Summary
The microbiota plays a central role in human health and disease by shaping immune development, immune responses and metabolism, and by protecting from invading pathogens. Technical advances that allow comprehensive characterization of microbial communities by genetic sequencing have sparked the hunt for disease‐modulating bacteria. Emerging studies in humans have linked the increased abundance of Prevotella species at mucosal sites to localized and systemic disease, including periodontitis, bacterial vaginosis, rheumatoid arthritis, metabolic disorders and low‐grade systemic inflammation. Intriguingly, Prevotella abundance is reduced within the lung microbiota of patients with asthma and chronic obstructive pulmonary disease. Increased Prevotella abundance is associated with augmented T helper type 17 (Th17) ‐mediated mucosal inflammation, which is in line with the marked capacity of Prevotella in driving Th17 immune responses in vitro. Studies indicate that Prevotella predominantly activate Toll‐like receptor 2, leading to production of Th17‐polarizing cytokines by antigen‐presenting cells, including interleukin‐23 (IL‐23) and IL‐1. Furthermore, Prevotella stimulate epithelial cells to produce IL‐8, IL‐6 and CCL20, which can promote mucosal Th17 immune responses and neutrophil recruitment. Prevotella‐mediated mucosal inflammation leads to systemic dissemination of inflammatory mediators, bacteria and bacterial products, which in turn may affect systemic disease outcomes. Studies in mice support a causal role of Prevotella as colonization experiments promote clinical and inflammatory features of human disease. When compared with strict commensal bacteria, Prevotella exhibit increased inflammatory properties, as demonstrated by augmented release of inflammatory mediators from immune cells and various stromal cells. These findings indicate that some Prevotella strains may be clinically important pathobionts that can participate in human disease by promoting chronic inflammation.
Keywords: cytokines, inflammation, inflammatory disease, mucosa, Prevotella, T cells
Abbreviations
- APC
antigen‐presenting cell
- BV
bacterial vaginosis
- CCL
C‐C motif chemokine
- CCR
chemokine receptor
- CII
type II collagen
- COPD
chronic obstructive pulmonary disease
- CRA
chronic RA
- IFN
Interferon
- IL
interleukin
- LPS
lipopolysaccharide
- MIP‐1α
macrophage inflammatory protein‐1α
- NAFLD
non‐alcoholic fatty liver disease
- NASH
non‐alcoholic steatohepatitis
- NORA
new‐onset RA
- RA
rheumatoid arthritis
- Th
helper T cell
- TLR
Toll‐like receptor
- TNF
tumour necrosis factor
- TSLP
thymic stromal lymphopoietin
Introduction
Studies in germ‐free mice and the use of microbial reconstitution have underlined the importance of the commensal microbiota in shaping immune development and function, and hence the risk of inflammatory disease.1 The advances in next‐generation‐sequencing have allowed in‐depth characterization of non‐culturable bacterial communities.2 This has sparked significant interest in deciphering the health‐ and disease‐related microbiotas to identify bacteria and mechanisms that could play a part in disease aetiology and progression. Characterization of the healthy human microbiota has revealed distinct bacterial communities at different body sites (gastrointestinal, urogenital, skin, lung, oral and nasal) supporting the notion of microbial communities adapting to different ecological environments in the body.3 Interestingly, bacterial Prevotella species have been found to be prevalent commensal colonizers at mucosal sites; being the predominant genus in the respiratory system4, 5 and a central constituent in one of three gut bacterial enterotypes,6 as well as present in saliva and several oral sites.3
Prevotella species are anaerobic Gram‐negative bacteria of the Bacteroidetes phylum, which also includes the clinically important genera Bacteroides and Porphyromonas.7 Prevotella strains are classically considered commensal bacteria due to their extensive presence in the healthy human body and their rare involvement in infections. Only a few strains have been reported to give rise to opportunistic endogenous infections, including chronic infections, abscesses and anaerobic pneumonia.8, 9, 10 In light of the abundant Prevotella colonization and low pathogenicity it is likely that humans have co‐evolved with Prevotella, giving rise to a mutualistic relationship. However, emerging studies have linked increased Prevotella abundance and specific strains to inflammatory disorders, suggesting that at least some strains exhibit pathobiontic properties. The present review addresses the interaction between Prevotella and the immune system, and how Prevotella may promote inflammatory disease. Many studies have addressed the link between bacteria and inflammatory diseases using various methods (e.g. culture techniques and quantitative PCR for specific stains, genera, or phyla); however, this review will mainly focus on recent studies employing genomics‐based culture‐independent methods of in‐depth microbiota characterization (16S rRNA or metagenomic sequencing).
Periodontitis
The first link between Prevotella and chronic inflammatory disease was indicated as early as 1928 with the observation of black‐pigmented Gram‐negative anaerobes in periodontal disease.11 Indeed, later studies confirmed the presence of Prevotella in biofilms of gingivitis and periodontitis.12 It is well established that bacteria are a central driver of these diseases characterized by neutrophil recruitment, pro‐inflammatory cytokines and metalloproteinase expression mediating destruction of connective tissues and alveolar bone.13 Most mechanistic research has focused on the role of Porphyromonas gingivalis (a Gram‐negative anaerobic member of the Bacteroidetes phyla, similarly to Prevotella), because this species was thought to be the main driver of disease. However, metagenomic studies have revealed a more diverse dysbiotic bacterial community that collectively may shape disease progression.14 A recent study in mice has shown that Prevotella nigrescens, similarly to P. gingivalis, can drive periodontal disease – as demonstrated by maxillary alveolar bone loss following oral inoculation.15 It was found that infection promoted immune responses characterized by increased T helper type 17 (Th17) [i.e. interleukin (IL‐17)], suppressed Th2 (IL‐4, IL‐5 and IL‐9), and similar Th1 [interferon‐γ (IFN‐γ)] cytokine production by lymph node T cells compared with uninfected mice. Prevotella nigrescens was found to drive Th17 responses in vitro through the production of IL‐1 by bone‐marrow‐derived dendritic cells in a Toll‐like receptor 2 (TLR2) ‐dependent manner (Fig. 1). The role of Prevotella in driving Th17‐mediated immune responses in periodontitis is supported by studies linking IL‐1α and IL‐1β levels in crevicular fluid to Prevotella colonization.16
Figure 1.
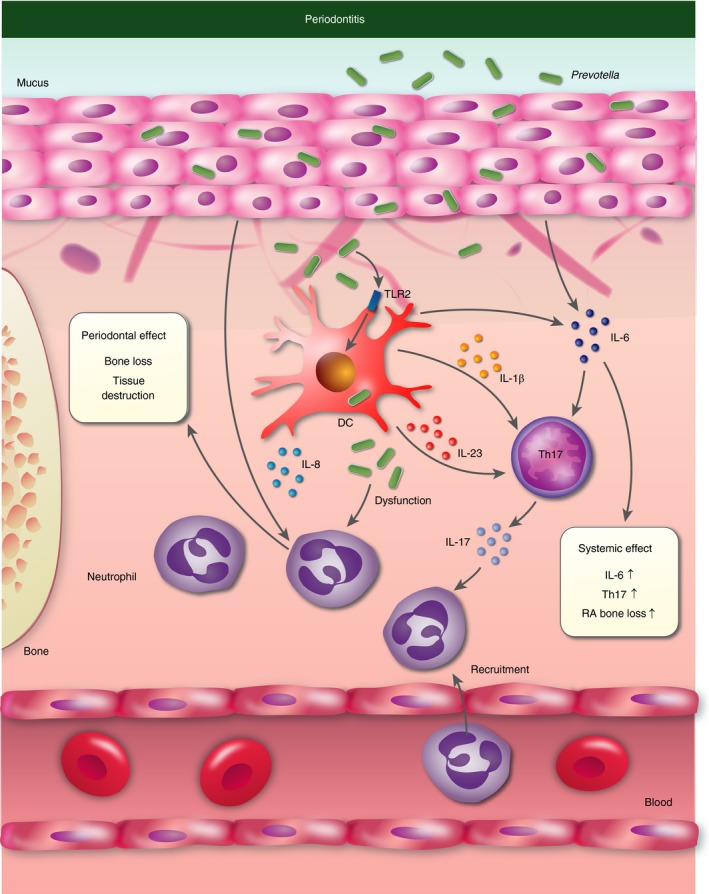
Prevotella‐mediated inflammation in periodontitis. Prevotella stimulate the release of interleukin‐1β (IL‐1β), IL‐6 and IL‐23 by dendritic cells (DC) through Toll‐like receptor 2 (TLR2), which in turn mediates IL‐17 production by T helper 17 (Th17) cells that activate neutrophils. Epithelial cells contribute to neutrophil recruitment and Th17 cell activation through the production of IL‐8 and IL‐6, respectively. Prevotella directly induce dysfunction in recruited neutrophils. Chronic inflammation, characterized Th17 immune responses and recruitment of neutrophils, leads to localized bone loss and tissue destruction characteristic of periodontitis. Local inflammation disseminates and affects systemic disease, including bone loss in rheumatoid arthritis (RA).
Only a few studies have compared the immunological properties of Prevotella to innocuous oral commensal bacteria, like Streptococcus and Lactobacillus species.17 In vitro studies using human monocyte‐derived dendritic cells,18 odontoblast‐like cell clones19 and a gingival epithelial cell line20 suggest that Prevotella exhibit an enhanced capacity to induce inflammatory mediators [IL‐6, IL‐8 and tumour necrosis factor‐α (TNF‐α)] when compared with strict commensal oral bacteria and even P. gingivalis. Interestingly, a murine study using a subcutaneous chamber model found that oral commensal Streptococcus mitis infection could readily be cleared, whereas infection with Prevotella intermedia was uncontrolled for more than 7 days.21 The Prevotella intermedia infection was found to induce increased host cell infiltration compared with S. mitis; however the infiltrating neutrophils were defective in terms of phagocytosis and reactive oxygen species production, and exhibited a necrotic morphology. Interestingly, neutrophil dysfunction is a prominent feature of periodontal disease.22 Combined, the studies suggest that Prevotella can promote periodontitis by driving neutrophil recruitment via Th17 immune responses. Chronic activation of the Th17 pathway may mediate tissue and bone destruction because recruited neutrophils are unable to clear the bacteria and promote resolution of tissue inflammation.
Rheumatoid arthritis
Epidemiological studies have linked periodontal disease to increased risk of systemic diseases, including rheumatoid arthritis (RA).14 It has been speculated that periodontal pathogens drive systemic inflammation or disseminate to affected tissue, thereby promoting localized inflammation. Indeed, increased specific IgG to periodontal pathogens, including Prevotella intermedia and P. gingivalis, has been reported in RA.23, 24, 25, 26, 27 Furthermore, the presence of DNA from periodontal pathogens has been found in serum and synovial fluid of patients with RA, indicating systemic dissemination of bacteria that directly promote localized synovial inflammation.28, 29, 30 However, the effect‐size and strength of association vary among bacterial species in these studies, suggesting a more complicated relationship that may depend on individual study design and clinical characteristics of the patient cohorts. Interestingly, a study employing 16S rRNA sequencing of the subgingival microbiota found that the presence of P. gingivalis correlated with periodontal disease but not RA, whereas Prevotella was associated with new‐onset RA (NORA) but not chronic RA (CRA) independent of periodontal disease.31 In mice, Prevotella nigrescens was found to induce periodontitis after oral inoculation.15 The infected mice exhibited accelerated onset and severity of experimental arthritis compared with control mice when immunized with type II collagen (CII). Importantly, systemic administration of 100‐fold heat‐killed bacteria was unable to emulate this effect, suggesting that establishment of chronic oral infection and periodontitis by Prevotella is needed to promote RA. It was found that mice with Prevotella‐mediated periodontitis promoted IL‐17 but not IFN‐γ production by CII‐specific T cells. The enhanced IL‐17 production by CII‐specific T cells was found to correlate with arthritic bone erosion, indicating a central role of Th17 responses in promoting RA pathology. Collectively, the present studies suggest that Prevotella‐mediated periodontitis can affect the progression of RA by modulating systemic immune responses; however, the relevance of oral Prevotella to human disease and specific patient groups requires further research.
Dysbiosis in the gut has been linked to RA, and suggested to be a risk factor responsible for the rise in disease incidence. Two recent studies performing 16S rRNA sequencing of faecal samples found dysbiosis associated with Prevotella strains closely related to Prevotella copri in patients with NORA.32, 33 Faecal matter from patients with RA and healthy controls was used to colonize the gut of arthritis‐prone SKG mice.33 The microbiota from patients with RA was found to induce increased numbers of intestinal IL‐17+ Th17 cells, but similar numbers of IFN‐γ + Th1 and FoxP3+ regulatory T cells compared with healthy microbiota. After triggering disease by zymosan administration, SKG mice colonized by the RA microbiota developed severe arthritis characterized by increased disease and histology scores, and increased serum rheumatoid factor levels. It was demonstrated that the severe arthritis was associated with increased IL‐17 (Th17) and unchanged IFN‐γ (Th1) production in response to the arthritis‐related autoantigen RPL23A. Similarly, oral administration of Prevotella melanogenica in humanized HLA‐DQ8 mice immunized with CII augmented RA onset and severity, which was associated with increased gut inflammation as demonstrated by shortening of intestinal villi and leucocyte infiltration.34 Interestingly, the same study reported suppression of experimental RA by Prevotella histicola through the expansion of regulatory T cells in the gut and suppression of systemic CII‐specific immune responses. These findings suggest that specific members of the Prevotella genus have different disease modulating properties, and highlight the importance of in‐depth characterization of the microbiota in the study of inflammatory disease.
In vitro studies comparing Prevotella copri to the gut commensal bacteria Bacteroides fragilis, Bifidobacterium bifidum, Lactobacillus acidophilus and Escherichia coli using bone‐marrow‐derived dendritic cells found that Prevotella copri was superior in inducing the Th17 driving cytokines IL‐6 and IL‐23.33 Hence, Prevotella copri‐stimulated bone‐marrow‐derived dendritic cells were able to prime naive Th cells to produce up to fivefold increased IL‐17 levels compared with the commensal bacteria. These findings suggest that Prevotella copri exhibits intrinsic Th17 promoting capability, which when present in the gut microbiota can promote RA.
A recent study has shed light on the possible immune modulatory role of Prevotella copri in human RA. Of RA patients (including NORA and CRA), 32% were found to have serum IgA or IgG antibodies specific for Prevotella copri, which was almost absent in healthy controls and patients with other arthritic diseases.35 In comparison, IgA or IgG antibodies specific for P. gingivalis were present at similar frequencies and levels in all patient groups and healthy controls, whereas antibodies to the gut commensals Bacteroides fragilis and Escherichia coli were largely absent. These findings indicate that the immune responses to Prevotella copri exclusively develop in RA patients and may contribute to disease initiation or progression in some patients. Interestingly, Prevotella copri‐specific IgA but not IgG levels were associated with systemic levels of innate [macrophage inflammatory protein 1α (MIP‐1α) and MIP‐1β], Th1 (IFN‐γ and IL‐12), and Th17 (IL‐23, IL‐22, IL‐17A, IL‐17E and IL‐17F) cytokines.35 Patients with RA had either IgA, IgG or no specific antibodies, indicating that different immune responses to Prevotella copri can develop within the individual patient, which in turn may have implications for disease risk and outcomes.
Bacterial vaginosis
Bacterial vaginosis (BV) is associated with poor health outcomes for women, including pre‐term birth, and increased risk of acquiring HIV or other infections. The disease is characterized by the loss of a commensal Lactobacillus‐rich microbiota and the blooming of anaerobic bacteria in the vaginal tract. Although earlier reports have implicated Prevotella bivia in BV,36 a couple of very recent studies found that Prevotella abundance increased with severity of BV, which inversely correlated with the presence of Lactobacillus.37, 38, 39 Prevotella in the vaginal microbiota was associated with increased innate (IL‐1α, IL‐1β, IL‐8, and TNF‐α) cytokines, and production of Th17 (IL‐23 and IL‐17) and Th1 (IL‐12p70 and IFN‐γ) related cytokines in cervicovaginal fluid.37, 38 These findings are in line with a previous study linking increased IL‐1β and IL‐8 levels in cervicovaginal fluid to Prevotella bivia colonization.40 Increased numbers of activated CCR5+ HLA‐DR+ CD38+ Th cells were associated with the presence of Prevotella in the vaginal mucosa.38 No apparent change in mucosal antigen‐presenting cell (APC) numbers (CD11c+ dendritic cells or CD14+ monocytes/macrophages) was observed in the same study; however, APCs from Lactobacillus‐rich compared with Prevotella‐rich vaginal mucosas exhibited distinct transcriptional profiles.37 Prevotella‐rich mucosa APCs showed a profile similar to APCs activated by lipopolysaccharide (LPS), and expressed cytokine genes known to promote Th17 immune responses (IL23A, IL6, IL1A and IL1B). These findings indicate that Prevotella in the vaginal tract contributes to activation of a Th17 immune response via APCs, leading to the recruitment and activation of Th cells in the inflamed vaginal mucosa (Fig. 2). Important in relation to women's health, the recruitment of CCR5+ Th cells into the vaginal mucosa may be a central underlying risk factor for increased HIV transmission in BV.38
Figure 2.
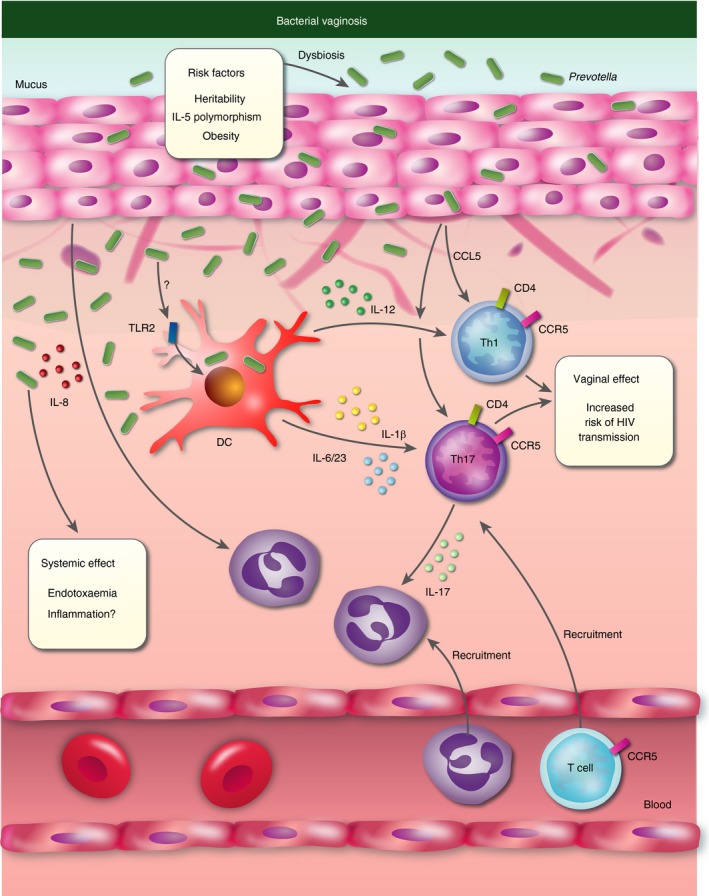
Prevotella‐mediated inflammation in bacterial vaginosis. Prevotella stimulate release of interleukin‐1β (IL‐1β), IL‐6 and IL‐23 by dendritic cells (DC), which in turn mediates IL‐17 production by T helper 17 (Th17) cells that activate neutrophils. DCs also produce IL‐12, which mediates the activation of Th1 cells. Epithelial cells contribute to neutrophil and Th cell recruitment through the production of IL‐8 and CCL5, respectively. Genetic background and obesity predispose to Prevotella‐rich dysbiosis in bacterial vaginosis. Dysbiosis leads to systemic release of lipopolysaccharides (endotoxaemia), and possibly systemic inflammation. Increased numbers of mucosal CCR5‐positive Th cells are associated with increased risk of HIV transmission.
Cervicovaginal epithelial cultures stimulated with Prevotella (Prevotella bivia or Prevotella amnii) compared with vaginal commensal Lactobacillus (Lactobacillus crispatus, Lactobacillus iners and/or Lactobacillus acidophilus) were reported to induce higher levels of IL‐1α, IL‐1β, IL‐6, IL‐8, CCL20, CXCL2, CXCL3 and CCL5.37, 38, 41, 42, 43 This cytokine/chemokine profile suggests that Prevotella has a high intrinsic capacity to promote vaginal Th17‐mediated immune responses and neutrophil recruitment through epithelial cells. The enhanced inflammatory property of Prevotella is supported by studies in mice, which found increased numbers of activated CD44+ and CCR5+ Th cells in the vaginal tract of germ‐free mice following inoculation with Prevotella bivia compared with Lactobacillus crispatus.38
One of the recent studies linking Prevotella to BV39 addressed the influence of host genetics on the vaginal microbiota in a twin‐family cohort. It was reported that microbiota composition and Prevotella abundance were largely determined by host genetics, but influenced by environmental factors, including menopause, hormone therapy, human papillomavirus infection and obesity. Gene candidate analysis found that the minor allele group with polymorphism in the IL‐5 gene had increased abundance of Prevotella melaninogenica. To address a potential causal effect of obesity in driving vaginal dysbiosis, the microbiota of obese mice on a high‐fat diet was compared with that of lean mice on a control diet.39 Obesity in mice was found to induce vaginal dysbiosis linked to increased abundance of Prevotella, and vaginal transfer of the dysbiotic microbiota to lean mice increased plasma LPS levels in recipient mice. These findings underline that complex gene–environment interactions shape the risk of acquiring a Prevotella‐rich vaginal microbiota. However, once acquired, Prevotella may drive chronic inflammation associated with BV that in turn has systemic effects on other diseases.
Gut dysbiosis triggered by HIV
Persistent chronic inflammation is a central hallmark of HIV infection even after successful antiviral therapy. Low‐grade systemic inflammation characterized by activated T cells, inflammatory cytokines, endotoxaemia and gut bacteria translocation has been linked to poor disease outcomes and increased mortality.44, 45 Studies have demonstrated that HIV infection is associated with intestinal dysbiosis characterized by increased Prevotella and reduction in Bacteroides.46, 47, 48, 49 Recent studies suggest that increased Prevotella in HIV is a driver for persistent inflammation in the gut leading to mucosal dysfunction and systemic inflammation.50, 51, 52 Colon biopsies from untreated HIV‐infected individuals showed increased Prevotella colonization, and Prevotella abundance was specifically associated with the elevated numbers of activated HLA‐DR+ CD38+ Th and Tc cells, as well as higher CD40 expression on HLA‐DR+CD1chigh myeloid dendritic cells in colon mucosa.50 A later study from the same group reported that colon dysbiosis (linked to Prevotella copri and Prevotella stercorea) in HIV‐infected individuals was associated with elevated CD40 expression on a CD1c+ subset of HLA‐DR+ CD11chigh myeloid dendritic cells, which in turn correlated with systemic levels of activated HLA‐DR+ CD38+ Th and Tc cells.52 Furthermore, CD40 expression levels on CD1c+ myeloid dendritic cells correlated with Th17 (IL‐1β, IL‐5, IL‐23 and IL‐17), Th1 (IFN‐γ) and innate (TNF‐α and IL‐10) cytokines in colonic tissue. Another study confirmed the link between Prevotella‐rich gut dysbiosis in HIV and increased systemic levels of activated T cells.51 The study additionally reported correlations between dysbiosis and increased systemic high‐sensitivity C‐reactive protein and LPS levels. However, no specific correlations to gut Prevotella abundance were investigated. Combined, these studies indicate that HIV‐induced Prevotella accumulation leads to dysfunctional immune responses in the gut mucosa driving bacterial translocation, endotoxaemia and systemic inflammation (Fig. 3). A recent interventional study administering prebiotics to HIV patients found that short dietary supplementation attenuated gut dysbiosis and systemic inflammation.53 However, longitudinal studies in humans and experimental work in mice are needed to delineate a causal relationship between HIV‐induced Prevotella‐rich dysbiosis, inflammation and adverse disease outcomes. Studies addressing Prevotella and gut dysbiosis independent of HIV are reviewed below.
Figure 3.
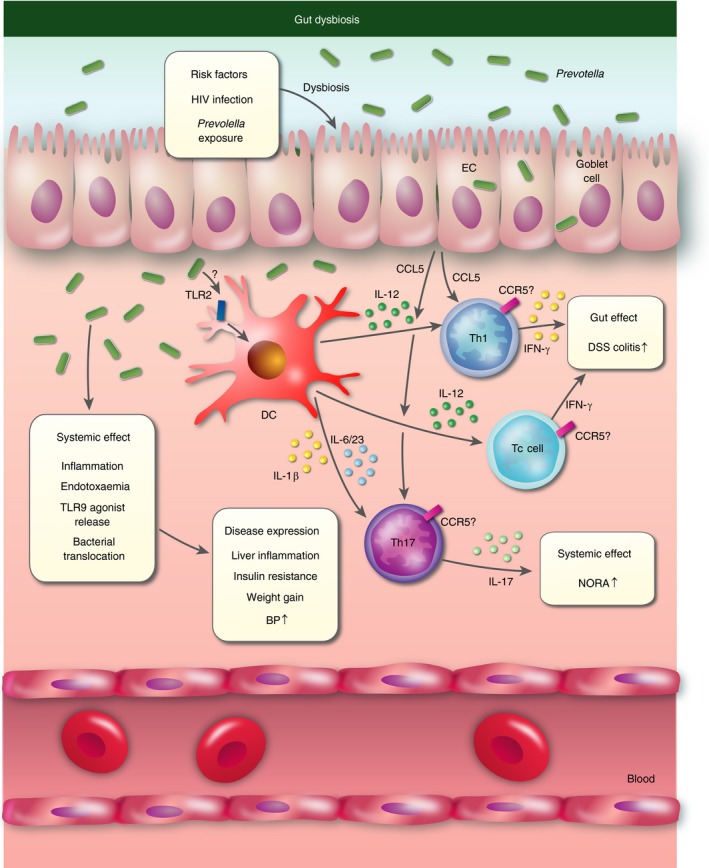
Gut inflammation associated Prevotella‐rich dysbiosis. Prevotella stimulates release of interleukin‐1β (IL‐1β), IL‐6 and IL‐23 by dendritic cells (DC), which in turn mediate IL‐17 production by T helper 17 (Th17) cells that activate neutrophils. DCs also produce IL‐12, which mediates the activation of Th1 and cytotoxic T (Tc) cells. Epithelial cells may contribute to recruitment of CCR5‐positive T cells through the production of CCL5. HIV infection and exposure to Prevotella are risk factors for Prevotella‐rich dysbiosis in the gut. Dysbiosis leads to systemic release of inflammation, bacteria, lipopolysaccharides (endotoxaemia) and Toll‐like receptor 9 (TLR9) agonists, which in turn mediates systemic disease expression, including liver inflammation, insulin resistance, weight gain and increased blood pressure (BP). Dysbiosis‐associated increase in Th17 immune responses may affect new‐onset rheumatoid arthritis (NORA). Dysbiosis increases Th1‐mediated inflammation in dextran sulphate sodium (DSS)‐induced experimental colitis.
Gut dysbiosis and metabolic syndrome
Metabolic syndrome is a collection of risk factors (obesity, insulin resistance, high blood pressure and increased blood cholesterol/triglycerides) predisposing to the development of diabetes, cardiovascular disease and non‐alcoholic fatty liver disease (NAFLD). These diseases are highly interrelated, and a common feature is low‐grade systemic inflammation. Gut dysbiosis is associated with disease, but reports on the involvement of Prevotella have been inconsistent. Increased abundance of Prevotella was found to be associated with insulin‐resistance in a non‐diabetic cohort,54 in a cohort of morbidly obese patients,55 and was linked to obesity,56, 57 hypertension,58 and NAFLD56, 59 in case–control studies. Yet other studies found no such association in type 2 diabetes,60, 61, 62 obesity,59, 63 and ischaemic stroke patients,64 although a comprehensive study found an association with increased Paraprevotella (member of the Prevotellaceae family) in type 2 diabetes.60 These discrepancies may be due to the complex interrelatedness of the diseases, making patient selection and stratification important study parameters. Furthermore, different bacterial species could be involved in expression of the same disease features. Indeed, increased abundance of both Prevotella copri and Bacteroides vulgatus was associated with insulin‐resistance, but the species were mutually exclusive in the gut.54 Adding further to the complexity, a study showed that individuals with improved glucose metabolism following a high‐fibre dietary intervention had a Prevotella‐rich gut microbiota,65 suggesting interaction of diet on the outcome of specific disease features.
Studies in mice indicate that Prevotella can drive features of metabolic syndrome. Colonization of germ‐free mice with a Prevotella‐rich microbiota from patients with hypertension induced higher blood pressure compared with mice receiving microbiota from a normotensive donor.58 Prevotella copri colonization in mice on a high‐fat diet promoted increased insulin‐resistance.54 Furthermore, the role of Prevotella‐rich dysbiosis in NAFLD and obesity was studied using the transfer of dysbiotic microbiota enriched with Prevotella, unknown Prevotellaceae and TM7 from mice with a deficient inflammasome pathway (Asc knockout or IL‐18 knockout) to wild‐type mice by co‐housing.66 The presence of a Prevotella‐rich gut microbiota exacerbated methionine‐choline‐deficient diet‐induced non‐alcoholic steatohepatitis (NASH) characterized by increased liver steatosis and inflammation, and elevated liver‐enzymes alanine aminotransferase and aspartate aminotransferase in blood. The study found that Prevotella‐rich dysbiosis was associated with the presence of black‐pigmented bacteria in colonic epithelial cells and macrophages. Furthermore, the study indicated that NASH disease propagation was driven by epithelium‐derived CCL5‐dependent intestinal inflammation giving rise to systemic release of bacterial TLR4 and TLR9 agonists promoting TNF‐α‐dependent inflammation and pathology in the liver. Additionally, transfer of Prevotella‐rich microbiota from Asc knockout mice caused increased weight‐gain in both wild‐type mice on high‐fat diet, and obesity‐prone ob/ob mice.66 However, the transfer did not affect insulin resistance, underlining a heterogenic effect of the microbiota in metabolic disease phenotypes. Although these findings are compelling, additional investigations of immune mechanisms in metabolic disease are needed in humans.
Gut dysbiosis and inflammatory bowel disease
An interesting line of research has demonstrated a central role of the Nucleotide‐binding and oligomerization domain‐Like Receptor P6 (NLRP6)‐inflammasome in maintaining gut homeostasis and protection from Prevotella‐rich dysbiosis, which can promote experimental colitis in mice. NLRP6‐deficiency was found to cause goblet cell dysfunction and reduced mucus secretion on intestinal surfaces leading to increased susceptibility to Citrobacter rodentium infection.67 The NLRP6‐inflammasome in intestinal epithelial cells was found to sense microbiota‐derived metabolites leading to IL‐18‐dependent production of antimicrobial peptides, which in turn shapes microbiota composition under homeostatic conditions.68 Homeostatic epithelial cell‐derived IL‐18 has previously been shown to mediate Foxp3+ regulatory T cell function, and directly suppress pathogenic Th17 cell function in the intestine.69 This finding provides a possible model for immunological maintenance of gut homeostasis, where another layer of microbial protection involving regulatory T cell‐stimulated IgA production could further shape the gut microbiota.70, 71 Indeed, gut bacteria known to promote colitis have been shown to be a target of T‐cell‐dependent IgA,72 although a protective role of Prevotella‐specific IgA remains to be formally demonstrated.
Transfer of Prevotella‐rich dysbiotic gut microbiota from Asc knockout or NLRP6 knockout mice to wild‐type mice was found to promote dextran sulphate sodium‐induced experimental colitis characterized by increased weight loss, tissue pathology and death in recipient mice. Increased susceptibility to experimental colitis was dependent on CCL5, and increased CCL5 levels were associated with intestinal recruitment of conventional T cells, B cells and APCs in NLRP6 knockout mice.73 Furthermore, the study indicated that Prevotella‐rich dysbiosis drive decreased NLRP6‐dependent IL‐18 production in intestinal epithelial cells. Interestingly, this was supported by a later study demonstrating that transfer of Prevotella‐rich dysbiotic microbiota would subvert IL‐18‐dependent antimicrobial peptide production through the production of metabolites antagonizing NLRP6 function, which in turn promoted establishment of the dysbiotic microbial community in the gut.68 Colonization of antibiotic‐treated C57BL/6 mice with Prevotella copri enhanced dextran sulphate sodium‐induced colitis compared with control mice or mice colonized by commensal Bacteroides thetaiotamicron.32 The enhanced colitis was associated with increased IFN‐γ production by lamina propria Th cells from Prevotella copri colonized mice, suggesting that Prevotella promote Th1 immune responses in experimental colitis.
Although a role for Prevotella in inflammatory bowel disease is compelling from studies of experimental colitis in mice, currently no studies have provided an association between increased Prevotella abundance and disease in humans. In fact, a study indicated reduced Prevotella in paediatric Crohn's disease.74 Furthermore, the most comprehensive study to date75 found no association between Prevotella and new‐onset Crohn's before treatment.76 Rather, Crohn's disease was associated with outgrowth of Enterobacteriaceae, Pasteurellaceae, Veillonellaceae and Fusobacteriaceae, which is in line with earlier microbiome studies of Crohn's disease and ulcerative colitis.77, 78 The mechanisms by which Prevotella promote disease in mice (subversion of gut homeostasis and initiation of intestinal inflammation) may be shared with other bacterial species linked to human disease. Furthermore, human inflammatory bowel disease is highly heterogeneous and specific bacteria may be involved in different disease phenotypes and immune mechanisms,79 suggesting a need for larger prospective cohort studies to delineate causal relationships.
Asthma and COPD
The healthy lung has traditionally been viewed as sterile due to the absence of culturable bacteria in the absence of clinical respiratory infection. However, a study in 20104 reported a low‐density, but distinct microbial community dominated by Prevotella in the lung. This finding has subsequently been confirmed by later studies controlling for potential sources of contamination.5, 80, 81, 82 Intriguingly, Prevotella abundance was reported to be reduced in patients with asthma and with COPD, which instead presented with outgrowth of pathogenic proteobacteria.4 Lung colonization by proteobacteria has previously been linked to increased risk of developing asthma in childhood,83 exacerbation episodes,84 as well as increased neutrophilia and IL‐8 levels in patients with asthma.85 Similarly, patients with COPD present with predominant proteobacterial colonization during both stable disease and excerbations.86, 87, 88 This has led to speculation that proteobacteria take part in disease development and progression in COPD.89 This hypothesis is supported by studies associating increased bacteria loads to increased airway inflammation90, 91 and accelerated decline in lung function.92
A study compared the inflammatory properties of Prevotella associated with healthy lungs (Prevotella melaninogenica, Prevotella nanceiensis and Prevotella salivae) with proteobacteria associated with asthma and COPD (Haemophilus influenzae B, non‐typeable Haemophilus influenzae and Moraxella catarrhalis).93 Prevotella was found to induce similar levels of CD83, CD86 and CD40 activation‐maker surface expression, but reduced production of IL‐12p70, IL‐23 and IL‐10 cytokines in monocyte derived dendritic cells when compared with proteobacteria. This lower inflammatory capacity of Prevotella compared with proteobacteria was further demonstrated in mice reporting decreased MIP‐2a (IL‐8), TNF‐α and thymic stromal lymphopoietin production by lung stromal cells, and decreased levels of TNF‐α production by lung immune cells.94 Titration experiments indicated that the lower stimulatory capacity of Prevotella was due to intrinsic differences in composition of pathogen‐associated molecular patterns. It was hypothesized93, 94 that the difference could be ascribed to alternate LPS structures as Prevotella produce penta‐acylated LPS whereas Haemophilus influenzae and Moraxella catarrhalis produce hexa‐acylated and hepta‐acylated LPS, respectively. Indeed, an analysis of the LPS synthesis pathway in publicly available genomes found that only gammaproteobacteria have the genetic capacity to produce hexa‐acylated LPS (the prototypic LPS commonly isolated from Escherichia coli), which exhibit 100‐fold stimulatory capacity on TLR4 compared with penta‐acylated LPS.95 Non‐typeable Haemophilus influenzae was found to induce severe lung neutrophilia in mice accompanied by increased levels of MIP‐2a (IL‐8), CCL20 and IL‐1β in lung tissue compared with Prevotella nanceiensis.94 Furthermore, non‐typeable Haemophilus influenzae induced severe immune pathology in lung tissue, whereas no pathology could be observed in response to Prevotella nanceiensis when compared with control mice. The diminished lung inflammatory capacity of Prevotella nanceiensis was dependent on TLR2, whereas the inflammation mediated by non‐typeable Haemophilus influenzae was TLR2‐independent. These findings support that Prevotella exhibit limited TLR4‐stimulating capacity as this genus cannot produce hexa‐acylated LPS. Furthermore, proteobacteria may specifically participate in driving inflammatory features of asthma and COPD, whereas Prevotella in comparison may be well tolerated in the lung.
A possible homeostatic role for Prevotella in the healthy lung remains largely unknown. Induction of COPD‐like lung inflammation and pathology in mice by LPS/elastase inhalation was found to decrease Prevotella abundance, and mediate Pseudomonas and Lactobacillus outgrowth.96 This suggests that reduced Prevotella in human asthma and COPD4 may not be a risk factor before disease, as disease‐related inflammation may directly drive decreased Prevotella abundance by creating a microenvironment not suitable for survival. Studies suggest that the low‐density lung microbiota is transmitted from the oral microbiota by microaspiration and continuously eliminated.80, 81 It could therefore be speculated that the limited immune stimulatory potential of Prevotella may drive its own elimination by mediating a low‐grade inflammatory process, which in turn may protect from invading respiratory pathogens and chronic disease under homeostatic conditions.80 Alternatively, a study found that early establishment of a Bacteroidetes‐rich lung microbiota in neonatal mice drives expansion of regulatory T cells protecting from allergic airway disease in response to house‐dust‐mite.97 Hence Prevotella (a member of the Bacteroidetes phyla) may participate in establishing tolerance in the lung, although a role for Bacteroides species with known immune regulatory properties98 cannot be ruled out. Combined, these studies show that many speculations can be made as to the role of Prevotella in the healthy lung, and so there is a need for more experimental work for clarification.
Concluding remarks
Emerging studies are linking Prevotella abundance and specific strains to inflammatory disease mediated by Th17‐related immune responses. Indeed, at least some Prevotella strains seem to be inflammophilic pathobionts that thrive in an inflammatory environment, and exhibit superior intrinsic capacity to stimulate Th17‐mediated inflammation compared with strict commensal bacteria. There is compelling mechanistic and causal evidence in mice that Prevotella can promote inflammatory disease features. However, there is a need for more studies in humans to ascertain a causal and potential disease‐triggering role for Prevotella. Inflammatory diseases are highly heterogeneous and develop through the complex interactions between host genetic risk factors and environmental exposures.99 Prevotella may only play a part in certain disease endotypes, and larger cohort studies are needed to delineate causal relationships. Additionally, Prevotella may not be the only genus participating in inflammatory disease, and specific Prevotella species may exhibit different properties. A recent comprehensive study comparing several bacterial species suggests that membership of a specific phylum does not predict immunological properties, underlining the importance of characterizing properties at species level.100 Furthermore, studies indicate that Prevotella is a genus with high genetic diversity within and between species.101, 102 This may explain why Prevotella is abundant in the healthy microbiota, and suggests that only certain strains may exhibit pathobiontic properties. Species heterogeneity also underlines the need to continue in‐depth metagenomic characterization of the microbiota in inflammatory disease to reveal disease‐modulating properties. Intriguingly, some Prevotella species could have evolved immune escape mechanisms, including induction of neutrophil dysfunction,21, that may lead to chronic inflammation due to defective clearance. Deciphering the genetic and mechanistic basis of immune escape by Prevotella may in the future reveal disease‐modifying drug targets.
Disclosures
The author declares that there is no conflict of interest.
Acknowledgement
The author wishes to thank Alison Schroeer for her artistic contributions in preparing the illustrations that accompany this manuscript.
References
- 1. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009; 9:313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Medini D. Microbiology in the post‐genomic era. Nat Rev Microbiol 2008; 6:419–30. [DOI] [PubMed] [Google Scholar]
- 3. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486, 207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C et al Disordered microbial communities in asthmatic airways. PLoS One 2010; 5:e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A et al Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med 2011; 184:957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR et al Enterotypes of the human gut microbiome. Nature 2011; 473:174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jousimies‐Somer HR. Update on the taxonomy and the clinical and laboratory characteristics of pigmented anaerobic gram‐negative rods. Clin Infect Dis 1995; 20(Suppl 2):S187–91. [DOI] [PubMed] [Google Scholar]
- 8. Nagy E. Anaerobic infections: update on treatment considerations. Drugs 2010; 70:841–58. [DOI] [PubMed] [Google Scholar]
- 9. Brook I. Anaerobic pulmonary infections in children. Pediatr Emerg Care 2004; 20:636–40. [DOI] [PubMed] [Google Scholar]
- 10. Brook I. Microbiology of common infections in the upper respiratory tract. Prim Care 1998; 25:633–48. [DOI] [PubMed] [Google Scholar]
- 11. Dahlén GG. Black‐pigmented gram‐negative anaerobes in periodontitis. FEMS Immunol Med Microbiol 1993; 6:181–92. [DOI] [PubMed] [Google Scholar]
- 12. Berezow AB, Darveau RP. Microbial shift and periodontitis. Periodontol 2011; 2000:36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cekici A, Kantarci A, Hasturk H, Van Dyke TE. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol 2014; 2000:57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol 2014; 15:30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Aquino SG, Abdollahi‐Roodsaz S, Koenders MI, de van Loo FAJ, Pruijn GJM, Marijnissen RJ et al Periodontal pathogens directly promote autoimmune experimental arthritis by inducing a TLR2‐ and IL‐1‐driven Th17 response. J Immunol 2014; 192:4103–11. [DOI] [PubMed] [Google Scholar]
- 16. Schincaglia GP, Hong BY, Rosania A, Barasz J, Thompson A, Sobue T et al Clinical, immune, and microbiome traits of gingivitis and peri‐implant mucositis. J Dent Res 2017; 96:47–55. [DOI] [PubMed] [Google Scholar]
- 17. Greer A, Irie K, Hashim A, Leroux BG, Chang AM, Curtis MA et al Site‐specific neutrophil migration and CXCL2 expression in periodontal tissue. J Dent Res 2016; 95:946–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cury PR, Carmo JP, Horewicz VV, Santos JN, Barbuto JA. Altered phenotype and function of dendritic cells in individuals with chronic periodontitis. Arch Oral Biol 2013; 58:1208–16. [DOI] [PubMed] [Google Scholar]
- 19. Horst OV, Tompkins KA, Coats SR, Braham PH, Darveau RP, Dale BA. TGF‐β1 Inhibits TLR‐mediated odontoblast responses to oral bacteria. J Dent Res 2009; 88:333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ji S, Kim Y, Min B‐M, Han SH, Choi Y. Innate immune responses of gingival epithelial cells to nonperiodontopathic and periodontopathic bacteria. J Periodontal Res 2007; 42:503–10. [DOI] [PubMed] [Google Scholar]
- 21. Matsui A, Jin J‐O, Johnston CD, Yamazaki H, Houri‐Haddad Y, Rittling SR. Pathogenic bacterial species associated with endodontic infection evade innate immune control by disabling neutrophils. Infect Immun 2014; 82:4068–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uriarte SM, Edmisson JS, Jimenez‐Flores E. Human neutrophils and oral microbiota: a constant tug‐of‐war between a harmonious and a discordant coexistence. Immunol Rev 2016; 273:282–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kimura Y, Yoshida S, Takeuchi T, Kimura M, Yoshikawa A, Hiramatsu Y et al Periodontal pathogens participate in synovitis in patients with rheumatoid arthritis in clinical remission: a retrospective case–control study. Rheumatology (Oxford) 2015; 54:2257–63. [DOI] [PubMed] [Google Scholar]
- 24. Goh CE, Kopp J, Papapanou PN, Molitor JA, Demmer RT. Association between serum antibodies to periodontal bacteria and rheumatoid factor in the Third National Health and Nutrition Examination Survey. Arthritis Rheumatol. (Hoboken, N.J.) 2016; 68:2384–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moen K, Brun JG, Madland TM, Tynning T, Jonsson R. Immunoglobulin G and A antibody responses to Bacteroides forsythus and Prevotella intermedia in sera and synovial fluids of arthritis patients. Clin Diagn Lab Immunol 2003; 10:1043–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okada M, Kobayashi T, Ito S, Yokoyama T, Komatsu Y, Abe A et al Antibody responses to periodontopathic bacteria in relation to rheumatoid arthritis in Japanese adults. J Periodontol 2011; 82:1433–41. [DOI] [PubMed] [Google Scholar]
- 27. Ogrendik M, Kokino S, Ozdemir F, Bird PS, Hamlet S. Serum antibodies to oral anaerobic bacteria in patients with rheumatoid arthritis. MedGenMed 2005; 7:2. [PMC free article] [PubMed] [Google Scholar]
- 28. Martinez‐Martinez RE, Abud‐Mendoza C, Patiño‐Marin N, Rizo‐Rodríguez JC, Little JW, Loyola‐Rodríguez JP. Detection of periodontal bacterial DNA in serum and synovial fluid in refractory rheumatoid arthritis patients. J Clin Periodontol 2009; 36:1004–10. [DOI] [PubMed] [Google Scholar]
- 29. Reichert S, Haffner M, Keyßer G, Schäfer C, Stein JM, Schaller H‐G et al Detection of oral bacterial DNA in synovial fluid. J Clin Periodontol 2013; 40:591–8. [DOI] [PubMed] [Google Scholar]
- 30. Moen K, Brun JG, Valen M, Skartveit L, Eribe EKR, Olsen I et al Synovial inflammation in active rheumatoid arthritis and psoriatic arthritis facilitates trapping of a variety of oral bacterial DNAs. Clin Exp Rheumatol 2006; 24:656–63. [PubMed] [Google Scholar]
- 31. Scher JU, Ubeda C, Equinda M, Khanin R, Buischi Y, Viale A et al Periodontal disease and the oral microbiota in new‐onset rheumatoid arthritis. Arthritis Rheum 2012; 64:3083–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C et al Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013; 2:e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maeda Y., Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K et al Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. (Hoboken, N.J.) 2016; 68:2646–61. [DOI] [PubMed] [Google Scholar]
- 34. Marietta EV, Murray JA, Luckey DH, Jeraldo PR, Lamba A, Patel R et al Suppression of inflammatory arthritis by human gut‐derived Prevotella histicola in humanized mice. Arthritis Rheumatol. (Hoboken, N.J.) 2016; 68:2878–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pianta A, Arvikar S, Strle K, Drouin EE, Wang Q, Costello CE et al Evidence for immune relevance of Prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol 2017; 69: 964–75 https://doi.org/10.1002/art.40003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Onderdonk AB, Delaney ML, Fichorova RN. The human microbiome during bacterial vaginosis. Clin Microbiol Rev 2016; 29:223–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M et al Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity 2015; 42:965–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu‐Ali G, Bowman BA et al Lactobacillus‐deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity 2017; 46:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Si J, You HJ, Yu J, Sung J, Ko G. Prevotella as a hub for vaginal microbiota under the influence of host genetics and their association with obesity. Cell Host Microbe 2017; 21:97–105. [DOI] [PubMed] [Google Scholar]
- 40. Kyongo JK, Crucitti T, Menten J, Hardy L, Cools P, Michiels J et al Cross‐sectional analysis of selected genital tract immunological markers and molecular vaginal microbiota in sub‐Saharan African women, with relevance to HIV risk and prevention. Clin Vaccine Immunol 2015; 22:526–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fichorova RN, Yamamoto HS, Delaney ML, Onderdonk AB, Doncel GF. Novel vaginal microflora colonization model providing new insight into microbicide mechanism of action. MBio 2011; 2:e00168–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Doerflinger SY, Throop AL, Herbst‐Kralovetz MM. Bacteria in the vaginal microbiome alter the innate immune response and barrier properties of the human vaginal epithelia in a species‐specific manner. J Infect Dis 2014; 209:1989–99. [DOI] [PubMed] [Google Scholar]
- 43. Zalenskaya IA, Joseph T, Bavarva J, Yousefieh N, Jackson SS, Fashemi T et al Gene expression profiling of human vaginal cells in vitro discriminates compounds with pro‐inflammatory and mucosa‐altering properties: novel biomarkers for preclinical testing of HIV microbicide candidates. PLoS One 2015; 10:e0128557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hunt PW. HIV and inflammation: mechanisms and consequences. Curr. HIV/AIDS Rep. 2012; 9:139–47. [DOI] [PubMed] [Google Scholar]
- 45. Marchetti G, Tincati C, Silvestri G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev 2013; 26:2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ et al Alterations in the gut microbiota associated with HIV‐1 infection. Cell Host Microbe 2013; 14:329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ling Z, Jin C, Xie T, Cheng Y, Li L, Wu N. Alterations in the fecal microbiota of patients with HIV‐1 infection: an observational study in a Chinese population. Sci Rep 2016; 6:30673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C et al A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog 2014; 10:e1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang L, Poles MA, Fisch GS, Ma Y, Nossa C, Phelan JA et al HIV‐induced immunosuppression is associated with colonization of the proximal gut by environmental bacteria. AIDS 2016; 30:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK et al An altered intestinal mucosal microbiome in HIV‐1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol 2014; 7:983–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vázquez‐Castellanos JF, Serrano‐Villar S, Latorre A, Artacho A, Ferrús ML, Madrid N et al Altered metabolism of gut microbiota contributes to chronic immune activation in HIV‐infected individuals. Mucosal Immunol 2015; 8:760–72. [DOI] [PubMed] [Google Scholar]
- 52. Dillon SM, Lee EJ, Kotter CV, Austin GL, Gianella S, Siewe B et al Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T‐cell activation in untreated HIV‐1 infection. Mucosal Immunol 2016; 9:24–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Serrano‐Villar S, Vázquez‐Castellanos JF, Vallejo A, Latorre A, Sainz T, Ferrando‐Martínez S. The effects of prebiotics on microbial dysbiosis, butyrate production and immunity in HIV‐infected subjects. Mucosal Immunol 2016;. https://doi.org/10.1038/mi.2016.122. [DOI] [PubMed] [Google Scholar]
- 54. Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BAH et al Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016; 535:376–81. [DOI] [PubMed] [Google Scholar]
- 55. Moreno‐Indias I, Sánchez‐Alcoholado L, García‐Fuentes E, Cardona F, Queipo‐Ortuño MI, Tinahones FJ. Insulin resistance is associated with specific gut microbiota in appendix samples from morbidly obese patients. Am J Transl Res 2016; 8:5672–84. [PMC free article] [PubMed] [Google Scholar]
- 56. Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD et al Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 2013; 57:601–9. [DOI] [PubMed] [Google Scholar]
- 57. Hu H‐J, Park S‐G, Jang HB, Choi M‐G, Park K‐H, Kang JH et al Obesity alters the microbial community profile in Korean adolescents. PLoS One 2015; 10:e0134333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G et al Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017; 5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Michail S, Lin M, Frey MR, Fanter R, Paliy O, Hilbush B et al Altered gut microbial energy and metabolism in children with non‐alcoholic fatty liver disease. FEMS Microbiol Ecol 2015; 91:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S et al Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015; 528:262–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F et al A metagenome‐wide association study of gut microbiota in type 2 diabetes. Nature 2012; 490:55–60. [DOI] [PubMed] [Google Scholar]
- 62. Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B et al Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013; 498:99–103. [DOI] [PubMed] [Google Scholar]
- 63. Nakayama J, Yamamoto A, Palermo‐Conde LA, Higashi K, Sonomoto K, Tan J et al Impact of westernized diet on gut microbiota in children on Leyte Island. Front Microbiol 2017; 8:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yamashiro K, Tanaka R, Urabe T, Ueno Y, Yamashiro Y, Nomoto K et al Gut dysbiosis is associated with metabolism and systemic inflammation in patients with ischemic stroke. PLoS One 2017; 12:e0171521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kovatcheva‐Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T et al Dietary fiber‐induced improvement in glucose metabolism is associated with increased abundance of Prevotella . Cell Metab 2015; 22:971–82. [DOI] [PubMed] [Google Scholar]
- 66. Henao‐Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T et al Inflammasome‐mediated dysbiosis regulates progression of NAFLD and obesity. Nature 2012; 482:179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wlodarska M, Thaiss CA, Nowarski R, Henao‐Mejia J, Zhang J‐P, Brown EM et al NLRP6 inflammasome orchestrates the colonic host‐microbial interface by regulating goblet cell mucus secretion. Cell 2014; 156:1045–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Levy M, Thaiss CA, Zeevi D, Dohnalová L, Zilberman‐Schapira G, Mahdi JA et al Microbiota‐modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 2015; 163:1428–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Harrison OJ, Srinivasan N, Pott J, Schiering C, Krausgruber T, Ilott NE et al Epithelial‐derived IL‐18 regulates Th17 cell differentiation and Foxp3+ Treg cell function in the intestine. Mucosal Immunol 2015; 8:1226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell‐IgA response to the intestinal microbiota. Proc Natl Acad Sci USA 2009; 106:19256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang L, Ray A, Jiang X, Wang J, Basu S, Liu X et al T regulatory cells and B cells cooperate to form a regulatory loop that maintains gut homeostasis and suppresses dextran sulfate sodium‐induced colitis. Mucosal Immunol 2015; 8:1297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L et al Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014; 158:1000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Elinav E, Strowig T, Kau AL, Henao‐Mejia J, Thaiss CA, Booth CJ et al NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011; 145:745–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D et al Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe 2015; 18:489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gevers D, Kugathasan S, Knights D, Kostic AD, Knight R, Xavier RJ. A microbiome foundation for the study of Crohn's disease. Cell Host Microbe 2017; 21:301–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gevers D, Kugathasan S, Denson LA, Vázquez‐Baeza Y, Van Treuren W, Ren B et al The treatment‐naive microbiome in new‐onset Crohn's disease. Cell Host Microbe 2014; 15:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV et al Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012; 13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D et al Non‐invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One 2012; 7:e39242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ray A, Dittel BN. Interrelatedness between dysbiosis in the gut microbiota due to immunodeficiency and disease penetrance of colitis. Immunology 2015; 146:359–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bassis CM, Erb‐Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB et al Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 2015; 6:e00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dickson RP, Erb‐Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB et al Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc 2015; 12:821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Park H, Shin JW, Park S‐G, Kim W. Microbial communities in the upper respiratory tract of patients with asthma and chronic obstructive pulmonary disease. PLoS One 2014; 9:e109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bønnelykke K et al Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med 2007; 357:1487–95. [DOI] [PubMed] [Google Scholar]
- 84. Bisgaard H, Hermansen MN, Bønnelykke K, Stokholm J, Baty F, Skytt NL et al Association of bacteria and viruses with wheezy episodes in young children: prospective birth cohort study. BMJ 2010; 341:c4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wood LG, Simpson JL, Hansbro PM, Gibson PG. Potentially pathogenic bacteria cultured from the sputum of stable asthmatics are associated with increased 8‐isoprostane and airway neutrophilia. Free Radic Res 2010; 44:146–54. [DOI] [PubMed] [Google Scholar]
- 86. Murphy TF, Sethi S, Niederman MS. The role of bacteria in exacerbations of COPD. A constructive view. Chest 2000; 118:204–9. [DOI] [PubMed] [Google Scholar]
- 87. Monsó E, Rosell A, Bonet G, Manterola J, Cardona PJ, Ruiz J et al Risk factors for lower airway bacterial colonization in chronic bronchitis. Eur Respir J Off J Eur Soc Clin Respir Physiol 1999; 13:338–42. [DOI] [PubMed] [Google Scholar]
- 88. Zalacain R, Sobradillo V, Amilibia J, Barrón J, Achótegui V, Pijoan JI et al Predisposing factors to bacterial colonization in chronic obstructive pulmonary disease. Eur Respir J Off J Eur Soc Clin Respir Physiol 1999; 13:343–8. [DOI] [PubMed] [Google Scholar]
- 89. Papi A, Luppi F, Franco F, Fabbri LM. Pathophysiology of exacerbations of chronic obstructive pulmonary disease. Proc Am Thorac Soc 2006; 3:245–51. [DOI] [PubMed] [Google Scholar]
- 90. Sethi S, Maloney J, Grove L, Wrona C, Berenson CS. Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2006; 173:991–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hill AT, Campbell EJ, Hill SL, Bayley DL, Stockley RA. Association between airway bacterial load and markers of airway inflammation in patients with stable chronic bronchitis. Am J Med 2000; 109:288–95. [DOI] [PubMed] [Google Scholar]
- 92. Wilkinson TMA, Patel IS, Wilks M, Donaldson GC, Wedzicha JA. Airway bacterial load and FEV1 decline in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2003; 167:1090–5. [DOI] [PubMed] [Google Scholar]
- 93. Larsen JM, Steen‐Jensen DB, Laursen JM, Søndergaard JN, Musavian HS, Butt TM et al Divergent pro‐inflammatory profile of human dendritic cells in response to commensal and pathogenic bacteria associated with the airway microbiota. PLoS One 2012; 7:e31976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Larsen JM, Musavian HS, Butt TM, Ingvorsen C, Thysen AH, Brix S. Chronic obstructive pulmonary disease and asthma‐associated Proteobacteria, but not commensal Prevotella spp., promote Toll‐like receptor 2‐independent lung inflammation and pathology. Immunology 2015; 144:333–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Brix S, Eriksen C, Larsen JM, Bisgaard H. Metagenomic heterogeneity explains dual immune effects of endotoxins. J Allergy Clin Immunol 2015; 135:277–80. [DOI] [PubMed] [Google Scholar]
- 96. Yadava K, Pattaroni C, Sichelstiel AK, Trompette A, Gollwitzer ES, Salami O et al Microbiota promotes chronic pulmonary inflammation by enhancing IL‐17a and autoantibodies. Am J Respir Crit Care Med 2016; 193:975–87. [DOI] [PubMed] [Google Scholar]
- 97. Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD et al Lung microbiota promotes tolerance to allergens in neonates via PD‐L1. Nat Med 2014; 20:642–7. [DOI] [PubMed] [Google Scholar]
- 98. Neff CP, Rhodes ME, Arnolds KL, Collins CB, Donnelly J, Nusbacher N et al Diverse intestinal bacteria contain putative zwitterionic capsular polysaccharides with anti‐inflammatory properties. Cell Host Microbe 2016; 20:535–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Renz H, von Mutius E, Brandtzaeg P, Cookson WO, Autenrieth IB, Haller D. Gene–environment interactions in chronic inflammatory disease. Nat Immunol 2011; 12:273–7. [DOI] [PubMed] [Google Scholar]
- 100. Geva‐Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz‐Lopez A et al Mining the human gut microbiota for immunomodulatory organisms. Cell 2017; 168:928–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhu A, Sunagawa S, Mende DR, Bork P. Inter‐individual differences in the gene content of human gut bacterial species. Genome Biol 2015; 16:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ley RE. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat Rev Gastroenterol Hepatol 2016; 13:69–70. [DOI] [PubMed] [Google Scholar]


