Abstract
The genetic component of Immunoglobulin-A (IgA) vasculitis is still far to be elucidated. To increase the current knowledge on the genetic component of this vasculitis we performed the first genome-wide association study (GWAS) on this condition. 308 IgA vasculitis patients and 1,018 healthy controls from Spain were genotyped by Illumina HumanCore BeadChips. Imputation of GWAS data was performed using the 1000 Genomes Project Phase III dataset as reference panel. After quality control filters and GWAS imputation, 285 patients and 1,006 controls remained in the datasets and were included in further analysis. Additionally, the human leukocyte antigen (HLA) region was comprehensively studied by imputing classical alleles and polymorphic amino acid positions. A linkage disequilibrium block of polymorphisms located in the HLA class II region surpassed the genome-wide level of significance (OR = 0.56, 95% CI = 0.46–0.68). Although no polymorphic amino acid positions were associated at the genome-wide level of significance, P-values of potential relevance were observed for the positions 13 and 11 of HLA-DRB1 (P = 6.67E-05, P = 1.88E-05, respectively). Outside the HLA, potential associations were detected, but none of them were close to the statistical significance. In conclusion, our study suggests that IgA vasculitis is an archetypal HLA class II disease.
Introduction
Immunoglobulin-A (IgA) vasculitis, also known as Henoch-Schöenlein purpura (HSP), is the most common type of primary small-sized blood vessel leukocytoclastic vasculitis in children, although it may also develop in adults1. Although the classic clinical triad of IgA vasculitis consists of palpable purpura (involving the lower extremities), joints and the gastrointestinal tract, renal complications may also develop in affected individuals2. In this regard, the outcome of IgA vasculitis patients is related to the presence of glomerulonephritis, which may lead to chronic renal failure1, 2.
IgA vasculitis has a multifactorial etiology in which both environmental and genetic factors seem to contribute to the predisposition and clinical phenotype of the disease1, 3. However, the genetic component of this type of vasculitis remains poorly understood, as only a few candidate gene studies have been performed to date4, 5.
Unlike the candidate gene approach, genome-wide association studies (GWAS) imply a hypothesis free analysis of hundreds of thousands of single-nucleotide polymorphisms (SNPs) across the whole genome6. This strategy has proven to be a powerful tool to unravel the genetic component of complex diseases during the last decade, including primary vasculitides such as Takayasu Arteritis, Behçet disease, and antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis (AAV)7.
This study aimed at conducting the first GWAS of IgA vasculitis using the largest series of IgA vasculitis patients of European ancestry ever assessed for a genetic study.
Patients and Methods
Study population
A series of 308 patients diagnosed with IgA vasculitis and 1,018 unaffected and unrelated controls were genotyped in this study. The total number of individuals that passed the quality control (QC) filters mentioned below and were finally included in further analysis was 1,291 (285 and 1,006 for IgA vasculitis patients and controls, respectively). All subjects were from Spain and had European ancestry. IgA vasculitis condition was diagnosed accordingly with both the guidelines included in Michel et al.8 and the American College of Rheumatology classification criteria for this form of vasculitis9. A description of the main clinical features of the IgA vasculitis patients and controls analyzed after QC filters is shown in Supplementary Table S1. For experiments involving humans and the use of human blood samples, all the methods were carried out in accordance with the approved guidelines and regulations, according to the Declaration of Helsinki. All experimental protocols were approved by the Ethics Committees of clinical research of the Spanish regions of Galicia, Cantabria, Madrid, Andalucía, and País Vasco. All participants or their parents signed an informed consent form before being enrolled in the study.
Genotyping and quality controls
Genomic DNA was extracted from peripheral blood samples using standard methods. Genotyping was conducted using the GWAS platform “Infinium® HumanCore Beadchip” in an iScan System (Illumina, Inc) and following the manufacturer’s protocol.
Raw data were subjected to the following QC filters using PLINK v.1.0710: (1) SNPs with cluster separation <0.4, call rates <0.98, minor allele frequencies (MAF) <0.01, and those deviating from Hardy-Weinberg equilibrium (HWE; P < 0.001) were excluded; (2) samples with call rates <0.95, and those with identity by descent >0.4 were also removed. Sex chromosomes were not analyzed. The number of IgA vasculitis patients and controls that remained after each QC filter is shown in Supplementary Table S2.
Imputation of GWAS data
SNP genotype imputation throughout the genome was performed after initial QC using the 1000 Genomes Project (1KG) Phase III dataset as reference panel (www.1000genomes.org) and the software IMPUTE v.211. For that, we set the strand orientation, chromosome position, and SNP nomenclature accordingly with the build 37 (HG19) of the 1KG using PLNK. Imputation was carried out in individual chunks of 50,000 Mb covering whole-genome regions with a probability threshold for merging genotypes of 0.9 to maximize the quality of imputed variants. Imputed data were also subjected to the above mentioned QC filters in PLINK. Singletons were removed. Finally, possible population sub-stratification was controlled by principal component (PC) analyses using PLINK and the gcta64 and R-base software under GNU Public license v2. To identify outliers, we calculated and plotted the ten first PCs of each individual, and those deviating >4 standard deviations from the cluster centroid were excluded. PC analysis for the first three PCs for each individual are plotted in Supplementary Fig. The total number of SNPs that passed the QC and were finally analyzed was 1,909,910 (2,581,927 and 2,185,351 for IgA vasculitis patients and controls, respectively). The number of polymorphisms that remained after each QC filter is shown in Supplementary Table S2.
Human leukocyte antigen (HLA) imputation
Considering that IgA vasculitis is an immune-mediated condition, a more comprehensive analysis of the HLA region was conducted. With that aim, we extracted the extended HLA region (29,000,000 to 34,000,000 bp in chromosome 6) from the non-imputed data and imputed SNPs, classical HLA alleles at two- and four-digits, and polymorphic amino acid positions as described12–16. In brief, to impute this genomic region, we used the SNP2HLA method with the Beagle software package and the Type 1 Diabetes Genetics Consortium (T1DGC) reference panel comprised of 5,225 individuals of European origin with genotyping data of 8,961 common SNPs and indel polymorphisms across the xMHC region, and four digits genotyping data of the HLA class I and II molecules12–16. Imputed HLA data were also filtered with PLINK with the following thresholds: success call rate >0.95 for alleles and amino acids, deviation from HWE (P < 0.001) for SNPs, and >0.95 total call rate for individuals. Information of a total of 7,179 SNPs, 423 classical HLA alleles (126 at two-digit and 297 at four-digit resolution) of the HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, HLA-DQA1, HLA-DPB1 and HLA-DPA1 genes, and 1,275 amino acidic variants of the HLA system remained after the filters.
Statistical analyses
An estimation of the statistical power of the final cohort (285 IgA vasculitis patients/1,006 healthy controls) was obtained with CaTS Power Calculator for Genetic Studies software (Supplementary Table S3).
To test for association, we compared the genotype frequencies of every SNP between cases and controls by logistic regression on the best-guess genotypes assuming an additive model in PLINK. The ten first PCs were included as covariates. In the case of the HLA region, we tested SNPs, classical HLA alleles and all possible combinations of amino acid residues per position. A likelihood ratio test of amino acid positions was also conducted, as described12.
P-values, odds ratios (OR), and 95% confidence intervals (CI) were then calculated. The statistical threshold was set at the genome-wide level of significance (P < 5E-08). In the HLA analysis, despite not interrogating the whole genome but a specific region of chromosome 6, we decided to maintain the statistical threshold at the genome-wide level of significance (P < 5E-08) to avoid possible false positive results.
Results
Figure 1 summarizes the overall results of the study. Several association signals in high linkage disequilibrium (LD, r2 > 0.8) at the genome-wide level of significance were disclosed within the HLA region at chromosome 6. The strongest signal corresponded to a disequilibrium block of polymorphisms (OR = 0.56, 95% CI = 0.46–0.68) (Table 1), which we refer to as rs9275260, that maps to an intergenic region in HLA class II between HLA-DQA1 and HLA-DQB1. To confirm these results, we obtained direct genotypes of the Spanish cohort using a TaqMan probe for rs9275260. The overall concordance reached after comparing TaqMan types with the corresponding imputed data was 99.84%. Outside the HLA, some potential signals located in different intronic and intergenic regions were observed (Fig. 1), but none of them reached the statistical level of significance (Supplementary Table S4).
Figure 1.
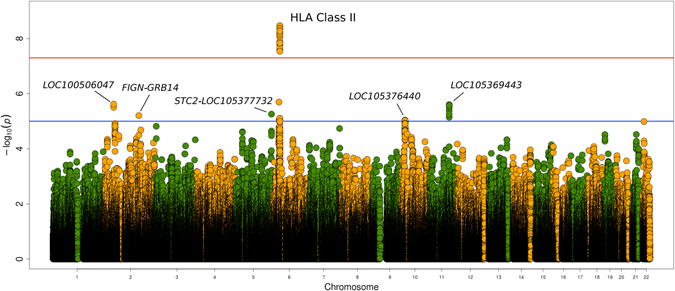
Manhattan plot representation of the results of this study. The −log10 of the p values are plotted against their physical chromosomal position. The red line represents the genome-wide level of significance (P < 5E-08). A less stringent threshold (p < 1E-05) is highlighted in blue.
Table 1.
Signals within HLA associated with IgA susceptibility at the GWAS significance level-P < 5E-08-after imputation of GWAS data.
| SNP | Position in chr 6 (GRCh37) | Reference allele | P | OR [CI 95%] |
|---|---|---|---|---|
| rs9275260 | 32.661.575 | C | 3.42E-09 | 0.56 [0.46–0.68] |
| rs9275259 | 32.661.572 | C | 3.42E-09 | 0.56 [0.46–0.68] |
| rs9275284 | 32.663.073 | C | 4.30E-09 | 0.56 [0.46–0.68] |
| rs9275285 | 32.663.080 | A | 4.30E-09 | 0.56 [0.46–0.68] |
| rs9275286 | 32.663.143 | T | 4.30E-09 | 0.56 [0.46–0.68] |
| rs9275288 | 32.663.203 | A | 4.30E-09 | 0.56 [0.46–0.68] |
| rs9275292 | 32.663.289 | C | 4.30E-09 | 0.56 [0.46–0.68] |
| rs9275244 | 32.660.881 | G | 4.92E-09 | 0.56 [0.46–0.68] |
| rs5000633 | 32.663.610 | C | 5.20E-09 | 0.56 [0.46–0.68] |
| rs2395522 | 32.664.722 | A | 5.25E-09 | 0.56 [0.46–0.68] |
| rs9275279 | 32.662.843 | G | 5.32E-09 | 0.56 [0.46–0.68] |
| rs9275281 | 32.662.920 | G | 5.32E-09 | 0.56 [0.46–0.68] |
| rs4248168 | 32.659.743 | G | 5.46E-09 | 0.56 [0.47–0.68] |
| rs9275224 | 32.659.878 | A | 5.46E-09 | 0.56 [0.47–0.68] |
| rs4713580 | 32.659.994 | C | 5.46E-09 | 0.56 [0.47–0.68] |
| rs4713584 | 32.660.237 | C | 5.46E-09 | 0.56 [0.47–0.68] |
| rs9275225 | 32.660.262 | G | 5.46E-09 | 0.56 [0.47–0.68] |
| rs5002704 | 32.659.279 | T | 5.67E-09 | 0.56 [0.46–0.68] |
| rs4713581 | 32.660.023 | T | 6.08E-09 | 0.56 [0.47–0.68] |
| rs4713583 | 32.660.153 | T | 6.08E-09 | 0.56 [0.47–0.68] |
| rs9275228 | 32.660.347 | G | 6.12E-09 | 0.56 [0.47–0.68] |
| rs9275227 | 32.660.337 | C | 6.12E-09 | 0.56 [0.47–0.68] |
| rs9275295 | 32.663.391 | A | 6.19E-09 | 0.56 [0.46–0.68] |
| rs9275277 | 32.662.677 | G | 7.33E-09 | 0.57 [0.47–0.69] |
| rs9275276 | 32.662.676 | T | 7.33E-09 | 0.57 [0.47–0.69] |
| rs9275245 | 32.660.943 | A | 7.89E-09 | 0.57 [0.47–0.69] |
| rs5002708 | 32.659.357 | T | 8.00E-09 | 0.57 [0.47–0.69] |
| rs5002707 | 32.659.337 | T | 8.00E-09 | 0.57 [0.47–0.69] |
| rs67838634 | 32.662.128 | G | 8.89E-09 | 1.76 [1.45–2.13] |
| rs9275222 | 32.659.516 | T | 1.28E-08 | 1.75 [1.44–2.12] |
| rs6457617 | 32.663.851 | C | 1.44E-08 | 0.57 [0.47–0.70] |
| rs6457620 | 32.663.999 | G | 1.44E-08 | 0.57 [0.47–0.70] |
| rs5002702 | 32.659.158 | G | 1.47E-08 | 0.57 [0.47–0.70] |
| rs9275226 | 32.660.311 | C | 1.49E-08 | 0.57 [0.47–0.70] |
| rs9275230 | 32.660.442 | A | 1.49E-08 | 0.57 [0.47–0.70] |
| rs5002705 | 32.659.319 | C | 1.60E-08 | 0.57 [0.47–0.70] |
| rs4713582 | 32.660.051 | T | 1.63E-08 | 0.57 [0.47–0.70] |
| rs4711304 | 32.660.170 | T | 1.65E-08 | 0.58 [0.47–0.70] |
| rs9275246 | 32.661.003 | C | 2.11E-08 | 0.58 [0.48–0.70] |
| rs4713587 | 32.659.535 | G | 2.36E-08 | 0.58 [0.48–0.70] |
| rs9275247 | 32.661.015 | T | 2.87E-08 | 0.58 [0.48–0.70] |
| rs9275231 | 32.660.505 | C | 2.81E-08 | 1.73 [1.42–2.09] |
HLA: Human leukocyte antigen; IgA: Immunoglobulin-A; GWAS: genome-wide association study; SNP: single nucleotide polymorphism; chr: chromosome; OR: odds ratio; CI: confidence interval.
We tried to narrow down the HLA association with IgA vasculitis by inferring SNPs, classical HLA alleles, and polymorphic amino acid positions using as reference the T1DGC panel. Accordingly, association signals at the genome-wide level of significance were disclosed (Fig. 2A). The genetic variant rs9275224 represented the strongest peak (P = 5.74E-09, OR = 0.56, 95% CI = 0.46–0.68) (Supplementary Table S5). The polymorphism rs9275260 (and SNPs in high linkage disequilibrium with it) observed in the genome-wide data analysis was not detected in the analysis of the HLA region since 1KG Phase III dataset was not used as reference panel in this analysis. Nevertheless, rs9275224 was in complete LD (r2 = 1) with rs9275260 (and, consequently, with all the SNPs of the same disequilibrium block) observed in the genome-wide data analysis, meaning that these polymorphisms represent the same signal. Although no polymorphic amino acid positions were associated at the genome-wide significance level, P-values of potential relevance were observed for the HLA-DRB1 positions 13 and 11 (P = 6.67E-05 and P = 1.88E-05, respectively) (Supplementary Table S6). Conditional logistic regression analyses of the HLA data indicated that rs9275224 explained most of the HLA associated variants in HLA class II (Fig. 2B). Regarding HLA class I, a potential signal in HLA-B was observed (rs2523650, P = 1.10E-05, OR = 1.59, 95% CI = 1.29–1.96).
Figure 2.
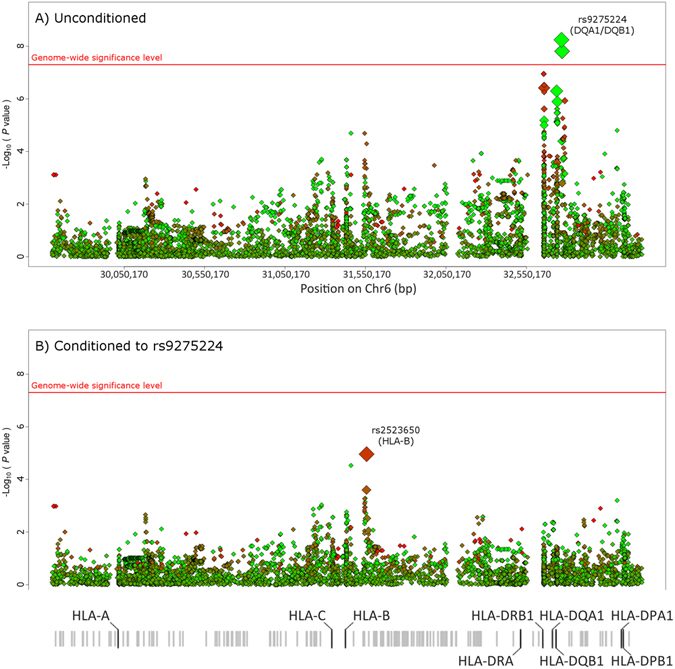
Manhattan plot representation of the step-wise conditional logistic regression of the HLA region. (A) Unconditioned test of the HLA region. (B) Results of the HLA region after controlling for rs9275224. The −log10 of the p values are plotted against their physical chromosomal position. A red/green color gradient was used to represent the effect size of each analyzed variant (red for risk and green for protection). The red line represents the genome-wide level of significance (P < 5E-08).
Discussion
This study represents the first GWAS of IgA vasculitis. Consistent with the results obtained in a former study5, our data suggest the involvement of HLA class II region in the pathophysiology of IgA vasculitis, thus supporting the high relevance of the immune system in the development of this disease and suggesting that IgA vasculitis may be related to other class II vasculitides such as giant cell arteritis (GCA)12 or AAV17. The strongest signal mapped to the HLA-DQA1/DQB1 region, which is in high LD with the HLA-DRB1 gene. Consequently, the associated polymorphisms may be tagging a putative aetiologic variant at HLA-DRB1. Regarding polymorphic amino acid positions, none of the signals reached the genome-wide level of significance. Nevertheless, likewise rheumatoid arthritis13 and GCA12, the HLA-DRB1 positions 13 and 11 were amongst the strongest signals, which support the notion that IgA vasculitis may share immunopathogenic pathways with these conditions. On the other hand, after performing the conditional analysis on the HLA data, a potential signal that maps to HLA-B was observed, although it did not reach the genome-wide level of significance. This result could be indicating a potential effect of HLA class I in the pathogenesis of IgA vasculitis, as previously proposed4. In addition, no consistent associations with IgA vasculitis susceptibility were detected outside the HLA region, probably due to an insufficient statistical power to detect risk variants with a moderate effect.
Vasculitides constitute a heterogeneous group of diseases that often have overlapping clinical and pathological manifestations18. Nevertheless, differences between them in molecular terms have been described7. In this regard, the results derived from our study classify IgA vasculitis as a HLA class II condition linking it to GCA and AAV. Nonetheless, it is important to keep in mind that the number of cases recruited in our study was not high and replication was not carried out. Because of that, further confirmatory studies in independent populations should be performed to validate our data.
In summary, our results suggest that IgA vasculitis is an archetypal HLA class II disease.
Electronic supplementary material
Acknowledgements
We wish to thank all the patients with IgA vasculitis and controls for their participation in this study. We want to specially thank Patricia Fuentevilla Rodríguez, María Del Camino Villa Llamazares and María Eugenia CuadradoMantecón for their technical assistance. This study was supported by European Union FEDER funds and “Fondo de Investigaciones Sanitarias” (grant PI12/00193) from ‘Instituto de Salud Carlos III’ (ISCIII, Health Ministry, Spain). RL-M was supported by the Miguel Servet I programme of the Spanish Ministry of Economy and Competitiveness through the grant CP16/00033. FDC was supported by the Ramón y Cajal programme of the Spanish Ministry of Economy and Competitiveness through the grant RYC-2014-16458. FG was recipient of a Sara Borrell postdoctoral fellowship from the “Instituto Carlos III de Salud” at the Spanish Ministry of Health (Spain) (CD15/00095). SR-M and BU were supported by funds from the RETICS Program (RIER) (RD16/0012/0009 and RD12/0009/0013, respectively).
Author Contributions
R.L.-M. and F.D.C. conceptualized and designed the study, drafted the initial manuscript, and approved the final manuscript as submitted. S.C., F.G., S.R.-M., B.S.-P., N.O.-C. and J.L. carried out the initial analyses, reviewed and revised the manuscript, and approved the final manuscript as submitted. B.U. and V.M. performed the initial analyses, drafted the manuscript and read and approved the final version of the manuscript. T.P., J.A.M.-F., A.N.P., D.A., M.A., E.R., M.L.L., J.M.B.-M., E.G.-A. and D.J. Jayne carried out the initial analyses, reviewed and revised the manuscript, and approved the final manuscript as submitted. R.B., J.M. and M.A.G.-G. designed the data collection instruments, and coordinated and supervised data collection, critically reviewed the manuscript, and approved the final manuscript as submitted.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Raquel López-Mejías and F. David Carmona contributed equally to this work. Ricardo Blanco, Javier Martín and Miguel A. González-Gay jointly supervised this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-03915-2
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gonzalez-Gay MA, Garcia-Porrua C. Epidemiology of the vasculitides. Rheum Dis Clin North Am. 2001;27:729–49. doi: 10.1016/S0889-857X(05)70232-5. [DOI] [PubMed] [Google Scholar]
- 2.Calvo-Río V, et al. Henoch-Schonlein purpura in northern Spain: clinical spectrum of the disease in 417 patients from a single center. Medicine (Baltimore). 2014;93:106–13. doi: 10.1097/MD.0000000000000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyatt RJ, Julian BA. IgA nephropathy. N Engl J Med. 2013;368:2402–14. doi: 10.1056/NEJMra1206793. [DOI] [PubMed] [Google Scholar]
- 4.López-Mejías R, et al. Association of HLA-B*41:02 with Henoch-Schonlein Purpura (IgA Vasculitis) in Spanish individuals irrespective of the HLA-DRB1 status. Arthritis Res Ther. 2015;17:102. doi: 10.1186/s13075-015-0622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.López-Mejías R, et al. HLA-DRB1 association with Henoch-Schonlein purpura. Arthritis Rheumatol. 2014;67:823–7. doi: 10.1002/art.38979. [DOI] [PubMed] [Google Scholar]
- 6.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 7.Carmona FD, Martín J, González-Gay MA. Genetics of vasculitis. CurrOpinRheumatol. 2015;27:10–7. doi: 10.1097/BOR.0000000000000124. [DOI] [PubMed] [Google Scholar]
- 8.Michel BA, Hunder GG, Bloch DA, Calabrese LH. Hypersensitivity vasculitis and Henoch-Schonlein purpura: a comparison between the 2 disorders. J Rheumatol. 1992;19:721–8. [PubMed] [Google Scholar]
- 9.Mills JA, et al. The American College of Rheumatology 1990 criteria for the classification of Henoch-Schonlein purpura. Arthritis Rheum. 1990;33:1114–21. doi: 10.1002/art.1780330809. [DOI] [PubMed] [Google Scholar]
- 10.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoSGenet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carmona FD, et al. A large-scale genetic analysis reveals a strong contribution of the HLA class II region to giant cell arteritis susceptibility. Am J Hum Genet. 2015;96:565–80. doi: 10.1016/j.ajhg.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raychaudhuri S, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–6. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia X, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayes MD, et al. Immunochip analysis identifies multiple susceptibility loci for systemic sclerosis. Am J Hum Genet. 2014;94:47–61. doi: 10.1016/j.ajhg.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ortiz-Fernández L, et al. Genetic Analysis with the Immunochip Platform in Behçet Disease. Identification of Residues Associated in the HLA Class I Region and New Susceptibility Loci. PLoS One. 2016;11:e0161305. doi: 10.1371/journal.pone.0161305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyons PA, et al. Genetically distinct subsets within ANCA-associated vasculitis. N Engl J Med. 2012;367:214–23. doi: 10.1056/NEJMoa1108735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jennette JC, et al. 2012 revised International Chapel Hill Consensus Conference Nomenclature of Vasculitides. Arthritis Rheum. 2013;65:1–11. doi: 10.1002/art.37715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


