Summary
Sgf73, a core component of SAGA, is the yeast orthologue of ataxin‐7, which undergoes CAG–polyglutamine repeat expansion leading to the human neurodegenerative disease spinocerebellar ataxia type 7 (SCA7). Deletion of SGF73 dramatically extends replicative lifespan (RLS) in yeast. To further define the basis for Sgf73‐mediated RLS extension, we performed ChIP‐Seq, identified 388 unique genomic regions occupied by Sgf73, and noted enrichment in promoters of ribosomal protein (RP)‐encoding genes. Of 388 Sgf73 binding sites, 33 correspond to 5′ regions of genes implicated in RLS extension, including 20 genes encoding RPs. Furthermore, half of Sgf73‐occupied, RLS‐linked RP genes displayed significantly reduced expression in sgf73Δ mutants, and double null strains lacking SGF73 and a Sgf73‐regulated, RLS‐linked RP gene exhibited no further increase in replicative lifespan. We also found that sgf73Δ mutants display altered acetylation of Ifh1, an important regulator of RP gene transcription. These findings implicate altered ribosomal protein expression in sgf73Δ yeast RLS and highlight altered acetylation as a pathway of relevance for SCA7 neurodegeneration.
Keywords: genome‐wide occupancy, longevity gene, Neurodegeneration, replicative lifespan, Sgf73, yeast
Introduction
The SAGA (Spt‐Ada‐Gcn5 Acetyltransferase) complex is a major transcriptional coactivator complex responsible for regulation of a large number of yeast genes, the majority of which are stress‐induced (Huisinga & Pugh, 2004). Additionally, SAGA regulates many actively transcribed yeast genes, and the cognate orthologous STAGA (Spt3‐Taf9‐Gcn5 acetyltransferase) complex regulates the expression of numerous mammalian genes (Bonnet et al., 2014), suggesting its broad significance for transcription regulation. The SAGA/STAGA complex is recruited to its target genes by many transcription factors, and through both its Gcn5/GCN5 acetylation and Ubp8/USP22 deubiquitination activities at promoters, it facilitates transcription activation and elongation [reviewed in (Koutelou et al., 2010)]. The SAGA complex has four distinct modules responsible for acetyltransferase activity, deubiquitination activity, complex recruitment, and architecture of the complex. Sgf73 is a subunit of SAGA (Helmlinger et al., 2004; Timmers & Tora, 2005), which acts in the Ubp8 deubiquitinase module (DUBm), linking the core SAGA Gcn5‐mediated acetylation activity to the Ubp8 DUBm (Kohler et al., 2008; Zhao et al., 2008; Lee et al., 2009; Morgan et al., 2016).
Deletion of SGF73 results in a dramatic extension in yeast RLS (McCormick et al., 2014). RLS is defined as the number of daughter cells that a mother cell produces before senescence. SGF73 is required for Ubp8‐mediated histone H2B deubiquitination, with sgf73Δ strains having elevated H2BK123 ubiquitination, as is seen in ubp8Δ strains (Kohler et al., 2008). This increase in ubiquitinated H2BK123 contributes to the RLS extension of sgf73Δ mutants, as strains that lack UBP8 are also long‐lived, and strains harboring the H2B‐K123R mutation, which cannot be ubiquitinated, are short‐lived (McCormick et al., 2014).
The RLS extension in sgf73Δ mutants is Sir2 dependent, and sgf73Δ strains have altered Sir2‐related activities (McCormick et al., 2014). SIR2 encodes a class III NAD+‐dependent deacetylase that has been implicated in both the regulation of lifespan and the benefits of caloric restriction in a variety of species, making it a central focus of aging research. Overexpression of SIRT1, the closest mammalian orthologue of SIR2, has varying effects on senescence depending on the cell type, further underscoring the important role of the sirtuins in aging (Langley et al., 2002; Chua et al., 2005). Additionally, yeast Sir2 is required for telomeric and ribosomal DNA (rDNA) silencing, and suppression of homologous recombination within the rDNA repeats [reviewed in (Kueng et al., 2013)]. Inappropriate rDNA recombination results in the accumulation of rDNA circles, which is one factor correlated with aging in yeast (reviewed in (Blander & Guarente, 2004). SAGA has been shown to play a role in the asymmetric retention of rDNA circles in the mother cell by tethering the rDNA circles to nuclear pore complexes. SGF73 deletion reduces rDNA circle retention (Denoth‐Lippuner et al., 2014).
Ribosomal biogenesis is another key determinant of aging. In particular, a reduction in 60S ribosomal subunits promotes lifespan extension in yeast (Steffen et al., 2008), and a reduction in both the 40S and 60S ribosomal subunits in Caenorhabditis elegans yields longer lifespan in this metazoan model organism (Chen et al., 2007; Curran & Ruvkun, 2007; Hansen et al., 2007). Multiple signaling pathways regulate ribosomal biogenesis in response to nutrients and stress. One of these is the nutrient‐sensing target of rapamycin (TOR) signaling pathway that regulates the interacting coactivator Ifh1 (Interacts with Forkhead) and corepressor Crf1 (Co‐Repressor with FHL1) of the Forkhead‐like transcription factor (Fhl1) (Martin et al., 2004). The dynamic process of ribosomal protein gene transcription is regulated by the interaction of Ifh1 with Fhl1, which is bound at RP gene promoters (Warner, 1999). Ifh1 is first recruited to ribosomal gene promoters upon nutrient stimulus, with acetylation being a key regulator of Ifh1 activity (Schawalder et al., 2004). Ifh1 activity is decreased upon Gcn5‐mediated acetylation, whereas Sir2 and another sirtuin, Hst1, deacetylate Ifh1, resulting in increased activity. It is this on–off cycle that contributes to proper regulation of RP gene transcription (Downey et al., 2013).
The human counterpart of Sgf73 is ataxin‐7, and the gene encoding this protein causes the autosomal dominant neurodegenerative disorder spinocerebellar ataxia type 7 (SCA7), upon CAG–polyglutamine repeat expansion. Ataxin‐7 is a member of the mammalian transcription coactivator complex STAGA, where it plays a similar role to its yeast DUBm counterpart (Martinez et al., 2001; Helmlinger et al., 2004). Ataxin‐7 has an amino‐terminal CAG/polyglutamine tract that normally ranges from four to 35 repeats. However, when this tract expands to repeat lengths greater than 37, humans develop SCA7 (David et al., 1997; Stevanin et al., 2000). SCA7 is thus one of the nine heritable CAG–polyglutamine repeat expansion disorders that all result in expanded polyglutamine tracts in different proteins. Accumulation of polyglutamine‐expanded ataxin‐7 protein leads to the dysfunction and death of neurons in the retina, cerebellum, and brainstem, causing blindness, progressive loss of coordination, and premature death (Lebre & Brice, 2003).
To probe the molecular mechanisms of Sgf73/ataxin‐7 function, we used a ChIP‐Seq approach, identifying 388 genomic regions bound by Sgf73. Notably, we found an enrichment of binding in the 5′ promoter regions of genes encoding ribosomal proteins (RPs). Of the Sgf73‐occupied yeast genomic regions, 33 corresponded to the 5′ regions of genes previously linked to RLS extension, with the majority of these genes encoding RPs. We found that deletion of SGF73 altered the expression of Sgf73‐occupied RP genes at baseline and upon rapamycin stress. Furthermore, we found that double null strains of yeast lacking SGF73 and a Sgf73‐regulated RLS‐linked RP gene did not exhibit any further increase in RLS. Finally, we noted altered acetylation of the TOR target Ifh1 in sgf73Δ mutants. Our results reveal Sgf73 target genes that are integral to pathways of aging regulation in yeast and may highlight pathways regulating mammalian aging and SCA7 neurodegeneration in humans.
Results
Sgf73 ChIP‐Seq analysis yields 388 unique genomic occupancy sites
To perform ChIP on Sgf73, we integrated a carboxy‐terminal 13‐Myc epitope. Integration was verified by molecular genetic analysis, while tagged protein production and ChIP‐tagged protein recovery were confirmed by immunoblotting (Fig. S1). In addition, functionality of the tagged protein was confirmed using standard growth assays that demonstrated that the 13‐Myc tag did not interfere with Sgf73 function (Fig. S2). Sgf73 ChIP'd DNA was processed into libraries, and single‐end sequencing was performed to generate ~17 million 50‐bp reads per sample. Raw sequence reads were aligned to the Saccharomyces cerevisiae S288C genome 3 (sacCer3). Significant peaks were identified using HOMER (Hypergeometric Optimization of Motif EnRichment) v4.2 (Heinz et al., 2010). These were defined as a collection of sequence reads, collectively mapping to genomic locations at a significantly higher density than background and surrounding genomic areas (Fig. 1). Our Sgf73 ChIP‐Seq analysis identified 389 significant peaks (Table S1), with two significant peaks positioned 5′ to the same transcriptional start site (TSS), resulting in 388 unique occupancy sites. The tag counts, which correspond to the number of unique reads mapping to the indicated genomic region, were used to rank the peaks.
Figure 1.
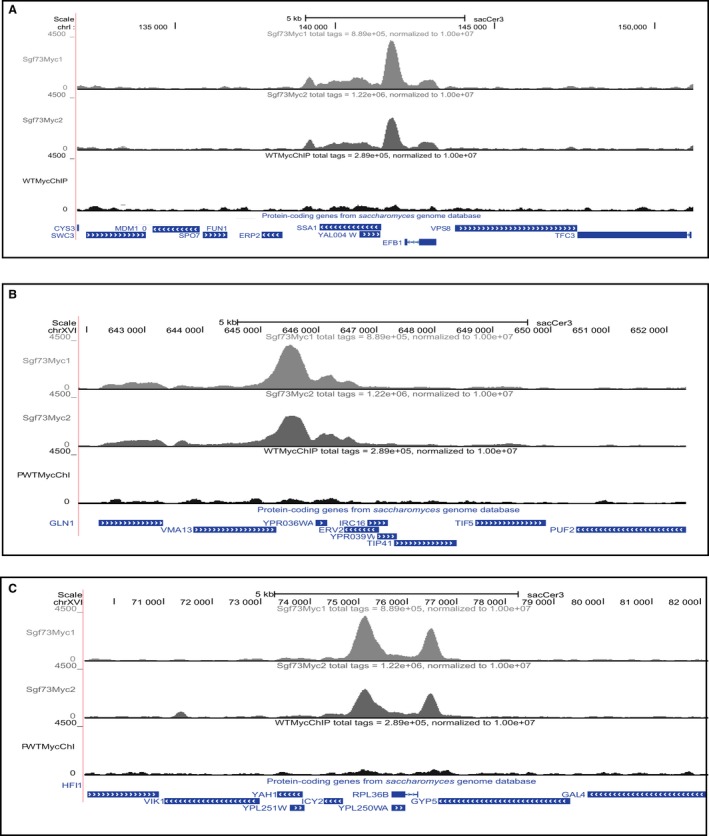
Representative genome tracks of Sgf73‐bound peaks. Visual inspection of genome tracks of reads from significant peaks shows correct assignment of called peaks, uniformity between Sgf73 ChIP replicates, and significance over background. The three top‐ranked peaks (highest peak scores) are presented: (A) SSA1, (B) YPR036W‐A, and (C) ICY2. Note that in the ICY2 track image (C), the significant peak for RPL36B occurs 3′ to ICY2.
Genes encoding ribosomal proteins (RPs) are enriched for Sgf73 occupancy
To understand the most significant genes and pathways affected by Sgf73, we performed enrichment analyses using WebGestalt (Zhang et al., 2005; Wang et al., 2013), followed by Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa & Goto, 2000; Kanehisa et al., 2012, 2014) analysis to reveal gene families and pathways that were enriched in the set of Sgf73‐occupied genes (Fig. S3A). Most striking from this analysis is the enrichment of ‘ribosome’ in GO terms and, significantly, ‘ribosome’ is the only enriched KEGG pathway (P = 1.96e‐34), with 57 of the 388 peaks (~14.5%) displaying Sgf73 occupancy proximal to a RP gene (Table 1). Survey of the ChIP‐Seq tracks confirmed the presence of Sgf73 RP occupancy peaks (Fig. 2). Of the 57 ribosomal protein genes, 36 encode large ribosomal subunits, 20 encode small ribosomal subunits, and one encodes a ribosomal stalk protein. It is noteworthy that numerous studies have linked ribosomal biogenesis to aging and have established that reduced expression of large ribosomal subunits can yield dramatically increased RLS (Kaeberlein et al., 2005; Steffen et al., 2008). Our findings thus indicate that Sgf73 loss of function may promote increased RLS by altering ribosome regulation.
Table 1.
Ribosomal protein genes with 5′ occupancy by Sgf73
| Gene symbol | Gene name | Ensembl gene stable ID | Entrez ID |
|---|---|---|---|
| RPL1B | 60S ribosomal protein L1 | YGL135W | 852742 |
| RPL2A | 60S ribosomal protein L2 | YFR031C‐A | 850590 |
| RPL5 | 60S ribosomal protein L5 | YPL131W | 855972 |
| RPL6A | 60S ribosomal protein L6‐A | YML073C | 854902 |
| RPL6B | 60S ribosomal protein L6‐B | YLR448W | 851169 |
| RPL7A | 60S ribosomal protein L7‐A | YGL076C | 852804 |
| RPL8A | 60S ribosomal protein L8‐A | YHL033C | 856352 |
| RPL10 | 60S ribosomal protein L10 | YLR075W | 850764 |
| RPL13B | 60S ribosomal protein L13‐B | YMR142C | 855173 |
| RPL14A | 60S ribosomal protein L14‐A | YKL006W | 853864 |
| RPL16B | 60S ribosomal protein L16‐B | YNL069C | 855655 |
| RPL17A | 60S ribosomal protein L17‐A | YKL180W | 853674 |
| RPL19A | 60S ribosomal protein L19 | YBR084C‐A | 852379 |
| RPL19B | 60S ribosomal protein L19 | YBL027W | 852254 |
| RPL20A | 60S ribosomal protein L20‐A | YMR242C | 855283 |
| RPL20B | 60S ribosomal protein L20 | YOR312C | 854489 |
| RPL22A | 60S ribosomal protein L22‐A | YLR061W | 850750 |
| RPL23A | 60S ribosomal protein L23 | YBL087C | 852191 |
| RPL23B | 60S ribosomal protein L23 | YER117W | 856853 |
| RPL26A | 60S ribosomal protein L26‐A | YLR344W | 851058 |
| RPL27B | 60S ribosomal protein L27‐B | YDR471W | 852082 |
| RPL28 | 60S ribosomal protein L28 | YGL103W | 852775 |
| RPL30 | 60S ribosomal protein L30 | YGL030W | 852853 |
| RPL31A | 60S ribosomal protein L31‐A | YDL075W | 851484 |
| RPL33A | 60S ribosomal protein L33‐A | YPL143W | 855960 |
| RPL34B | 60S ribosomal protein L34‐B | YIL052C | 854759 |
| RPL35A | 60S ribosomal protein L35 | YDL191W | 851336 |
| RPL36B | 60S ribosomal protein L36‐B | YPL249C‐A | 855826 |
| RPL37B | 60S ribosomal protein L37‐B | YDR500C | 852111 |
| RPL39 | 60S ribosomal protein L39 | YJL189W | 853250 |
| RPL40A | 60S ribosomal protein L40; Ubiquitin | YIL148W | 854658 |
| RPL41A | 60S ribosomal protein L41 | YDL184C | 851344 |
| RPL41B | 60S ribosomal protein L41 | YDL133C‐A | 851422 |
| RPL42A | 60S ribosomal protein L42 | YNL162W | 855560 |
| RPL43A | 60S ribosomal protein L43 | YPR043W | 856156 |
| RPL43B | 60S ribosomal protein L43 | YJR094W‐A | 853557 |
| RPP2A | 60S acidic ribosomal protein P2‐alpha | YOL039W | 854118 |
| RPS0A | 40S ribosomal protein S0‐A | YGR214W | 853128 |
| RPS0B | 40S ribosomal protein S0‐B | YLR048W | 850737 |
| RPS1A | YLR441C | YLR441C | 851162 |
| RPS3 | 40S ribosomal protein S3 | YNL178W | 855543 |
| RPS5 | 40S ribosomal protein S5 | YJR123W | 853587 |
| RPS7A | 40S ribosomal protein S7‐A | YOR096W | 854263 |
| RPS7B | 40S ribosomal protein S7‐B | YNL096C | 855628 |
| RPS8B | 40S ribosomal protein S8 | YER102W | 856839 |
| RPS9A | 40S ribosomal protein S9‐A | YPL081W | 856024 |
| RPS9B | 40S ribosomal protein S9‐B | YBR189W | 852487 |
| RPS11B | 40S ribosomal protein S11 | YBR048W | 852337 |
| RPS12 | 40S ribosomal protein S12 | YOR369C | 854551 |
| RPS14A | 40S ribosomal protein S14‐A | YCR031C | 850397 |
| RPS16B | 40S ribosomal protein S16 | YDL083C | 851476 |
| RPS17B | 40S ribosomal protein S17‐B | YDR447C | 852058 |
| RPS18B | 40S ribosomal protein S18 | YML026C | 854982 |
| RPS19B | 40S ribosomal protein S19‐B | YNL302C | 855414 |
| RPS21B | 40S ribosomal protein S21‐B | YJL136C | 853305 |
| RPS24A | 40S ribosomal protein S24 | YER074W | 856805 |
| RPS31 | 40S ribosomal protein S31; Ubiquitin | YLR167W | 850864 |
Figure 2.
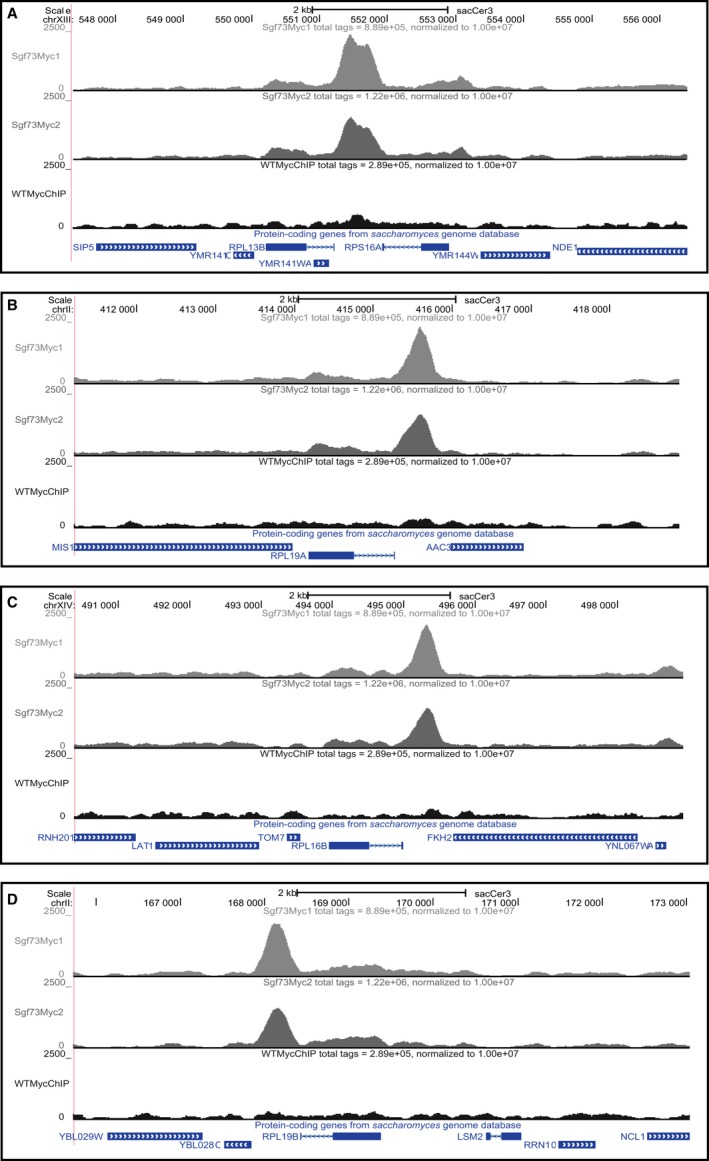
Representative genome tracks of RLS‐linked RP Sgf73 occupancy peaks. Many Sgf73‐occupied peaks that are linked to genes involved in RLS extension upon deletion map 5′ to RP genes. Shown are genome tracks of representative peaks of the following RLS‐linked RP genes: (A) RPL13B, (B) RPL19A, (C) RPL16B, and (D) RPL19B.
Genes encoding large ribosomal subunits implicated in RLS extension studies are enriched for Sgf73 binding
Deletion of SGF73 results in a dramatic extension in RLS. As a core component of the SAGA transcription coactivator complex, Sgf73 loss of function may alter the expression of genes catalogued to promote RLS extension. We considered the results of a recent study in which all nonessential yeast deletion mutants were subjected to RLS analysis (McCormick et al., 2015). Comparison of Sgf73 genomic occupancy sites with these 238 yeast genes, found to significantly promote RLS upon deletion, revealed 33 uniquely shared genes (Table 2; Fig. S3B). Among these 33 genes (McCormick et al., 2015), 20 encode RPs, of which 18 of the 20 RP genes encode large ribosomal subunits. To confirm that these promoters are SAGA DUBm‐targeted, we performed ChIP analysis of Ubp8 and observed that Ubp8 co‐occupied 14 of the 19 RLS‐linked RP regions enriched for Sgf73 binding (Table S1 and Fig. S4). However, it should be noted that Sgf73 is known to bind DNA through its zinc finger domain (Bonnet et al., 2010), whereas there is no evidence for Ubp8 directly binding to DNA, which is supported by our detection of only 174 Ubp8 peaks, of which 171 are also Sgf73‐occupied (Table S2; Gene Expression Omnibus accession #GSE76461). As the yeast RLS extension single‐gene deletion survey yielded a total of 26 RP genes encoding large ribosome subunits (McCormick et al., 2015), and 18 of these 26 large ribosome unit genes (i.e., ~69%) exhibit Sgf73 occupancy, altered regulation of large ribosomal subunit gene expression may contribute to the extreme RLS extension phenotype observed in sgf73Δ yeast strains.
Table 2.
Sgf73 occupied genes implicated in replicative lifespan extension
| Chr # | Start | End | Peak score | bp to TSS | Promoter ID | Gene name | Gene description |
|---|---|---|---|---|---|---|---|
| chrXIII | 388 438 | 388 553 | 2899.699951 | 235 | YMR058W | FET3 | Ferro‐02‐oxidoredutase |
| chrXVI | 679 114 | 679 269 | 2307.100098 | 240 | YPR064W | YPR064W | |
| chrlV | 765 359 | 765 481 | 2291.649902 | −265 | YDR151C | CTH1 | Cth1p |
| chrXIII | 551 398 | 551 502 | 2158.399902 | −243 | YMR142C | RPL13B | Ribosomal 60S subunit protein L13B |
| chrII | 415 546 | 415 648 | 2027.050049 | −336 | YBR084C‐A | RPL19A | Ribosomal 60S subunit protein L19A |
| chrXIV | 495 272 | 495 401 | 1984.149902 | −335 | YNL069C | RPL16B | Ribosomal 60S subunit protein L16B |
| chrII | 168 066 | 168 211 | 1955 | −285 | YBL027W | RPL19B | Ribosomal 60S subunit protein L19B |
| chrVII | 23 427 | 23 542 | 1886.400024 | −451 | YGL253W | HXK2 | Hexokinase 2 |
| chrVII | 254 368 | 254 492 | 1819.050049 | −211 | YGL135W | RPL1B | Ribosomal 60S subunit protein L1B |
| chrIV | 321 879 | 321 984 | 1733.550049 | −295 | YDL075W | RPL31A | Ribosomal 60S subunit protein L31A |
| chrXIV | 559 344 | 559 470 | 1728.550049 | −405 | YNL037C | IDH1 | Isocitrate dehydrogenase [NAD(+)] |
| chrXV | 253 915 | 254 041 | 1710.600098 | −319 | YOL039W | RPP2A | Ribosomal protein P2A |
| chrIX | 257 361 | 257 495 | 1688.699951 | −365 | YIL052C | RPL34B | Ribosomal 60S subunit protein L34B |
| chrXIII | 124 367 | 124 483 | 1627.349976 | −253 | YML073C | RPL6A | Ribosomal 60S subunit protein L6A |
| chrIV | 117 431 | 117 534 | 1530.599976 | −182 | YDL191W | RPL35A | Ribosomal 60S subunit protein L35A |
| chrXV | 901 425 | 901 527 | 1482.649902 | −282 | YOR312C | RPL20B | Ribosomal 60S subunit protein L20B |
| chrXII | 1 028 449 | 1 028 574 | 1472.050049 | −343 | YLR448W | RPL6B | Ribosomal 60S subunit protein L6B |
| chrXIII | 754 426 | 754 526 | 1431.550049 | 179 | YMR242C | RPL20A | Ribosomal 60S subunit protein L20A |
| chrV | 153 026 | 153 167 | 1352.5 | −424 | YER001W | MNN1 | Mnn1p |
| chrXII | 125 194 | 125 303 | 1343.649902 | −286 | YLL012W | YEH1 | Yeh1p |
| chrII | 60 934 | 61 043 | 1330.300049 | −249 | YBL087C | RPL23A | Ribosomal 60S subunit protein L23A |
| chrVII | 366 078 | 366 180 | 1292.449951 | −133 | YGL076C | RPL7A | Ribosomal 60S subunit protein L7A |
| chrXV | 1 028 799 | 1 028 904 | 1232.300049 | −226 | YOR369C | RPS12 | Ribosomal 40S subunit protein S12 |
| chrX | 525 859 | 525 976 | 1129.050049 | −418 | YJR048W | CYC1 | Cyc1p |
| chrIV | 1 450 918 | 1 451 035 | 1095.849976 | −123 | YDR500C | RPL37B | Ribosomal 60S subunit protein L37B |
| chrVII | 1 050 308 | 1 050 489 | 1042.550049 | −440 | YGR279C | SCW4 | Scw4p |
| chrXVI | 280 095 | 280 222 | 954.099976 | −322 | YPL144W | POC4 | Poc4p |
| chrXII | 818 597 | 818 710 | 822.75 | −659 | YLR344W | RPL26A | Ribosomal 60S subunit protein L26A |
| chrXII | 514 958 | 515 073 | 775.25 | −247 | YLR180W | SAM1 | Methionine adenosyltransferase |
| chrX | 607 973 | 608 113 | 763.5 | −262 | YJR094W‐A | RPL43B | Rpl43 bp |
| chrXII | 263 015 | 263 122 | 756.400024 | −126 | YLR061W | RPL22A | Ribosomal 60S subunit protein L22A |
| chrII | 697 587 | 697 702 | 604.550049 | −342 | YBR238C | YBR238C | Hypothetical protein |
| chrVII | 976 969 | 977 114 | 599.450012 | −295 | YGR243W | FMP43 | Fmp43p |
RP‐occupied genes Sgf73 have reduced expression in sgf73Δ mutants
To determine whether RP‐encoding genes identified as Sgf73 genomic occupancy sites undergo changes in gene expression upon SGF73 deletion, we measured RNA expression levels of RP‐encoding genes that promote RLS extension in the sgf73Δ mutant. We detected reduced levels of gene expression for all tested RP‐encoding genes, with significant reductions in gene expression documented for nine of these RP‐encoding genes (Fig. 3A). We selected an additional 12 Sgf73‐occupied, non‐RLS‐promoting genes that encode ribosomal subunits for qRT–PCR analysis, as not all RPs may result in significant longevity on their own but could contribute in conjunction with other factors, and we similarly observed reduced expression levels for all 12 RP‐encoding genes, with the majority showing significant reductions (Fig. 3B). Reduced expression of genes encoding ribosomal subunits further supports a role for altered transcriptional regulation of RP‐encoding genes in the dramatic RLS phenotype documented in sgf73Δ mutants.
Figure 3.
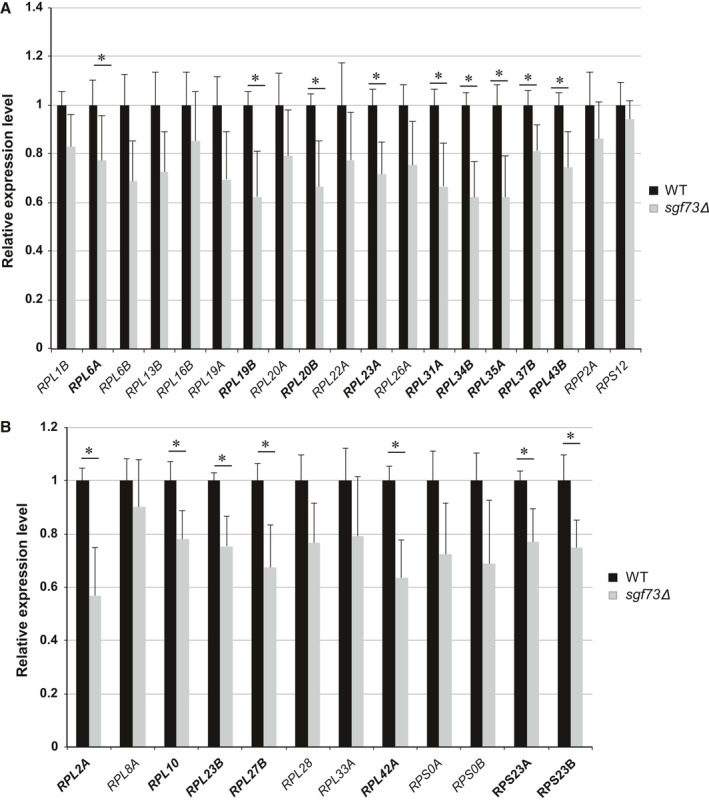
Ribosomal protein subunit genes, subject to Sgf73 occupancy and linked to replicative lifespan extension, display reduced expression in sgf73Δ mutants. RNA expression analysis of yeast ribosome protein (RP) subunit genes linked to replicative lifespan extension (A), and of yeast RP subunit genes not linked to replicative lifespan extension (B). Results represent qRT–PCR analysis of indicated RP subunit gene after normalization to the internal control SCR1. For each gene, expression in control WT (wild‐type) yeast was arbitrarily set to 1. Bold text indicates significant alterations. *P < 0.05, t‐test; n = 6 isolates/group; error bars = s.e.m.
The sgf73Δ mutant exhibits blunted transcriptional repression in response to rapamycin
One well‐documented response to various stress stimuli is a reduction in the transcription of RP‐encoding genes [reviewed in (Xiao & Grove, 2009)]. To test whether downregulation of RP gene transcription is defective upon stress induction in sgf73Δ mutants, we treated WT and sgf73Δ yeast strains with rapamycin. Rapamycin inhibits TOR, resulting in the repression of RP gene expression, due to a shift toward binding with the repressive Fhl1 cofactor Crf1, as Ifh1 is constitutively bound at RP promoters and its cofactor binding partners determine RP transcriptional levels (Martin et al., 2004; Xiao & Grove, 2009). After treatment with 200 ng mL−1 rapamycin or ethanol vehicle control, we examined the level of RP gene expression in WT and sgf73Δ cells by qRT–PCR analysis. Although all tested RP genes displayed decreased expression upon rapamycin treatment in both WT and sgf73Δ yeast, we documented smaller reductions in RP gene expression in sgf73Δ strains compared to WT (Fig. 4). Strikingly, nine of the RLS‐linked RP genes (45%) retained significantly higher transcript levels in the sgf73Δ mutant in the face of rapamycin stress (Fig. 4A), whereas only two (16%) of the non‐RLS‐linked RP genes exhibited such blunted repression (Fig. 4B). Thus, despite maintaining lower RP gene expression levels in unchallenged conditions, sgf73Δ strains did not fully repress RP gene expression when treated with rapamycin. This implies defective RP regulation in both normal and stress conditions in the sgf73Δ mutant. Thus, in unchallenged situations, levels are beneficially lower, but during stress, they are higher—allowing cellular processes to persist.
Figure 4.
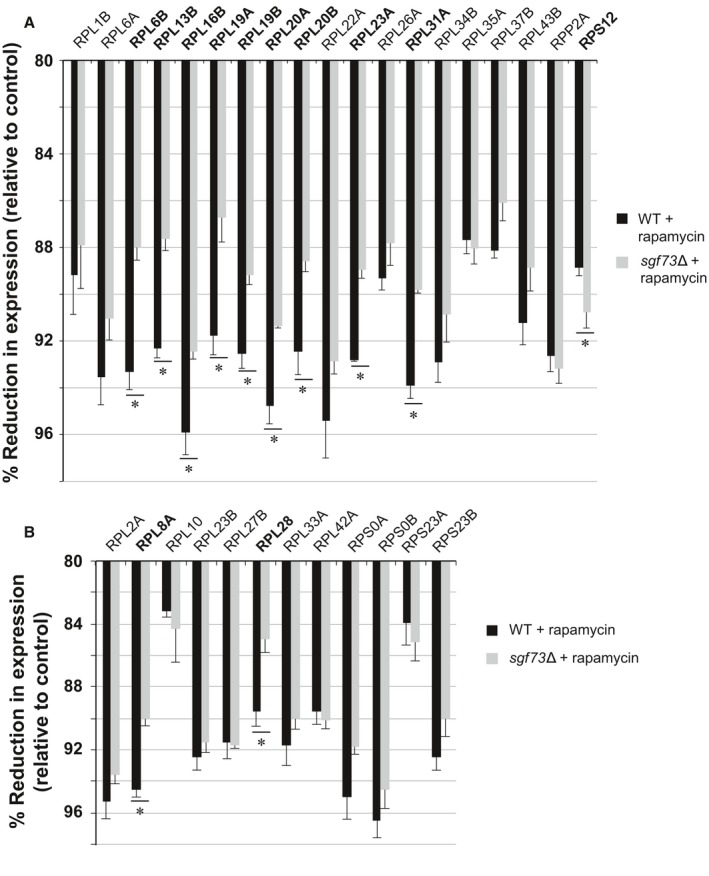
Rapamycin treatment elicits decreased transcriptional repression in sgf73Δ mutants. RNA expression analysis of yeast ribosomal protein subunit genes linked to replicative lifespan extension (A), and of yeast RP subunit genes not linked to replicative lifespan extension (B), after rapamycin treatment or exposure to ethanol (control). Results represent qRT–PCR analysis of indicated ribosomal protein subunit gene after normalization to internal control SCR1, and then after determining the ratio of expression in rapamycin‐treated yeast to expression in control‐treated yeast. The resultant ratio value was then subtracted from 100% to yield the % reduction in expression. Bold text indicates significant alterations. *P < 0.05, t‐test; n = 6 isolates/group; error bars = s.e.m.
To assess whether this transcription dysregulation is a general phenomenon or is restricted to RP‐encoding genes, we quantified the expression levels of non‐RP genes that display Sgf73 occupancy, including genes that promote RLS when deleted. Although significant differences were seen between WT and sgf73Δ strains, their expression did not follow the pattern observed for RP genes (Fig. 5A). This suggests that altered RP gene regulation under rapamycin stress conditions is specific to this set of transcripts, and is especially pronounced among RLS‐linked RP genes.
Figure 5.
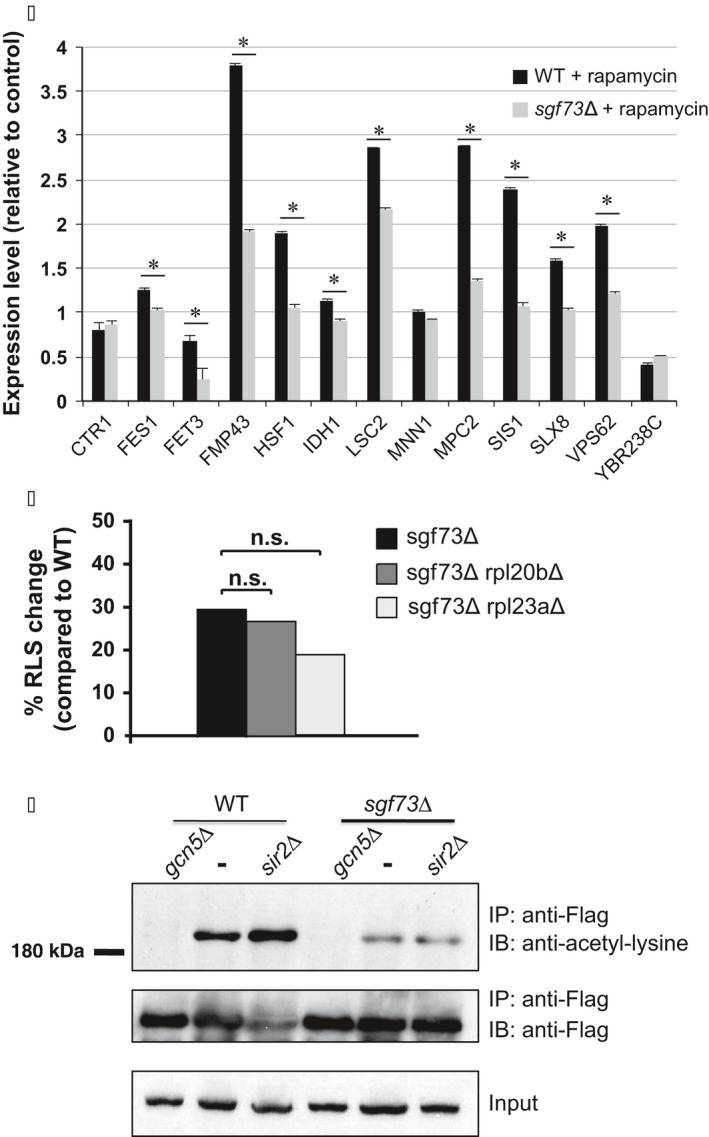
Sgf73 regulation of RLS‐linked RP genes is functionally linked to extreme lifespan extension in sgf73Δ yeast. (A) RNA expression analysis of Sgf73‐occupied yeast genes linked to replicative lifespan extension, but not encoding ribosomal protein subunits or protein translation regulators, after rapamycin treatment or exposure to ethanol (control). Results represent qRT–PCR analysis of indicated gene after normalization to the internal control SCR1, and then after determining the ratio of expression in rapamycin‐treated yeast to expression in control‐treated yeast. Unlike the response of ribosomal protein subunit genes upon rapamycin treatment, none of these genes exhibited blunted repression in sgf73Δ yeast in comparison with WT. *P < 0.05, t‐test; n = 6 isolates/group; error bars = s.e.m. (B) Double null yeast strains lacking SGF73 and either RPL20B or RPL23A were created, and individual yeast cells from each double null strain, from the sgf73Δ yeast strain, and from a control wild‐type (WT) strain were subjected to RLS analysis by counting the number of daughter cells per 80 individual yeast cells/strain. Results are shown as the % increase in RLS compared to the WT strain. For sgf73Δ vs. sgf73Δ rpl20bΔ, P = 0.865, t‐test. For sgf73Δ vs. sgf73Δ rpl23aΔ, P = 0.765, t‐test. (C) Yeast strains were transformed as indicated with Ifh1‐Flag, and then after anti‐Flag antibody immunoprecipitation (IP: anti‐Flag), immunoblots were probed with anti‐acetylated lysine (IB: anti‐acetyl‐lysine). Top: Note increased acetyl‐lysine signal for Ifh1 in sir2Δ compared to WT yeast (–), and markedly reduced acetyl‐lysine signal for Ifh1 in sgf73Δ yeast without a second deletion mutation (–) compared to WT yeast without a deletion mutation (–). Also, note that the sgf73Δ sir2Δ double‐mutant strain does not exhibit increased acetyl‐lysine signal for Ifh1 when compared to the sgf73Δ yeast strain, indicating that Sir2‐catalyzed deacetylation of Ifh1 is impaired in the sgf73Δ yeast strain. Anti‐Flag immunoblots of anti‐Flag immunoprecipitated material (middle) and of protein lysates from input‐transformed yeast strains (bottom) confirm success of the immunoprecipitation and equivalent expression of transformed Ifh1‐Flag. Yeast lacking GCN5 acetyltransferase (gcn5Δ) were used as a negative control.
Sgf73 acts on the same pathway as ribosomal subunit genes to regulate replicative lifespan
Our results indicate that Sgf73 is regulating a subset of RP genes, and loss of Sgf73 regulation of these RP genes accounts for the extreme replicative lifespan phenotype in the sgf73∆ mutant. To determine whether Sgf73 and RLS‐linked RP genes are acting on the same pathway, we created Sgf73 double null strains with either of two Sgf73‐regulated RLS‐linked RP genes (Rpl20b and Rpl23a) and then performed replicative lifespan analysis in comparison with the corresponding single‐gene null yeast strains. In both cases, we observed no further increase in replicative lifespan (Fig. 5B), indicating that Sgf73 and these RP genes are acting on the same pathway to regulate yeast lifespan.
Acetylation of Ifh1 is reduced in the sgf73∆ mutant
We considered the possibility that Sgf73 may affect RP gene expression through Ifh1, which is present at all RP promoters (Cai et al., 2013). Ifh1 is acetylated by Gcn5, which affects its binding to Fhl1, thereby modulating RP gene expression (Cherel & Thuriaux, 1995). Opposing activities of SAGA and Sir2 dictate the acetylation status of Ifh1, thereby linking metabolic state to RP gene expression. In cells lacking Gcn5 and the SAGA structural subunit Spt7, Ifh1 acetylation is greatly reduced, whereas disruption of SIR2 results in higher acetylation levels of Ifh1 (Cai et al., 2013; Downey et al., 2013). As a member of SAGA, Sgf73 may determine the acetylation levels of Ifh1 by affecting Gcn5 activity or by recruiting the SAGA complex to the regulatory regions of RP genes. Alternatively, Sgf73 may alter Ifh1 acetylation levels through its physical interaction with Sir2 (McCormick et al., 2014). To evaluate the role of Ifh1 acetylation in Sgf73 mutants, we immunoprecipitated Ifh1‐Flag from WT and sgf73∆ strains and immunoblotted with an anti‐acetyl‐lysine antibody. We observed reduced Ifh1 acetylation levels in the sgf73∆ strain (Fig. 5C), although global histone H3K9/14 acetylation levels were not altered in the sgf73∆ cells (Fig. S5), as previously reported (Lee et al., 2009). This lack of global disruption of SAGA target acetylation in the sgf73∆ strain indicates that loss of Sgf73 function specifically affects SAGA‐mediated acetylation of Ifh1, resulting in increased Ifh1 deacetylation. When SIR2 function is lost, acetylation of Ifh1 markedly increases (Fig. 5C), consistent with Sir2 deacetylation of Ifh1, as shown previously (Downey et al., 2013). To determine whether deletion of SIR2 leads to increased Ifh1 acetylation levels in the sgf73∆ mutant, we constructed the sgf73∆ sir2∆ double mutant and measured Ifh1 acetylation. Strikingly, Ifh1 acetylation levels were not higher in the sgf73∆ sir2∆ double mutant, compared to the sgf73∆ mutant alone (Fig. 5C). These results indicate that Sir2‐dependent deacetylation of Ifh1 is already impaired in the absence of Sgf73, suggesting that Sgf73 is important for Sir2 targeting, an observation consistent with our previous findings (McCormick et al., 2014). When we compared Ifh1 acetylation in the sgf73∆ sir2∆ double mutant to the sir2∆ mutant alone, we observed reduced Ifh1 acetylation, indicating that a defect in SAGA complex‐mediated acetylation of Ifh1 is present in the sgf73∆ mutant. Taken together, these results demonstrate that proper acetylation regulation of Ifh1 is lost in the sgf73∆ strain through a disruption of both normal Gcn5 acetylation and Sir2 deacetylation.
Discussion
Sgf73 is a transcriptional adaptor linking the core of the SAGA transcriptional coactivator complex to the Ubp8 DUB module. Previous studies have highlighted the importance of Sgf73 for Ubp8 DUB activity, showing that deletion of SGF73 is sufficient to completely abolish Ubp8‐regulated deubiquitination of histone H2BK123 (Kohler et al., 2008). Furthermore, we have documented an interaction between Sgf73 and the deacetylase Sir2 and demonstrated that sgf73Δ cells have enhanced Sir2‐dependent activity (McCormick et al., 2014). These findings implicate Sgf73 as a coordinator of gene regulation between the Sir2 and Ubp8 chromatin modifiers, which is key to lifespan extension in sgf73Δ cells. In this study, we sought to identify the genomic regions bound by Sgf73 and thus determine how Sgf73 may coordinate epigenetic modifications to modulate gene expression in order to gain additional insight into mechanisms of aging and the human neurodegenerative disease SCA7.
Deletion of SGF73 results in dramatic RLS extension, increasing median RLS by 65% and maximum RLS by 53% (McCormick et al., 2014). Our previous studies indicated that two major contributing factors to extended RLS in sgf73Δ mutants are increased H2BK123 ubiquitination and altered Sir2‐dependent deacetylation. In this study, we asked whether transcription dysregulation may also directly contribute to RLS extension in sgf73 mutants. To define mechanisms of Sgf73 loss‐of‐function RLS extension, we performed Sgf73 ChIP‐Seq and identified 388 unique chromatin regions of Sgf73 occupancy. Most striking was the enriched occupancy of Sgf73 at positions 5′ to ribosomal subunit genes (57 genes; P = 1.96e‐34). Among these genes, 20 are known to extend RLS upon their deletion. In total, we found Sgf73 occupancy 5′ of 33 genes known to extend RLS upon deletion, including 20 ribosomal‐related genes.
With the finding of enrichment for Sgf73 occupancy at ribosomal genes and deep connections between ribosomal gene deletion and RLS extension, we asked whether loss of Sgf73 elicited transcriptional changes in these genes, and documented reduced RP gene expression upon SGF73 deletion. We then constructed double null yeast strains lacking SGF73 and either of two Sgf73‐regulated RLS‐linked RP genes and observed no further extension in RLS in such double null strains. Thus, Sgf73 appears to have a previously unrecognized role in ribosome biogenesis, and impaired ribosome biogenesis, resulting in reduced protein translation, is likely a major contributor to the extreme RLS extension observed in the sgf73Δ mutant. As reduced protein translation has been shown to promote lifespan extension in mice (Selman et al., 2009), our findings reinforce the importance of protein production balance as a deeply conserved factor in determining organismal lifespan.
A role for Sgf73 in ribosomal biogenesis is perhaps not unexpected, as SAGA is a stress‐induced transcriptional coactivator complex (Huisinga & Pugh, 2004), and previous studies have identified SAGA binding at RP genes (Cai et al., 2011), as well as binding of Gcn5 at RP genes (Venters et al., 2011). Gcn5 acetylates the RP gene transcription coactivator Ifh1 to reduce RP gene expression, and Ifh1 is also subject to Sir2 deacetylation (Cai et al., 2013; Downey et al., 2013). Indeed, Sgf73 appears to play a role in ribosomal biogenesis under treatment with rapamycin, a TOR inhibitor. Upon treatment with rapamycin, RP gene expression is reduced in WT cells through altered TOR regulation of Ifh1. Although we also observed reduced RP gene expression in sgf73Δ cells, this was not to the same extent as in WT cells, identifying Sgf73 as a key target of Ifh1 regulation. Our previous studies identified Sir2 as a key player in sgf73Δ RLS extension and documented a physical interaction between Sgf73 and Sir2 (McCormick et al., 2014). As ribosome protein biogenesis is highly dependent on Ifh1, with its recruitment and acetylation state affecting the degree to which RP genes are expressed (Downey et al., 2013), we considered a role for altered Ifh1 acetylation in the transcriptional dysregulation of RP‐encoding genes in the sgf73Δ mutant. Both SAGA and Sir2 influence acetylation of Ifh1, with Gcn5 promoting acetylation and Sir2 mediating deacetylation (Downey et al., 2013). Current models of RP gene expression regulation posit that a cycle of rapid acetylation and deacetylation of Ifh1 permits a burst of RP gene expression when nutrients become plentiful. According to this model, Sir2‐mediated Ifh1 deacetylation resets the system so that increased RP gene transcription can occur, and this increased transcription is then restrained upon Ifh1 acetylation (Downey et al., 2013). When we examined the acetylation levels of Ifh1 in the sgf73Δ mutant, we observed reduced Ifh1 acetylation. In the sgf73Δ sir2Δ double‐mutant strain, we found that Ifh1 acetylation levels were comparable to the sgf73Δ mutant, but were reduced compared to the sir2Δ mutant strain, indicating that both SAGA acetylation and Sir2 deacetylation of Ifh1 are impaired in the sgf73Δ strain. Thus, in sgf73Δ mutants, dynamic regulation of Ifh1 acetylation is lost, resulting in reduced RP gene expression at baseline and a failure to properly repress RP gene expression under stress. These findings support a model whereby Sgf73 is necessary for Sir2 deacetylation of Ifh1, and Sgf73 is also required for optimal Gcn5 acetylation of Ifh1. Reduced ribosomal biogenesis during progressive aging in sgf73Δ mutants is likely a factor in the extended RLS, as reduction in 60S ribosome subunits extends RLS. Taken together, these results highlight altered acetylation dynamics of target proteins as an upstream event in transcriptional dysregulation of ribosome biogenesis, which in the case of yeast aging appears to be beneficial. Whether ataxin‐7, the mammalian orthologue of Sgf73, is similarly important for acetylation‐mediated regulation of transcription and how polyglutamine expansion of ataxin‐7 in the human neurological disorder SCA7 may alter this acetylation regulation to promote neurodegeneration are important questions for future studies.
Materials and methods
Yeast strain construction and dilution assay
Tagging was performed as described, using the pFA6a‐13‐Myc‐kanMX6 plasmid and C‐terminal tagging primers (Longtine et al., 1998). In brief, the 13‐Myc‐KanMX6 sequence was amplified using primers with overhangs corresponding to 50 bp 5′ to the SGF73 or UBP8 stop codon, as well as 50 bp 3′ to the stop codon. Competent WT cells were prepared and transformed with the amplified product to integrate the tag by homologous recombination. Positive clones were selected on G418. Molecular genotyping was used to confirm the tags’ presence; positive strains were backcrossed to WT cells. Cells were grown overnight in 3 mL of YPD, then 1 A600 equivalent of cells was spun down, spent media were removed, and cells were resuspended in 1 mL of sterile water. 1:5 serial dilutions were then pinned onto control, selective, and/or drug plates.
Chromatin immunoprecipitation
A 5 mL of YPD culture from a single yeast colony grown overnight at 30 °C was diluted in 100 mL of YPD for harvest at ~A600 L mL−1. Cells were fixed with 0.86% formaldehyde at room temperature (RT) for 25 min. The reaction was quenched with 125 mm glycine and 0.2% NH4OH for 5 min at RT. Cells were collected by centrifugation for 15 min. The pellet was washed three times with 20 mL of cold phosphate‐buffered saline (PBS). Washed cells were resuspended in the remaining wash buffer, moved to a 2‐mL tube, spun, and additional liquid removed. Cells were lysed in 1.5 mL of FA‐lysis buffer (50 mm HEPES‐KOH pH 7.5, 140 mm NaCl, 1 mm EDTA pH 8.0 0.1% sodium deoxycholate, 1% Triton X‐100, and 1x Roche complete proteinase inhibitors) with additional detergents (0.5% NP‐40 and 0.1% SDS) and divided between three 1.5‐mL tubes. Acid‐washed glass beads were then added to the meniscus, and cells were lysed by bead beating at 4 °C for 1 h. Lysate was transferred to a new 1.5‐mL tube. Chromatin was then sheared in a precooled horn sonicator (Sonic Dismembrator FB‐505, Fisher Scientific, Waltham, Massachusetts, USA), using pulse cycles of 20 s on and 30 s off at 80% amp for a total time of 30 min to achieve fragments of 250–400 bp. After sonication, tubes were spun at 4 °C for 5 min and the supernatant was transferred to low‐bind tubes (Lifetech, Carlsbad, California, USA AM12450). An equal volume of FA‐lysis buffer was added, and lysate was precleared with 10 μL of protein A:G magnetic beads for 30 min at 4 °C. A sample from each preparation was taken after preclearing to analyze input chromatin and protein. 100 μL of anti‐c‐Myc EZview resin (Sigma, St. Louis, Missouri, USA E6654‐1ML) was added to the samples and rotated overnight at 4 °C. Postbinding beads were washed twice with 1 mL FA‐lysis buffer, twice with 1 mL of wash buffer (10 mm Tris pH 8, 0.25M LiCl, 0.5% NP‐40, 0.5% sodium deoxycholate, 1 mm EDTA), and once with 1 mL of Tris‐EDTA (TE). Samples were de‐cross‐linked with 100 μL of TE + 1% SDS (with 10 μL taken for protein analysis) and incubated at 65 °C for 6 h. Following cross‐link reversal, 8 μL of TE and 2 μL of RNase were added and incubated at 37 °C for 1 h. 1.5 μL of proteinase K (20 mg mL−1) was added followed by overnight incubation at 37 °C. DNA was purified using the MinElute PCR Kit (Qiagen, Hilden, Germany 28006) and eluted with 13 μL of nuclease‐free water. Fragment size was analyzed on 6% acrylamide gels.
ChIP‐Seq analysis
Libraries for sequencing were generated using the ChIP'd fragments and the Ovation ultralow DR multiplex 1–8 library system (NuGEN—0330). Library amplification was performed using 18 cycles of 94 °C—30 s, 60 °C—30 s, 72 °C—1 min, with a 2‐min 72 °C predenaturation step and 5‐min 72 °C extension step added. After library construction, fragments were size‐selected. Libraries were separated on 6% TBE‐acrylamide using Invitrogen's 25‐bp ladder, and DNA in the 150‐ to 400‐bp range was excised. DNA was recovered in 350 μL of elution buffer (300 mm NaCl, 10 mm Tris pH 7.6). The mixture was transferred to a SpinX tube (Sigma CLS1862) to remove gel from the DNA in suspension. DNA was precipitated in 1 mL cold 100% ethanol, 35 μL of sodium acetate (3M pH 5.2), and 2 μL glycogen (5 mg mL−1). Libraries were resuspended in 15 μL of Qiagen EB, and 1 μL was run on a 2100 Bioanalyzer DNA 100 chip (Agilent, Santa Clara, California, USA 5067‐1504). Sequencing was performed at Beijing Genomics Institute on the Illumina Hi‐seq 2000 platform. Single‐end 50‐bp read sequencing of the pooled libraries. Postsequencing, all reads were sorted by barcode and low‐quality sequence reads were removed. All ChIP‐Seq data are deposited and publicly available via Gene Expression Omnibus accession #GSE76461.
Bioinformatics analysis and validation
To analyze the sequencing reads, we employed homer (Hypergeometric Optimization of Motif EnRichment) v4.2 http://biowhat.ucsd.edu/homer/. Raw sequence reads were evaluated with fastqc (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) for quality control. Sequence duplication was compensated for using only uniquely mapping reads. The S. cerevisiae S288c genome assembly from the Saccharomyces Genome Database (GCA_000146055.2) was used for all alignments and annotations. Oligonucleotide sequences for evaluation of ChIP and validation of occupancy will be furnished upon request.
Whole‐cell lysate preparation and anti‐Flag immunoprecipitations
Protocol was adapted from Downey et al., 2013. Between 90 and 100 A600 equivalents of cells were collected by centrifugation, washed with phosphate‐buffered saline, and stored at −80 °C until lysis. Pellets were resuspended in 500 μL cold lysis buffer (0.5% Triton, 200 mm NaCl, 50 mm Tris–HCl pH 7.5, and 1 mm EDTA), supplemented with 10 mm sodium butyrate, 10 mm nicotinamide, 5 mm sodium fluoride, 1 mm dithiothreitol, and a cocktail of protease inhibitors including phenylmethylsulfonyl fluoride, leupeptin, benzamidine HCl, pepstatin, and tosyl phenylalanyl chloromethyl ketone (lysis buffer supplemented or LBS). Glass lysis beads were added to the meniscus in 50‐mL conical tubes and lysed at 4 °C. Six 1‐min durations of vortexing were carried out with 1 min on ice in between. Lysates were recovered by pipette, and the beads were washed with LBS. This wash was combined with the lysate. Extracts were clarified by centrifugation at 4 °C. Anti‐Flag M2 beads (Sigma‐Aldrich, St. Louis, Missouri, USA) were added to the lysate and incubated for 2–3 h on cold room nutator. The beads were washed three times with LBS. Following washes, remaining liquid was aspirated with an 18‐gauge needle. Immunoprecipitated proteins were eluted with 100 μL 2xUSB (8 M urea, 4% SDS, 10% ß‐mercaptoethanol, 125 mm Tris, pH 6.8) at 65 °C for 10 min. Prior to loading onto an 8% SDS‐PAGE gel, eluates were boiled for 5 min. Typically 50% of immunoprecipitated material was analyzed for each anti‐acetyl‐lysine (Chemicon, Temecula, California, USA) immunoblot.
Conflict of interest
None declared.
Funding
This study was supported by funds from the NIH: R01 EY014061 and R01 AG033082 (A.R.L.S.), R01 GM090177 (L.P.), T32 GM008666 (A.G.M.), T32 AG000266 (M.A.M.), and F32 GM089101 (R.M.G.).
Supporting information
Fig. S1 Validation of C‐terminal Myc‐tagged Sgf73 constructs and Myc ChIP.
Fig. S2 Confirmation of Sgf73‐Myc function and Sgf73/Ubp8 ChIP for expected occupancy sites.
Fig. S3 Gene ontology analysis of Sgf73 ChIP‐Seq data and comparison of Sgf73‐occupied genes with replicative lifespan promoting genes.
Fig. S4 Ubp8 shares occupancy with Sgf73 RLS‐linked RP peaks.
Fig. S5 Overall acetylation activity is not diminished in sgf73Δ yeast.
Table S1 Sgf73 occupancy peaks from ChIP‐Seq analysis
Table S2 Sgf73 and Ubp8 shared occupancy peaks from ChIP‐Seq analysis
Acknowledgments
We thank members of the La Spada, Kennedy, and Pillus laboratories for technical assistance and useful discussion. We also thank Benjamin P. Tu for the Ifh1 yeast strains, and Manuel Ares for his advice on the rapamycin experiments.
Contributor Information
Lorraine Pillus, Email: lpillus@ucsd.edu.
Albert R. La Spada, Email: alaspada@ucsd.edu
References
- Blander G, Guarente L (2004) The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435. [DOI] [PubMed] [Google Scholar]
- Bonnet J, Wang YH, Spedale G, Atkinson RA, Romier C, Hamiche A, Pijnappel WW, Timmers HT, Tora L, Devys D, Kieffer B (2010) The structural plasticity of SCA7 domains defines their differential nucleosome‐binding properties. EMBO Rep. 11, 612–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J, Wang CY, Baptista T, Vincent SD, Hsiao WC, Stierle M, Kao CF, Tora L, Devys D (2014) The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 28, 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP (2011) Acetyl‐CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42, 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, McCormick MA, Kennedy BK, Tu BP (2013) Integration of multiple nutrient cues and regulation of lifespan by ribosomal transcription factor Ifh1. Cell Rep. 4, 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Pan KZ, Palter JE, Kapahi P (2007) Longevity determined by developmental arrest genes in Caenorhabditis elegans . Aging Cell 6, 525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherel I, Thuriaux P (1995) The IFH1 gene product interacts with a fork head protein in Saccharomyces cerevisiae . Yeast 11, 261–270. [DOI] [PubMed] [Google Scholar]
- Chua KF, Mostoslavsky R, Lombard DB, Pang WW, Saito S, Franco S, Kaushal D, Cheng HL, Fischer MR, Stokes N, Murphy MM, Appella E, Alt FW (2005) Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 2, 67–76. [DOI] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G (2007) Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 3, e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, Abbas N, Stevanin G, Durr A, Yvert G, Cancel G, Weber C, Imbert G, Saudou F, Antoniou E, Drabkin H, Gemmill R, Giunti P, Benomar A, Wood N, Ruberg M, Agid Y, Mandel JL, Brice A (1997) Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat. Genet. 17, 65–70. [DOI] [PubMed] [Google Scholar]
- Denoth‐Lippuner A, Krzyzanowski MK, Stober C, Barral Y (2014) Role of SAGA in the asymmetric segregation of DNA circles during yeast ageing. Elife 3, e03790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M, Knight B, Vashisht AA, Seller CA, Wohlschlegel JA, Shore D, Toczyski DP (2013) Gcn5 and sirtuins regulate acetylation of the ribosomal protein transcription factor ifh1. Curr. Biol. 23, 1638–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C (2007) Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans . Aging Cell 6, 95–110. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage‐determining transcription factors prime cis‐regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Hardy S, Sasorith S, Klein F, Robert F, Weber C, Miguet L, Potier N, Van‐Dorsselaer A, Wurtz JM, Mandel JL, Tora L, Devys D (2004) Ataxin‐7 is a subunit of GCN5 histone acetyltransferase‐containing complexes. Hum. Mol. Genet. 13, 1257–1265. [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF (2004) A genome‐wide housekeeping role for TFIID and a highly regulated stress‐related role for SAGA in Saccharomyces cerevisiae . Mol. Cell 13, 573–585. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK (2005) Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310, 1193–1196. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M (2012) KEGG for integration and interpretation of large‐scale molecular data sets. Nucleic Acids Res. 40, D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E (2008) Yeast Ataxin‐7 links histone deubiquitination with gene gating and mRNA export. Nat. Cell Biol. 10, 707–715. [DOI] [PubMed] [Google Scholar]
- Koutelou E, Hirsch CL, Dent SY (2010) Multiple faces of the SAGA complex. Curr. Opin. Cell Biol. 22, 374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Oppikofer M, Gasser SM (2013) SIR proteins and the assembly of silent chromatin in budding yeast. Annu. Rev. Genet. 47, 275–306. [DOI] [PubMed] [Google Scholar]
- Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T (2002) Human SIR2 deacetylates p53 and antagonizes PML/p53‐induced cellular senescence. EMBO J. 21, 2383–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebre AS, Brice A (2003) Spinocerebellar ataxia 7 (SCA7). Cytogenet. Genome Res. 100, 154–163. [DOI] [PubMed] [Google Scholar]
- Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL (2009) Yeast Sgf73/Ataxin‐7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenetics Chromatin 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR‐based gene deletion and modification in Saccharomyces cerevisiae . Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Martin DE, Soulard A, Hall MN (2004) TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119, 969–979. [DOI] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG (2001) Human STAGA complex is a chromatin‐acetylating transcription coactivator that interacts with pre‐mRNA splicing and DNA damage‐binding factors in vivo . Mol. Cell. Biol. 21, 6782–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick MA, Mason AG, Guyenet SJ, Dang W, Garza RM, Ting MK, Moller RM, Berger SL, Kaeberlein M, Pillus L, La Spada AR, Kennedy BK (2014) The SAGA histone deubiquitinase module controls yeast replicative lifespan via Sir2 interaction. Cell Rep. 8, 477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick MA, Delaney JR, Tsuchiya M, Tsuchiyama S, Shemorry A, Sim S, Chou AC, Ahmed U, Carr D, Murakami CJ, Schleit J, Sutphin GL, Wasko BM, Bennett CF, Wang AM, Olsen B, Beyer RP, Bammler TK, Prunkard D, Johnson SC, Pennypacker JK, An E, Anies A, Castanza AS, Choi E, Dang N, Enerio S, Fletcher M, Fox L, Goswami S, Higgins SA, Holmberg MA, Hu D, Hui J, Jelic M, Jeong KS, Johnston E, Kerr EO, Kim J, Kim D, Kirkland K, Klum S, Kotireddy S, Liao E, Lim M, Lin MS, Lo WC, Lockshon D, Miller HA, Moller RM, Muller B, Oakes J, Pak DN, Peng ZJ, Pham KM, Pollard TG, Pradeep P, Pruett D, Rai D, Robison B, Rodriguez AA, Ros B, Sage M, Singh MK, Smith ED, Snead K, Solanky A, Spector BL, Steffen KK, Tchao BN, Ting MK, Vander Wende H, Wang D, Welton KL, Westman EA, Brem RB, Liu XG, Suh Y, Zhou Z, Kaeberlein M, Kennedy BK (2015) A comprehensive analysis of replicative lifespan in 4,698 single‐gene deletion strains uncovers conserved mechanisms of aging. Cell Metab. 22, 895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MT, Haj‐Yahya M, Ringel AE, Bandi P, Brik A, Wolberger C (2016) Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 351, 725–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schawalder SB, Kabani M, Howald I, Choudhury U, Werner M, Shore D (2004) Growth‐regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432, 1058–1061. [DOI] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al‐Qassab H, Carmignac D, Ramadani F, Woods A, Robinson IC, Schuster E, Batterham RL, Kozma SC, Thomas G, Carling D, Okkenhaug K, Thornton JM, Partridge L, Gems D, Withers DJ (2009) Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326, 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M (2008) Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133, 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanin G, Durr A, Brice A (2000) Clinical and molecular advances in autosomal dominant cerebellar ataxias: from genotype to phenotype and physiopathology. Eur. J. Hum. Genet. 8, 4–18. [DOI] [PubMed] [Google Scholar]
- Timmers HT, Tora L (2005) SAGA unveiled. Trends Biochem. Sci. 30, 7–10. [DOI] [PubMed] [Google Scholar]
- Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck‐Walsh C, Pugh BF (2011) A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell 41, 480–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, Zhang B (2013) WEB‐based gene set analysis toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41, W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440. [DOI] [PubMed] [Google Scholar]
- Xiao L, Grove A (2009) Coordination of ribosomal protein and ribosomal RNA gene expression in response to TOR signaling. Curr. Genomics 10, 198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J (2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schule R, Takeyama K, Kato S, Tora L, Devys D (2008) A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell 29, 92–101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Validation of C‐terminal Myc‐tagged Sgf73 constructs and Myc ChIP.
Fig. S2 Confirmation of Sgf73‐Myc function and Sgf73/Ubp8 ChIP for expected occupancy sites.
Fig. S3 Gene ontology analysis of Sgf73 ChIP‐Seq data and comparison of Sgf73‐occupied genes with replicative lifespan promoting genes.
Fig. S4 Ubp8 shares occupancy with Sgf73 RLS‐linked RP peaks.
Fig. S5 Overall acetylation activity is not diminished in sgf73Δ yeast.
Table S1 Sgf73 occupancy peaks from ChIP‐Seq analysis
Table S2 Sgf73 and Ubp8 shared occupancy peaks from ChIP‐Seq analysis


