Abstract
Hypotheses on the existence of a universal “Root Economics Spectrum” (RES) have received arguably the least attention of all trait spectra, despite the key role root trait variation plays in resource acquisition potential. There is growing interest in quantifying intraspecific trait variation (ITV) in plants, but there are few studies evaluating (i) the existence of an intraspecific RES within a plant species, or (ii) how a RES may be coordinated with other trait spectra within species, such as a leaf economics spectrum (LES). Using Coffea arabica (Rubiaceae) as a model species, we measured seven morphological and chemical traits of intact lateral roots, which were paired with information on four key LES traits. Field collections were completed across four nested levels of biological organization. The intraspecific trait coefficient of variation (cv) ranged from 25 to 87% with root diameter and specific root tip density showing the lowest and highest cv, respectively. Between 27 and 68% of root ITV was explained by site identity alone for five of the seven traits measured. A single principal component explained 56.2% of root trait covariation, with plants falling along a RES from resource acquiring to conserving traits. Multiple factor analysis revealed significant orthogonal relationships between root and leaf spectra. RES traits were strongly orthogonal with respect to LES traits, suggesting these traits vary independently from one another in response to environmental cues. This study provides among the first evidence that plants from the same species differentiate from one another along an intraspecific RES. We find that in one of the world’s most widely cultivated crops, an intraspecific RES is orthogonal to an intraspecific LES, indicating that above and belowground responses of plants to managed (or natural) environmental gradients are likely to occur independently from one another.
Keywords: agroecology, agroforestry, Coffea arabica, functional traits, intraspecific trait variation, leaf economics spectrum, root economics spectrum, trait coordination
Introduction
Elucidating the key dimensions of functional trait variation among plant species has been critical for understanding, predicting, and managing terrestrial ecological responses to environmental or land-use change. Supporting the application of functional trait-based approaches to terrestrial ecology is arguably one unifying framework: evaluating how traits covary or trade-off to form distinct trait “spectra” (Grime, 1979; Lambers and Poorter, 1992; Reich et al., 1997; Westoby et al., 2002; Reich, 2014; Diaz et al., 2016). In addition to early mentions of plant strategies, trait suites, and trait trade-offs (Grime, 1979; Givnish, 1988; Lambers and Poorter, 1992; Chapin et al., 1993), the concept of trait spectra was popularized in the 2000s, most notably with the publication of the “Leaf Economics Spectrum” (LES) (Wright et al., 2004). Shortly before and since the LES was published, nearly all groups of functional traits have been hypothesized to covary along a distinct spectrum that describes functional variation among plant species (Westoby et al., 2002; Chave et al., 2009; Reich, 2014; Diaz et al., 2016). Evidence on trait spectra in plants has been critical for evaluating the evolutionary pressures driving trait variation among species (e.g., Shipley et al., 2006), and understanding how differences in species traits influence ecosystem structure and function (e.g., Diaz et al., 2004; Cornwell et al., 2008).
Among all traits, hypotheses on the existence of a universal “Root Economics Spectrum” (RES) has received arguably the least attention (Reich, 2014), despite the key role root trait variation plays in resource acquisition potential (Lambers et al., 2008; Cahill et al., 2010; Bardgett et al., 2014), and other ecosystem functions such as soil stability (e.g., Rillig et al., 2015). Evidence for the existence of a community-level RES has emerged, with certain traits covarying along a spectrum from resource acquiring to resource conserving root traits. Distinct root morphological and chemical traits [i.e., high specific root length (SRL), specific root area (SRA), specific root tip density (SRTD) (or branching intensity, fine root tip density, and root tip abundance), and root N concentration] designate resource acquisition while large root diameter and high root C:N signify resource conservation (Prieto et al., 2015; Fort et al., 2016; Weemstra et al., 2016). However, studies testing for the presence of a RES across species have produced mixed results. For example, recent studies have found evidence for a RES in herbaceous species, but analyses suggest woody species show drastically different patterns (Larson and Funk, 2016; Roumet et al., 2016; Weemstra et al., 2016).
Even less well resolved, is whether or not there exists a within-species RES, unrelated to genetic diversity but driven by environmental factors. The expression of root plasticity, and thus measurable root intraspecific trait variation (ITV), may be owing to within-species genetic variability or the integration of signals from the rooting environment (Miner et al., 2005). Yet, artificial selection during domestication can limit root trait variation, such that ITV from genetic sources are constrained. Here, we employ two highly related varieties from the economically important tree-crop breeding program for Coffea arabica. Given this, our study focuses near solely on phenotypic plasticity derived from the effects of environmental conditions on root trait values and root-leaf trait coordination. Recent analyses of ITV point to within-species trait spectra (Albert et al., 2010; Siefert et al., 2015) as having a key role in determining plant community composition (Laughlin et al., 2012), plant responses to environmental change (Jung et al., 2014; Moran et al., 2016), and rates of ecosystem function (Lecerf and Chauvet, 2008; Gagliardi et al., 2015) (but see Jackson et al., 2013). However, to date this literature has almost exclusively focused on leaf traits. In comparison, to our knowledge there is only one study that directly tests for the presence of a within-species RES. Hajek et al. (2013) found that certain root traits covaried strongly along a single axis of variation that, when coupled with relative growth rate, leaf size, and specific leaf area (SLA), described nearly 70% of the variation in root chemistry and morphology among Populus trema individuals. Ostonen et al. (2007) illustrated the relationships between SRL and fertilization within tree species through meta-analysis. Beyond this, there is no evidence indicating whether or not a RES applies to individuals of other species.
Documenting systematic responses of traits to environmental conditions has long been a main theme when evaluating inter- and intraspecific variation in traits (e.g., Givnish, 1988; Chapin et al., 1993). Understanding how plants integrate signals from highly heterogeneous soil environments in order to modify root traits, is arguably more challenging as compared to other aboveground traits. Unlike well-described relationships between light availability and rates of C assimilation and associated traits in leaves (e.g., Rijkers et al., 2000), root traits respond to a range of regional and localized soil chemical resources including macro- and micro-nutrients, soil moisture regimes, and soil pH, as well as multiple physical properties including structure, texture, and aggregation (reviewed by McCormack et al., 2015; Weemstra et al., 2016). These factors would be expected to vary most widely across broad growing regions, as compared to individual-plant scales; an assumption that is widely reflected in many process-based models of agricultural yield and ecosystem services, where trait values are generalized across growing regions (e.g., Bouman and van Laar, 2006; van Oijen et al., 2010). In this sense quantifying the primary sources of variation in traits, especially root traits that are difficult to collect, is key for ensuring that trait values are actually reflective of the plants growing in a particular region, site, or management scenario, and for designing trait sampling strategies that adequately capture the main aspects of ITV (Carmona et al., 2015; Martin et al., 2017).
Another key question in trait-based research is whether or not different trait spectra are “parallel” vs. “orthogonal” (independent of) to one another (Baraloto et al., 2010; Weemstra et al., 2016). Studies on interspecific trait variation have found the LES is orthogonal to suites of whole plant (Diaz et al., 2016), stem (Baraloto et al., 2010), and leaf hydraulic traits (Li et al., 2015). With respect to root traits, studies have found evidence of parallel coordination between RES and other spectra, but these results have been limited to herbaceous species (Craine et al., 2005; Tjoelker et al., 2005), or have been observed only when evaluating trait differences among plant communities (de la Riva et al., 2016).
To date, hypotheses on trait spectra orthogonality within species have been only weakly tested, especially for root traits. One study did find evidence of orthogonality among LES and leaf hydraulic traits in a Eucalyptus genotype (Blackman et al., 2016), while the analysis by Hajek et al. (2013) indicates that only certain leaf traits (i.e., leaf area and SLA) covary along a within-species RES, while other leaf traits (i.e., leaf N) do not. Drawing on literature from trees and tree-crops, we expect root ITV to occur along environmental gradients, namely soil moisture (Moser et al., 2010; Padovan et al., 2015) and edaphic conditions (Isaac et al., 2014; Defrenet et al., 2016); gradients that may occur independently of those driving aboveground ITV, such as air temperature, light, and CO2 availability (Gagliardi et al., 2015; Niinemets, 2015; Blackman et al., 2016). Additionally, mycorrhizal associations, a key biotic aspect of soils, may lead to systematic differences in ITV of root traits as compared to other trait spectra (Collins et al., 2016).
Testing hypotheses on trait coordination or orthogonality has applied implications for both agroecosystem models and crop biology. From a modeling perspective, if above- and belowground traits coordinate along single axes of resource acquisition/conservation, data on aboveground traits (which are generally easier to collect) could be used to approximate root trait data (that are generally more difficult to ascertain). Alternatively, evidence of orthogonality among root and leaf traits within species would indicate that root trait values are essential when describing, or modeling the impacts of, independent belowground dimensions of plant functional biology such as nutrient capture and retention (Meister et al., 2014). From a crop science perspective, researchers have pointed to the difficulties in artificially selecting crops for desirable root traits (Meister et al., 2014). If root and leaf show coordinated patterns of ITV, managing the environmental conditions that are known to influence leaf traits, would also be expected to influence root traits. Alternatively, if suites of traits vary independently from one another, multiple environmental gradients would have to be managed in order to reach desired functional trait profiles of crops of crop assemblages.
Our study was designed to evaluate patterns of intraspecific root trait variation within an agroecosystem. We use individuals of C. arabica, grown in four climatically different sites across two management systems (monoculture and agroforestry) in a nested design to determine primary sources of trait variation. As one of the world’s most widespread tree-crops, ITV of C. arabica LES traits have long been a focus on agronomists (e.g., DaMatta, 2004; Matos et al., 2009), and have more recently been evaluated to test hypotheses regarding ITV. For example, Martin et al. (2017) report that while C. arabica plants differ along the LES, high-resource agricultural environments lead this species to express weakened patterns of LES trait covariation (when compared to wild plants). Similarly, an analysis by Gagliardi et al. (2015) found that the position of coffee plants along an “intraspecific LES” was associated with plant yield. So while evidence indicates C. arabica follows along the LES, and that this variation has implications for agroecosystem functioning, tests on the patterns of C. arabica root trait covariation are considerably less common (van Kanten et al., 2005; Dias et al., 2007).
Our objectives were to (1) describe the magnitude of root trait variation within individuals of the same genotype across a range of organizational levels (site, management, and individuals); (2) evaluate which nested level of biological organization best explain root trait variation in C. arabica; (3) determine if root traits coordinate along a single axis of resource acquisition to resource conservation, an intraspecific RES; and (4) test whether or not an intraspecific RES is related to other well-known dimensions of ITV. Based on prior studies of root trait variation, as well as studies in intraspecific leaf trait variation in coffee, we hypothesized that root traits would vary most widely across sites (Gagliardi et al., 2015; Martin et al., 2017), which represent major climatic gradients of coffee growing conditions. Based on the emerging literature on trait ITV and within-species economic spectra (Niinemets, 2015; Martin et al., 2017), we also hypothesized that root traits would covary within coffee, along a single primary intraspecific RES; more specifically, we anticipated that root traits associated with resource acquisition trade-off with traits associated with resource conservation. Lastly, since leaves and roots respond differently to environmental gradients, we hypothesized that intraspecific root and leaf economic spectra in coffee would show little coordination among one another, and instead show patterns of orthogonality.
Materials and Methods
Sampling Design
Our study employed a nested design in order to quantify root and leaf trait variation in C. arabica plants across four different hierarchical scales: (1) individuals within blocks, (2) among blocks within management treatments (i.e., full sun and agroforestry); (3) among management treatments within a site; and (4) among sites. This design resulted in traits measured on 64 coffee plants, which were collected from n = 4 coffee plants per block, within n = 2 blocks per management treatment, within n = 2 management treatments per site, and across n = 4 sites.
We identified four sites in the coffee growing regions of Costa Rica and Nicaragua where closely related varieties of C. arabica (var. Caturra and var. Pacas, respectively, both derived from var. Bourbon) are grown (hereafter referred to as, collectively, C. arabica). While C. arabica var. Pacas is technically a different variety, this genotype is long-recognized only as a mutation of the same genetic strain as C. arabica var. Caturra (Bertrand et al., 1999). Given this mutation, there is a possibility of slight genetic variability between the two highly related varieties (but see Table 5 for evidence of no systematic or consistent differences between varieties). These sites ranged in latitude from 9 to 11°N and elevation of 455–1500 m (covering a range of coffee growing altitudes) with a mean annual temperature (MAT) from 18.7 to 24°C and mean annual precipitation (MAP) from 1386 to 3200 mm (Table 1). These four sites represented four broad coffee growing regions as outlined in Table 1.
Table 5.
Mean (± s.d.) of root traits among the four sites.
| Trait | Cool and wet (CW) | Hot and wet (HW) | Cool and dry (CD) | Hot and dry (HD) |
|---|---|---|---|---|
| D | 1.12 (0.13)a | 1.04 (0.14)a | 1.47 (0.30)b | 1.05 (0.31)a |
| SRL | 4.01 (2.06)a | 2.98 (1.29)a | 1.05 (0.80)b | 3.56 (2.62)a |
| SRA | 329.1 (156.2)ab | 289.0 (109.7)ab | 131.8 (63.5)a | 367.5 (379.7)b |
| SRTD | 607.4 (426.0)ab | 378.5 (195.3)ab | 160.6 (121.3)a | 458.9 (404.0)b |
| RLD | 1.74 (0.82)a | 2.22 (1.16)a | 0.30 (0.22)b | 1.90 (1.10)a |
| Nroot | 1.96 (0.44)a | 1.94 (0.33)a | 1.34 (0.34)b | 1.59 (0.31)b |
| CNroot | 24.61 (4.52)a | 21.36 (3.17)a | 37.23 (9.85)b | 31.02 (6.83)b |
| PCA 1 score | -0.31 (0.39)a | 0.26 (0.29)a | 0.66 (0.38)b | -0.11 (0.63)a |
| PCA 2 score | 0.05 (0.61)a | -0.45 (0.45)b | 0.30 (0.26)a | 0.11 (0.68)a |
Values followed by the same letter within a row are significantly different based on Tukey’s HSD test (n = 16).
Table 1.
Site and management information for farms employed in this study.
| Site | Country and Coordinates | Elevation | MAP/MAT | Variety/shade tree (density) | Management treatment | Soil N | Soil C | Soil P | Soil pH | Soil moisture |
|---|---|---|---|---|---|---|---|---|---|---|
| Cool and wet (CW) | Costa Rica | 1020 | 3014/19.5 | Caturra/E. poeppigiana (6300) | Full sun | 1.07 ± 0.04 | 10.53 ± 0.39 | 4.95 ± 0.59 | 4.23 ± 0.10 | 71.46 ± 2.90 |
| 09°56′19″ N | ||||||||||
| 83°43′46″ W | ||||||||||
| Agroforestry | 0.96 ± 0.08 | 9.47 ± 0.88 | 9.63 ± 1.93 | 4.00 ± 0.10 | 71.85 ± 7.30 | |||||
| Hot and wet (HW) | Costa Rica | 685 | 3200/23.4 | Caturra/E. poeppigiana (5000) | Full sun | 0.43 ± 0.03 | 4.38 ± 0.38 | 11.19 ± 1.12 | 4.38 ± 0.08 | 35.01 ± 1.38 |
| 09°53′44″ N | ||||||||||
| 83°40′7″ W | ||||||||||
| Agroforestry | 0.37 ± 0.03 | 4.02 ± 0.30 | 7.74 ± 1.33 | 4.55 ± 0.13 | 36.45 ± 2.70 | |||||
| Cool and dry (CD) | Costa Rica | 1500 | 1491/18.7 | Caturra/E. poeppigiana (6860) | Full sun | 0.26 ± 0.02 | 2.83 ± 0.25 | 4.98 ± 1.59 | 4.66 ± 0.08 | 13.39 ± 1.39 |
| 09°40′03″ N | ||||||||||
| 83°06′32″ W | ||||||||||
| Agroforestry | 0.41 ± 0.01 | 4.66 ± 0.17 | 6.56 ± 1.60 | 4.68 ± 0.05 | 17.16 ± 2.74 | |||||
| Hot and dry (HD) | Nicaragua | 455 | 1386/24.0 | Pacas/I. laurina (4000) | Full sun | 0.54 ± 0.05 | 6.10 ± 0.55 | 4.39 ± 1.05 | 4.51 ± 0.15 | 22.84 ± 1.54 |
| 11°53′54″ N | ||||||||||
| 86°08′56″ W | ||||||||||
| Agroforestry | 0.57 ± 0.02 | 6.40 ± 0.20 | 3.35 ± 1.13 | 4.39 ± 0.19 | 23.28 ± 1.41 | |||||
We show qualitative classifications of the different study sites that coarsely approximate the range of Coffea arabica growing conditions, country and coordinates, elevation (m), mean annual precipitation (MAP, mm)/mean annual temperature (MAT, °C), C. arabica variety, shade tree (Erythrina poeppigiana or Inga laurina) and density (stems ha-1), and soil metrics at the management treatment (full sun or agroforestry) level [soil N (%), soil C (%), soil P (mg kg-1), soil pH and soil moisture (%); n = 16)].
Coffea arabica is cultivated in two main management systems: monoculture (hereafter “full sun” management) and in agroforestry systems where coffee is intercropped with regularly pruned N2-fixing shade tree (hereafter “agroforestry”). For the agroforestry management system, coffee is specifically intercropped with Erythrina poeppigiana (Fabaceae) at three of these sites, and Inga laurina (Fabaceae) at one site. Within each site-by-management combination, we delineated two 25-m2 blocks that were spaced at minimum 20 m from one another to ensure spatial interspersion of sampling. Within each block, we selected four sample plants for collection of traits where n = 64 coffee plants total distributed equally among sites. In sum, all of the traits measured on coffee plants were sampled and associated with the following information: (i) roots/leaves within plants; (ii) plants within blocks; (iii) blocks within management treatments; and (iv) management treatments within sites.
At all of the sites coffee plants are stump pruned approximately every 5–7 years leading to uneven-aged canopies of coppiced resprouts, with generally 2–3 resprouts per plant (Charbonnier et al., 2013). All sampled plants were at reproductive maturity, were between 140 and 235 cm in height with a resprout basal diameter between 14.4 and 34.6 mm. In the monoculture treatments, coffee plants were at minimum 20 m away from the nearest shade tree, a distance that is excess of the zone of influence for certain physiological processes including N transfer (Meylan, 2012). In the agroforestry treatments, all sample plants were between 0.5 and 9.6 m from the nearest shade tree.
Root Trait Collection
We excavated one complete lateral root from each individual C. arabica plant in our study within 1 week in order to minimize any growing season effects. The C. arabica root system is typically composed of a primary taproot and lateral roots in the upper soil horizon as well as four to eight axial roots (Garriz, 1978). We followed the main aerial stump to the taproot until the upper-most lateral roots could be isolated. A lateral root was then excavated in its entirety, which included all feeder and fine roots and any gravitropically positive roots. Multiple levels of standardization were used in our collection of lateral roots. Specifically, we: (1) standardized all plants by genotype (described above); (2) standardized all plants by age (based on known relationships between diameter at sprout and age); (3) standardized all intact lateral roots by depth at which they were collected (top 20 cm); and (4) standardized all intact lateral roots by originating diameter class (<4 mm). Roots were then brushed, stored intact in freezer bags, and stored at -18°C until processing. We also collected an intact soil core (5 cm diameter) from the top 20 cm of the soil profile, bagged the collected soil and roots and stored at -18°C until further processing.
In the lab, intact lateral roots were rinsed by hand using deionized water. We restricted analysis to absorptive fine roots <2 mm in diameter (Perez-Harguindeguy et al., 2013), however, given our standardization in sampling protocol described above, we included higher ordered roots (4th order but <2mm) from our intact lateral root sample. (Note – we also replicated all of our analysis on a subset of roots that fall under the “absorptive” functional classification as described by McCormack et al. (2015), which includes only 1st, 2nd, and 3rd order roots. Our results were robust with respect to this data subsetting). Samples were then scanned using a flatbed scanner at 600 dpi. Root image analysis was then conducted using WinRhizo (Regents Instruments, Montreal, QC, Canada), to generate information on total root length, total root area, total tip number, and mean root diameter. Roots from the soil cores were extracted in a water bath with tweezers and scanned for length. All roots were then oven-dried at 60°C for 48 h and weighed, and then transported to the University of Toronto Scarborough. Chemical trait analysis was conducted on the intact roots of <2 mm, which were first ground into a homogeneous powder using a ball mill (Retsch Ltd., Haan, Germany). Root C and N concentrations were then measured on approximately 0.1 g of dried sample using a CN628 elemental analyzer (LECO Instruments, Mississauga, ON, Canada).
Based on these analyses, we derived data on seven root traits in total for each plant including two traits associated with resource conservation – average root diameter (D; mm) and root carbon:N ratios (CNroot), four traits associated with resource acquisition – SRL (root length divided by root dry mass; m g-1), SRA (root area divided by root dry mass; m2 kg-1), SRTD (number of root tips divided by root dry mass; tips g-1), root N concentrations (Nroot; %), and one trait derived from soil cores, root length density (RLD, total root length in a known soil volume; cm cm-3 soil). In sum, we present one trait on a standardized volume basis (RLD), three traits as a mean of <2 mm fine roots (D, Nroot, and CNroot), and three traits on a standardized mass basis for <2 mm fine roots (SRL, SRA, and SRTD).
Soil Conditions
Across all four sites, soil samples to a depth of 15 cm were collected and analyzed for soil properties (n = 64; 16 samples per site). Fresh soil samples were homogenized and divided into three for determination of soil moisture content (%), soil pH, and soil nutrients. For one of the subsamples, wet mass was measured, soils were then oven-dried at 105°C for 72 h, and soil moisture content was calculated as the difference between wet and dry mass divided by dry mass. Soil pH was measured with one of the subsamples in a 1:5 soil to water solution with a pH meter (Mettler Toledo pH meter, Mississauga, ON, Canada). The final subsample was air-dried and transported to the University of Toronto for total C and N concentrations (%), as well as available plant-available phosphorus (P, mg kg-1 soil). Total soil C and N concentrations were measured on approximately 1 g of dried sample using a CN628 elemental analyzer (LECO Instruments ULC, Mississauga, ON, Canada). For plant-available P determinations, samples were air-dried and sieved to 2 mm, and 4 g were extracted using 20 mL of Brays 1 and filtered through #1 Whatman filter paper. Plant-available P was then determined colorimetrically using a QuikChem8500 flow injection analyzer (Lachat Instruments, Loveland, CO, United States). Soil total N ranged from 0.34 to 1.02%, soil C ranged from 3.75 to 10.03%, soil available P ranged from 3.9 to 9.5 mg kg-1, soil pH ranged from 4.12 to 4.67, and gravimetric soil moisture content ranged from 15.3 to 71.6% (Table 1).
Leaf Trait Data
To test the hypothesis that root and leaf trait represent independent axes of variation among coffee plants, we used a modified version of the leaf trait dataset collected by Martin et al. (2017). In short, their study was designed to quantify intraspecific variation in LES traits among the same coffee plants that were sampled here for root traits. Their dataset entailed eight traits measured on 384 coffee leaves, which corresponds to leaf traits being measured on six leaves for each coffee plant in our sample (Martin et al., 2017). To merge their dataset with ours, a plant-level average value of four leaf traits [maximum photosynthetic rates on a mass basis (Amass, μmol CO2 g-1 s-1); leaf mass per area (LMA, g m-2); leaf N concentrations (Nleaf, %); leaf tissue density (g cm-3)] was calculated for analysis here.
Statistical Analysis – Intraspecific Root Trait (Co)variation
All statistical analyses were performed using R v. 3.0.2 (R Foundation for Statistical Computing, Vienna, Austria). In total, our dataset entailed 62 observations for each root trait (two observations were removed as outliers). For each root trait, we first described the magnitude of intraspecific variation by calculating coefficients of variation (cv) across the entire dataset. We then employed a maximum likelihood approach to fit both normal and log-normal distributions to each root trait dataset, and compared models based on log-likelihood ratios. Where traits were best-described by log-normal distributions, log-transformed data was used in further analyses.
We used standardized major axis (SMA) regression models performed using the ‘lmodel2’ R package (Legendre, 2014) to examine bivariate relationships between all seven root traits (where n = 62 in all SMA models). For these pairwise tests, SMA were employed since we were primarily interested in the slopes of the relationships between any two given traits, all variables were assumed to be measured with error, and we did not have a priori hypotheses regarding the causal relationship between any pair of traits (Warton et al., 2006). Lastly, to evaluate root trait relationships in multivariate trait space, we performed a principal component analysis (PCA) using all seven root traits with the ‘vegan’ R package (Oksanen et al., 2016). Based on these analyses, we calculated PCA axis 1 and 2 scores for each root, and included these in analyses on variance decomposition (detailed below).
Statistical Analysis – Causes of Root Trait Variation
We used a nested analysis of variance (ANOVA) coupled with variance partitioning techniques, to evaluate how categorical environmental or management (full sun or agroforestry) factors explained variation in coffee root traits. This was done by first using the ‘lme4’ R package (Bates et al., 2014) to fit a linear mixed model for each trait individually, as well as both PCA axis 1 and 2 scores. In these models, all three nested levels (i.e., block within management within site) were entered as sequential random effects, and the intercept was the only fixed effect. We then used the ‘varcomp’ function in the ‘ape’ R package (Paradis et al., 2004) to calculate the variance components associated with each nested level. These analyses were based on log-transformed data for all root traits except Nroot and PCA axis 1 and 2 scores (Table 2).
Table 2.
Intraspecific variation in roots traits of C. arabica.
| Root trait | Log-likelihood values | Descriptive statistics | |||
|---|---|---|---|---|---|
| Normal | Log-normal | Mean (± s.d.) | Range | Intraspecific cv (%) | |
| D | -10.56 | -5.86 | 1.17 (0.29) | 0.67–1.93 | 24.69 |
| SRL | -133.56 | -121.78 | 2.87 (2.10) | 0.37–9.97 | 73.23 |
| SRA | -423.43 | -395.88 | 277.2 (255.6) | 63.6–1538.9 | 81.39 |
| SRTD | -449.68 | -428.67 | 397.1 (344.5) | 29.6–1987.2 | 86.76 |
| RLD | -96.59 | -91.22 | 1.53 (1.16) | 0.09–5.17 | 75.55 |
| Nroot | -35.78 | -37.65 | 1.71 (0.43) | 0.87–2.87 | 25.46 |
| CNroot | -223.47 | -216.70 | 28.58 (8.97) | 14.14–53.69 | 31.38 |
The maximum likelihood-based model that best-describes the distribution of each trait is highlighted in bold (where n = 62 in all cases). Root traits are: root diameter (D; mm), specific root length (SRL; m g-1), specific root area (SRA; m2 kg1), specific root tip density (SRTD; tips g1), root length density (RLD; cm cm-3), root nitrogen concentrations (Nroot; %), and root carbon:N ratios (CNroot).
For each trait, we then performed an additional mixed model analysis that included soil pH, soil N, soil C, soil P, and soil moisture as fixed effects, and our three nested categorical factors (site, management, and block) as random effects. For these models we then calculated the proportion of intraspecific root trait variation explained by the fixed effects alone (i.e., the “fixed effects r2”) and the proportion of ITV explained by both the fixed and random factors combined (i.e., the “fixed effects + random effects r2”) (Nakagawa and Schielzeth, 2013). These values were calculated using the ‘sem.model.fits’ function in the ‘piecewiseSEM’ R package (Lefcheck, 2016).
Statistical Analysis – Intraspecific Root and Leaf Trait Coordination
To test if root and leaf traits represent independent axes of variation in C. arabica, we used a MFA: a multivariate ordination method that tests if different groups of variables form independent structures within a dataset. This analysis follows previous studies testing the independence of trait spectra across species (c.f. Baraloto et al., 2010), but in our case, the MFA tests if root traits covary and leaf traits covary independently from one another within C. arabica. The test statistic derived from our MFA is the RV coefficient that describes the relationship among root and leaf trait spectra; values closer to zero indicate little correlation among leaf and root traits, and coefficients approaching an absolute value of 1 indicate stronger relationships among leaf and root trait dimensions. We used a permutation test to generate a one-tailed significance value for our RV coefficient. Specifically, root and leaf traits were determined to vary independently from one another, if our observed RV coefficient fell within the lower 95% percentile of the distribution of RV coefficients, derived from n = 10000 permuted datasets. In order to test if our observed patterns of coordination/decoupling among leaf and root trait spectra were heavily influence by a single root trait that did not strongly align with an RES (see results below), this MFA analysis and permutation test was also performed on a dataset with RLD removed. All MFA tests were performed using the “FactoMineR” R package (Le et al., 2008).
Results
Root Trait (Co)variation
All C. arabica root traits exhibited considerable intraspecific variation, with cvs ranging from approximately 25–87% (Table 2). The two root chemical traits, Nroot and CNroot, exhibited low variation at 25.46 and 31.38%, respectively, while root D also exhibited low intraspecific variation (cv = 24.69 %). All other traits varied by over an order of magnitude within C. arabica, with cvs ≥ 73%. In particular, SRL ranged from 0.37 to 9.97 m g-1, SRA from 63.6 to 1538.9 m2 kg-1, and RLD from 0.09 to 5.17 cm cm-3 (Table 2). Specific root tip density exhibited the highest intraspecific variation (cv = 86.76%), with SRTD values ranging over almost two orders of magnitude from 29.6 to 1987.2 tips g-1 (Table 2).
Bivariate Root Trait Correlations
Pairwise relationships between all C. arabica root traits were strong and highly significant (Table 3). Specifically, three root morphological traits that are associated with resource acquisition (i.e., SRL, SRA, and SRTD) were strongly and significantly positively related with one another (r2 = 0.479–0.804, p < 0.001 for all three tests), while root D was negatively related with SRL (r2 = 0.645; p < 0.001), SRA (r2 = 0.497; p < 0.001), and SRTD (r2 = 0.501; p < 0.001). The root chemical trait associated with resource acquisition, Nroot, was significantly positively related with SRL, SRA, SRTD, and RLD, (r2 = 0.172–0.431; p < 0.001), and significantly negatively related with root D (r2 = 0.226; p < 0.001).
Table 3.
Bivariate relationships among seven root traits in C. arabica.
| log-D | log-SRL | log-SRA | log-SRTD | log-RLD | Nroot | log-CNroot | |
|---|---|---|---|---|---|---|---|
| log-D | — | -0.212(-0.341, -0.251) | -0.361(-0.434, -0.301) | -0.273(-0.328, -0.228) | -0.229(-0.288, -0.183) | -1.846(-2.312, -1.473) | 0.8008(0.644, 0.995) |
| log-SRL | 0.645 (<0.001) | — | 0.809(0.700, 0.934) | 0.935(0.834, 1.048) | 0.784(0.626, 0.982) | 1.845(1.528, 2.250) | -2.740(-3.323, -2.260) |
| log-SRA | 0.497 (<0.001) | 0.687 (<0.001) | — | 0.756(0.628, 0.910) | 0.634(0.503, 0.800) | 0.667(0.538, 0.824) | -2.216(-2.731, -1.798) |
| log-SRTD | 0.501 (<0.001) | 0.804 (<0.001) | 0.479 (<0.001) | — | 0.839(0.658, 1.068) | 1.983(1.617, 2.431) | -2.930(-3.582, -2.397) |
| log-RLD | 0.209 (<0.001) | 0.227 (<0.001) | 0.180 (<0.001) | 0.105 (0.005) | — | 2.364(1.873, 2.984) | -3.494(-4.394, -2.778) |
| Nroot | 0.226 (<0.001) | 0.431 (<0.001) | 0.318 (<0.001) | 0.369 (<0.001) | 0.172 (<0.001) | — | -1.478(-1.630, -1.340) |
| log-CNroot | 0.279 (<0.001) | 0.436 (<0.001) | 0.335 (<0.001) | 0.387 (<0.001) | 0.198 (<0.001) | 0.856 (<0.001) | — |
All relationships are based on standardized major axis regression analysis, where n = 62 for all tests. The upper right portions of the table represent the slopes of bivariate trait relationships with associated 95% confidence limits (in brackets). The bottom left portion represents r2 values and associated p-values (in brackets).
Multivariate Root Trait (Co)variation
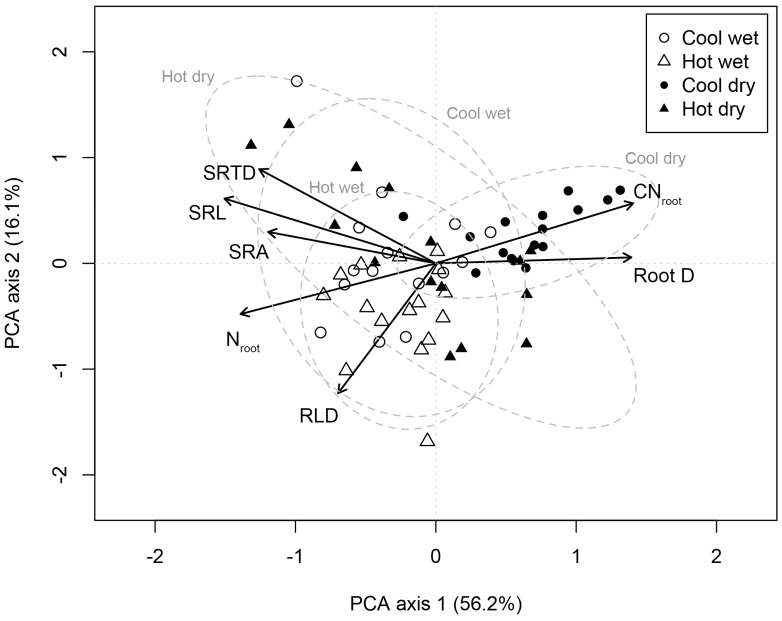
The first two PCA axes explained 72.3% of the variation in the seven root traits among C. arabica plants (Figure 1). PCA axis 1 explained the majority of this variation (56.2%) and was significantly positively associated with D (p < 0.001) and CNroot (p < 0.001) while negatively associated to root traits associated with resource acquisition, SRL (p < 0.001), SRA (p < 0.001), SRTD (p < 0.001), RLD (p = 0.001), and Nroot (p < 0.001) (Figure 1). PCA axis 1 highlights the trade-offs between traits associated with resource acquisition and traits associated with resource conservation; PCA axis 1 scores were also significantly (p < 0.001) related to site category. PCA axis 2 explained a further 16.1% of the variation in the seven root traits among C. arabica plants (Figure 1).
FIGURE 1.
Principal components analysis of intraspecific variation in root functional traits in Coffea arabica. The PCA is based on root traits measured on 62 individual-plants, sampled across four different sites (open circles = CW, open triangles = HW, closed circles = CD, and closed triangles = HD). Dashed lines correspond to 95% confidence ellipses for C. arabica roots sampled at each site. PCA axis 1 scores were significantly different (p < 0.001) for CD as compared to CW, HW, and HD.
Variance Partitioning
For all root traits, the site in which a C. arabica plant was growing consistently explained the majority of intraspecific variation among plants (Table 4). Specifically, among morphological traits, site identity explained 29.1% of the variance in root D, 68.3% of the variance in RLD, and 36.1% of the variance in SRL (Table 4). Intraspecific variation in root chemical traits was also best explained by site (Nroot = 27.4%; CNroot = 42.1%), as compared to management treatment or block. Management treatment did explain the largest proportion of variance in SRTD (35.4%) and SRA (29.1%), but site identity explained an additional 21.4 and 16.9% of the variation, respectively. Management treatment also explained between ∼15 and 28% of the variation in SRL, Nroot, and CNroot (Table 4). Site and management also explained root trait variation in multivariate trait space. Specifically, 33.7 and 31.7% of the variation in PCA score 1 was explained by site and management, respectively, while 4.1 and 39.4% of the variation in PCA score 2 was explained by site and management, respectively (Table 4). Within the exception of Nroot, block identity explained <11% of the intraspecific variation in any coffee root trait (Table 4).
Table 4.
Sources of intraspecific variation in root traits of C. arabica.
| Root trait | Variance decomposition | Mixed effects model | ||||
|---|---|---|---|---|---|---|
| Site | Management | Block | Error | Fixed effects r2 | Fixed effects + random effects r2 | |
| log-D | 29.1 | 18.7 | 8.4 | 43.9 | 0.0463 | 0.6158 |
| log-SRL | 36.1 | 27.5 | 3.2 | 33.2 | 0.0721 | 0.6864 |
| log-SRA | 16.9 | 29.7 | 10.9 | 42.4 | 0.0660 | 0.6337 |
| log-SRTD | 21.4 | 35.4 | 2.0 | 41.2 | 0.1045 | 0.6259 |
| log-RLD | 68.3 | 6.7 | 3.2 | 21.8 | 0.0181 | 0.7944 |
| Nroot | 27.4 | 15.6 | 16.2 | 40.8 | 0.0434 | 0.7351 |
| log-CNroot | 42.1 | 24.1 | 2.7 | 31.1 | 0.0245 | 0.7479 |
| PCA1 | 33.7 | 31.7 | 5.4 | 2.9 | 0.0148 | 0.7432 |
| PCA2 | 4.1 | 39.4 | 5.4 | 51.1 | 0.0846 | 0.4697 |
Variance decomposition was based on a nested analysis of variance, which for each trait was based on n = 62 individual-plants. The nested level explaining the highest percentage of variation in a given trait is highlighted in bold. Also presented are the explained variance associated with continuous soil variables (soil N, soil C, soil P, soil pH, and soil moisture) measured within these nested levels (“Fixed effects r2”), and the explained variance associated with both the fixed effects and random effects.
Continuous soil parameters (soil N, soil C, soil P, soil pH, and soil moisture) measured within the nested levels also explained very little of the variance in any root traits (Table 4). Specifically, these continuous variables explained only between 1.5 and 10.5% of the variation in any root trait, indicating that continuous soil variables did not systematically explain intraspecific root trait variation. In comparison, the addition of the nested categorical levels to mixed models increased the explanatory power to 47–79% (Table 4).
Given that the nested level “site” strongly explained variance in root traits, we conducted an ANOVA of root traits among sites (Table 5). Root D, SRL, and RLD were significantly different at the CD site as compared to the other three sites (Table 5); larger root D was paired with lower SRL and SRA at the CD site. Similarly, Nroot and CNroot were significantly different at this site, expressing lower concentrations of N and higher C:N ratios (Table 5).
Root and Leaf Trait Coordination
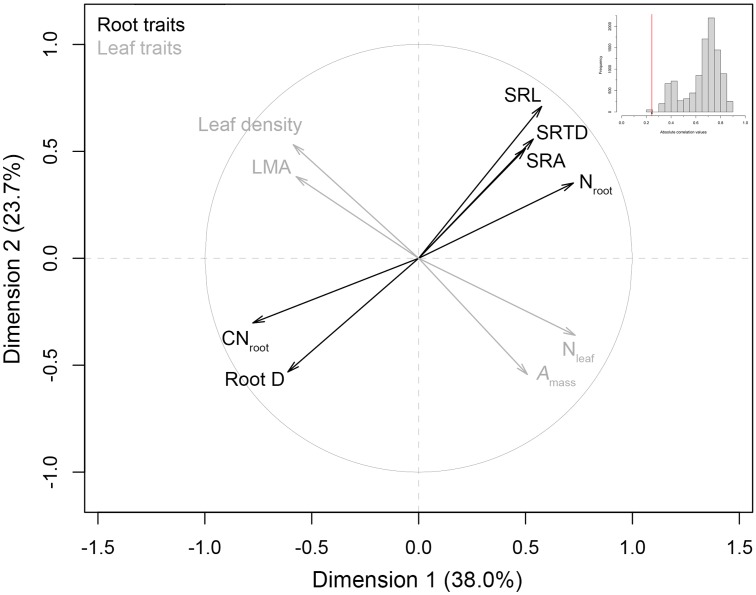
Multiple factor analysis indicated a significant lack of intraspecific coordination among C. arabica root traits and leaf traits (Figure 2). These two trait spectra were strongly orthogonal to one another, with an observed RV correlation coefficient of 0.245 that ranked as the lowest observed RV value in our randomized distribution of RV coefficients (randomization test p < 0.001, Figure 2 inset). These MFA results were robust with respect to the inclusion of RLD: the one root trait that is measured on a per soil volume basis and aligns most closely with axis 2 of our RES PCA (see Figure 1). Specifically, when RLD is included in the analysis, the MFA RV = 0.244 (p = 0.02) (data not shown). Qualitatively, Amass, Nleaf, LMA, and leaf tissue density covaried along an LES, while root traits covaried along an independent trait spectrum (Figure 2).
FIGURE 2.
Multiple factor analysis of root traits and leaf economics traits in C. arabica. Leaf traits were based on tree-level values derived from Martin et al. (2017), and a priori multiple factor analysis (MFA) groupings of root traits [with the removal of root length density (RLD)] and leaf traits, are denoted by gray and black coloring, respectively. A histogram of the randomized absolute correlation values (RV values) between these trait groups (where n = 10, 000 randomizations) is also presented as an inset. The red line indicates the observed correlation (RV) between root and leaf groups, relative to the distribution expected if traits were randomly assigned to groups.
Discussion
The Existence an Intraspecific Root Economics Spectrum
Our findings contribute to the growing literature evaluating how trait spectra commonly used to describe interspecific differences in functional biology, may also describe the ecological variability among plants within a species (Hajek et al., 2013; Gagliardi et al., 2015; Niinemets, 2015). Although there remains limited evidence that species in fact differentiate across a universal RES (Weemstra et al., 2016), we did find strong support for a single, well-defined RES in C. arabica. Specifically, this intraspecific RES describes bi- and multivariate trade-offs among resource acquiring traits on one hand – namely high values of SRL, SRA, SRTD, RLD, and Nroot – and resource conservation traits on the other – namely high values of CNroot and root D (Figure 1).
The relationship among root D and specific root length and area measures must be tempered with an inherent autocorrelation to SRL and SRA (Weemstra et al., 2016), and controversy in regards to the meaningfulness of these relationships (van Kanten et al., 2005). However, the trade-off between root D with SRTD and Nroot as well as SRL, SRA, and RLD with CNroot provides substantive evidence of a singular axis. We indicate strong covariation across root acquisition and conservative traits, with coffee plants assembling along a hypothesized resource acquisitive-conservative spectrum. Our results also suggest that when evaluating the traits comprising a RES, including all traits that are strongly tied to resource acquisition potential (c.f. Cahill et al., 2010), may increase the ability to detect a RES within species.
The Role of Abiotic Gradients and Biotic Interactions in Forming an RES
In our study, site identity explained the largest portion of variance for most of our root traits for five of the seven traits, while management systems (full sun versus agroforestry) explained the largest portion of variance for two of the seven traits (SRA and SRTD) (Table 4). Multivariate trait syndromes differed most consistently according to site-level differences. We show that PCA axis 1 scores were significantly different for the cold and dry site as compared to the other three sites (p < 0.001; Figure 1 and Table 5). Generally, root traits for plants growing at the CD site were significantly different than those at the other sites (Table 5), tending toward the resource conservative end of the intraspecific RES (notably, higher D, lower Nroot, higher CNroot); differences that appear largely attributable to the significantly lower soil moisture content at the CD site (Table 1), but could be due to a range of abiotic factors not captured in this study. This conforms to previous suggestions that plant traits will hinge on environmental gradients, highlighting the need to replace species mean traits with distributions that can be used to describe the breadth of ITV (Albert et al., 2010).
Significant relationships between root traits and soil fertility metrics have been demonstrated at the plant community level (Fort et al., 2016) and across land-use types (Prieto et al., 2015), though results are not consistent; for example, SRL has been observed to both increase (Fort et al., 2016) and decrease (Prieto et al., 2015) with increasing soil fertility. Similar inconsistencies have been shown in other studies on interspecific variation in root trait: Holdaway et al. (2011) found that species grown in P limited environments exhibited high SRL, low root D, and high root branching, while Ostonen et al. (2007) found that SRL decreases with increasing fertilization. Although Tobner et al. (2013) illustrate that intraspecific variation in certain root traits, namely root D, may indicate environmental change, our continuous soil variables (soil N, soil C, soil P, soil pH, and soil moisture) embedded within our nested levels explained little ITV (1.5–10.5%) in C. arabica roots (Table 4), and were not sufficient to capture the full range of explanatory soil variables. Highly dynamic soil metrics, such as N mineralization rates or hydraulic conductivity may be more well suited to predict root ITV at the site- through to the individual-plant level, however, this is beyond the scope of this study.
Unmeasured components of the root economy in ecosystems may also be more adept in explaining ITV in roots. For example, Valverde-Barrantes et al. (2016) suggest an ‘alternative adaptation’ to resource acquisition leading to higher colonization space for arbuscular mycorrhizal fungi (AMF), which results in root traits that defy strict trade-offs along a RES. Furthermore, associations to AMF versus ectomycorrhizal fungi among woody species may differentially impact root morphology, root elongation, and tip density (Chen et al., 2016). In our study systems, the most significant feature differentiating the two management systems is the presence or absence of N2-fixing Fabaceae trees, which consequently results in alternate pathways for N deposition (Munroe and Isaac, 2014). Whereas the presence of an N2-fixing tree has a strong influence on C. arabica leaf chemical traits (Martin et al., 2017), but not roots (Table 4), much remains unknown about the impact of N2-fixing trees on the N economy of neighboring plants as well as fungal communities within these systems, two potential drivers of root trait form and function.
Intraspecific RES and LES Orthogonality
Our findings suggest that orthogonality of trait spectra that has been observed across species (Baraloto et al., 2010; Li et al., 2015), also describe the relationship between an intraspecific RES and LES in C. arabica (Figure 2). These two axes of variation are independent of one another, suggesting that within our study species, root traits vary in response to environmental conditions that do not necessarily result in commensurate changes in leaf form and function. These results support recent work on the lack of coordination between a RES and other trait spectra across species (Weemstra et al., 2016), but it is important to note that interspecific studies on root trait coordination/orthogonality with other known dimensions of plant functional specialization, have produced conflicting results (e.g., Liu et al., 2010; de la Riva et al., 2016).
One explanation for the decoupling of RES and LES traits within a species observed here, is that root traits are less constrained than leaves in terms of ‘phenotypic morphospace’ (Donovan et al., 2014). Individual phenotypic leaf plasticity is constrained while root trait expression exhibits larger variability. These differences are despite strong phylogenetic constraints on root traits that should limit their variability (Cornwell et al., 2014; Kramer-Walter et al., 2016). C. arabica plants exhibited relatively low variation in leaf morphological (LMA cv = 17.3%) and chemical traits (leaf C and N cv ≤ 10.1%, Martin et al., 2017), as compared to analogous root traits (SRL cv = 73.23%; SRA cv = 81.39%; Nroot cv = 25.46%; Table 2). This may be due to greater differentiation as a result of the multifunctional nature of roots for resource acquisition in a highly heterogeneous space as well as structural support. Furthermore, this lack of constrained ITV may be particularly strong in agroecosystems, given management practices that increase resource (water and nutrients) stability and thus allow for greater niche differentiation.
An alternative explanation engages the role of artificial selection (Milla et al., 2015; Martin et al., 2017). Given luxury resource availability in agroecosystems, artificial selection has been hypothesized to shift crop trait values toward the resource acquisition end of any trait spectra (Milla et al., 2014; Gagliardi et al., 2015). Additional studies testing hypotheses on how root trait syndromes have evolved across species are needed (Valverde-Barrantes and Blackwood, 2016). It is possible that selection has resulted in trait trade-offs in C. arabica that are less strong as compared to those observed in wild plants, but comparative analyses (c.f. Figure 2 in Martin et al., 2017) would be needed to test this expectation.
Conclusion
Observations that an intraspecific RES in C. arabica is orthogonal to an intraspecific LES, contributes to our understanding of the causes of whole plant ITV and will advance our predictive ability of C. arabica productive vigor and response to environmental change. Recent analyses point to a lack of root functional trait data as a major limitation when predicting the effects of climate change on yield or other agroecosystem functions (Rosenzweig et al., 2014). On one hand, our results indicate that root trait data cannot be reliably approximated based on correlations with other traits that might be more easily measured. However, our results here suggest root trait data from different sites or management-systems, may indeed capture the large majority of ITV in root traits, and could be incorporated into local-level models of agroecosystem function. For example in C. arabica, certain models (e.g., van Oijen et al., 2010) incorporate CNroot data as a predictor of yield and C storage; our results show that nearly 70% of the total variation in this particular trait can be accounted for by sampling C. arabica roots across sites and management-systems. As quantifying ITV in crops remains a key data requirement for many of the world’s most widely employed crop models (Bouman and van Laar, 2006), new site-specific observations of root traits will be central in refining such models.
As C. arabica is one of the most economically important tree-crops globally, providing evidence-based criteria to manage such systems across climatic and management conditions is undoubtedly needed for the success of coffee producers, particularly smallholder farmers. Extending our results to other important crop or wild plant species could have important implications for predicting ecosystem structure and function.
Author Contributions
MI designed and coordinated the study, conducted data collection and analysis, and drafted the manuscript. AM assisted in study design, participated in data collection, data analysis, and manuscript writing. EdM, BR, OR, and KV contributed to site establishment, study design, and manuscript writing. All authors gave final approval for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to sincerely thank Mirna Barrios, Sanjeeb Bhattarai and Francisco Estrada for invaluable logistical support and assistance in the field, and to Serra Buchanan, Luzianne Reid, and Keane Tirona for their assistance with lab analyses. We would like to thank three journal reviewers for their insightful and constructive comments. CoffeeFlux observatory (http://www.umrecosols.fr/index.php/fr/implantations/costa-rica) was developed by CIRAD (Centre de Coopération Internationale en Recherche Agronomique pour le Développement) and CATIE (Centro Agronómico Tropical de Investigación y Enseñanza) with thanks to Cafetalera Aquiares farm. Coffee-Flux was supported by the AIRD-SAFSE project and the MACACC project (ANR-13-AGRO-0005).
Footnotes
Funding. This research was undertaken thanks to funding from the Canada Research Chairs program and a Natural Sciences and Engineering Research Council of Canada Discovery Grant to MI.
References
- Albert C. H., Thuiller W., Yoccoz N. G., Douzet R., Aubert S., Lavorel S. (2010). A multi-trait approach reveals the structure and the relative importance of intra- vs. interspecific variability in plant traits. Funct. Ecol. 24 1192–1201. 10.1111/j.1365-2435.2010.01727.x [DOI] [Google Scholar]
- Baraloto C., Paine C. E. T., Poorter L., Beauchene J., Bonal D., Domenach A. M., et al. (2010). Decoupled leaf and stem economics in rain forest trees. Ecol. Lett. 13 1338–1347. 10.1111/j.1461-0248.2010.01517.x [DOI] [PubMed] [Google Scholar]
- Bardgett R. D., Mommer L., De Vries F. T. (2014). Going underground: root traits as drivers of ecosystem processes. Trends Ecol. Evol. 29 692–699. 10.1016/j.tree.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Bates D., Maechler M., Bolker B., Walker S. (2014). lme4: linear mixed-effects models using Eigen and S4. R Package Version 1 1–23. [Google Scholar]
- Bertrand B., Aguilar G., Santacreo R., Anzueto F. (1999). “El mejoramiento genético en América central,” in Desafíos de la Caficultura Centroamericana, eds Bertrand B., Rapidel B. (San José: Inter-American Institute for Cooperation on Agriculture; ), 407–456. [Google Scholar]
- Blackman C. J., Aspinwall M. J., Resco De Dios V., Smith R. A., Tissue D. T. (2016). Leaf photosynthetic, economics and hydraulic traits are decoupled among genotypes of a widespread species of eucalypt grown under ambient and elevated CO2. Funct. Ecol. 30 1491–1500. 10.1111/1365-2435.12661 [DOI] [Google Scholar]
- Bouman B. A. M., van Laar H. H. (2006). Description and evaluation of the rice growth model ORYZA2000 under nitrogen-limited conditions. Agric. Syst. 87 249–273. 10.1016/j.agsy.2004.09.011 [DOI] [Google Scholar]
- Cahill J. F., Mcnickle G. G., Haag J. J., Lamb E. G., Nyanumba S. M., Clair C. C. S. (2010). Plants integrate information about nutrients and neighbors. Science 328 1657–1657. 10.1126/science.1189736 [DOI] [PubMed] [Google Scholar]
- Carmona C. P., Rota C., Azcarate F. M., Peco B. (2015). More for less: sampling strategies of plant functional traits across local environmental gradients. Funct. Ecol. 29 579–588. 10.1111/1365-2435.12366 [DOI] [Google Scholar]
- Chapin F. S., Autumn K., Pugnaire F. (1993). Evolution of suites of traits in response to environmental stress. Am. Nat. 142 S78–S92. 10.1086/285524 [DOI] [Google Scholar]
- Charbonnier F., Le Maire G., Dreyer E., Casanoves F., Christina M., Dauzat J., et al. (2013). Competition for light in heterogeneous canopies: application of MAESTRA to a coffee (Coffea arabica L.) agroforestry system. Agric. For. Meteorol. 181 152–169. 10.1016/j.agrformet.2013.07.010 [DOI] [Google Scholar]
- Chave J., Coomes D., Jansen S., Lewis S. L., Swenson N. G., Zanne A. E. (2009). Towards a worldwide wood economics spectrum. Ecol. Lett. 12 351–366. 10.1111/j.1461-0248.2009.01285.x [DOI] [PubMed] [Google Scholar]
- Chen W. L., Koide R. T., Adams T. S., Deforest J. L., Cheng L., Eissenstat D. M. (2016). Root morphology and mycorrhizal symbioses together shape nutrient foraging strategies of temperate trees. Proc. Natl. Acad. Sci. U.S.A. 113 8741–8746. 10.1073/pnas.1601006113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C. G., Wright S. J., Wurzburger N. (2016). Root and leaf traits reflect distinct resource acquisition strategies in tropical lianas and trees. Oecologia 180 1037–1047. 10.1007/s00442-015-3410-7 [DOI] [PubMed] [Google Scholar]
- Cornwell W. K., Cornelissen J. H. C., Amatangelo K., Dorrepaal E., Eviner V. T., Godoy O., et al. (2008). Plant species traits are the predominant control on litter decomposition rates within biomes worldwide. Ecol. Lett. 11 1065–1071. 10.1111/j.1461-0248.2008.01219.x [DOI] [PubMed] [Google Scholar]
- Cornwell W. K., Westoby M., Falster D. S., Fitzjohn R. G., O’meara B. C., Pennell M. W., et al. (2014). Functional distinctiveness of major plant lineages. J. Ecol. 102 345–356. 10.1111/1365-2745.12208 [DOI] [Google Scholar]
- Craine J. M., Lee W. G., Bond W. J., Williams R. J., Johnson L. C. (2005). Environmental constraints on a global relationship among leaf and root traits of grasses. Ecology 86 12–19. 10.1890/04-1075 [DOI] [Google Scholar]
- DaMatta F. M. (2004). Ecophysiological constraints on the production of shaded and unshaded coffee: a review. Field Crop Res. 86 99–114. 10.1016/j.fcr.2003.09.001 [DOI] [Google Scholar]
- de la Riva E. G., Tosto A., Pérez-Ramos I. M., Navarro-Fernández C. M., Olmo M., Anten N. P. R., et al. (2016). A plant economics spectrum in Mediterranean forests along environmental gradients: is there coordination among leaf, stem and root traits? J. Veg. Sci. 27 187–199. 10.1111/jvs.12341 [DOI] [Google Scholar]
- Defrenet E., Roupsard O., Van Den Meersche K., Charbonnier F., Perez-Molina J. P., Khac E., et al. (2016). Root biomass, turnover and net primary productivity of a coffee agroforestry system in Costa Rica: effects of soil depth, shade trees, distance to row and coffee age. Ann. Bot. 118 833–851. 10.1093/aob/mcw153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias P. C., Araujo W. L., Moraes G. A. B. K., Barros R. S., Damatta F. M. (2007). Morphological and physiological responses of two coffee progenies to soil water availability. J. Plant Physiol. 164 1639–1647. 10.1016/j.jplph.2006.12.004 [DOI] [PubMed] [Google Scholar]
- Diaz S., Hodgson J. G., Thompson K., Cabido M., Cornelissen J. H. C., Jalili A., et al. (2004). The plant traits that drive ecosystems: evidence from three continents. J. Veg. Sci. 15 295–304. 10.1111/j.1654-1103.2004.tb02266.x [DOI] [Google Scholar]
- Diaz S., Kattge J., Cornelissen J. H. C., Wright I. J., Lavorel S., Dray S., et al. (2016). The global spectrum of plant form and function. Nature 529 167–171. 10.1038/nature16489 [DOI] [PubMed] [Google Scholar]
- Donovan L. A., Mason C. M., Bowsher A. W., Goolsby E. W., Ishibashi C. D. A. (2014). Ecological and evolutionary lability of plant traits affecting carbon and nutrient cycling. J. Ecol. 102 302–314. 10.1111/1365-2745.12193 [DOI] [Google Scholar]
- Fort F., Cruz P., Lecloux E., De Oliveira L. B., Stroia C., Theau J. P., et al. (2016). Grassland root functional parameters vary according to a community-level resource acquisition-conservation trade-off. J. Veg. Sci. 27 749–758. 10.1111/jvs.12405 [DOI] [Google Scholar]
- Gagliardi S., Martin A. R., Virginio E. D., Rapidel B., Isaac M. E. (2015). Intraspecific leaf economic trait variation partially explains coffee performance across agroforestry management regimes. Agric. Ecosyst. Environ. 200 151–160. 10.1016/j.agee.2014.11.014 [DOI] [Google Scholar]
- Garriz P. I. (1978). Distribución radicular de tres cultivares de Coffea arabica L. en un suelo limo-arcilloso. Agron. Trop. 29 91–103. [Google Scholar]
- Givnish T. J. (1988). Adaptation to sun and shade - a whole-plant perspective. Aust. J. Plant Physiol. 15 63–92. 10.1071/PP9880063 [DOI] [Google Scholar]
- Grime J. P. (1979). Plant Strategies and Vegetation Processes. New York, NY: John Wiley and Sons. [Google Scholar]
- Hajek P., Hertel D., Leuschner C. (2013). Intraspecific variation in root and leaf traits and leaf-root trait linkages in eight aspen demes (Populus tremula and P. tremuloides). Front. Plant Sci. 4:415 10.3389/fpls.2013.00415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdaway R. J., Richardson S. J., Dickie I. A., Peltzer D. A., Coomes D. A. (2011). Species- and community-level patterns in fine root traits along a 120 000-year soil chronosequence in temperate rain forest. J. Ecol. 99 954–963. 10.1111/j.1365-2745.2011.01821.x [DOI] [Google Scholar]
- Isaac M. E., Anglaaere L. C. N., Borden K., Adu-Bredu S. (2014). Intraspecific root plasticity in agroforestry systems across edaphic conditions. Agric. Ecosyst. Environ. 185 16–23. 10.1016/j.agee.2013.12.004 [DOI] [Google Scholar]
- Jackson B. G., Peltzer D. A., Wardle D. A. (2013). The within-species leaf economic spectrum does not predict leaf litter decomposability at either the within-species or whole community levels. J. Ecol. 101 1409–1419. 10.1111/1365-2745.12155 [DOI] [Google Scholar]
- Jung V., Albert C. H., Violle C., Kunstler G., Loucougaray G., Spiegelberger T. (2014). Intraspecific trait variability mediates the response of subalpine grassland communities to extreme drought events. J. Ecol. 102 45–53. 10.1111/1365-2745.12177 [DOI] [Google Scholar]
- Kramer-Walter K. R., Bellingham P. J., Millar T. R., Smissen R. D., Richardson S. J., Laughlin D. C. (2016). Root traits are multidimensional: specific root length is independent from root tissue density and the plant economic spectrum. J. Ecol. 104 1299–1310. 10.1111/1365-2745.12562 [DOI] [Google Scholar]
- Lambers H., Poorter H. (1992). Inherent variation in growth-rate between higher-plants - a search for physiological causes and ecological consequences. Adv. Ecol. Res. 23 187–261. 10.1016/S0065-2504(08)60148-8 [DOI] [Google Scholar]
- Lambers H., Raven J. A., Shaver G. R., Smith S. E. (2008). Plant nutrient-acquisition strategies change with soil age. Trends Ecol. Evol. 23 95–103. 10.1016/j.tree.2007.10.008 [DOI] [PubMed] [Google Scholar]
- Larson J. E., Funk J. L. (2016). Seedling root responses to soil moisture and the identification of a belowground trait spectrum across three growth forms. New Phytol. 210 827–838. 10.1111/nph.13829 [DOI] [PubMed] [Google Scholar]
- Laughlin D. C., Joshi C., Van Bodegom P. M., Bastow Z. A., Fule P. Z. (2012). A predictive model of community assembly that incorporates intraspecific trait variation. Ecol. Lett. 15 1291–1299. 10.1111/j.1461-0248.2012.01852.x [DOI] [PubMed] [Google Scholar]
- Le S., Josse J., Husson F. (2008). FactoMineR: an R package for multivariate analysis. J. Stat. Softw. 25 1–18. 10.18637/jss.v025.i01 [DOI] [Google Scholar]
- Lecerf A., Chauvet E. (2008). Intraspecific variability in leaf traits strongly affects alder leaf decomposition in a stream. Basic Appl. Ecol. 9 598–605. 10.1016/j.baae.2007.11.003 [DOI] [Google Scholar]
- Lefcheck J. S. (2016). piecewiseSEM: piecewise structural equation modeling in R for ecology, evolution, and systematics. Methods Ecol. Evol. 7 573–579. 10.1111/2041-210X.12512 [DOI] [Google Scholar]
- Legendre P. (2014). lmodel2: Model II Regression. R Package Version 1.7–2. Available at: http://CRAN.R-project.org/package=lmodel2 [Google Scholar]
- Li L., Mccormack M. L., Ma C. G., Kong D. L., Zhang Q., Chen X. Y., et al. (2015). Leaf economics and hydraulic traits are decoupled in five species-rich tropical-subtropical forests. Ecol. Lett. 18 899–906. 10.1111/ele.12466 [DOI] [PubMed] [Google Scholar]
- Liu G. F., Freschet G. T., Pan X., Cornelissen J. H. C., Li Y., Dong M. (2010). Coordinated variation in leaf and root traits across multiple spatial scales in Chinese semi-arid and arid ecosystems. New Phytol. 188 543–553. 10.1111/j.1469-8137.2010.03388.x [DOI] [PubMed] [Google Scholar]
- Martin A. R., Rapidel B., Roupsard O., Van Den Meersche K., De M., Virginio Filho E., et al. (2017). Intraspecific trait variation across multiple scales: the leaf economics spectrum in coffee. Funct. Ecol. 31 604–612. 10.1111/1365-2435.12790 [DOI] [Google Scholar]
- Matos F. S., Wolfgramm R., Goncalves F. V., Cavatte P. C., Ventrella M. C., Damatta F. M. (2009). Phenotypic plasticity in response to light in the coffee tree. Environ. Exp. Bot. 67 421–427. 10.1111/j.1399-3054.2011.01525.x [DOI] [Google Scholar]
- McCormack M. L., Dickie I. A., Eissenstat D. M., Fahey T. J., Fernandez C. W., Guo D. L., et al. (2015). Redefining fine roots improves understanding of below-ground contributions to terrestrial biosphere processes. New Phytol. 207 505–518. 10.1111/nph.13363 [DOI] [PubMed] [Google Scholar]
- Meister R., Rajani M. S., Ruzicka D., Schachtman D. P. (2014). Challenges of modifying root traits in crops for agriculture. Trends Plant Sci. 19 779–788. 10.1016/j.tplants.2014.08.005 [DOI] [PubMed] [Google Scholar]
- Meylan L. (2012). Design of Cropping Systems Combining Production and Ecosystem Services: Developing a Methodology Combining Numerical Modeling and Participation of Farmers. Application to a coffee-Based Agroforestry in Costa Rica. Ph.D. thesis, Montpellier SupAgro, Montpellier. [Google Scholar]
- Milla R., Morente-Lopez J., Alonso-Rodrigo J. M., Martin-Robles N., Chapin F. S. (2014). Shifts and disruptions in resource-use trait syndromes during the evolution of herbaceous crops. Proc. R. Soc. B Biol. Sci. 281:20141429 10.1098/rspb.2014.1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milla R., Osborne C. P., Turcotte M. M., Violle C. (2015). Plant domestication through an ecological lens. Trends Ecol. Evol. 30 463–469. 10.1016/j.tree.2015.06.006 [DOI] [PubMed] [Google Scholar]
- Miner B. G., Sultan S. E., Morgan S. G., Padilla D. K., Relyea R. A. (2005). Ecological consequences of phenotypic plasticity. Trends Ecol. Evol. 20 685–692. 10.1016/j.tree.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Moran E. V., Hartig F., Bell D. M. (2016). Intraspecific trait variation across scales: implications for understanding global change responses. Glob. Change Biol. 22 137–150. 10.1111/gcb.13000 [DOI] [PubMed] [Google Scholar]
- Moser G., Leuschner C., Roderstein M., Graefe S., Soethe N., Hertel D. (2010). Biomass and productivity of fine and coarse roots in five tropical mountain forests stands along an altitudinal transect in southern Ecuador. Plant Ecol. Divers. 3 151–164. 10.1080/17550874.2010.517788 [DOI] [Google Scholar]
- Munroe J. W., Isaac M. E. (2014). N2-fixing trees and the transfer of fixed-N for sustainable agroforestry: a review. Agron. Sustain. Dev. 34 417–427. 10.1007/s13593-013-0190-5 [DOI] [Google Scholar]
- Nakagawa S., Schielzeth H. (2013). A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol. Evol. 4 133–142. 10.1111/j.2041-210x.2012.00261.x [DOI] [Google Scholar]
- Niinemets U. (2015). Is there a species spectrum within the world-wide leaf economics spectrum? Major variations in leaf functional traits in the Mediterranean sclerophyll Quercus ilex. New Phytol. 205 79–96. 10.1111/nph.13001 [DOI] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’hara R. B., et al. (2016). vegan: Community Ecology Package in R version 2.3–5. Available at: https://CRAN.R-project.org/package=vegan [Google Scholar]
- Ostonen I., Puttsepp U., Biel C., Alberton O., Bakker M. R., Lohmus K., et al. (2007). Specific root length as an indicator of environmental change. Plant Biosyst. 141 426–442. 10.1080/11263500701626069 [DOI] [Google Scholar]
- Padovan M. P., Cortez V. J., Navarrete L. F., Navarrete E. D., Deffner A. C., Centeno L. G., et al. (2015). Root distribution and water use in coffee shaded with Tabebuia rosea Bertol. and Simarouba glauca DC. compared to full sun coffee in sub-optimal environmental conditions. Agroforest. Syst. 89 857–868. 10.1007/s10457-015-9820-z [DOI] [Google Scholar]
- Paradis E., Claude J., Strimmer K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20 289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- Perez-Harguindeguy N., Diaz S., Garnier E., Lavorel S., Poorter H., Jaureguiberry P., et al. (2013). New handbook for standardised measurement of plant functional traits worldwide. Aust. J. Bot. 61 167–234. 10.1071/BT12225 [DOI] [Google Scholar]
- Prieto I., Roumet C., Cardinael R., Dupraz C., Jourdan C., Kim J. H., et al. (2015). Root functional parameters along a land-use gradient: evidence of a community-level economics spectrum. J. Ecol. 103 361–373. 10.1111/1365-2745.12351 [DOI] [Google Scholar]
- Reich P. B. (2014). The world-wide ‘fast-slow’ plant economics spectrum: a traits manifesto. J. Ecol. 102 275–301. 10.1111/1365-2745.12211 [DOI] [Google Scholar]
- Reich P. B., Walters M. B., Ellsworth D. S. (1997). From tropics to tundra: global convergence in plant functioning. Proc. Natl. Acad. Sci. U.S.A. 94 13730–13734. 10.1073/pnas.94.25.13730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers T., Pons T. L., Bongers F. (2000). The effect of tree height and light availability on photosynthetic leaf traits of four neotropical species differing in shade tolerance. Funct. Ecol. 14 77–86. 10.1046/j.1365-2435.2000.00395.x [DOI] [Google Scholar]
- Rillig M. C., Aguilar-Trigueros C. A., Bergmann J., Verbruggen E., Veresoglou S. D., Lehmann A. (2015). Plant root and mycorrhizal fungal traits for understanding soil aggregation. New Phytol. 205 1385–1388. 10.1111/nph.13045 [DOI] [PubMed] [Google Scholar]
- Rosenzweig C., Elliott J., Deryng D., Ruane A. C., Muller C., Arneth A., et al. (2014). Assessing agricultural risks of climate change in the 21st century in a global gridded crop model intercomparison. Proc. Natl. Acad. Sci. U.S.A. 111 3268–3273. 10.1073/pnas.1222463110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumet C., Birouste M., Picon-Cochard C., Ghestem M., Osman N., Vrignon-Brenas S., et al. (2016). Root structure-function relationships in 74 species: evidence of a root economics spectrum related to carbon economy. New Phytol. 210 815–826. 10.1111/nph.13828 [DOI] [PubMed] [Google Scholar]
- Shipley B., Lechowicz M. J., Wright I., Reich P. B. (2006). Fundamental trade-offs generating the worldwide leaf economics spectrum. Ecology 87 535–541. 10.1890/05-1051 [DOI] [PubMed] [Google Scholar]
- Siefert A., Violle C., Chalmandrier L., Albert C. H., Taudiere A., Fajardo A., et al. (2015). A global meta-analysis of the relative extent of intraspecific trait variation in plant communities. Ecol. Lett. 18 1406–1419. 10.1111/ele.12508 [DOI] [PubMed] [Google Scholar]
- Tjoelker M. G., Craine J. M., Wedin D., Reich P. B., Tilman D. (2005). Linking leaf and root trait syndromes among 39 grassland and savannah species. New Phytol. 167 493–508. 10.1111/j.1469-8137.2005.01428.x [DOI] [PubMed] [Google Scholar]
- Tobner C. M., Paquette A., Messier C. (2013). Interspecific coordination and intraspecific plasticity of fine root traits in North American temperate tree species. Front. Plant Sci. 4:242 10.3389/fpls.2013.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde-Barrantes O. J., Blackwood C. B. (2016). Root traits are multidimensional: specific root length is independent from root tissue density and the plant economic spectrum: commentary on Kramer-Walter et al. (2016). J. Ecol. 104 1311–1313. 10.1111/1365-2745.12605 [DOI] [Google Scholar]
- Valverde-Barrantes O. J., Horning A. L., Smemo K. A., Blackwood C. B. (2016). Phylogenetically structured traits in root systems influence arbuscular mycorrhizal colonization in woody angiosperms. Plant Soil 404 1–12. 10.1007/s11104-016-2820-6 [DOI] [Google Scholar]
- van Kanten R., Schroth G., Beer J., Jimenez F. (2005). Fine-root dynamics of coffee in association with two shade trees in Costa Rica. Agroforest. Syst. 63 247–261. 10.1007/s10457-005-4163-9 [DOI] [Google Scholar]
- van Oijen M., Dauzat J., Harmand J. M., Lawson G., Vaast P. (2010). Coffee agroforestry systems in Central America: II. Development of a simple process-based model and preliminary results. Agroforest. Syst. 80 361–378. 10.3389/fmicb.2015.00091 [DOI] [Google Scholar]
- Warton D. I., Wright I. J., Falster D. S., Westoby M. (2006). Bivariate line-fitting methods for allometry. Biol. Rev. 81 259–291. 10.1017/S1464793106007007 [DOI] [PubMed] [Google Scholar]
- Weemstra M., Mommer L., Visser E. J. W., Van Ruijven J., Kuyper T. W., Mohren G. M. J., et al. (2016). Towards a multidimensional root trait framework: a tree root review. New Phytol. 211 1159–1169. 10.1111/nph.14003 [DOI] [PubMed] [Google Scholar]
- Westoby M., Falster D. S., Moles A. T., Vesk P. A., Wright I. J. (2002). Plant ecological strategies: some leading dimensions of variation between species. Annu. Rev. Ecol. Syst. 33 125–159. 10.1146/annurev.ecolsys.33.010802.150452 [DOI] [Google Scholar]
- Wright I. J., Reich P. B., Westoby M., Ackerly D. D., Baruch Z., Bongers F., et al. (2004). The worldwide leaf economics spectrum. Nature 428 821–827. 10.1038/nature02403 [DOI] [PubMed] [Google Scholar]