Abstract
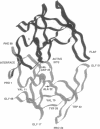
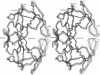
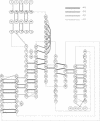
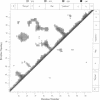
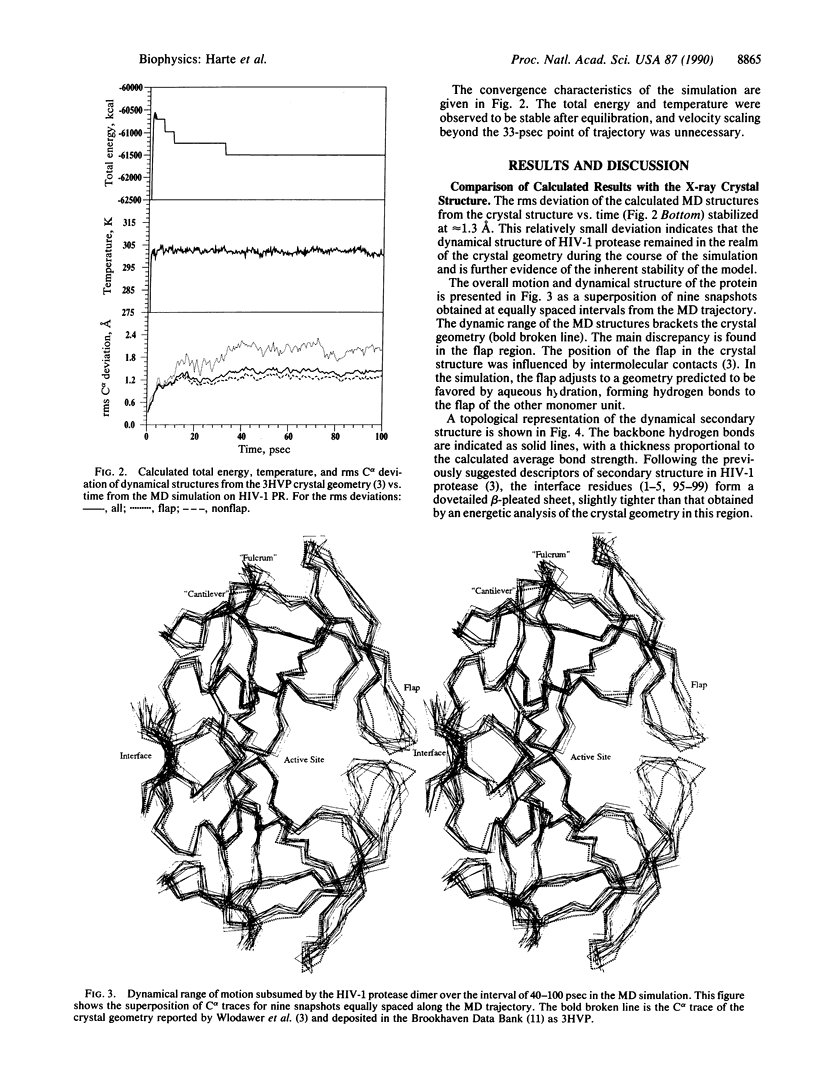
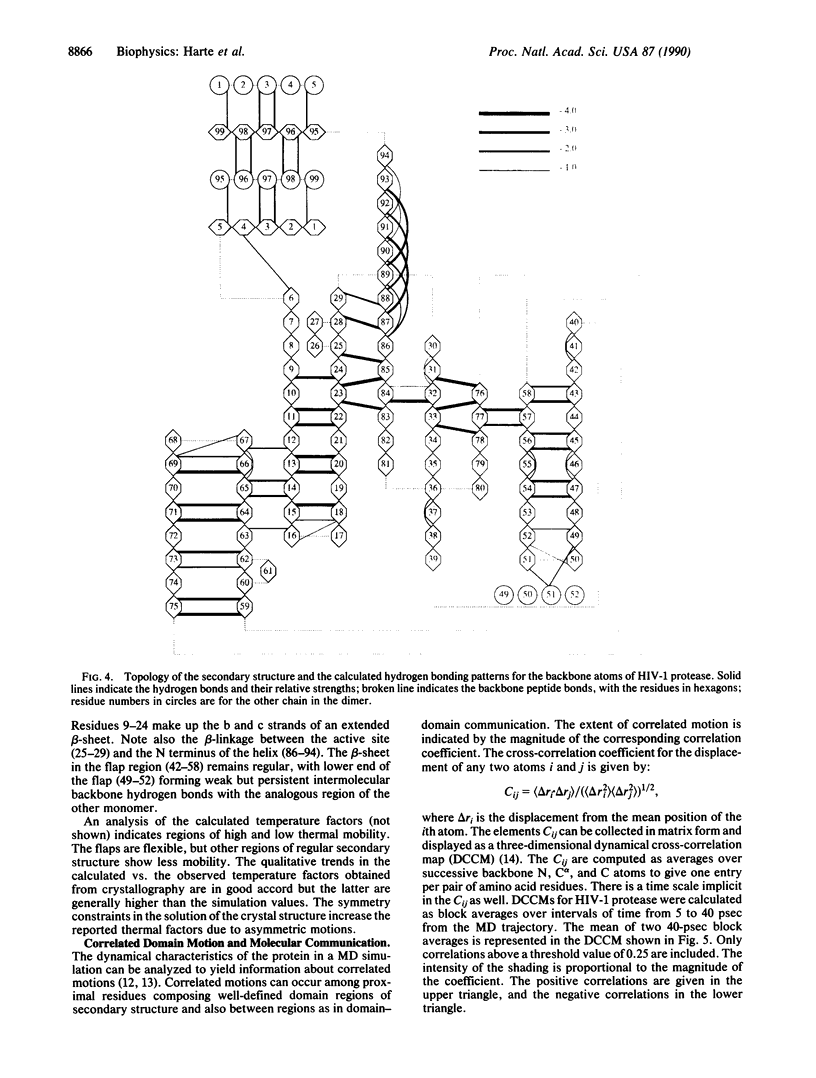
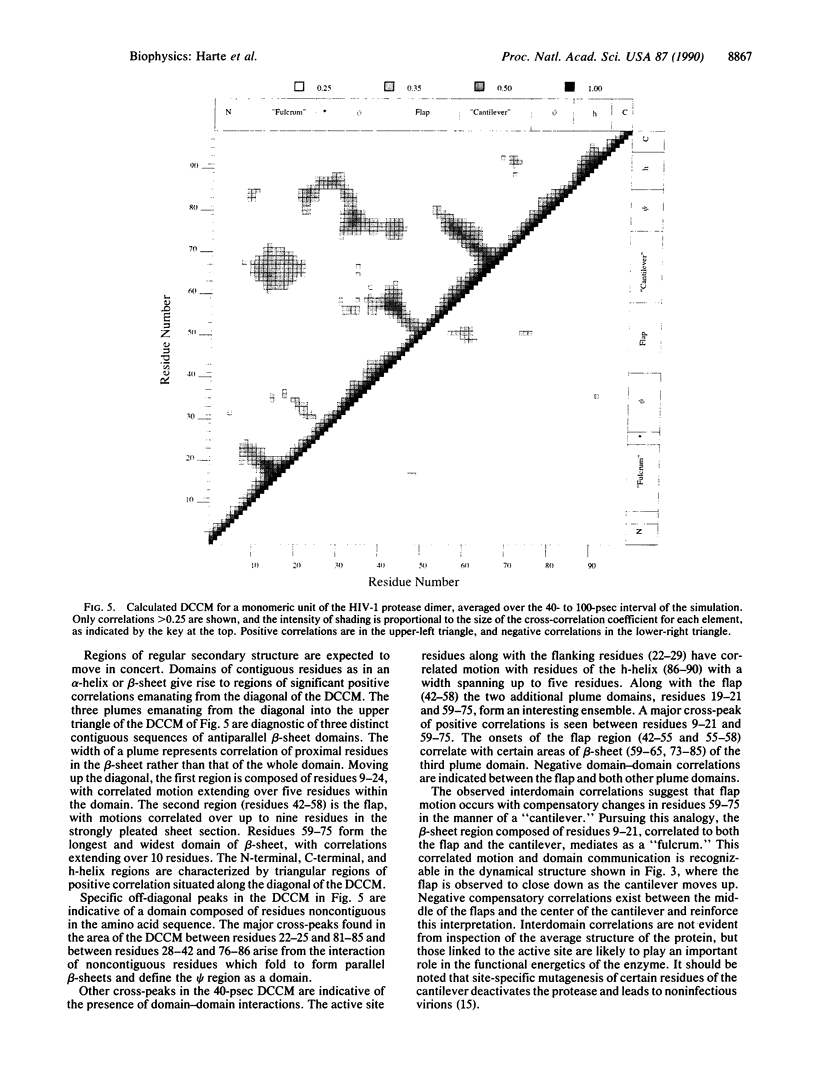
A dynamical model for the structure of the human immunodeficiency virus 1 (HIV-1) protease dimer in aqueous solution has been developed on the basis of molecular dynamics simulation. The model provides an accurate account of the crystal geometry and also a prediction of the structural reorganization expected to occur in the protein in aqueous solution compared to the crystalline environment. Analysis of the results by means of dynamical cross-correlation coefficients for atomic displacements indicates that domain-domain communication is present in the protein in the form of a molecular "cantilever" and is likely to be involved in enzyme function at the molecular level. The dynamical structure also suggests information that may ultimately be useful in understanding and further development of specific inhibitors of HIV-1 protease.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Beveridge D. L., DiCapua F. M. Free energy via molecular simulation: applications to chemical and biomolecular systems. Annu Rev Biophys Biophys Chem. 1989;18:431–492. doi: 10.1146/annurev.bb.18.060189.002243. [DOI] [PubMed] [Google Scholar]
- Blundell T., Pearl L. Retroviral proteinases. A second front against AIDS. Nature. 1989 Feb 16;337(6208):596–597. doi: 10.1038/337596a0. [DOI] [PubMed] [Google Scholar]
- Kohl N. E., Emini E. A., Schleif W. A., Davis L. J., Heimbach J. C., Dixon R. A., Scolnick E. M., Sigal I. S. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapatto R., Blundell T., Hemmings A., Overington J., Wilderspin A., Wood S., Merson J. R., Whittle P. J., Danley D. E., Geoghegan K. F. X-ray analysis of HIV-1 proteinase at 2.7 A resolution confirms structural homology among retroviral enzymes. Nature. 1989 Nov 16;342(6247):299–302. doi: 10.1038/342299a0. [DOI] [PubMed] [Google Scholar]
- Loeb D. D., Swanstrom R., Everitt L., Manchester M., Stamper S. E., Hutchison C. A., 3rd Complete mutagenesis of the HIV-1 protease. Nature. 1989 Aug 3;340(6232):397–400. doi: 10.1038/340397a0. [DOI] [PubMed] [Google Scholar]
- Miller M., Schneider J., Sathyanarayana B. K., Toth M. V., Marshall G. R., Clawson L., Selk L., Kent S. B., Wlodawer A. Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution. Science. 1989 Dec 1;246(4934):1149–1152. doi: 10.1126/science.2686029. [DOI] [PubMed] [Google Scholar]
- Navia M. A., Fitzgerald P. M., McKeever B. M., Leu C. T., Heimbach J. C., Herber W. K., Sigal I. S., Darke P. L., Springer J. P. Three-dimensional structure of aspartyl protease from human immunodeficiency virus HIV-1. Nature. 1989 Feb 16;337(6208):615–620. doi: 10.1038/337615a0. [DOI] [PubMed] [Google Scholar]
- Pearl L. H., Taylor W. R. A structural model for the retroviral proteases. Nature. 1987 Sep 24;329(6137):351–354. doi: 10.1038/329351a0. [DOI] [PubMed] [Google Scholar]
- Skalka A. M. Retroviral proteases: first glimpses at the anatomy of a processing machine. Cell. 1989 Mar 24;56(6):911–913. doi: 10.1016/0092-8674(89)90621-1. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Miller M., Jaskólski M., Sathyanarayana B. K., Baldwin E., Weber I. T., Selk L. M., Clawson L., Schneider J., Kent S. B. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989 Aug 11;245(4918):616–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]
- Yarchoan R., Mitsuya H., Broder S. AIDS therapies. Sci Am. 1988 Oct;259(4):110–119. doi: 10.1038/scientificamerican1088-110. [DOI] [PubMed] [Google Scholar]