Abstract
The safe and effective delivery of therapeutic genes into target cell interiors is of great importance in gene therapy. Chitosan has been extensively studied as a gene delivery carrier due to its good biocompatibility and biodegradability. Understanding the atomic interaction mechanism between chitosan and DNA is important in the design and application of chitosan-based drug and gene delivery systems. In this work, the interactions between single-stranded polynucleotides and different types of chitosan were systematically investigated by using molecular dynamics (MD) simulation. Our results demonstrate that the functional groups of chitosan, the types of base and length of polynucleotides regulate the interaction behavior between chitosan and polynucleotides. The encapsulation capacity of polynucleotide by chitosan is mainly balanced by two factors: the strength of polynucleotide binding to chitosan and the tendency of self-aggregation of polynucleotide in the solution. For –NH3 + chitosan, due to the strong electrostatic interaction, especially the H-bond between –NH3 + groups in chitosan and phosphate groups in polynucleotide, the aggregation effect could be partially eliminated. The good dispersal capacity of polynucleotides may improve the encapsulation of polynucleotides by chitosan, and hence increase the delivery and transfection efficiency of chitosan-based gene carrier.
Introduction
The safe and effective delivery of therapeutic genes into target cell interiors is of great importance in gene therapy1–5. Although the traditional viral vectors can offer better efficiency of gene delivery, due to its side effect (e.g., endogenous virus recombination, oncogenic effects, unexpected immune response etc.) on clinical applications6–8, it has been gradually replaced by non-viral vectors (i.e., typically based on polymeric micelles, dendrimers, nanoparticles, liposomes etc.)9–11.
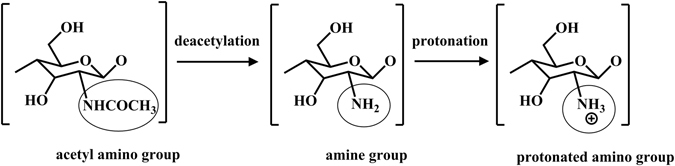
As a member of non-viral vectors, much attention have been given to chitosan and chitosan derivatives due to their biodegradability, biocompatibility and their mucoadhesive as well as high positive charge density in low pH solution6, 12–14. As shown in Fig. 1, chitosan can be obtained by deacetylation of chitin, and the primary amines in the chitosan backbone can subsequently become positively charged in low pH solution. Its positively charged character enables chitosan binding strongly to several mammalian cells15. Moreover, chitosan forms polyelectrolyte complexes with negatively charged DNA, in which the DNA becomes better protected against nuclease degradation16. These beneficial characters give chitosan the abilities to be a competent gene carrier. However, the gene delivery and transfection efficiency of chitosan is relatively low, thus their clinical applications were greatly limited2.
Figure 1.
The schemetic of chitosan’s versatility with different functional groups.
Further studies also showed that the transfection efficiency of chitosan is significantly influenced by a series of chitosan-based formulation parameters, such as the molecular weight (MW) of chitosan, its degree of deacetylation (DD), the charge ratio of chitosan to DNA/siRNA etc.17–21. For instance, the study of Lavertu et al.21 has shown that the maximum transfection efficiency could be obtained by simultaneously increasing the MW and decreasing the DD, indicating a multi-factor effect (i.e., particle stability and electrostatic binding of chitosan-DNA complexes) in determining transfection efficiency. There are also some studies reported the modification of chitosan for the purpose of improving transfection efficiency22, 23. Gao et al.22 prepared an arginine-chitosan gene carrier where the arginine was linked through NH2 group. They concluded that modification of functional groups on chitosan could improve chitosan’s water solubility and enhance its gene transfection efficiency in HEK 293 and COS-7 cells. Guţoaia et al.23 recommended that fine-tuned PEGylation of chitosan can be used to generate PEG-chitosan/siRNA delivery systems with maximum bioactivity. These experiments greatly enhanced our understanding of the property of chitosan as a gene carrier. In addition, all of these experiments, including chitosan-based formulation parameters and chitosan modification show that the purposeful changing of functional group in the backbone of chitosan may provide an effective strategy in improving the transfection efficiency. However, the interaction mechanism between chitosan and DNA is still unclear at molecular level. In particular, the effect of functional group in the backbone of chitosan (e.g., –NH2, –NH3 + and –NHCOCH3, as shown in Fig. 1) on DNA delivery is still obscure.
In fact, with the goal of improving the transfection efficiency and realizing real application of chitosan-based gene systems, it is of great importance to investigate the effect of functional group on DNA delivery and underlying mechanism in details. Recently, with the help of fast-growing computational power, molecular dynamics (MD) simulation may offer plenty of useful information on this challenge by shedding light onto the atomic level details of chitosan-based drug/gene delivery systems24, 25. Recently, MD simulation have been widely used to study the critical steps in gene delivery, including nanoparticle formation dynamics with various carriers, complexation of carriers with polynucleotides, carrier conformations in endosome stages, and release of polynucleotides from carriers etc.26–32. For example, Zhan et al.31 reported that the hydrophobic/hydrophilic modifications of linear polycations may bring improvement on its properties as gene carrier. Ding et al.32 found that the property of polyelectrolyte chains grafted to nanovector and DNA molecules can have important impacts on the endocytosis.
In this study, the mechanism of single-stranded polynucleotide encapsulation by chitosan and interaction between different types of polynucleotide and chitosan with atomic details were investigated by MD simulations. Herein, three extreme cases for chitosan with 10 units were considered for simplicity: 0% deacetylated structure (designated as –NHCOCH3 chitosan), fully deacetylated structure in neutral solution (–NH2 chitosan), and fully deacetylated structure in low pH solution (–NH3 + chitosan). Besides, four types of single-stranded polynucleotide models were used (3 or 10 units of polycytidylic acid and polyguanylic acid) to understand the effect of base and chain length of polynucleotide on the encapsulation by chitosan. These polynucleotides were represented as C3, G3, C10 and G10 in the following sections of the paper. The choice of single-stranded polynucleotides in this work mainly based on two facts: (1) the importance of single-stranded oligonucleotides in gene editing and gene therapy; (2) single-stranded polynucleotide is one of the simplest DNA structure, and it was selected to understand the fundament mechanism of interaction between chitosan and DNA via molecular dynamics simulation. The structure of chitosan and single-stranded polynucleotides with atomic details can be found in Figs S1 and S2 in Supplementary Information.
Simulation methods
Firstly, the structure and topology of polyglucose with 10 units were generated by LEaP module from the AMBER33 suite of programs. Secondly, the initial structures of chitosans with 10 units were generated by replacing the glucose unit –OH group bonded with C2 into –NH2, –NH3 + and –NHCOCH3 groups in our home made script (see Fig. S1 in Supplementary Information). The geometry of unit of chitosan with different groups was optimized with HF/6–31G+ by Gaussian03 software34. Then the force field parameters of –OH group in glucose topology were replaced by the parameters (including the atom charges, bonded and non-bonded parameters) of –NH2, –NH3 + and –NHCOCH3 groups in this home made script. These force field parameters of chitosan unit with different groups were derived from the general AMBER force field (GAFF) and the standard AMBER 2003 force field, respectively, by using AmberTools33. The parameters for bonds, angles and dihedral angles between different units of chitosan were obtained from the reference35. The partial charges were derived by the calculation of chitosan unit with different functional groups using restrained electrostatic potential (RESP) method at the HF/6-31G+ level in Gaussian 03.
The initial structure of single-stranded polynucleotides was obtained from one chain of standard B-form double-stranded DNA constructed by the nucleic acid database in the Hyperchem software (Version 7.0, Hypercube, Inc). In B-form dsDNA, there is ~10.5 bp in one turn, and extends 34 Å per 10 bp of sequence. The inclination of base pair to axis is around −1.2° and mean propeller twist is +16°. The parameters of polynucleotides were derived from the Amber99sb-ildn force field36. All simulations were performed by GROMACS 5.0.7 package37. In each system, certain number of chitosans and polynucleotides chains with different ratio (shown in Table 1) were solvated in a box of TIP3P38 water molecules by packmol39 software. Counter-ions (Na+ or Cl−) were added into the solution to neutralize the systems. Periodic boundary conditions and a time step of 2 fs were applied. The cut-off of the van der Waals (vdW) interaction was set to be 1.2 nm. The LINCS algorithm40 was used to constrain all the bond lengths. The particle mesh Ewald (PME) method41 was used to calculate the electrostatic interactions with a cut-off of 1.2 nm. The Lorentz-Berthelot rule42 was employed to calculate the Lennard-Jones (LJ) potential of cross interaction between chitosan and polynucleotide. Temperature coupling using velocity rescaling43 with a stochastic term was used to keep the temperature at 300 K. The Berendsen method44 was used to control the pressure at 1 bar with an isothermal compressibility of 4.5 × 10−5 bar−1. In each system, a 50000 step energy minimization was firstly performed by steepest descent method45. Subsequently, a 200 ps pre-equilibration under an NVT ensemble was performed. Finally, a 100 ns MD simulation in the NPT ensemble was carried out. Visual molecular dynamics (VMD) graphics software was used for molecular visualization46.
Table 1.
All performed simulations in this study.
| MD simulations | |||||
|---|---|---|---|---|---|
| Name of system | Number of chitosan | Number of polynucleotides | Type of counter-ions | Number of counter-ions | Simulation time (ns) |
| NH3 +_C3 | 6 | 20 | Cl− | 20 | 100 |
| NH2_C3 | 6 | 20 | Na+ | 40 | 100 |
| NHCOCH3_C3 | 6 | 20 | Na+ | 40 | 100 |
| NH3 +_G3 | 6 | 20 | Cl− | 20 | 100 |
| NH2_G3 | 6 | 20 | Na+ | 40 | 100 |
| NHCOCH3_G3 | 6 | 20 | Na+ | 40 | 100 |
| NH3 +_C10 | 6 | 6 | Cl− | 6 | 100 |
| NH2_C10 | 6 | 6 | Na+ | 54 | 100 |
| NHCOCH3_C10 | 6 | 6 | Na+ | 54 | 100 |
| NH3 +_G10 | 6 | 6 | Cl− | 6 | 100 |
| NH2_G10 | 6 | 6 | Na+ | 54 | 100 |
| NHCOCH3_G10 | 6 | 6 | Na+ | 54 | 100 |
Results and Discussion
Effect of chitosan’s functional groups on DNA encapsulation
Figure 2(a) shown the radial distribution functions (RDFs) of C3 and G3 around different functional groups of chitosan (with the ratio of chitosan:C3/G3 = 3:10, see Table 1). The RDFs was calculated between the center of mass of chitosan’s functional groups (e.g., –NH2, –NH3 + and –NHCOCH3, as shown in Fig. 1) and any atoms of single-stranded polynucleotides. It could be used to quantitatively estimate the distribution of polynucleotides around chitosan and evaluated the effect of chitosan’s functional groups on polynucleotides distribution. The peak height of g(r) of –NH3 + chitosan/C3 system (solid blue line) is around 9.1, and it is highest comparing with other systems. It indicates that C3 tends to tightly absorbed on the –NH3 + chitosan, due to the strong electrostatic attraction between negatively charged C3 and protonated amines of –NH3 + chitosan. Besides, one could find that the peak height of g(r) is about 8.9 in –NH2 chitosan/C3 system (solid red line). It is relatively high and very close to that of –NH3 + chitosan system, with slightly narrow distribution of C3 within 2 nm around –NH2 chitosan. The peak height of g(r) is around 6.9 in –NHCOCH3 chitosan/C3 system (solid black line), and it is much lower than that of other two types of chitosan complexed with C3. The encapsulation of polynucleotides was also characterized by monitoring the number of intimate contact atoms between chitosan and C3/G3. Herein, an intimate contact between chitosan and C3/G3 was considered if any C3/G3 atoms are within the distance of 6.0 Å of the center of mass of chitosan’s functional groups (e.g., –NH2, –NH3 + and –NHCOCH3, as shown in Fig. 1). The time evolution of the number of contact atoms between chitosan and C3/G3 was shown in Fig. 2(b), where one could find more C3 atoms around –NH3 + chitosan systems. The contact curve have quite similar trend with the results of RDFs curve. Moreover, these differences can also be found in the typical snapshots of these systems at the end of 100 ns MD simulation displayed in Fig. 3. The trajectory and snapshot showed that C3 dispersed into –NH2 chitosan, leading to a sandwich-like conformation, while –NHCOCH3 chitosan prefer to aggregate together and free C3 can be found distribute around the big condensates of –NHCOCH3 chitosan. Figures S3–S8 displayed the polynucleotide encapsulation process in chitosan-C3/G3 systems. These conformations of different chitosan are in accordance with the results of RDFs curve and contact curve, and C3 can be well encapsulated in both –NH3 + and –NH2 chitosan systems.
Figure 2.
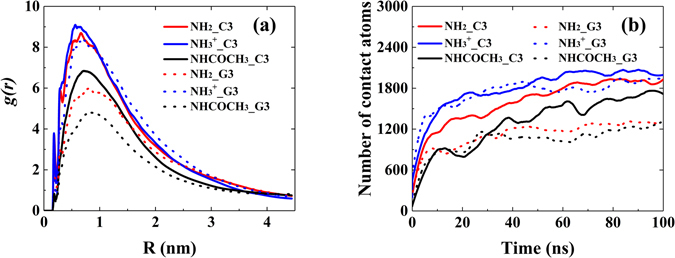
(a) The RDFs of C3 and G3 around different functional groups in chitosan (b) Number of contact atoms between chitosan and C3/G3 as a function of simulation time, with the ratio chitosan:C3/G3 = 3:10.
Figure 3.
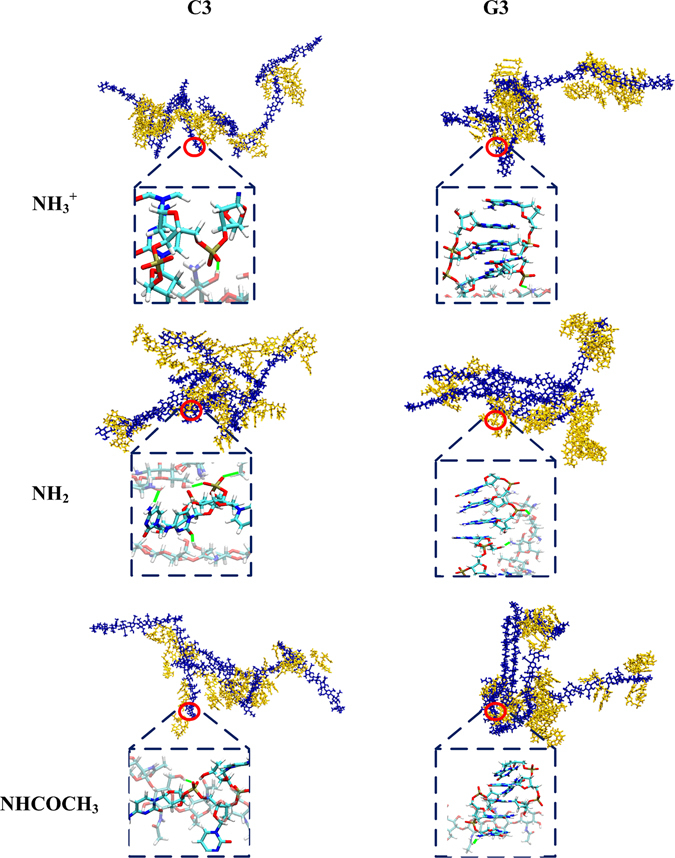
The snapshots of C3 and G3 encapsulated by different types of chitosan at the end of 100 ns MD simulation (blue licorice model: chitosan, yellow licorice model: C3 or G3). Water molecules were omitted for clarity. Enlarged atomic pictures of interaction between chitosan and single-stranded polynucleotides were displayed in blue dashed box (transparent licorice model: chitosan, non-transparent licorice model: C3 or G3, blue: nitrogen, white: hydrogen, red: oxygen, cyan: carbon and orange: phosphorus). The hydrogen bonds formed between chitosan and C3/G3 were marked by solid green line.
To better understand the effect of functional group on the encapsulation of polynucleotides, the mechanism of the C3 adsorption onto different types of chitosan was studied. As claimed in several experimental and theoretical studies, hydrogen bond is the main driving force in chitosan structural changes47, 48. Herein, the number of inter-molecular H-bond in C3 system was calculated to address the effect of functional groups. Figure 4(a) showed that the number of H-bond formed between C3 and different functional groups increased as a function of simulation time and slightly fluctuated at the end of the simulation. One could find that the system of –NH3 + chitosan/C3 has the largest H-bond numbers among three systems, which indicates that the strength of H-bond between C3 and –NH3 + chitosan is strongest. It could be attribute to the strong electrostatic interaction between positive charged –NH3 + group in chitosan and negatively charged –PO4 2− group in C3, as shown in Fig. 5(a). With tight contact between –NH3 + group in chitosan and –PO4 2− in C3, the chance of forming H-bond is high. Figure 5(a) also showed that the electrostatic interaction dominates the interaction between C3 and different types of chitosan. In the AMBER force field, there is no special term that is specific to hydrogen bonds. The hydrogen bond energy still arises from the dipole–dipole interaction of the donor and acceptor groups, and it is added to the electrostatic potential. Therefore, H-bond has great contribution to the electrostatic interaction and total interaction energy. It has large effect on the interaction strength between C3 and chitosan, and thus affects the distribution of C3 around chitosan.
Figure 4.
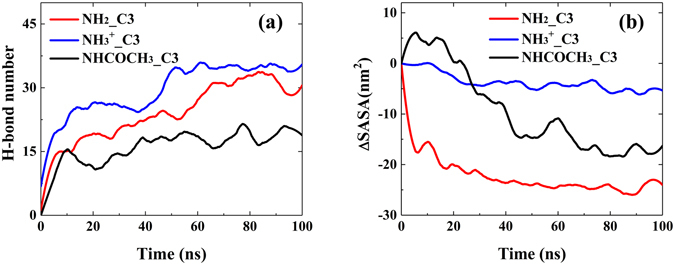
(a) The number of inter-molecular H-bonds between different functional groups and C3. (b) The SASA change of different types of chitosan in chitosan/C3 systems versus the simulation time.
Figure 5.
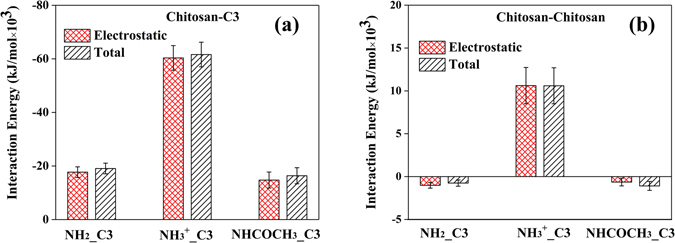
The average electrostatic and total interaction energies of (a) Chitosan–C3 and (b) Chitosan–chitosan in the systems with the ratio chitosan: C3 = 3:10. Data were taken from the last 50 ns trajectories of the total 100 ns in the MD simulations. The error bars represent the standard deviations of the data.
In Fig. 4(b), the solvent accessible surface area (SASA) change of different types of chitosan versus the simulation time was displayed. Comparing with other two systems, –NH3 + chitosan has smallest change of SASA. Due to the inter-molecular and intra-molecular repulsive electrostatic interaction between –NH3 + groups, –NH3 + chitosans have more stretched chain conformations and less aggregate structure. Both –NH2 and –NHCOCH3 chitosan has lower SASA value at the end of simulation, and the notable change of SASA in these two systems suggests that they could form more aggregated and condensed structure. To confirm this argument and provide a reference for dynamics of aggregation of –NH2 and –NHCOCH3 chitosan, the SASA change of different types of chitosan without polynucleotides during 100 ns MD simulation were plotted in Fig. S9. It was found that without polynucleotide the SASA of both of –NH2 and –NHCOCH3 chitosan remarkably decreased. These results consist with the fact that deacetylation could increase the solubility of chitosan, and the solubility of chitosan at low pH (most of the –NH2 groups are protonated) is much higher49.
The difference of the RDFs curve for –NH2 and –NHCOCH3 chitosan systems in Fig. 2(a) can also be explained by comparing the H-bond numbers and ∆SASA curve. Note that C3 in –NH2 chitosan/C3 system can form much higher number of inter-molecular H-bond with –NH2 chitosan than that with –NHCOCH3 chitosan/C3 system, and this could be attributed to the fact that –NH2 group have more H-donors than –NHCOCH3 group. Therefore, –NHCOCH3 chitosan have weaker interaction energy with C3 comparing with –NH2 chitosan (see Fig. 5). In addition, the strong tendency of –NH2 chitosan’s self-assembly will tightly encapsulate C3, leads to a sandwitch-like morphology of –NH2 chitosan/C3 system and increase the distribution of C3 around –NH2 chitosan. On the basis of strong interaction (including H-bond) between C3 and –NH2 chitosan, although –NH2 chitosan could form stronger aggregated structure than –NH3 + chitosan, the distribution of C3 around –NH2 chitosan is slightly lower than that around –NH3 + chitosan.
Effect of base type on DNA encapsulation
As reported in several studies50, 51, the nature of polynucleotides has great influence on their interaction with gene carriers. For instance, compared with DNA, the siRNA was able to establish stronger interactions with carriers due to its more flexible structure50. Herein, for the purpose of understanding the effect of base types on DNA encapsulation, the process of G3 encapsulation into different types of chitosan with the ratio chitosan: G3 = 3:10 were also investigated (see Table 1).
As shown in Fig. 2(a), the peak height of RDF in –NH3 + chitosan/G3 system (blue dot line) is around 8.2, and it is a little bit lower than that in –NH3 + chitosan/C3 system. However, in the case of –NH2 (red dot line) and –NHCOCH3 (black dot line) chitosan/G3 systems, the peak height of the RDFs curve of G3 is much lower, which indicates that the distribution of G3 around these two types of chitosan is much lower than that of C3. These results can also be found by comparing the difference of number of contact atoms in these systems, as shown in Fig. 2(b). For further comparison, the typical snapshots of different types of chitosan complexed with C3 and G3 at the end of 100 ns MD simulation were also displayed in Fig. 3. It suggested that in –NH2 and –NHCOCH3 chitosan/G3 systems, the aggregation of G3 is more obvious than that of C3 in solution. It is not surprising that guanine has larger conjugated ring than that of cytosine, and the aggregation of G3 in solution is stronger. To better address the effect of different bases, 20 C3 and G3 molecules in solution were separately simulated for 80 ns in absence of chitosan, and the typical snapshots during the aggregation process for two polynucleotides were displayed in Fig. S10 (see Supplementary Information). Figure S10 suggested that G3 have stronger tendency of aggregating comparing with C3. This is the reason that in both –NH2 and –NHCOCH3 systems, G3 has less distribution around chitosan. However, due to the strong electrostatic interaction (including high strength of H-bonds) between –NH3 + groups in chitosan and –PO4 2− groups in nucleotides, the aggregation of both C3 and G3 could be highly suppressed, and both of them could well disperse around the positively charged –NH3 + chitosan. In other words, the electrostatic interaction is the driving force for the interaction between –NH3 + chitosan and polynucleotides.
Since the polynucleotides are composed of nitrogenous base, five-carbon sugar and phosphate group, the H-bond number between the functional groups in three chitosan and different part of polynucleotides were calculated to extend the analysis with more chemical detail, as shown in Fig. 6. Due to the aggregation of G3, the number of H-bond decreased for all three systems comparing with the systems containing C3. Moreover, one could find that the number of H-bond between G3 and –NH2/–NHCOCH3 chitosan is quite small, which may also cause by strong aggregation of G3. The higher number of H-bond between G3 and –NH3 + chitosan primarily arise from the H-bond formed between –NH3 + group and phosphate group. This fact confirms the significance of electrostatic interaction in polynucleotide encapsulation by chitosan.
Figure 6.
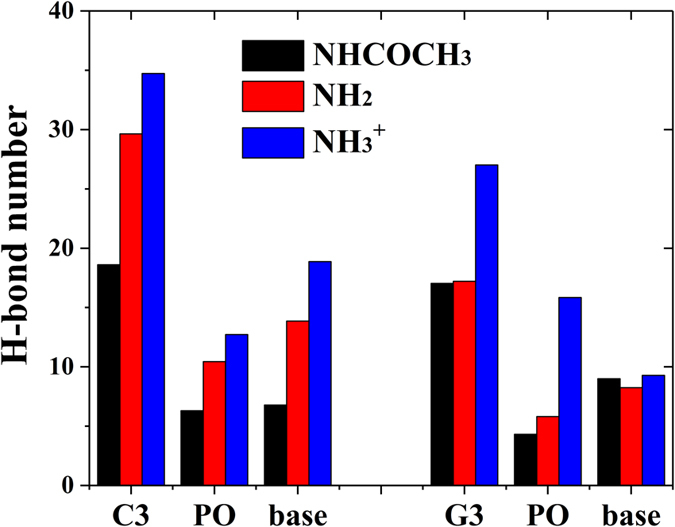
The average number of inter-molecular H-bond between different functional groups and different part of C3 and G3 during the MD simulations (C3/G3: total number of H-bond formed between chitosan and polynucleotides; PO: phosphate; base: guanine or cytosine).
In short summary, compared with C3, due to stronger stacking ability of base, G3 have stronger tendency of aggregating in the solution, and this property may decrease the polynucleotide encapsulation capacity by chitosan and following delivery efficiency. For –NH3 + chitosan, due to the strong electrostatic interaction, especially the H-bond between –NH3 + group in chitosan and phosphate group in polynucleotide, the aggregation effect could be partially eliminated. Thus the encapsulation capacity of polynucleotide by chitosan is mainly balanced by two factors: the strength of polynucleotide binding to chitosan and the tendency of self-aggregation of polynucleotide.
Effect of chain length on polynucleotide encapsulation
In this section, the effect of chain length on polynucleotide encapsulation was investigated. Six systems with polynucleotide chain length of 10 (C10 or G10) and different types of chitosan were constructed and simulated (see Table 1). The molecular ratio of chitosan: C10/G10 = 1:1.
Figure 7(a) showed the RDFs of C10/G10 around different types of chitosan. For –NH3 + chitosan system, increasing the chain length of polynucleotide has very little effect on the distribution of polynucleotide around chitosan: C3 (see Fig. 2(a)) has slightly higher density and closer distance around –NH3 + chitosan than C10. However, for –NH2 and –NHCOCH3 chitosan, chain length has large effect on the polynucleotide encapsulation. For example, both C3 and G3 have larger and closer distribution around –NH2 chitosan (see Fig. 2(a)), comparing with C10 and G10. The number of contact atoms in these systems, as shown in Fig. 2(b) and Fig. 7(b), also confirmed the results derived from RDFs. If one carefully check the equilibrated structure of –NH2 chitosan-polynucleotide complexes, it was interesting that part of the long chain polynucleotide could stack to each other or even keep certain distance from chitosan, as shown in Fig. 7(d). Figure S11–S16 showed the polynucleotide encapsulation process in chitosan-C10/G10 systems. In the cases of –NH2 chitosan systems, the capability of C10 and G10 dispersed along the chitosan chain was highly decreased. This phenomenon probably origins from (1) relative flexible conformation of C10 and G10 and (2) more base-base stacking sites of C10 and G10. Similar to G3 aggregation, the stacking of polynucleotide with chain length of 10 would decrease the interaction strength between polynucleotide and chitosan, and thus decrease the distribution of polynucleotide around chitosan. This could also be reflected by H-bond number between C10/G10 and functional groups in chitosan, as shown in Fig. 7(c). The number of H-bond between –NH2 groups and C3/G3 (see Fig. 6) greatly decreased with chain length increase, which indicates the decreased interaction strength. Meanwhile, the number of H-bonds between –NH3 + chitosan and polynucleotide with different chain length has little difference, which consist with the RDFs curves. This phenomenon could attribute to the fact that strong interaction between positively charged –NH3 + and negatively charged phosphate groups could minimized the effect of base stacking of C10 and G10.
Figure 7.
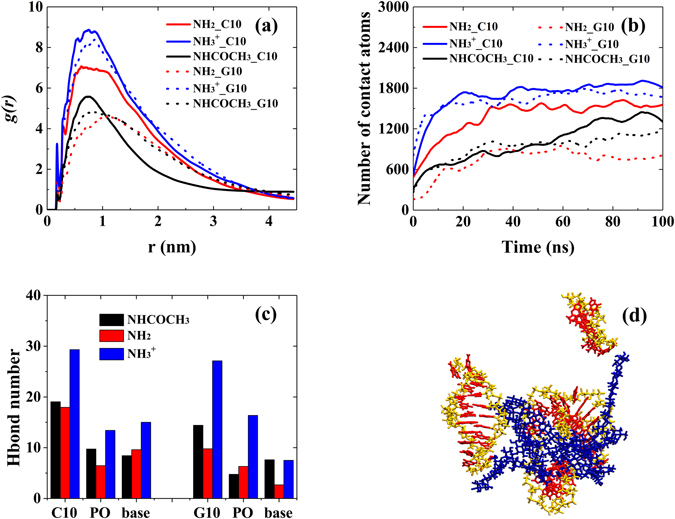
(a) The RDFs of C10/G10 around different types of chitosan. (b) Number of contact atoms between chitosan and C10/G10 as a function of simulation time. (c) Inter-molecular H-bond numbers between functional groups in chitosan and different parts of nucleotide (C10/G10: total number of H-bond formed between chitosan and C10/G10; PO: phosphate; base: guanine or cystosine). (d) The snapshots of –NH2 chitosan/G10 system at the end of 100 ns MD simulation (blue: chitosan, yellow: G10, red: base). All bases in G10 were displayed in red to better understand the role of base-base interaction in aggregation of G10. Water molecules were omitted for clarity.
We need add a cautious remark that in this study we only considered three extreme cases for chitosan: 0% deacetylated structure, fully deacetylated structure in neutral solution, and fully deacetylated structure in low pH solution. Although the model is relatively simple, it is the early stage of investigating chitosan-DNA interaction at the molecular level by molecular simulation. Further consideration of structure of double-stranded DNA, different degree of deacetylation of chitosan, different pH environment and hydrophilic/hydrophobic modification of chitosan etc. will be taken in our future studies to help understand the gene/drug encapsulation and delivery by chitosan-based delivery systems.
Conclusion
In this study, molecular dynamics simulations were performed to investigate the effects on single-stranded polynucleotide encapsulation by chitosan and the underlying interaction mechanism between polynucleotide and different types of chitosan with atomic details. It was found that the functional groups of chitosan, the types of base and length of polynucleotides regulate the interaction behavior between chitosan and polynucleotides. The encapsulation capacity of polynucleotide by chitosan is mainly balanced by two factors: the strength of polynucleotide binding to chitosan and the tendency of self-aggregation of polynucleotide in the solution. For –NH3 + chitosan, due to the strong electrostatic interaction, especially the H-bond between –NH3 + group in chitosan and phosphate group in polynucleotide, the aggregation effect could be partially eliminated. Therefore, both C3/G3 and C10/G10 in –NH3 + chitosan have better dispersal capacity than that in –NH2 and –NHCOCH3 chitosan. The good dispersal capacity of polynucleotides may improve the encapsulation of polynucleotides by chitosan, and hence increase the delivery and transfection efficiency of gene carrier. Our results revealed the atomic interaction mechanism between polynucleotide and various types of chitosan, and may provide novel ideas for the design and application of chitosan-based drug and gene delivery systems.
Electronic supplementary material
Acknowledgements
We acknowledge financial support by the National Natural Science Foundation of China (Grant Nos 21674032, 21403049, 21503186), Qianjiang Talents Program of Zhejiang Province (Nos QJD1602011) and National Undergraduate Training Programs for Innovation and Entrepreneurship.
Author Contributions
J.S., L.Z. and L.L. conceived the study, J.S., J.L., Z.Z. and G.P. conducted the MD simulation and analysis, J.S. and J.L. wrote the paper. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Jia-Wei Shen and Jiachen Li contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05197-0
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jia-Wei Shen, Email: shen.jiawei@hotmail.com.
Li Zhang, Email: lizhang@zstu.edu.cn.
Lijun Liang, Email: michael.lijunl@gmail.com.
References
- 1.Guo X, Huang L. Recent Advances in Nonviral Vectors for Gene Delivery. Acc. Chem. Res. 2012;45:971–979. doi: 10.1021/ar200151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mintzer MA, Simanek EE. Nonviral Vectors for Gene Delivery. Chem. Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 3.Whitehead KA, Langer R, Anderson DG. Knocking Down Barriers: Advances in siRNA Delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuckerman JE, Davis ME. Clinical Experiences with Systemically Administered siRNA-based Therapeutics in Cancer. Nat. Rev. Drug Discov. 2015;14:843–856. doi: 10.1038/nrd4685. [DOI] [PubMed] [Google Scholar]
- 5.Chiappini C, et al. Biodegradable Silicon Nanoneedles Delivering Nucleic Acids Intracellularly Induce Localized in Vivo Neovascularization. Nat. Mater. 2015;14:532–539. doi: 10.1038/nmat4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao S, Sun W, Kissel T. Chitosan-Based Formulations for Delivery of DNA and siRNA. Adv. Drug Deliv. Rev. 2010;62:12–27. doi: 10.1016/j.addr.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Yue Y, Wu C. Progress and Perspectives in Developing Polymeric Vectors for in Vitro Gene Delivery. Biomat. Sci. 2013;1:152–170. doi: 10.1039/C2BM00030J. [DOI] [PubMed] [Google Scholar]
- 8.Molla MR, Levkin PA. Combinatorial Approach to Nanoarchitectonics for Nonviral Delivery of Nucleic Acids. Adv.Mater. 2016;28:1159–1175. doi: 10.1002/adma.201502888. [DOI] [PubMed] [Google Scholar]
- 9.Duan X, et al. Smart pH-sensitive and Temporal-Controlled Polymeric Micelles for Effective Combination Therapy of Doxorubicin and Disulfiram. ACS Nano. 2013;7:5858–5869. doi: 10.1021/nn4010796. [DOI] [PubMed] [Google Scholar]
- 10.Jiang S, Eltoukhy AA, Love KT, Langer R, Anderson DG. Lipidoid-Coated Iron Oxide Nanoparticles for Efficient DNA and siRNA Delivery. Nano Lett. 2013;13:1059–1064. doi: 10.1021/nl304287a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Junquera E, Aicart E. Recent Progress in Gene Therapy to Deliver Nucleic Acids with Multivalent Cationic Vectors. Adv. Colloid Interfac. 2016;233:161–175. doi: 10.1016/j.cis.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Shu XZ, Zhu KJ. The Influence of Multivalent Phosphate Structure On the Properties of Ionically Cross-Linked Chitosan Films for Controlled Drug Release. Eur. J. Pharm. Biopharm. 2002;54:235–243. doi: 10.1016/S0939-6411(02)00052-8. [DOI] [PubMed] [Google Scholar]
- 13.Lee M, et al. The Use of Chitosan as a Condensing Agent to Enhance Emulsion-Mediated Gene Transfer. Biomaterials. 2005;26:2147–2156. doi: 10.1016/j.biomaterials.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Taranejoo S, Chandrasekaran R, Cheng W, Hourigan K. Bioreducible PEI-functionalized Glycol Chitosan: A Novel Gene Vector with Reduced Cytotoxicity and Improved Transfection Efficiency. Carbohydr. Polym. 2016;153:160–168. doi: 10.1016/j.carbpol.2016.07.080. [DOI] [PubMed] [Google Scholar]
- 15.Katas H, Alpar HO. Development and Characterisation of Chitosan Nanoparticles for siRNA Delivery. J. Controlled Release. 2006;26:216–225. doi: 10.1016/j.jconrel.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 16.Richardson SW, Kolbe HJ, Duncan R. Potential of Low Molecular Mass Chitosan as a DNA Delivery System: Biocompatibility, Body Distribution and Ability to Complex and Protect DNA. Int. J. Pharm. 1999;178:231–243. doi: 10.1016/S0378-5173(98)00378-0. [DOI] [PubMed] [Google Scholar]
- 17.Köping-Höggård M, et al. Chitosan as a Nonviral Gene Delivery System. Structure-Property Relationships and Characteristics Compared with Polyethylenimine in Vitro and After Lung Administration in Vivo. Gene Ther. 2001;8:1108–1121. doi: 10.1038/sj.gt.3301492. [DOI] [PubMed] [Google Scholar]
- 18.Sato T, Ishii T, Okahata Y. In Vitro Gene Delivery Mediated by Chitosan. Effect of pH, Serum, and Molecular Mass of Chitosan On the Transfection Efficiency. Biomaterials. 2001;22:2075–2080. doi: 10.1016/S0142-9612(00)00385-9. [DOI] [PubMed] [Google Scholar]
- 19.Kiang T, Wen J, Lim HW, Leong KW. The Effect of the Degree of Chitosan Deacetylation On the Efficiency of Gene Transfection. Biomaterials. 2004;25:5293–5301. doi: 10.1016/j.biomaterials.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 20.Strand SP, Danielsen S, Christensen BE, Vårum KM. Influence of Chitosan Structure On the Formation and Stability of DNA− Chitosan Polyelectrolyte Complexes. Biomacromolecules. 2005;6:3357–3366. doi: 10.1021/bm0503726. [DOI] [PubMed] [Google Scholar]
- 21.Lavertu M, Methot S, Tran-Khanh N, Buschmann MD. High Efficiency Gene Transfer Using chitosan/DNA Nanoparticles with Specific Combinations of Molecular Weight and Degree of Deacetylation. Biomaterials. 2006;27:4815–4824. doi: 10.1016/j.biomaterials.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 22.Gao Y, et al. Arginine-chitosan/DNA Self-Assemble Nanoparticles for Gene Delivery: In Vitro Characteristics and Transfection Efficiency. Int. J. Pharm. 2008;359:241–246. doi: 10.1016/j.ijpharm.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 23.Guţoaia A, et al. Fine-Tuned PEGylation of Chitosan to Maintain Optimal siRNA-nanoplex Bioactivity. Carbohydr. Polym. 2016;143:25–34. doi: 10.1016/j.carbpol.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Meneksedag-Erol D, Tang T, Uludağ H. Molecular Modeling of Polynucleotide Complexes. Biomaterials. 2014;35:7068–7076. doi: 10.1016/j.biomaterials.2014.04.103. [DOI] [PubMed] [Google Scholar]
- 25.Ramezanpour M, et al. Computational and Experimental Approaches for Investigating Nanoparticle-Based Drug Delivery Systems. Biochim. Biophys. Acta Biomembr. 2016;1858:1688–1709. doi: 10.1016/j.bbamem.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 26.Korolev N, Lyubartsev AP, Laaksonen A, Nordenskiöld L. A Molecular Dynamics Simulation Study of Oriented DNA with Polyamine and Sodium Counterions: Diffusion and Averaged Binding of Water and Cations. Nucleic Acids Res. 2003;31:5971–5981. doi: 10.1093/nar/gkg802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziebarth J, Wang Y. Molecular Dynamics Simulations of DNA-polycation Complex Formation. Biophys. J. 2009;97:1971–1983. doi: 10.1016/j.bpj.2009.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun C, Tang T, Uludağ H, Cuervo JE. Molecular Dynamics Simulations of DNA/PEI Complexes: Effect of PEI Branching and Protonation State. Biophys. J. 2011;100:2754–2763. doi: 10.1016/j.bpj.2011.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun C, Tang T, Uludaǧ H. Molecular Dynamics Simulations for Complexation of DNA with 2 kDa PEI Reveal Profound Effect of PEI Architecture On Complexation. J. Phys. Chem. B. 2012;116:2405–2413. doi: 10.1021/jp211716v. [DOI] [PubMed] [Google Scholar]
- 30.Zheng M, et al. Targeting the Blind Spot of Polycationic Nanocarrier-Based siRNA Delivery. ACS Nano. 2012;6:9447–9454. doi: 10.1021/nn301966r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhan B, et al. Coarse-Grained Simulation of polycation/DNA-like Complexes: Role of Neutral Block. Mol. Pharm. 2015;12:2834–2844. doi: 10.1021/mp500861c. [DOI] [PubMed] [Google Scholar]
- 32.Ding H, Ma Y. Design Maps for Cellular Uptake of Gene Nanovectors by Computer Simulation. Biomaterials. 2013;34:8401–8407. doi: 10.1016/j.biomaterials.2013.06.067. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Wang W, Kollman PA, Case DA. Antechamber, An Accessory Software Package For Molecular Mechanical Calculations. J. Comput. Chem. 2005;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [Google Scholar]
- 34.Frisch, A. et al. Gaussian 03, Revision C.02. Gaussian, Inc., Wallingford (2004).
- 35.Ferreira ML, Pedroni VI, Alimenti GA, Gschaider ME, Schulz PC. The Interaction Between Water Vapor and Chitosan II: Computational Study. Colloids Surf. A: Physicochem. Eng. Aspects. 2008;315:241–249. doi: 10.1016/j.colsurfa.2007.08.006. [DOI] [Google Scholar]
- 36.Lindorff Larsen K, et al. Improved Side‐Chain Torsion Potentials for the Amber ff99SB Protein Force Field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Der Spoel D, et al. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 38.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 39.Martínez L, Andrade R, Birgin EG, Martínez JM. PACKMOL: A Package for Building Initial Configurations for Molecular Dynamics Simulations. J. Comput. Chem. 2009;30:2157–2164. doi: 10.1002/jcc.21224. [DOI] [PubMed] [Google Scholar]
- 40.Hess B, Bekker H, Berendsen HJ, Fraaije JG. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 41.Darden T, York D, Pedersen L. Particle Mesh Ewald: An N-Log (N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 42.Curtiss, C. F. & Bird, R. B. Molecular Theory of Gases and Liquids, Wiley New York, 581 (1954).
- 43.Bussi G, Donadio D, Parrinello M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007;126:14101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 44.Berendsen, H. J., Postma, J. V., van Gunsteren, W. F., DiNola, A. & Haak, J. R. J. Chem. Phys. 81, 3684–3690 (1984).
- 45.Fletcher R, Powell MJ. A Rapidly Convergent Descent Method for Minimization. Comput. J. 1963;6:163–168. doi: 10.1093/comjnl/6.2.163. [DOI] [Google Scholar]
- 46.Humphrey W, Dalke A, Schulten K. VMD: Visual Molecular Dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 47.Franca EF, Freitas LC, Lins RD. Chitosan Molecular Structure as a Function of N‐Acetylation. Biopolymers. 2011;95:448–460. doi: 10.1002/bip.21602. [DOI] [PubMed] [Google Scholar]
- 48.Borca CH, Arango CA. Molecular Dynamics of a Water-Absorbent Nanoscale Material Based on Chitosan. J. Phys. Chem. B. 2016;120:3754–3764. doi: 10.1021/acs.jpcb.5b11230. [DOI] [PubMed] [Google Scholar]
- 49.Pillai C, Paul W, Sharma CP. Chitin and Chitosan Polymers: Chemistry, Solubility and Fiber Formation. Prog. Polym. Sci. 2009;34:641–678. doi: 10.1016/j.progpolymsci.2009.04.001. [DOI] [Google Scholar]
- 50.Zheng M, et al. Targeting the Blind Spot of Polycationic Nanocarrier-Based siRNA Delivery. ACS Nano. 2012;6:9447–9454. doi: 10.1021/nn301966r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pavan GM, et al. Computational Insights Into the Interactions Between DNA and siRNA with “Rigid” and “Flexible” Triazine Dendrimers. Biomacromolecules. 2010;11:721–730. doi: 10.1021/bm901298t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


