Abstract
Humans have exploited natural resources for a variety of applications. Chitin and its derivative chitin oligosaccharides (CHOS) have potential biomedical and agricultural applications. Availability of CHOS with the desired length has been a major limitation in the optimum use of such natural resources. Here, we report a single domain hyper-transglycosylating chitinase, which generates longer CHOS, from Enterobacter cloacae subsp. cloacae 13047 (EcChi1). EcChi1 was optimally active at pH 5.0 and 40 °C with a Km of 15.2 mg ml−1, and k cat/Km of 0.011× 102 mg−1 ml min−1 on colloidal chitin. The profile of the hydrolytic products, major product being chitobiose, released from CHOS indicated that EcChi1 was an endo-acting enzyme. Transglycosylation (TG) by EcChi1 on trimeric to hexameric CHOS resulted in the formation of longer CHOS for a prolonged duration. EcChi1 showed both chitobiase and TG activities, in addition to hydrolytic activity. The TG by EcChi1 was dependent, to some extent, on the length of the CHOS substrate and concentration of the enzyme. Homology modeling and docking with CHOS suggested that EcChi1 has a deep substrate-binding groove lined with aromatic amino acids, which is a characteristic feature of a processive enzyme.
Introduction
Chitin [(C8H13O5N) n»1], is a linear homopolymer of N-acetyl glucosamine (GlcNAc) units linked through β (1 → 4) glycosidic bonds. It is an abundant renewable natural resource next to cellulose in the biosphere. Chitin is a primary structural component of the fungal cell wall, insects, molluscs, squid, internal shells of cephalopods and the exoskeletons of arthropod, exists in three biological forms i.e. α-chitin, β-chitin and γ-chitin. The presence of acetamide group (NH–CO–CH3) allows increased hydrogen bonding between adjacent polymers, giving increased strength of chitin – polymer matrix. Chitin oligosaccharides (CHOS) generated from polymeric chitin with specific composition and length have potential applications. The remarkable properties of CHOS i.e. bio-degradability, bio-compatibility, and non-toxicity suit wider industrial application1, 2. Production of CHOS by chemical methods has been challenging due to disadvantages like non-specific random hydrolysis, and difficulty to remove the acidity of the CHOS oligomers3. Chitinases could be possible alternatives for production of long chain CHOS, especially for biological applications.
Chitinases (EC 3.2.1.14) hydrolyze chitin to chitin monomer (DP1; Degree of polymerization 1), chitin dimer (DP2) and CHOS of shorter length. Chitinases, classified into two glycoside hydrolase (GH) families i.e. GH18 and GH19 could be exochitinases that act terminally and endochitinases that cleave randomly at internal sites of the chitin, eventually producing a variety of low molecular mass or short length CHOS4. Chitinases that act on crystalline polysaccharides need to associate and disrupt the polymer from packing and also direct the travelling of single polymer chain into the catalytic center through a catalytic groove, a mechanism called processivity5, 6. The aromatic amino acids lining the substrate-binding site, facilitate processivity by functioning as a flexible and hydrophobic sheet and allow the polymer chain to slide7, 8.
A few of the GH18 chitinases exhibit an uncommon transglycosylation (TG), in addition to chitin hydrolysis, by introducing new glycosidic bonds between donor and acceptor saccharides9. The GH18 chitinases gained special interest due to potential applications for the enzymatic production of CHOS by TG from chitin10. Bacterial chitinases like chitinase-D from Serratia proteamaculans (SpChiD) exhibited hyper-TG with chitotriose to chitohexaose (DP3–6) substrates generating products longer than chitohexaose (DP7–13)11. Similarly, chitinase A from Stenotrophomonas maltophilia 12 and ChiCW from Bacillus cereus 28–9 synthesized chitohexaose (DP6) from chitotetraose (DP4)13. A close relationship between bacterial chitinases and their pathogenicity may exist14, 15, and the molecular targets of chitinases in chitinous or non-chitinous hosts remain unclear. Hence, detailed understanding of chitinases at the biochemical or functional level may help in understanding their role in pathogenicity.
Enterobacter cloacae subsp. cloacae (Ec), a nosocomial pathogen, was originally isolated from the human cerebrospinal fluid by Edwin Oakes Jordan in 189016. Analysis of the genome sequence of E. cloacae subsp. cloacae revealed that four chitinases and two N-acetyl-glucosaminidases could be involved in chitin degradation. In addition, E. cloacae genome has genes that code for one CBM-33 lytic polysaccharide monooxygenase (LPMO-GenBank: ADF60226.1) and one polysaccharide deacetylase (PDA-GenBank: ADF62202.1), which may have a crucial role in depolymerization of chitin. Of the four chitinases, two belong to family GH18 (EcChi1-GenBank: ADF62010.1 and EcChi2 (GenBank: ADF62328.1) and other two belong to GH19 (EcChi3-GenBank: ADF62326.1 and EcChi4-GenBank: ADF61237.1). Here, the EcChi1 with only the catalytic GH18 domain and no auxiliary domains displayed an unprecedented hyper-TG. We show that EcChi1 has structural features of a hyper-TG chitinase similar to SpChiD of Serratia proteamaculans.
Results
Cloning, heterologous expression, purification and dot blot assay
Full-length DNA sequence coding for EcChi1 consisted of 1257 nucleotides and an ORF of 418 amino acid residues with a predicted isoelectric point of 6.15. A 1.2-kb segment of EcChi1 gene was amplified with specific primers using gDNA of E. cloacae subsp. cloacae as a template, cloned into pET-28a (+), and transformed E. coli Rosetta-gami for expression. Extracellular protein was isolated from the induced E. coli Rosetta-gami culture pellet and purified EcChi1 through Ni-NTA affinity chromatography. The molecular mass of EcChi1 (42.5 kDa) calculated from amino acid sequence without the signal peptide was in agreement with experimentally observed molecular weight as obtained by 12% SDS-PAGE (Supplementary Fig. S1A). Dot - blot assay revealed that, EcChi1 exhibited clear zone on water-soluble glycol chitin containing gel confirming the chitinase activity (Supplementary Fig. S1B).
Steady state kinetics of EcChi1 on colloidal chitin and chitobiose
The EcChi1 was active in mildly acidic to a neutral range of pH 5.0 to 7.0 with the optimum activity in 50 mM sodium citrate pH 5.0 (Fig. 1A). However, the activity decreased sharply below pH 5.0 and above 7.0, with diminished or no activity below pH 4.0 and above 9.0. EcChi1 was optimally active at 40 °C (Fig. 1B) and retained 60% activity at 50 °C, that was decreased with increase of temp up to 80 °C, while 70% activity was retained at 30 °C. The effect of metal ions on the activity of EcChi1 was also evaluated using colloidal chitin as substrate. The activity of EcChi1 in presence of Cu+2 was similar to control but was partially inhibited in the presence of urea, Ca+2, Mg+2, Zn+2, Mn+2, EDTA, and Fe+2. The presence of Hg+2 and SDS inhibited the activity by 80% and 90%, respectively. There was no increase in the activity of EcChi1 in the presence of test cations. The kinetic parameters of EcChi1 were determined with colloidal chitin and chitobiose as the substrate. The velocity measurements fitted to Michaelis-Menten kinetics revealed that the Km, k cat, and k cat/Km values were 15.2 mg ml−1, 0.16 × 102 min−1 and 0.011 × 102 mg−1 ml min−1 for colloidal chitin (Fig. 1C) and 213.2 μM, 1.41 min−1 and 0.6 × 10−2 for chitobiose (Supplementary Fig. S2).
Figure 1.
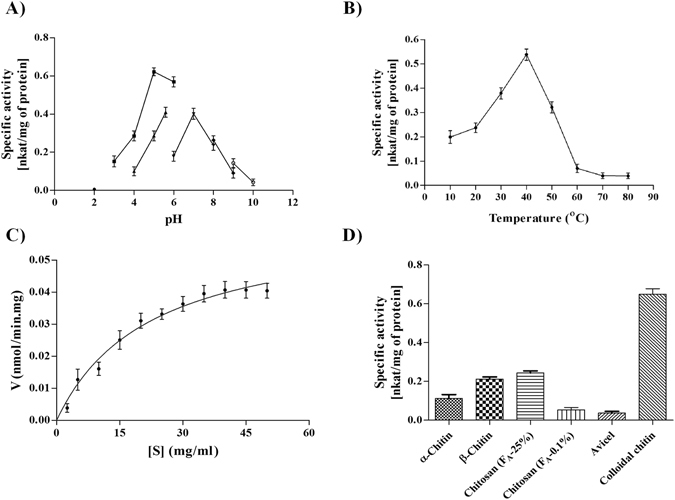
pH, temperature optima, kinetic parameters and activity on different insoluble substrates of EcChi1. The pH and temperature optima were determined by incubating 30 mg/ml colloidal chitin and 30 μg of EcChi1 for 1 h at 40 °C in various buffers at different pH values, as follows: (A) 50 mM glycine-HCl (pH 2.0, ▼), 50 mM sodium citrate (pH 3.0–6.0, ■), 50 mM sodium acetate (pH 4.0–6.0, ▲), 50 mM sodium phosphate (pH 6.0–8.00, ●), 50 mM Tris-HCl (pH 8.0–9.0, ♦) and 50 mM glycine-NaOH (pH 9.0–10.0, ○) and at 10 °C to 80 °C (B), Different concentrations of colloidal chitin (2.5–60 mg/ml) and 30 μg of EcChi1 in 50 mM sodium citrate buffer, pH 5.0. The average of the triplicate data was fitted to the Michaelis-Menten equation by a nonlinear regression function of graphpad prism version 6.0 (C), The reaction mixture containing 30 μg EcChi1 and 1 mg ml−1 of one of the polymeric substrates, was incubated at pH 5.0 and 40 °C for 1 h. The reaction mixture was centrifuged, and products were quantified using a reducing group assay (D). The error bars represent the standard deviations from three individual experiments.
Activity of EcChi1 on different insoluble substrates
EcChi1 exhibited maximum activity on colloidal chitin followed by chitosan, β-chitin, α-chitin and avicel. The activity of EcChi1 was 41% on chitosan (FA-25%), 35% on β-chitin, 17% on α-chitin, 8% on chitosan (FA-0.1%), and 5.7% on avicel.
Binding study with soluble substrates
Binding of EcChi1 to the soluble substrates was analysed using native PAGE with or without polysaccharides embedded in the gel. In presence of laminarin, CM-cellulose and glycol chitin there was a decrease in electrophoretic mobility of EcChi1 (Fig. 2). Among the three soluble substrates, the retardation in the mobility of EcChi1 was more by soluble glycol chitin than CM- cellulose, and laminarin suggesting specificity of EcChi1 towards chitinous substrates.
Figure 2.
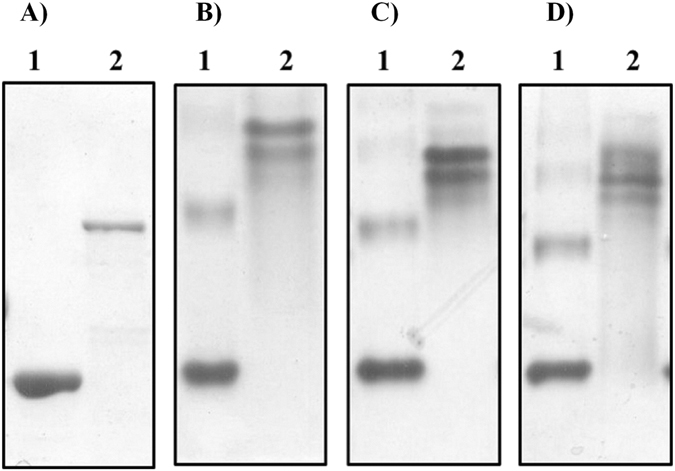
Binding of EcChi1 with the soluble polymeric substrates. Affinity non-denaturating electrophoresis was performed in 8% native PAGE gels by preparing with or without substrate. (A) Proteins (EcChi1 or non-interacting BSA) without substrate, while (B) with 0.1% (wt/vol) substrate glycol chitin, (C) CM-cellulose and (D) laminarin incorporated in gels. Proteins were visualized by Coomassie blue G-250 staining after electrophoresis. Lane 1. BSA, Lane 2. EcChi1.
Time-course of colloidal chitin and oligosaccharide degradation
EcChi1 released DP1 to DP4 hydrolytic products on polymeric colloidal chitin substrate with DP2 and DP1 as the major products (Fig. 3A and B). The concentration of DP1 increased with the extension of incubation time. EcChi1 hydrolyzed DP2 into DP1 after 240 min, and complete hydrolysis occurred by 420 min (Supplementary Fig. S3). Chitobiose was the major hydrolytic product from both DP3 and DP4 substrate (Fig. 4A and B). With DP5 substrate, a very little DP1 (1.4%) formed from 120 min. After prolonged incubation, DP1 was the dominant product with 37.5% followed by DP2 with 28% at 720 min (Fig. 5A and C). The hydrolysis of DP6 by EcChi1 was rapid with only 2.2% of DP6 at 120 min (Fig. 5B and D).
Figure 3.
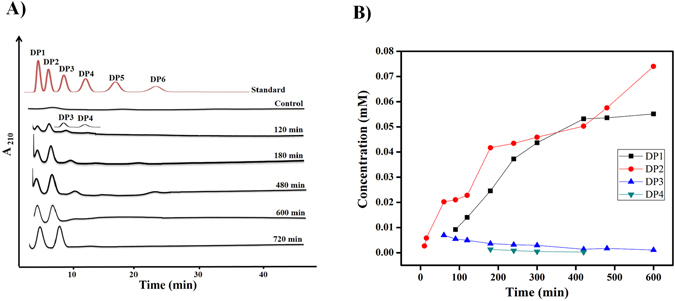
Hydrolysis of colloidal chitin by EcChi1. The EcChi1 (0.8 μM) was incubated with 30 mg/ml of the substrate in 900 μL reaction mixture for different time periods from 0 to 720 min at 40 °C. The reaction products were analyzed by binary gradient HPLC. (A) The topmost profile shows a standard mixture of CHOS ranging from DP1 to DP6. The remaining profiles are the reactions at different incubation times. Control represents the substrate without EcChi1 while standard represents CHOS ranging from DP1 to DP6. (B) Overview of the quantifiable CHOS products generated during hydrolysis. Products were quantified from respective peak areas using standard calibration curves of CHOS ranging from DP1 to DP4.
Figure 4.
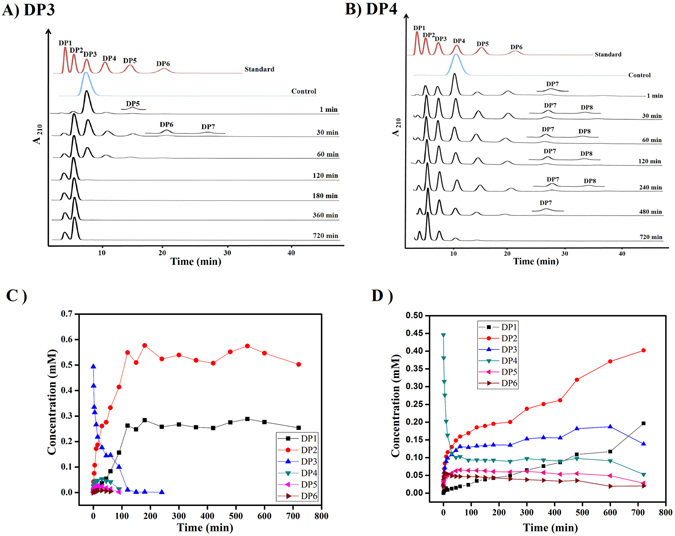
Time course hydrolysis and TG catalyzed by EcChi1 with DP3, DP4. The reaction mixture was incubated with 200 nM of EcChi1 and 1 mM of DP3/DP4 for different time periods from 0 to 720 min at 40 °C. Products were analyzed by binary gradient HPLC. (A and B) HPLC profiles of reaction products from DP3 (A) and DP4 (B) substrates. The topmost profile shows a standard mixture of CHOS ranging from DP1 to DP6 (A and B). The other profiles show the reaction products from DP3 and DP4 substrates at the indicated incubation times. The inset shows a magnified view of the low-peak-area products. Control represents the substrate without EcChi1. Overview of concentrations of CHOS products generated during reaction time courses with DP3 (C) and DP4 (D) substrates. Products were quantified from respective peak areas using standard calibration curves of CHOS ranging from DP1 to DP6.
Figure 5.
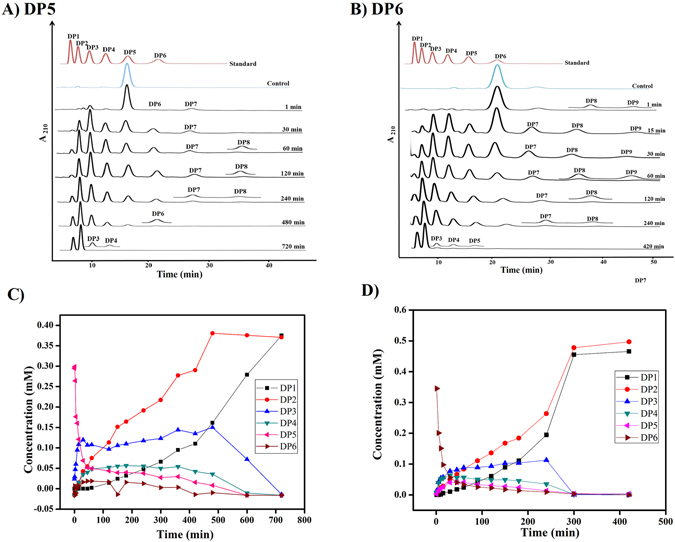
Time course hydrolysis and TG catalyzed by EcChi1 with DP5 and DP6. The reaction mixture products were analyzed by binary gradient HPLC. (A and B) HPLC profiles of reaction products from DP5 (A) and DP6 (B) substrates. The topmost profile shows a standard mixture of CHOS ranging from DP1 to DP6 (A and B). The other profiles show the reaction products from DP5 and DP6 substrates at the indicated incubation times. The inset shows a magnified view of the low-peak-area products. Control represents the substrate without EcChi1. (C and D) Overview of concentrations of CHOS products generated during reaction time courses with DP5 (C) and DP6 (D) substrates. Products were quantified from respective peak areas using standard calibration curves of CHOS ranging from DP1 to DP6.
EcChi1 exhibited high TG activity on chitotriose (DP3), chitotetraose (DP4), chitopentaose (DP5) and chitohexaose (DP6) substrates. With DP3 as starting substrate, TG products DP4-DP7 were detectable. DP4 and DP5 appeared from 1 min to 90 min, while DP6 and DP7 products appeared from 3 min to 60 min (Fig. 4A and C). Chitobiose was the major hydrolytic product from DP3 substrate. Use of DP4, DP5, and DP6 as starting substrates showed the formation of longer TG products (Figs 4B and 5A,B). With DP4 substrate, DP5-DP8 products formed due to TG, whereas DP9 was detectable as the longest TG product from DP5 and DP6 substrates (Figs 5B and 6). MALDI-TOF MS confirmed the products ≥DP6 of EcChi1. The products ranging from DP5 to DP9 and DP4 to DP9 were detectable with DP5 and DP6 substrates, respectively (Fig. 6A and B).
Figure 6.
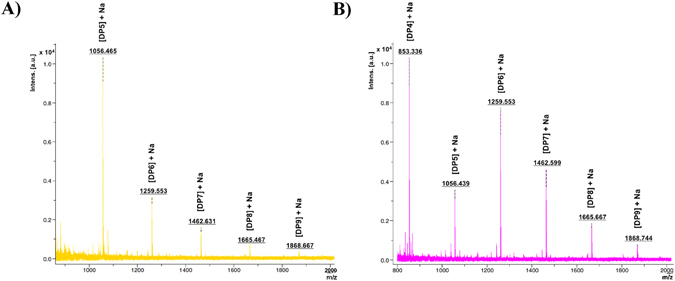
MALDI-TOF MS analysis of CHOS/TG products with DP5 and DP6 substrates catalyzed by EcChi1. The each 40 μL of reaction sample was concentrated under reduced pressure at 25 °C and dissolved in 10 μL of MilliQ H2O. The 2 μL of the reaction mixture was mixed with equal volume of 2,5-dihydroxybenzoic acid (2,5-DHB), and the resultant solution was subjected to mass measurements using an Ultraflex MALDI – TOF/TOF instrument. The masses shown here are with Na adducts.
Reaction with DP4 substrate accumulated TG products from 1 min and lasted for 480 min, while DP8 product was detectable until 240 min. The TG products DP5 and DP6 reached a maximum quantity of 5.3% and 4.4% at 60 and 5 min, respectively (Fig. 4B and D). With DP5 substrate, DP6-DP9 products formed due to TG. DP6 formed from 1 min that reached to a maximum (2.5%) by 45 min, that was hydrolyzed to shorter CHOS (Fig. 5A and C). The CHOS DP7-DP9 were detectable TG products with DP6 substrate (Fig. 5B and D). The TG products formed from 1 min onwards and remained until 240 min.
The hydrolytic and TG activity of EcChi1 were assayed at low substrate and enzyme concentrations with DP4 substrate. EcChi1 exhibited remarkable TG activity even with the low substrate and enzyme concentrations. A 10-fold lower concentration of the substrate (100 μM) and enzyme (20 nM) resulted in the formation of DP5-DP7 as TG products until 30 min that was eventually degraded (Supplementary Fig. S4A). When the substrate concentration decreased by 20 fold (50 μM), the TG products, DP5 and DP6 formed between 0–8 min, which gradually decreased over time (Supplementary Fig. S4B). These results indicated that the TG activity of EcChi1 was proportional to the substrate concentration.
Docking of EcChi1 with chitin tetramer
Sequence analysis revealed that EcChi1 is a single modular GH18 chitinase. The closest homologue of EcChi1 with a known crystal structure was chitinase II from Klebsiella pneumoniae (PDB: 3QOK; 86% sequence identity) which was used as the template to generate a 3D model. The stereo chemical quality of the generated model was checked for its quality through Ramachandran plot generated from PROCHECK analysis. The model showed 92.1% amino acids of modelled protein fell in the most favored region. Verify-3D graph also showed that 95.71% of the residues had an averaged 3D-1D score > = 0.2. Coordinates for chitin tetramer were extracted from the crystal structure of chitinase A from Serratia marcescens (PDB: 1NH6) and docked with modelled EcChi1 protein. EcChi1 complexed with DP4 substrate showed binding of DP4 from +2 to −2 subsites. Trp114 and Tyr154 and Arg278 and Trp395 are present at +2, +1 and −1, −2 positions, respectively. The residues forming interactions with ligand in the protein-ligand complex were Trp114, Tyr154, Arg278, and Trp395. Amino group in indole ring of Trp395 formed the hydrogen bond with a distance of 3.5 Å with the oxygen of acetamido group of non-reducing sugar moiety and a hydroxyl group on C6 of non-reducing sugar the formed hydrogen bond with 2.8 Å distance with the carboxylic group of Trp114, at −2 subsite (Fig. 7B). A terminal guanidium amino group of Arg278 and the amino group of Trp114 formed a hydrogen bond with 3.4 Å and 2.1 Å distances with the hydroxyl group of C6 and C4 of the penultimate non-reducing sugar moiety, respectively at −1 subsite. The hydroxyl group of Tyr154 formed a hydrogen bond with the hydroxyl group on C6 of sugar moiety at +1 subsite with 1.9 Å distance (Fig. 7B). EcChi1 has deep substrate binding cleft lined with aromatic residues that may play a major role in TG activity. The two aromatic amino acids Trp114 and Trp395 are located near to the catalytic center Glu153 (Fig. 7A and C).
Figure 7.
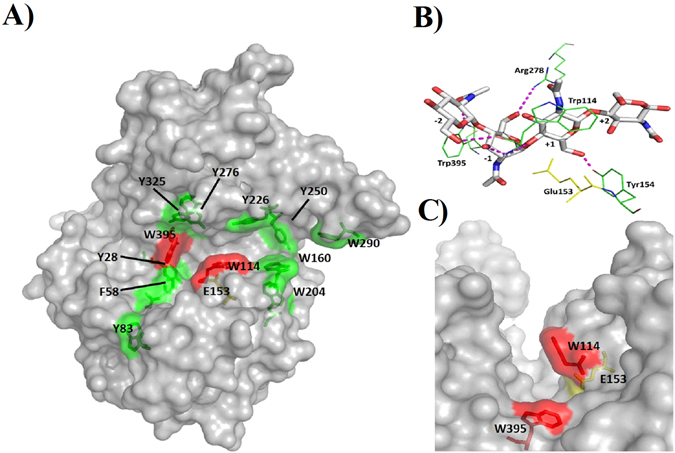
Substrate binding cleft of modelled EcChi1. (A) Aromatic residues lining along the substrate binding cleft (protein in surface representation and aminoacids in green and red sticks) and aminoacids interacting with chitin tetramer labeled in red colour. (B) Docked conformation of chitin tetramer (white sticks) and its possible interactions with residues (green lines) along with catalytic residue, Glu153 (yellow line) of EcChi1. (C) Deep active site cleft of EcChi1 showing catalytic (yellow colour) and aromatic (red colour) amino acids.
Discussion
The recent advances in whole genome sequencing technologies have paved the way to mine bacterial genomes for the genes and proteins of industrial importance. Search for new chitinases by utilizing the available genome information from diverse bacteria may provide chitinases with high TG activity for generating longer-chain CHOS. Phylogenetic analysis of chitinases from diverse bacterial members showed clustering of chitinases belonging to Enterobacteriaceae members along with a well characterized hyper-TG chitinase SpChiD with a few exceptions17. This observation suggested that mining Enterobacteriaceae members could be an option for novel TG enzyme. In the present study, we have identified and characterized a hyper-TG chitinase (EcChi1) from Enterobacter cloacae, a familiar representative species of family Enterobacteriaceae. Very few reports are available on chitinases and chitinolytic system of Enterobacter sp (Table 1). Among which, Enterobacter sp. G-118, E. aerogenes 19, and Enterobacter sp. NRG420 chitinases were characterized. ChiA was the first reported chitinase21 from an Enterobacter sp and shared 26% sequence homology with EcChi1. EcChi1 exhibited 87% sequence similarity with E. aerogenes chitinase, which did not have TG activity. There is no correlation between overall sequence similarity and TG as it depends on active site architecture of the enzyme and the orientation of water molecules at the catalytic centre which favors binding of incoming carbohydrate molecules, through strong interactions in the aglycon subsites22. The amino acid sequence of EcChi1 showed 87% identity to that of GH18 chitinase II (PDB ID: 3QOK) from Klebsiella pneumoniae for which no characterization was available. EcChi1 shared 75% identity with a hyper-TG chitinase of S. proteamaculans (SpChiD) (PDB ID: 4NZC)11. Nevertheless, many chitinases showing TG such as ChiA1 from B. circulans WI-1223, ChiA from Vibrio harveyi 24, and human chitotriosidase-125 showed only 31%, 26% and 29% sequence similarity to EcChi1, respectively.
Table 1.
Details of chitinases characterized from Enterobacter sp.
| Strain name | Chitinase | Mol. Wt (kDa) | Optimum pH | Optimum Temp (°C) | TG activity | Reference |
|---|---|---|---|---|---|---|
| Enterobacter agglomerans | ChiA | 61 | ND | ND | Not reported | 21 |
| Enterobacer sp.G-1 | N-acetyl glucosaminidase | 92 | 6.0 | 45 | Not reported | 18 |
| ChiA | 60 | ND | ND | Not reported | 61 | |
| E nterobacer sp. NRG4 | ChiA | 61 | 5.5 | 45 | Not reported | 20 |
| E. aerogenes | Chitinase | 42.5 | 6.0 | 55 | Not reported | 19 |
| E. cloacae subsp. cloacae | EcChi1 | 42.5 | 5 | 40 | Yes | Present study |
ND – Not determined.
Enzymes show a pH range at which they are most active. EcChi1 was optimally active at slightly acidic pH (pH 5.0–6.0) similar to the chitinases A, B, and D from S. proteamaculans 11, 26, chitinase B from S. marcescens 27, Paenibacillus sp. D128 and Enterobacter sp. NRG429. EcChi1 optimally active at 40 °C comparable to SpChiB, SpChiC26, SpChiD11 and Chitinibacter sp. GC7230. The optimum temperature of EcChi1 was higher than many other chitinases such as Vibrio sp31 and S. antarcticus 32. However, some chitinases from B. cereus IO833 and Paenibacillus barengoltzii 34 exhibited high optimal temperature than EcChi1.
Unlike pH and temperature optima, the kinetic parameters of EcChi1 were different from SpChiD. The Km and catalytic efficiency (k cat/Km) of EcChi1 were 6 fold decrease and nearly 2 fold increase than SpChiD, respectively11. Among all the kinetic parameters, there is a significant correlation between Km and TG, whereas no direct correlation between k cat and TG activity was observed in chitinases35. The single mutants of ChiA-D313N and ChiB-D142N from S. marcescens showed 2 to 3 fold lower Km and increased TG than the wild-type. The hydrophobicity of amino acids at substrate acceptor region and catalytic groove makes the enzyme efficiently bind to the substrates which resulted in less Km. The overall catalytic efficiency (k cat/Km) of EcChi1 was lower than ChiA, ChiB and, ChiC from S. proteamaculans 26 possible attributed to the lack of accessory domain (s) in EcChi1. The activity of EcChi1 on crystalline α-chitin and β-chitin was also low in comparison to colloidal chitin due to lack of accessory domains. The lining of aromatic amino acids in a substrate binding groove of EcChi1 (Fig. 7A) might responsible towards chitinous polymeric substrates. Similar findings were observed for SpChiD and StmChiA but not for StmChiB11, 12. EcChi1 had a very low activity on avicel (Fig. 1D). This could be due to the non-specific activity of EcChi1, as reported in a chitinase (ChiC) from Pseudoalteromonas sp. DL-636. Various cations, particularly metal ions influence the activity of chitinases37. Certain ions such Cu+2 and Zn+2 act as either stimulators or inhibitors on chitinases38, not in the case of EcChi1.
The highly conserved catalytic motif (DxxDxDxE) and chitin-binding motif (SXGG) are positioned between 146–153 and 110–113 amino acids, respectively. Single amino acid residue present immediate to the chitin-binding motif and presence of α/β fold insertion plays a pivotal role in processivity39. The two aromatic amino acids Trp97 and Trp220 were crucial for processivity in ChiB from S. marcescens 40. Presumably, corresponding aromatic amino acids Trp114 and Tyr226 might tightly bind with the polymeric substrate and steer the processive action of EcChi1, implying that EcChi1 might be a processive enzyme (Fig. 7A and C). In addition, homology modelling studies of EcChi1 showed the presence of deep substrate binding cleft lined with twelve aromatic amino acids, and α/β fold insertion further support the characteristic feature of a processive enzyme. Analysis of product profile for dominance of even number oligosaccharides and the ratio of DP2/DP1 have been employed to determine an apparent degree of processivity41, 42. For example, the ratio of initial DP2/DP1 for two processive enzymes SmChiA and SmChiB had 30.1 ± 1.5 and 24.3 ± 2.0, respectively42. Upon degradation of colloidal chitin by EcChi1, DP1 was not detectable up to 60 min indicating a possible processive mode (Fig. 3B). At 90 min the DP2/DP1 ratio was 1.45, which further decreased with increase in time possible because of chitobiase activity of EcChi1.
HPLC analysis revealed that DP2 and DP1 are the major end products with colloidal chitin substrate similar to SpChiD and StmChiA11, 12. Gradual formation of DP1 from DP2 indicated chitobiase activity and DP2 was not a suitable substrate for TG. EcChi1 exhibited marginally lower k cat when compared to SpChiD. The sequence analysis and modelling studies showed EcChi1 having a unique loop (Asn30 - Asp42) present at N- terminal region. The amino acids present at substrate binding cleft were occupied by the loop structure. The unique loop and aromatic residues highly conserved in single domain GH18 Enterobacteriaceae chitinases. The conserved amino acids Tyr28, Val35, and Thr36 might be responsible for chitobiase activity in EcChi1 similar to SpChiD17.
TG is a far-flung phenomenon in GH18 family chitinases, little known about the structural basis underlying this integral activity. EcChi1 showed significant TG activity on CHOS and produced long chain CHOS up to DP9. The product profile with DP3 substrate suggested that −1 to +2 or −2 to +1 subsites are possible for productive binding. DP3 was the minimum length of the substrate for EcChi1 to show TG activity. DP1-DP8 products were detected with DP4 substrate in which DP5-DP8 were TG products, whereas DP5-DP9 were TG products in SpChiD11. DP9 was the longest TG product detected with both DP5 and DP6 substrates. Similarly, incubation of 5 mM chitopentaose (DP5) with recombinant human chitotriosidase (HCT) resulted in the formation of the DP1-DP925. No TG activity was observed by StmChiA with DP5 substrate12. EcChi1 synthesized long-chain CHOS even with low concentrations of substrate and enzyme. Amino acid residues positioned at acceptor binding site are assumed to be crucial for TG activity43. The known chitinases with TG activity were occupied with aromatic amino acids at +n subsites. Phe396&Trp275 and Trp220&Trp97 were occupied by the position at +2 and +1 subsites of ChiA and ChiB from S. marcescens, respectively44, 45. Similarly, Trp114, Tyr154 at +1, +2 subsites might help further in delivering efficient TG activity in EcChi1. In addition, Trp285 and Trp164 are occupying in +2 and +1 subsites of ChiA from B. circulans 46. It was further supported by a non-processive endochitinase from S. marcescens, SmChiC2 which lacks TG activity and aromatic amino acids at +1, +2 subsites39. Thus bulk aromatic amino acids present at positive subsites and the lining of aromatic amino acids at substrate binding cleft might be increased the binding affinities for upcoming sugar acceptor molecule thereby promoted TG activity in EcChi1.
In addition to processivity, single amino acid residue, present next to this chitin-binding motif (SXGG), plays an important role in TG47, 48. SXGG motif was extended with tryptophan in other well-known TG chitinases, viz., HCT25, B. circulans ChiA46, ChiA and ChiB from S. marcescens 49 and SpChiD in S. proteamaculans 11. In non-TG and non-processive endo-chitinases, such as SmChiC2 and LlChi18A, this tryptophan was substituted with small amino acid alanine indicated its probable role in TG47, 50. Trp114 was the corresponding amino acid in EcChi1 which may contribute to TG and processivity. In addition, the highly conserved novel loop was identified among the GH18 single catalytic domain chitinases from Enterobacteriaceae, showing their divergence from other groups17. This unique loop (Asn30 - Asp42) is present in EcChi1 may favor the interaction with the incoming substrate molecule.
The degree of endo-activity is best assessed as a probability of endo-mode initiation where concentrations of both insoluble reducing groups (endo-initiation) and reducing end groups (exo-initiation) upon substrate degradation are determined51. Hence, the observation of CHOS with DP higher than DP2 is indicative of EcChi1 having endo-activity as observed for SmChiC. Homology modelling based structural analysis also suggests that EcChi1 has structural features of an endo-acting enzyme with an apparent processive mode of action. EcChi1 was the first reported chitinase from Enterobacter sp with high TG. The capability to hydrolyze chitin with high TG activity suggests EcChi1 could be a suitable enzyme for industrial production of long chain CHOS, while it would also be of interest to know the possible physiological role of EcChi1.
Methods
Chemicals and enzymes
Genomic DNA of E. cloacae subsp. cloacae ATCC 13047 was used as a template to amplify the EcChi1 gene. The plasmid pET-28a (+) and E. coli Rosetta-gami II (DE3) (Novagen, Madison, USA) were used for heterologous expression. E. coli was grown in Luria bertani (LB) broth (1% peptone, 0.5% yeast extract, 1% NaCl) at 37 °C. The antibiotics kanamycin (50 µg ml−1) and chloramphenicol (25 µg ml−1) were added to the medium as required. Oligonucleotide primers were designed based on the DNA sequence available in the database and procured from Integrated DNA Technologies (Bangalore, India). Restriction enzymes, T4 DNA ligase, and Q5 DNA polymerase were from New England Biolabs (NEB). Isopropyl-β-D-thiogalactoside (IPTG), kanamycin and all other chemicals were purchased from Calbiochem, Merck (Darmstadt, Germany) or from Hi-Media Labs (Mumbai, India). The polymeric substrates α-chitin (extracted from shrimp shells), β-chitin (extracted from squid pen), and high molecular weight chitosan substrates of a fraction of acetylation (FA) = 0.10% (Mahtani Chitosan Pvt. Ltd.) and FA = 25% (Sigma-Aldrich) were used for enzyme assay. Colloidal chitin and glycol chitin were prepared as described earlier52. CHOS with different DP were purchased from Seikagaku Corporation (Tokyo, Japan). Avicel was procured from Sigma-Aldrich (St. Louis, MO).
DNA manipulations and cloning of EcChi1
E. cloacae genomic DNA (gDNA) was isolated as described by53. A gradient PCR, with appropriate gene-specific forward primer: 5′-CTC CCA TGG GAC TGA TGT CCG TG-3′and reverse primer: 5′-GGC CTC GAG CTT CGC TAA CTG GTT-3′ was used to determine the suitable annealing temperature for maximum amplicon yield. The amplicon was purified using PCR cleanup kit (MACHEREY- NAGEL GmbH & Co., Germany) and cloned into the Nco I and Xho I sites of the pET-28a (+). The construct was used to transform E. coli Rosetta-gami-II (DE3) for expression of the cloned gene. Transformants were selected on kanamycin and chloramphenicol containing LB plates. The nucleotide sequence of the insert was confirmed by automated DNA sequencing (Scigenom, Kerala, India).
Expression and purification of EcChi1
Expression and purification of EcChi1 were done as described by ref. 52. The expressed protein was purified using Ni-nitrilotriacetic acid (Ni-NTA) affinity chromatography. Fractions of eluate having EcChi1were pooled and the protein purity was assessed by performing 12% SDS-PAGE. The purified EcChi1, in 50 mM sodium citrate buffer (pH 5.0), was stored at 4 °C for further characterization. The protein content of the sample was measured using a bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, USA) where bovine serum albumin (BSA) as a standard.
Dot blot assay
A dot blot assay was performed to detect the activity of purified EcChi1 as described by ref. 11. A composite gel mixed with 0.1% glycol chitin was prepared. Five micrograms of EcChi1 was spotted onto the gel and placed in the humid chamber at 37 °C overnight. After incubation, the gel was stained with 0.01% calcofluor white M2R in 0.5 M Tris-HCl (pH 8.9) for 10 min at 28 ± 2 °C. Finally, the brightener solution was removed. The gel was washed three times with distilled water for every 10 min at 28 ± 2 °C. The lytic zone was visualized on a UV transilluminator.
Reducing end assay
Chitinase activity was measured by a reducing-end assay as described by ref. 54 with slight modifications. Recombinant EcChi1 was incubated with colloidal chitin in 50 mM sodium citrate pH 5.0 at 40 °C, 200 rpm for 1 h, followed by centrifugation at 13,600 × g, 4 °C for 30 min. A 40 μL of the clear supernatant containing reducing groups was mixed with 300 μL of the freshly prepared color reagent (0.5 M sodium carbonate, 0.05% potassium ferricyanide) and boiled for 15 min at 120 °C. Optical density was measured for 200 μL of each reaction mixture at 420 nm. One unit was defined as the amount of enzyme that liberated 1 μmol of reducing sugar per min. All the reactions were performed in triplicate.
Optimum pH and temperature
To determine the optimum pH, the purified EcChi1, and colloidal chitin was incubated at 40 °C for 1 h in 50 mM buffers of different pH (2.0 to 10.0). Glycine-HCl buffer (pH 2.0), sodium citrate buffer (pH 3.0–6.0), sodium acetate buffer (pH 4.0–5.6), sodium phosphate buffer (pH 6.0–8.0), and glycine-NaOH buffer (pH 9.0 and 10.0) were used to determine optimum pH.
The effect of temperature on the activity of EcChi1 was assessed by incubating 3.5 μM enzyme with 30 mg ml−1 colloidal chitin in 50 mM citrate buffer, (pH 5.0) at 10, 20, 30, 40, 50, 60, 70 and 80 °C and measured chitinase activity.
Steady-state kinetics
Kinetic parameters of EcChi1 were determined using colloidal chitin and chitobiose as the substrate. The reaction mixture (200 μL) containing 0 to 60 mg ml−1 of colloidal chitin and 0 to 1000 μM chitobiose substrate with EcChi1 in 50 mM buffer was incubated at 40 °C for 1 h with constant shaking at 200 rpm for chitinase assay. The unit of chitinase was defined as the release of 1 μmol of GlcNAc per second under standard experimental conditions. Specific activity was calculated in nkat/mg of protein. Kinetic values were obtained from triplicates of data fitting to the Michaelis-Menten equation via Non-linear regression using GraphPad Prism version 5.0 (GraphPad Software Inc, San Diego, CA).
Substrate specificity
The substrate specificity of the purified EcChi1 was determined by replacing the colloidal chitin in the reaction mixture with different insoluble chitinous and non-chitinous substrates viz., α-chitin, β-chitin, colloidal chitin, glycol chitin (water-soluble chitin), chitosans (FA = 0.1 and 25%), or Avicel (microcrystalline cellulose). EcChi1 (3.5 μM) was incubated with different substrates (2.5% wt/vol) at 40 °C for 1 h followed by reducing end assay. For each experiment, we individually calculated the reducing ends present in substrate alone and deducted the initial number of reducing groups from the value obtained from the enzyme-substrate reaction. One unit was defined as the amount of chitinase that liberated 1 μmol of reducing sugar per min.
Soluble substrate binding
Soluble substrate binding was assessed as described by ref. 52. Different soluble polymeric substrates, viz., glycol chitin, laminarin, and CM-cellulose were incorporated in 8% polyacrylamide gels. BSA was loaded as a non-interacting protein. The electrophoresis was performed at 4 °C at a constant voltage of 80 V. The gels were stained with coomassie blue G-250 to detect the proteins. Binding was assessed visually or alternatively, the migration distances of the chitinases and protein marker were measured directly on the resolving gels.
Effect of various additives
To determine the effect of various additives viz., metal ions (Ca+2, Cu+2, Fe+2, Mg+2, Mn+2, Hg+2, Zn+2), chelator (EDTA) and denaturants (Urea, SDS), of 1 mM each were added to EcChi1 with colloidal chitin as a substrate. After a pre-incubation for 1 h at 40 °C at 200 rpm activity was measured by reducing end assay. For each additive, the residual activity was calculated, considering the activity without additive was 100%.
Time-course of colloidal chitin and oligosaccharide degradation
The hydrolytic activity of EcChi1 was assessed in a reaction mixture containing 1 mM substrate and 200 nM purified EcChi1 in 50 mM citrate buffer, (pH 5.0) at 40 °C over 720 min. The reaction was terminated by transferring 40 µL of the reaction mixture to an Eppendorf tube containing an equal amount of 70% acetonitrile and stored at −20 °C until analysis. Aliquots of 20 μL were injected into HPLC (Shimadzu, Tokyo, Japan) equipped with a ShodexAsahipack NH2P-504E column (4.6 mm [inner diameter] by 250 mm; Showa Denko K.K). Briefly, the mobile phase consisted of 70% acetonitrile and 30% MilliQ H2O, the flow rate was set to 0.7 ml min−1. The eluted CHOS were monitored at 210 nm. A CHOS mixture containing equal weights of oligomers ranging from DP1 to DP6 was used to generate standard chromatogram. Calibration curves were constructed separately for each CHOS in the mixture. The data points generated a linear curve for each CHOS with r2 values of 0.997 to 1.0.
MALDI-TOF MS analysis
The products of the reaction mixture with DP5 and DP6 CHOS substrates were analyzed by matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) after 45 min and 15 min, respectively. A 40 μL of the reaction mixture was concentrated and redissolved in 10 μL of HPLC-grade MilliQ H2O (Merck, Mumbai, India). Two microliters sample was mixed into the 2,5-dihydroxybenzoic acid (DHB) droplet and dried under a stream of air and analysis was done using Ultraflex MALDI-TOF/TOF (BrukerDaltonics GmbH, Bremen, Germany) with an autoflex 123 smart beam. The instrument was operated by the FlexControl 3.0 software package. All spectra were obtained using the reflectron mode with an acceleration voltage of 25 kV, a reflector voltage of 26, and pulsed ion extraction of 40 ns in the positive ion mode. The acquisition range was from m/z 800 to 2000. Peak lists were generated from the MS spectra using BrukerFlex Analysis software (version 3.0).
Docking of EcChi1 with chitin tetramer
The protein sequence of EcChi1 was retrieved from NCBI Database (http://www.ncbi.nlm.nih.gov/) with accession number ADF62010.1 for modeling. To select appropriate templates for constructing 3D structure models of the EcChi1 protein, The BLAST program was used for sequence search against known 3D structure available in the Protein Databank (PDB) (http://www.rcsb.org/), The homology model program MODELLER v9.1255 was employed to generate 3D models of EcChi1 based on the crystal coordinates of chitinase II from Klebsiella pneumoniae (PDB: 3QOK). The stereochemical quality of the modeled protein structure was checked in Ramachandran plot56 using PROCHECK57. The compatibility of the model with its sequence was measured by Verify-3D graph58, 59. Chitin tetramer ligand was extracted from the 1NH6 crystal structure, using Discovery studio 4.0 and used for docking studies using Autodock 4.260 via Auto Dock Tools (ADT) graphical user interface. Interactions with ligand were viewed in PyMol Molecular Graphics System 1.7.
Electronic supplementary material
Transglycosylation by a chitinase from <i>Enterobacter cloacae</i> subsp. <i>cloacae</i> generates longer chitin oligosaccharides
Acknowledgements
We thank the Department of Science and Technology, Government of India, Funds for Infrastructure in Science and Technology, Level II support, and Special Assistance Programme–University Grants Commission (SAP-UGC) to the Department of Plant Sciences. The authors also thank UGC-supported University with Potential for Excellence (UPE) (Phase II) and DBT sponsored Bioinformatics Infrastructure Facility (BIF), at University of Hyderabad. MKM and VPR thank Indian Council Medical Research (ICMR) and Department of Biotechnology (DBT), Government of India, respectively, for Senior Research Fellowship. ARP thanks DBT for Tata Innovation Fellowship.
Author Contributions
M.K.M., P.R.V., S.N.D and A.R.P. designed the work. M.K.M. constructed the clone of EcChi1, purified the recombinant protein, performed enzyme characterization and H.P.L.C. studies. B.B.C. conducted docking studies. The data was analyzed by M.K.M., P.R.V., SND and A.R.P. Manuscript was written by M.K.M. by taking critical inputs from S.N.D and A.R.P.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05140-3
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Das SN, et al. Biotechnological approaches for field applications of chitooligosaccharides (COS) to induce innate immunity in plants. Crit. Rev. Biotechnol. 2015;35:29–43. doi: 10.3109/07388551.2013.798255. [DOI] [PubMed] [Google Scholar]
- 2.Hamed I, Ozogul F, Regenstein JM. Industrial applications of crustacean by products (chitin, chitosan, and chitooligosaccharides): A review. Trends Food Sci. Technol. 2016;48:40–50. doi: 10.1016/j.tifs.2015.11.007. [DOI] [Google Scholar]
- 3.Gillard L, Tran AT, Boyer FD, Beau JM. Chitooligosaccharide Synthesis Using an Ionic Tag. European J. Org. Chem. 2016;2016:1103–1109. doi: 10.1002/ejoc.201501476. [DOI] [Google Scholar]
- 4.Dahiya N, Tewari R, Hoondal GS. Biotechnological aspects of chitinolytic enzymes: A review. Applied Microbiology and Biotechnology. 2006;71:773–782. doi: 10.1007/s00253-005-0183-7. [DOI] [PubMed] [Google Scholar]
- 5.Divne C, et al. The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science. 1994;265:524–8. doi: 10.1126/science.8036495. [DOI] [PubMed] [Google Scholar]
- 6.Rouvinen J, Bergfors T, Teeri T, Knowles JK, Jones TA. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science. 1990;249:380–6. doi: 10.1126/science.2377893. [DOI] [PubMed] [Google Scholar]
- 7.Divne C, Stahlberg J, Teeri TT, Jones TA. High-resolution crystal structures reveal how a cellulose chain is bound in the 50°A long tunnel of cellobiohydrolase I from Trichoderma reesei. J. Mol. Biol. 1998;275:309–25. doi: 10.1006/jmbi.1997.1437. [DOI] [PubMed] [Google Scholar]
- 8.Varrot A, et al. Structural basis for ligand binding and processivity in cellobiohydrolase Cel6A from Humicola insolens. Structure. 2003;11:855–864. doi: 10.1016/S0969-2126(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 9.Umekawa M, et al. Mutants of Mucor hiemalis endo beta N-acetyl glucosaminidase show enhanced transglycosylation and glycosynthase like activities. J. Biol. Chem. 2008;283:4469–4479. doi: 10.1074/jbc.M707137200. [DOI] [PubMed] [Google Scholar]
- 10.Taira T, et al. Transglycosylation reaction catalyzed by a class V chitinase from cycad, Cycas revoluta: A study involving site directed mutagenesis, HPLC, and real-time ESI-MS. Biochim. Biophys. Acta - Proteins Proteomics. 2010;1804:668–675. doi: 10.1016/j.bbapap.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 11.Purushotham P, Podile AR. Synthesis of long chain chitooligosaccharides by a hyper transglycosylating processive endochitinase of Serratia proteamaculans 568. J. Bacteriol. 2012;194:4260–4271. doi: 10.1128/JB.06473-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suma K, Podile AR. Chitinase A from Stenotrophomonas maltophilia shows transglycosylation and antifungal activities. Bioresour. Technol. 2013;133:213–220. doi: 10.1016/j.biortech.2013.01.103. [DOI] [PubMed] [Google Scholar]
- 13.Huang CJ, Chen CY. High level expression and characterization of two chitinases, ChiCH and ChiCW, of Bacillus cereus 28-9 in Escherichia coli. Biochem. Biophys. Res. Commun. 2005;327:8–17. doi: 10.1016/j.bbrc.2004.11.140. [DOI] [PubMed] [Google Scholar]
- 14.Frederiksen RF, et al. Bacterial chitinases and chitin binding proteins as virulence factors. Microbiology. 2013;159:833–47. doi: 10.1099/mic.0.051839-0. [DOI] [PubMed] [Google Scholar]
- 15.Prescott JF, Parreira VR, Mehdizadeh Gohari I, Lepp D, Gong J. The pathogenesis of necrotic enteritis in chickens: what we know and what we need to know: a review. Avian Pathol. 2016;45:288–94. doi: 10.1080/03079457.2016.1139688. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann H, et al. Reassignment of Enterobacter dissolvens to Enterobacter cloacae as E. cloacae sub species dissolvens comb. nov. and emended description of Enterobacter asburiae and Enterobacter kobei. Syst. Appl. Microbiol. 2005;28:196–205. doi: 10.1016/j.syapm.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Madhuprakash J, et al. Inverse relationship between chitobiase and transglycosylation activities of chitinase-D from Serratia proteamaculans revealed by mutational and biophysical analyses. Sci. Rep. 2015;5:15657. doi: 10.1038/srep15657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuo Y, et al. Purification, characterization and gene analysis of N-acetyl glucosaminidase from Enterobacter sp. G-1. Biosci. Biotechnol. Biochem. 1999;63:1261–8. doi: 10.1271/bbb.63.1261. [DOI] [PubMed] [Google Scholar]
- 19.Tang Y, Zhao J, Ding S, Liu S, Yang Z. Purification and properties of chitinase from Enterobacter aerogenes. Wei Sheng Wu Xue Bao. 2001;41:82–6. [PubMed] [Google Scholar]
- 20.Salam M, et al. Cloning, characterization and expression of the chitinase gene of Enterobacter sp. NRG4. Indian J. Microbiol. 2008;48:358–364. doi: 10.1007/s12088-008-0044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chernin LS, et al. Molecular cloning, structural analysis, and expression in Escherichia coli of a chitinase gene from Enterobacter agglomerans. Appl. Environ. Microbiol. 1997;63:834–9. doi: 10.1128/aem.63.3.834-839.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams SJ, Withers SG. Glycosyl fluorides in enzymatic reactions. Carbohydrate Research. 2000;327:27–46. doi: 10.1016/S0008-6215(00)00041-0. [DOI] [PubMed] [Google Scholar]
- 23.Sasaki C, et al. Comparative study of the reaction mechanism of family 18 chitinases from plants and microbes. J. Biochem. 2002;131:557–564. doi: 10.1093/oxfordjournals.jbchem.a003134. [DOI] [PubMed] [Google Scholar]
- 24.Suginta W, Vongsuwan A, Songsiriritthigul C, Svasti J, Prinz H. Enzymatic properties of wild type and active site mutants of chitinase A from Vibrio carchariae, as revealed by HPLC-MS. FEBS J. 2005;272:3376–3386. doi: 10.1111/j.1742-4658.2005.04753.x. [DOI] [PubMed] [Google Scholar]
- 25.Aguilera B, et al. Transglycosidase Activity of Chitotriosidase: Improved enzymatic assay for the human macrophage chitinase. J. Biol. Chem. 2003;278:40911–40916. doi: 10.1074/jbc.M301804200. [DOI] [PubMed] [Google Scholar]
- 26.Purushotham P, Sarma PVSRN, Podile AR. Multiple chitinases of an endophytic Serratia proteamaculans 568 generate chitin oligomers. Bioresour. Technol. 2012;112:261–269. doi: 10.1016/j.biortech.2012.02.062. [DOI] [PubMed] [Google Scholar]
- 27.Brurberg MB, Nes IF, Eijsink VGH. Comparative studies of chitinases A and B from Serratia marcescens. Microbiology. 1996;142:1581–1589. doi: 10.1099/13500872-142-7-1581. [DOI] [PubMed] [Google Scholar]
- 28.Singh AK, Chhatpar HS. Purification and characterization of chitinase from Paenibacillus sp. D1. Appl. Biochem. Biotechnol. 2011;164:77–88. doi: 10.1007/s12010-010-9116-8. [DOI] [PubMed] [Google Scholar]
- 29.Dahiya N, Tewari R, Tiwari RP, Hoondal GS. Chitinase from Enterobacter sp. NRG4: Its purification, characterization and reaction pattern. Electron. J. Biotechnol. 2005;8:134–145. doi: 10.2225/vol8-issue2-fulltext-6. [DOI] [Google Scholar]
- 30.Gao C, et al. Characterization of extracellular chitinase from Chitinibacter sp. GC72 and its application in GlcNAc production from crayfish shell enzymatic degradation. Biochem. Eng. J. 2015;97:59–64. doi: 10.1016/j.bej.2015.02.010. [DOI] [Google Scholar]
- 31.Bendt A, Huller H, Kammel U, Helmke E, Schweder T. Cloning, expression, and characterization of a chitinase gene from the Antarctic psychrotolerant bacterium Vibrio sp. strain Fi:7. Extremophiles. 2001;5:119–26. doi: 10.1007/s007920100179. [DOI] [PubMed] [Google Scholar]
- 32.Lee SG, et al. Expression of recombinant endochitinase from the Antarctic bacterium, Sanguibacter antarcticus KOPRI 21702 in Pichia pastoris by codon optimization. Protein Expr. Purif. 2010;71:108–114. doi: 10.1016/j.pep.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 33.Hammami I, et al. Partial purification and characterization of chiIO8, a novel antifungal chitinase produced by Bacillus cereus IO8. J. Appl. Microbiol. 2013;115:358–366. doi: 10.1111/jam.12242. [DOI] [PubMed] [Google Scholar]
- 34.Yang S, et al. Cloning, expression, purification and application of a novel chitinase from a thermophilic marine bacterium Paenibacillus barengoltzii. Food Chem. 2016;192:1041–8. doi: 10.1016/j.foodchem.2015.07.092. [DOI] [PubMed] [Google Scholar]
- 35.Zakarlassen H, et al. Aromatic residues in the catalytic center of chitinase A from Serratia marcescens affect processivity, enzyme activity, and biomass converting efficiency. J. Biol. Chem. 2009;284:10610–10617. doi: 10.1074/jbc.M900092200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, et al. Characterization of a cold-adapted and salt-tolerant exo-chitinase (ChiC) from Pseudoalteromonas sp. DL-6. Extremophiles. 2016;20:167–176. doi: 10.1007/s00792-016-0810-5. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Kopparapu NK, Yan Q, Yang S, Jiang Z. Purification and characterisation of a novel chitinase from persimmon (Diospyros kaki) with antifungal activity. Food Chem. 2013;138:1225–1232. doi: 10.1016/j.foodchem.2012.11.067. [DOI] [PubMed] [Google Scholar]
- 38.Yuli P, Suhartono MT, Rukayadi Y, Hwang JK, Pyun YR. Characteristics of thermostable chitinase enzymes from the indonesian Bacillus sp.13.26. Enzyme Microb. Technol. 2004;35:147–153. doi: 10.1016/j.enzmictec.2004.03.017. [DOI] [Google Scholar]
- 39.Payne CM, et al. Hallmarks of processivity in glycoside hydrolases from crystallographic and computational studies of the Serratia marcescens chitinases. J. Biol. Chem. 2012;287:36322–36330. doi: 10.1074/jbc.M112.402149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jana S, et al. Aromatic-Mediated Carbohydrate Recognition in Processive Serratia marcescens Chitinases. J. Phys. Chem. B. 2016;120:1236–1249. doi: 10.1021/acs.jpcb.5b12610. [DOI] [PubMed] [Google Scholar]
- 41.Sorbotten A, Horn SJ, Eijsink VGH, Varum KM. Degradation of chitosans with chitinase B from Serratia marcescens: Production of chito-oligosaccharides and insight into enzyme processivity. FEBS J. 2005;272:538–549. doi: 10.1111/j.1742-4658.2004.04495.x. [DOI] [PubMed] [Google Scholar]
- 42.Stockinger LW, et al. The effect of the carbohydrate binding module on substrate degradation by the human chitotriosidase. Biochim. Biophys. Acta - Proteins Proteomics. 2015;1854:1494–1501. doi: 10.1016/j.bbapap.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Eide KB, Lindbom AR, Eijsink VGH, Norberg AL, Sorlie M. Analysis of productive binding modes in the human chitotriosidase. FEBS Lett. 2013;587:3508–3513. doi: 10.1016/j.febslet.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Aronson NN, et al. Family 18 chitinase oligosaccharide substrate interaction: subsite preference and anomer selectivity of Serratia marcescens chitinase A. Biochem. J. 2003;376:87–95. doi: 10.1042/bj20030273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horn SJ, et al. Costs and benefits of processivity in enzymatic degradation of recalcitrant polysaccharides. Proc. Natl. Acad. Sci. USA. 2006;103:18089–94. doi: 10.1073/pnas.0608909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe T, et al. Aromatic residues within the substrate-binding cleft of Bacillus circulans chitinase A1 are essential for hydrolysis of crystalline chitin. Biochem. J. 2003;376:237–244. doi: 10.1042/bj20030419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Y, et al. Mutation of Trp137 to glutamate completely removes transglycosyl activity associated with the Aspergillus fumigatus AfChiB1. Glycoconj. J. 2009;26:525–534. doi: 10.1007/s10719-008-9203-z. [DOI] [PubMed] [Google Scholar]
- 48.Madhuprakash J, Tanneeru K, Purushotham P, Guruprasad L, Podile AR. Transglycosylation by Chitinase D from Serratia proteamaculans improved through altered substrate interactions. J. Biol. Chem. 2012;287:44619–44627. doi: 10.1074/jbc.M112.400879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horn SJ, et al. Endo/exo mechanism and processivity of family 18 chitinases produced by Serratia marcescens. FEBS J. 2006;273:491–503. doi: 10.1111/j.1742-4658.2005.05079.x. [DOI] [PubMed] [Google Scholar]
- 50.Vaaje Kolstad G, Bunaes AC, Mathiesen G, Eijsink VGH. The chitinolytic system of Lactococcus lactis ssp. lactis comprises a nonprocessive chitinase and a chitin-binding protein that promotes the degradation of α- And β-chitin. FEBS J. 2009;276:2402–2415. doi: 10.1111/j.1742-4658.2009.06972.x. [DOI] [PubMed] [Google Scholar]
- 51.Kurasin M, Kuusk S, Kuusk P, Sorlie M, Valjamae P. Slow Off-rates and Strong Product Binding Are Required for Processivity and Efficient Degradation of Recalcitrant Chitin by Family 18 Chitinases. J. Biol. Chem. 2015;290:29074–85. doi: 10.1074/jbc.M115.684977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neeraja C, Moerschbacher B, Podile AR. Fusion of cellulose binding domain to the catalytic domain improves the activity and conformational stability of chitinase in Bacillus licheniformis DSM13. Bioresour. Technol. 2010;101:3635–41. doi: 10.1016/j.biortech.2009.12.118. [DOI] [PubMed] [Google Scholar]
- 53.Sharma AD, Singh J. A nonenzymatic method to isolate genomic DNA from bacteria and actinomycete. Anal. Biochem. 2005;337:354–356. doi: 10.1016/j.ab.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 54.Imoto T, Yagishita K. A Simple Activity Measurement of Lysozyme. Agric. Biol. Chem. 1971;35:1154–1156. doi: 10.1080/00021369.1971.10860050. [DOI] [Google Scholar]
- 55.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 56.Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/S0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 57.Laskowski RA, MacArthur MA, Moss DS, Thornton JM. 20.19. PROCHECK - a program to check the stereochemical quality of protein structures. J. App. Cryst. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 58.Bowie JU, Luthy R, Eisenberg D. A method to identify protein sequences that fold into a known three dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 59.Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 60.Morris G, Huey R. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park J, et al. Purification and characterization of the chitinase (ChiA) from Enterobacter sp. G-1. Biosci Biotechnol Biochem. 1997;61:684–689. doi: 10.1271/bbb.61.684. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transglycosylation by a chitinase from <i>Enterobacter cloacae</i> subsp. <i>cloacae</i> generates longer chitin oligosaccharides


