation that Atp5g3 involves different pathways in different tissues in regulating alcoholism: The first finding is the difference in correlations between Atp5g3 and its partner genes in different tissues. The second finding is the difference in transcriptome maps that reveal the regulation of Atp5g3 expression in different tissues. However, these conclusions are purely speculative, not definitive connections between Atp5g3 and alcoholism and molecular pathways.
In conclusion, the function of Atp5g3 in the alcoholism pathway needs further investigation. In particular, future studies should focus on the genomic regulation of Atp5g3 genes and the evidence of its connection to alcoholism. Its role in obesity at present is unknown, although current data suggest that it may be involved. Any breakthrough on either an association with obesity or a potential molecular pathway will greatly improve current knowledge of the function of Atp5g3 in obesity.
Acknowledgments
The study was supported by grants from the National Institute on Alcohol Abuse and Alcoholism, the National Institutes of Health (#R01 AR51190 to WG), the State Key Development Program of Basic Research of China (#2009CB521905 to YJW), and the Veterans Administration Medical Center in Memphis, TN, USA.
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
References
- Boyle AE, Gill KJ. Confirmation of provisional quantitative trait loci for voluntary alcohol consumption: genetic analysis in chromosome substitution strains and F2 crosses derived from A/J and C57BL/6J progenitors. Pharmacogenet Genomics. 2008;18:1071–1082. doi: 10.1097/FPC.0b013e32831367f0. [DOI] [PubMed] [Google Scholar]
- Buscemi L, Turchi C. An overview of the genetic susceptibility to alcoholism. Med Sci Law. 2011;51(Suppl 1):S2–S6. doi: 10.1258/msl.2010.010054. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Wang J, Williams RW, et al. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat Neurosci. 2004;7:485–486. doi: 10.1038/nn0504-485. [DOI] [PubMed] [Google Scholar]
- Crabbe JC. Provisional mapping of quantitative trait loci for chronic ethanol withdrawal severity in BXD recombinant inbred mice. J Pharmacol Exp Ther. 1998;286:263–271. [PubMed] [Google Scholar]
- Cunningham CL. Localization of genes influencing ethanol-induced conditioned place preference and locomotor activity in BXD recombinant inbred mice. Psychopharmacology. 1995;120:28–41. doi: 10.1007/BF02246142. [DOI] [PubMed] [Google Scholar]
- Farber CR, Medrano JF. Dissection of a genetically complex cluster of growth and obesity QTLs on mouse chromosome 2 using subcongenic intercrosses. Mamm Genome. 2007;18:635–645. doi: 10.1007/s00335-007-9046-0. [DOI] [PubMed] [Google Scholar]
- Fernandez JR, Vogler GP, Tarantino LM, Vignetti S, et al. Sex-exclusive quantitative trait loci influences in alcohol-related phenotypes. Am J Med Genet. 1999;88:647–652. doi: 10.1002/(sici)1096-8628(19991215)88:6<647::aid-ajmg13>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- HTML. [Accessed August 23, 2007];HyperText Markup Language. 2012 Available at [ http://www.w3.org/TR/html401/]
- Jerez-Timaure NC, Kearney F, Simpson EB, Eisen EJ, et al. Characterization of QTL with major effects on fatness and growth on mouse chromosome 2. Obes Res. 2004;12:1408–1420. doi: 10.1038/oby.2004.177. [DOI] [PubMed] [Google Scholar]
- Kanis JA, Oden A, Johnell O, Johansson H, et al. The use of clinical risk factors enhances the performance of BMD in the prediction of hip and osteoporotic fractures in men and women. Osteoporos Int. 2007;18:1033–1046. doi: 10.1007/s00198-007-0343-y. [DOI] [PubMed] [Google Scholar]
- Kirstein SL, Davidson KL, Ehringer MA, Sikela JM, et al. Quantitative trait loci affecting initial sensitivity and acute functional tolerance to ethanol-induced ataxia and brain cAMP signaling in BXD recombinant inbred mice. J Pharmacol Exp Ther. 2002;302:1238–1245. doi: 10.1124/jpet.302.3.1238. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Ohno T, Hada N, Fujiyoshi M, et al. Genetic analysis of abdominal fat distribution in SM/J and A/J mice. J Lipid Res. 2010;51:3463–3469. doi: 10.1194/jlr.M009563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HS, Zhang JY, Thompson BS, Deng XY, et al. Rat mitochondrial ATP synthase ATP5G3: cloning and upregulation in pancreas after chronic ethanol feeding. Physiol Genomics. 2001;6:91–98. doi: 10.1152/physiolgenomics.2001.6.2.91. [DOI] [PubMed] [Google Scholar]
- Marti A, Ordovas J. Epigenetics lights up the obesity field. Obes Facts. 2011;4:187–190. doi: 10.1159/000329847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollah MB, Ishikawa A. Intersubspecific subcongenic mouse strain analysis reveals closely linked QTLs with opposite effects on body weight. Mamm Genome. 2011;22:282–289. doi: 10.1007/s00335-011-9323-9. [DOI] [PubMed] [Google Scholar]
- Overall RW, Kempermann G, Peirce J, Lu L, et al. Genetics of the hippocampal transcriptome in mouse: a systematic survey and online neurogenomics resource. Front Neurosci. 2009;3:55. doi: 10.3389/neuro.15.003.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero B, Ansah TA, et al. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav. 2010;9:129–159. doi: 10.1111/j.1601-183X.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Phillips TJ, Belknap JK, Finn DA, et al. Genetic analysis of the corticosterone response to ethanol in BXD recombinant inbred mice. Behav Neurosci. 1995;109:1199–1208. doi: 10.1037//0735-7044.109.6.1199. [DOI] [PubMed] [Google Scholar]
- Ruf C, Carosone-Link P, Springett J, Bennett B. Confirmation and genetic dissection of a major quantitative trait locus for alcohol preference drinking. Alcohol Clin Exp Res. 2004;28:1613–1621. doi: 10.1097/01.alc.0000145693.58448.95. [DOI] [PubMed] [Google Scholar]
- Slomko H, Heo HJ, Einstein FH. Minireview: Epigenetics of obesity and diabetes in humans. Endocrinology. 2012;153:1025–1030. doi: 10.1210/en.2011-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Gu J, Qi S, Lu L. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol. 2001;2:RESEARCH0046. doi: 10.1186/gb-2001-2-11-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]