Summary
We report a systematic RNAi longevity screen of 82 Caenorhabditis elegans genes selected based on orthology to human genes differentially expressed with age. We find substantial enrichment in genes for which knockdown increased lifespan. This enrichment is markedly higher than published genomewide longevity screens in C. elegans and similar to screens that preselected candidates based on longevity‐correlated metrics (e.g., stress resistance). Of the 50 genes that affected lifespan, 46 were previously unreported. The five genes with the greatest impact on lifespan (>20% extension) encode the enzyme kynureninase (kynu‐1), a neuronal leucine‐rich repeat protein (iglr‐1), a tetraspanin (tsp‐3), a regulator of calcineurin (rcan‐1), and a voltage‐gated calcium channel subunit (unc‐36). Knockdown of each gene extended healthspan without impairing reproduction. kynu‐1(RNAi) alone delayed pathology in C. elegans models of Alzheimer's disease and Huntington's disease. Each gene displayed a distinct pattern of interaction with known aging pathways. In the context of published work, kynu‐1, tsp‐3, and rcan‐1 are of particular interest for immediate follow‐up. kynu‐1 is an understudied member of the kynurenine metabolic pathway with a mechanistically distinct impact on lifespan. Our data suggest that tsp‐3 is a novel modulator of hypoxic signaling and rcan‐1 is a context‐specific calcineurin regulator. Our results validate C. elegans as a comparative tool for prioritizing human candidate aging genes, confirm age‐associated gene expression data as valuable source of novel longevity determinants, and prioritize select genes for mechanistic follow‐up.
Keywords: aging, Caenorhabditis elegans, comparative genetics, Homo sapiens, lifespan, transcriptomics
Introduction
Understanding which molecular processes contribute to aging is critical to developing interventions capable of extending healthy human lifespan and delaying onset of age‐associated diseases. A key step in this process is building a comprehensive model encompassing the range of genetic and environmental factors that influence lifespan and describing the complex interaction between these factors in an aging organism. Directly screening interventions for lifespan phenotypes in mammals is limited by long lifespans. Despite evolutionary distance and orders‐of‐magnitude differences in lifespan, processes that contribute to aging are sufficiently conserved that mechanistic knowledge gleaned from short‐lived invertebrates can be beneficially applied to mammalian systems. Genetic screens in the nematode, Caenorhabditis elegans, have identified hundreds of genes capable of influencing lifespan (Yanos et al., 2012; Sutphin & Korstanje, 2016). Pharmacological agents identified as prolongevity using invertebrate models – rapamycin, metformin, resveratrol – are now in clinical trials for treatment of age‐associated disease in humans (Kennedy & Pennypacker, 2015).
An approach that is tractable in humans is to characterize systemic changes that occur during normal aging. This approach identifies traits that change with age or during age‐associated disease and employs targeted studies to determine which play a causative role in aging. Early applications focused on easily measurable physiological traits, such as body weight or circulating molecules, but has now expanded into the ‘‐omics’ realm to provide systems‐level insight into molecular changes that occur with age. As part of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium, we published a large meta‐analysis of gene expression in human peripheral blood from 14,983 individuals representing ages across the adult lifespan (Peters et al., 2015). This study identified 1,497 genes with significantly different expression at different ages. Gene sets with a defined age‐associated expression pattern provide information about molecular processes with altered activity during aging and provide a valuable diagnostic tool for determining individual biological rate of aging and predicting risk of age‐associated disease, as demonstrated in follow‐up analyses (Peters et al., 2015). On a gene‐by‐gene basis, differential expression alone is insufficient to distinguish between genes that play a causative role in aging and genes that merely respond to the altered physiological environment in an aging organism.
In this study, we selected the human genes with the most significant differential expression with age from the CHARGE meta‐analysis and used RNAi to screen C. elegans orthologs for lifespan phenotypes. This selection criterion ensured that every gene identified in the lifespan screen was already of interest in the context of human aging. The short lifespan of C. elegans allowed genes capable of directly influencing lifespan to be rapidly identified and characterized. The resulting C. elegans candidate list was substantially enriched in genes for which knockdown extends lifespan. We selected the five genes with the greatest impact on lifespan for additional characterization and found that each gene produced a distinct pattern of influence on healthspan, reproduction, neurodegenerative pathology, and interaction with established aging pathways.
Results
Gene selection
We began by selecting the 125 genes with the most significant differential expression with age in human peripheral blood from the 1,497 identified in the CHARGE study (Peters et al., 2015). Each gene was replicated in independent samples with significant meta‐analysis P‐values ranging from 1.98 × 10−59 to 1.62 × 10−577 (Table S1, Supporting information). Our initial goal was to determine which genes were capable of directly influencing lifespan in C. elegans. We identified C. elegans orthologs for each human gene using the WORMHOLE ortholog prediction tool (Sutphin et al., 2016), yielding 88 C. elegans orthologs corresponding to 61 of the 125 human candidate genes (Tables 1 and S2, Supporting information). We obtained feeding RNAi bacterial strains with the correct target sequence for 82 of the 88 C. elegans genes, forming the ‘CHARGE gene set’. The CHARGE gene set includes 37 high‐confidence orthologs (HCOs) and 45 related proteins (RPs) (Table S2, Supporting information; see Experimental procedures). As a control, we selected 59 C. elegans genes (25 HCOs and 34 RPs) based on orthology to randomly selected human genes – the ‘Random gene set’ (Tables 1 and S3, Supporting information).
Table 1.
The CHARGE gene set is enriched in genes for which knockdown extends lifespan relative to the random gene set at 25 °C
| Orthologs included | All | HCO only | ||||||
|---|---|---|---|---|---|---|---|---|
| Temperature | 15 °C | 25 °C | 15 °C | 25 °C | ||||
| Gene set | CHARGE | Random | CHARGE | Random | CHARGE | Random | CHARGE | Random |
| Human genes (all) | 125 | 281 | 125 | 281 | 125 | 281 | 125 | 281 |
| Human genes with worm ortholog(s) | 61 | 63 | 61 | 63 | 45 | 39 | 45 | 39 |
| Worm orthologs identified | 88 | 160 | 88 | 160 | 39 | 37 | 39 | 37 |
| Worm orthologs testeda | 82 | 59 | 82 | 59 | 37 | 25 | 37 | 25 |
| # long‐lived RNAi | 5 | 7 | 36 | 4 | 3 | 2 | 15 | 1 |
| # not long‐lived RNAi | 77 | 52 | 46 | 55 | 34 | 23 | 22 | 24 |
| % long‐lived | 6.1% | 11.7% | 43.9% | 6.7% | 6.7% | 9.1% | 46.7% | 9.1% |
| Odds Ratiob | 0.49 | 10.79 | 1.01 | 15.78 | ||||
| P‐value (Fisher's Exact Test)b | 0.360 | 4.70E‐07 | 1.000 | 1.06E‐03 | ||||
HCO, high‐confidence ortholog.
Sequence‐verified RNAi available.
Enrichment for long‐lived RNAi in CHARGE vs. Random gene sets.
The charge gene set is enriched for determinants of lifespan at 25 °C
The efficacy of many longevity interventions is strongly dependent on environmental temperature (Lakowski & Hekimi, 1996; Gems et al., 1998; Lee & Kenyon, 2009; Leiser et al., 2011; Sutphin et al., 2012; Horikawa et al., 2015; Zhang et al., 2015). To capture genes that influence longevity across the temperature spectrum, we employed a two‐temperature screening strategy, measuring lifespan of wild‐type (N2) worms subjected to RNAi targeting each gene at 15 and 25 °C. We initially measured lifespan for ~105 worms per gene at each temperature and validated each RNAi that significantly extended lifespan relative to experiment‐matched empty vector (EV) RNAi through at least two additional rounds of lifespan measurement.
The CHARGE gene set was significantly enriched for RNAi capable of increasing lifespan relative to the Random gene set at 25 °C (P < 4.70 × 10−7), but not at 15 °C (P = 0.360) (Table 1). Of the 82 CHARGE genes, RNAi knockdown of 36 (43.9%) increased lifespan at 25 °C (Figs 1 and S2; Table S4, Supporting information), while RNAi knockdown of only five (6.1%) increased lifespan at 15 °C (Figs S1 and S2; Table S4, Supporting information). In contrast, RNAi knockdown of only four (6.7%) or seven (11.7%) of the 59 Random genes increased lifespan at 25 or 15 °C, respectively (Fig. S3, Supporting information; Tables 1 and S5, Supporting information). Of the 50 genes identified in either the CHARGE or Random gene sets, only 4 – atl‐1 (Suetomi et al., 2013), atm‐1 (Ventura et al., 2009), daf‐21 (Horikawa et al., 2015), and tag‐322 (Bell et al., 2009) – were previously reported to affect lifespan in C. elegans, while the remaining 46 are novel.
Figure 1.

RNAi knockdown of 36 of 82 genes in CHARGE gene set increases lifespan at 25 °C. Bars indicate percent change in mean lifespan for RNAi targeting CHARGE genes relative to experiment‐matched EV(RNAi) when experiments were pooled. Error bars are standard error. Blue bars indicate RNAi clones that increased lifespan by our significance criteria. For each Caenorhabditis elegans gene, the table to the left indicates the corresponding human ortholog, the ortholog confidence category (high‐confidence ortholog, HCO; related protein, RP), the life stage at which RNAi was started, and the direction of change in expression of each human gene with age in specified tissues [lymphoblastoid cell line, LCL; peripheral blood mononuclear cell, PBMC; assembled from Peters et al. (2015)].
To examine the effect of gene set on lifespan, we built a linear mixed model with target gene and experiment as nested random effects. Worms subjected to RNAi targeting CHARGE genes were significantly longer‐lived than worms subjected to RNAi targeting Random genes at 25 °C (β = 0.045, P < 0.0001), but not at 15 °C (β = −0.001, P = 0.927) (Fig. 2A,B; Table S6, Supporting information). The degree of lifespan extension at 15 °C vs. 25 °C correlated significantly for Random genes (R 2 = 0.161, P < 0.005), but not for CHARGE genes (R 2 = 0.016, P = 0.261) (Fig. 2C). Combined, these data show that our candidate selection strategy specifically enriched for genes that influence long‐term survival of worms at high temperature.
Figure 2.

CHARGE gene set is enriched for longevity determinants at 25 °C. Worms fed RNAi targeting CHARGE genes lived significantly longer than worms fed RNAi targeting Random genes at 25 °C (P < 0.0001) (A), but not at 15 °C (P = 0.927, linear mixed‐effects model) (B). Lines represent mean percent lifespan extension for candidate RNAi relative to experiment‐matched EV(RNAi). RNAi are rank‐ordered by mean lifespan extension (left to right). (C) Lifespan extension at 15 and 25 °C is significantly correlated for Random, but not CHARGE genes (R = Pearson correlation coefficient).
Knocking down genes with the greatest lifespan extension increases healthspan without impairing reproduction
To extend the most significant outcomes from our screen, we further characterized the genes with the greatest impact on lifespan. Knockdown of five genes increased lifespan by >20% at 25 °C (Figs 1 and 3A; Table S4, Supporting information) while maintaining or extending lifespan at 15 °C (Figs 3B and S1; Table S5, Supporting information): kynu‐1 (previously flu‐2) encodes the kynurenine pathway enzyme kynureninase (KYNU), iglr‐1 encodes a neuronal leucine‐rich repeat (NLRR) protein, tsp‐3 encodes a tetraspanin, rcan‐1 encodes a regulator of calcineurin, and unc‐36 encodes a voltage‐gated calcium channel subunit. We selected these genes for further analysis.
Figure 3.
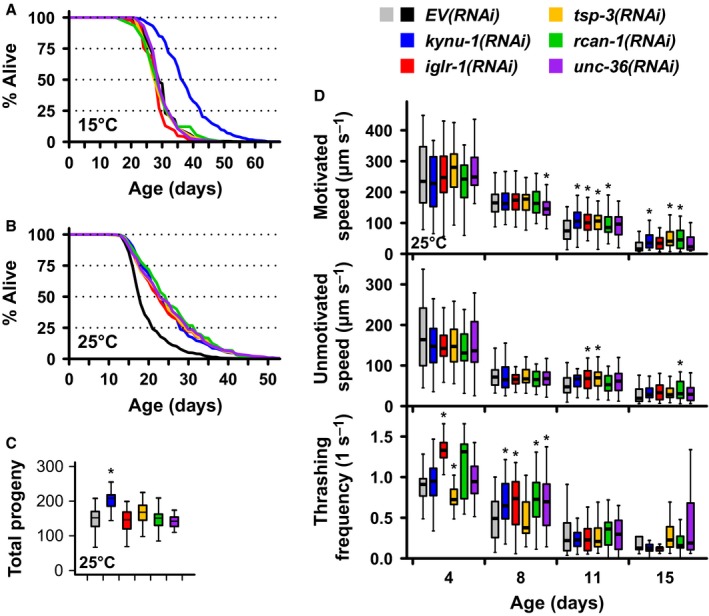
RNAi knockdown of selected genes differentially affects lifespan, reproduction, and healthspan. (A) RNAi knockdown of kynu‐1, iglr‐1, tsp‐3, rcan‐1, or unc‐36 extends lifespan at 25 °C. kynu‐1(RNAi) alone (B) extends lifespan at 15 °C and (C) increases total brood size at 25 °C. (D) RNAi knockdown of kynu‐1, iglr‐1, tsp‐3, or rcan‐1, but not unc‐36 delays decline in motivated speed with age (top), while unmotivated speed is largely unaffected (middle). RNAi knockdown of kynu‐1, iglr‐1, rcan‐1, or unc‐36, but not tsp‐3, increases thrashing rate in liquid early in life (bottom). *P < 0.05 vs. age‐matched EV(RNAi). For box and whisker plots, center line indicates median, boxes indicate 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles.
To validate the lifespan extension observed in the RNAi screen, we obtained null mutants for each gene. Each mutation extended lifespan to a similar degree as the corresponding RNAi (Fig. S4A–E; Table S7, Supporting information), with the exception of unc‐36. unc‐36 deletion impaired development, resulting in small adult body size and short lifespan (Fig. S4F; Table S7, Supporting information). Maintaining wild‐type worms on unc‐36(RNAi) for four generations prior to measuring lifespan partially replicated the unc‐36 mutant lifespan (Fig. S4G; Table S7, Supporting information), suggesting that functional UNC‐36 is required maternally or during early life (before larval stage 1) for normal development. kynu‐1 deletion resulted in a high‐penetrant temperature‐dependent developmental arrest. At 15 °C, kynu‐1 null worms that reached adulthood were small and long‐lived relative to wild‐type, but not as long‐lived as wild‐type worms subjected to kynu‐1(RNAi) from egg (Fig. S4B; Table S7, Supporting information). Maintaining wild‐type worms on kynu‐1(RNAi) for four generations replicated the lifespan phenotype of the mutant strain (Fig. S4H; Table S7, Supporting information) with a less penetrant developmental arrest, suggesting that KYNU‐1 is required for normal development at 15 °C. This is consistent with past studies that find enrichment for lifespan extension in response to RNAi knockdown of genes required for development (Chen et al., 2007; Curran & Ruvkun, 2007).
We next measured phenotypes commonly associated with increased lifespan. Antagonistic pleiotropy predicts that genes inferring increased late‐life mortality may be selected if they provide early‐life benefits. Accordingly, many C. elegans longevity mutants have defects in reproduction. Surprisingly, none of the candidate RNAi reduced the number of progeny produced, and kynu‐1(RNAi) even increased total brood size at 25 °C (Figs 3C and S4A; Table S8, Supporting information). This result does not discount antagonistic pleiotropy, as reproductive ability contributes to, but is not identical with, overall fitness.
Extending lifespan without delaying functional decline is undesirable. We measured the impact of each candidate RNAi on three healthspan phenotypes: (i) motivated movement on solid media (average speed following a plate tap), (ii) unmotivated movement on solid media (average speed without stimulation), and (iii) thrashing frequency in liquid. Each candidate RNAi significantly delayed the decline in motivated speed, thrashing frequency, or both, while unmotivated speed was largely unaffected (Fig. 3D; Table S9, Supporting information). Knocking down each gene thus allows worms to retain the ability to move later in life without affecting behavior.
Of the five candidate human/worm gene pairs, KYNU/kynu‐1, LRRN3/iglr‐1, RCAN3/rcan‐1, and CACNA2D2/unc‐36 have been implicated in neurological function and/or neurodegeneration. In C. elegans, neurodegenerative disease is modeled by expressing toxic aggregate‐prone peptides in body wall muscle, resulting in age‐dependent paralysis and aggregate formation. We measured age‐dependent paralysis in strains expressing amyloid‐beta (Aβ; modeling Alzheimer's disease) or a 35‐unit polyglutamine repeat (Q35; modeling Huntington's disease) at 25 °C. None of the candidate RNAi delayed paralysis in either model, except kynu‐1(RNAi), which slightly delayed paralysis in Aβ worms (Fig. S6A; Table S10, Supporting information).
Each candidate interacts with distinct combinations of established aging pathways
We next applied RNAi targeting our top candidate genes to mutant worms representing five established aging pathways to look for genetic interaction. For insulin signaling and the hypoxic response, we used mutant worms lacking the mediating transcription factors DAF‐16 and HIF‐1, respectively; for dietary restriction (DR), we used eat‐2 mutants, a genetic model of DR with reduced food intake; for mTOR signaling, we used worms lacking RSKS‐1, the ribosomal protein S6 kinase (S6K); and for sirtuins, we used worms lacking the C. elegans sirtuin sir‐2.1. Specifically, we asked whether each candidate RNAi was capable of extending lifespan in each aging mutant.
Each candidate gene displayed a distinct pattern of interaction with the aging pathway genes (Figs 4A and S7; Table S11, Supporting information). Lifespan extension from tsp‐3(RNAi) was completely prevented in daf‐16 mutants and slightly reversed in hif‐1 mutants, suggesting that tsp‐3 may influence lifespan, at least in part, by activating the hypoxic response (Fig. 4B). Both rcan‐1(RNAi) (Fig. 4C) and unc‐36(RNAi) (Fig. 4D) failed to extend lifespan of eat‐2 mutant worms, suggesting that both genes may extend lifespan in a manner similar to DR. Supporting this model – mTOR signaling is thought to mediate the beneficial effects of DR on lifespan – knockout of rsks‐1 partially repressed lifespan extension from rcan‐1(RNAi) and prevented lifespan extension from unc‐36(RNAi) (Fig. 4C,D).
Figure 4.

Each candidate aging gene interacts with known aging pathways in a distinct manner. (A) Summary of lifespan interaction between kynu‐1, iglr‐1, tsp‐3, rcan‐1, or unc‐36 and aging pathway mutants: insulin signaling (daf‐16), hypoxic response (hif‐1), DR (eat‐2), mTOR signaling (rsks‐1), and sirtuins (sir‐2.1). Each point represents the mean lifespan for an RNAi (x‐axis) applied to a specific strain (line color) for pooled data. Error bars are standard error. *P < 0.05 vs. EV(RNAi) (Wilcoxon rank sum test with Bonferroni multiple test correction). (B) Survival curves for tsp‐3(RNAi) in hif‐1 (left) or daf‐16 (right) mutants. (C) Survival curves for rcan‐1(RNAi) in eat‐2 (top) or rsks‐1 (bottom) mutants. (D) Survival curves for unc‐36(RNAi) in eat‐2 (top) or rsks‐1 (bottom) mutants.
iglr‐1(RNAi) failed to extend lifespan in any mutant background except sir‐2.1. In most cases, the lifespan of the aging pathway mutant fed iglr‐1(RNAi) was similar to or longer than wild‐type worms fed iglr‐1(RNAi) (Fig. 4A). This type of nonadditive interaction between lifespan‐extending interventions is difficult to interpret. The exception was the daf‐16 mutant, which had a short lifespan that was not extended by iglr‐1(RNAi), suggesting that iglr‐1 knockdown requires DAF‐16 to extend lifespan.
Among the candidates examined, knockdown of kynu‐1 alone significantly extended lifespan in all aging mutants (Fig. 4A). van der Goot et al. (2012) reported that RNAi knockdown of tdo‐2, encoding the upstream kynurenine pathway enzyme tryptophan (TRP) 2,3‐dioxygenase (TDO), increased lifespan and delayed neurodegenerative pathology in C. elegans. In contrast to kynu‐1(RNAi), lifespan extension from tdo‐2(RNAi) was largely dependent on daf‐16 and independent of eat‐2. They also report a protracted reproductive period for tdo‐2(RNAi), in contrast to the slightly increased brood size that we observed for kynu‐1(RNAi) (Figs 3C and S5A, Supporting information). To confirm that the RNAi was producing the expected metabolic perturbations (Fig. S8A, Supporting information) we measured kynurenine pathway metabolites and observed the expected increase in TRP in worms subjected to tdo‐2(RNAi), and in both kynurenine (KYN) and 3‐hydroxykynurenine (3HK) in worms subjected to kynu‐1(RNAi) (Fig. S8B, Supporting information). Our experiments with kynu‐1(RNAi) were conducted at 25 °C in a wild‐type background, while van der Goot et al. (2012) examined tdo‐2(RNAi) at 20 °C in a transgenic α‐synuclein background. To determine whether experimental context was responsible for the observed differences, we examined each phenotype using identical environmental conditions (15 or 25 °C) and genetic background (wild type).
kynu‐1(RNAi) and tdo‐2(RNAi) behaved similarly in several respects, robustly extending lifespan at both 15 and 25 °C (Fig. 5A; Table S12, Supporting information), extending all three measures of healthspan at 15 and 25 °C (Fig. S9; Table S9, Supporting information), and delaying paralysis in Aβ worms at 15 and 25 °C and in Q35 worms at 15 °C alone (Figs 5B and S6B; Table S10, Supporting information). Neither gene affected the number of aggregates that accumulated with age, but tdo‐2(RNAi) reduced total aggregate volume at all ages, while kynu‐1(RNAi) reduced aggregate volume only in middle‐aged worms (Fig. 5C). Starker differences became apparent when we examined reproduction and genetic interaction with established aging pathways. We confirmed the protracted reproductive period reported by van der Goot et al. (2012) at both temperatures in response to tdo‐2(RNAi) (Fig. S5B,C, Supporting information) and observed a reduction in total brood size at 15 °C (Fig. 5D; Table S8, Supporting information). In contrast, kynu‐1(RNAi) increased brood size at 25 °C, but did not otherwise substantively alter the reproductive profile (Figs 5D and S5B,C, Supporting information). As expected from prior studies (e.g. Gems et al., 1998; Petrella, 2014), brood size was markedly lower for all worms at 25 than 15 °C (Fig. 5D). Similar to 25 °C, kynu‐1(RNAi) significantly extended lifespan in all aging mutants at 15 °C (Fig. S7B; Table S11, Supporting information), while lifespan extension from tdo‐2(RNAi) was eliminated in daf‐16 mutants at 25 °C, in eat‐2 mutants at 15 °C, and in rsks‐1 mutants at 15 °C (Fig. 5E–G; Table S11, Supporting information). As van der Goot et al. (2012) found lifespan extension from tdo‐2(RNAi) to be dependent on daf‐16 and independent of eat‐2 at 20 °C, the environmental context at 20 °C is likely more similar to 25 °C than 15 °C.
Figure 5.

RNAi knockdown of kynu‐1 and tdo‐2 produces distinct aging phenotypes. (A) kynu‐1(RNAi) or tdo‐2(RNAi) robustly extends lifespan at 15 and 25 °C and (B) delays pathology in worms expressing amyloid‐beta (Aβ; modeling Alzheimer's disease) or a 35‐unit polyglutamine repeat (Q35; modeling Huntington's disease) in body wall muscle at 15 °C. (C) Neither kynu‐1(RNAi) nor tdo‐2(RNAi) affects the number of Q35::YFP aggregates that accumulate with age (top). tdo‐2(RNAi) significantly reduces total Q35::YFP volume per worm at all ages, while kynu‐1(RNAi) only does so in middle‐aged worms. *P < 0.05 vs. EV(RNAi) (Student's t‐test). (D) tdo‐2(RNAi) significantly reduces brood size at 15 °C, while kynu‐1(RNAi) significantly increases brood size at 25 °C. *P < 0.05 vs. EV(RNAi) (Student's t‐test). kynu‐1(RNAi), but not tdo‐2(RNAi), extends lifespan of (E) daf‐16 mutants at 25 °C, (F) eat‐2 mutants at 15 °C, and (G) rsks‐1 mutants at 15 °C. For box and whisker plots, center line indicates median, boxes indicate 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles.
In summary, we were largely able to confirm the observations of van der Goot et al. (2012) and conclude that (i) the mechanisms linking tdo‐2 to aging are strongly temperature dependent, and (ii) kynu‐1(RNAi) and tdo‐2(RNAi) have broadly similar effects on aging phenotypes, but achieve these outcomes through distinct mechanisms.
Discussion
In this study, we screened 82 C. elegans orthologs of 61 human genes that are differentially expressed with age in human blood. We report significant lifespan extension in response to RNAi knockdown of 40 of these genes. The CHARGE gene set was markedly enriched for genes capable of directly altering lifespan, suggesting a high degree of evolutionary conservation in genes influencing aging. Of the 40 pro‐aging genes in the CHARGE gene set and an additional 10 identified in the Random gene set, only four have been previously reported, while the remaining 46 are novel.
The percentage of candidate genes that directly affected lifespan at 25 °C (43.9%) was substantially higher than RNAi screens in C. elegans without a preselection criteria (<2%) (Lee et al., 2003b; Hamilton et al., 2005; Hansen et al., 2005; Samuelson et al., 2007) and within the range for screens that preselected candidates based on secondary C. elegans phenotypes, including developmental arrest (2–42%) (Chen et al., 2007; Curran & Ruvkun, 2007), reproductive senescence (16%) (Wang et al., 2014), thermal stress resistance (78%) (Munoz & Riddle, 2003), oxidative stress resistance (13–67%) (de Castro et al., 2004; Kim & Sun, 2007), and activation of the mitochondrial unfolded‐protein response (53%) (Bennett et al., 2014) (reviewed by Yanos et al., 2012 and Sutphin & Korstanje, 2016). Tacutu et al. (2012) found that knockdown of 27% of C. elegans orthologs of genes found in human or worm longevity protein–protein interaction networks increased lifespan. While differences in strain, environment, and criteria for identifying long‐lived candidates make comparison between screens necessarily qualitative, it is remarkable that preselection of C. elegans orthologs of human genes differentially expressed with age resulted in a similar enrichment for longevity determinants as preselection based on secondary phenotypes known to correlate with C. elegans longevity. Our results validate the use of C. elegans as a platform to directly screen human candidate aging genes identified through the application of ‘‐omics’ technologies and rapidly prioritize candidates for further study.
Environmental temperature is a key regulator of longevity in C. elegans
Temperature is consistently identified as a critical environmental factor in C. elegans aging. Previous work has detailed the role of temperature in specific aging interventions, such as reduced insulin signaling (Gems et al., 1998), reduced hypoxic signaling (Leiser et al., 2011), caffeine supplementation (Sutphin et al., 2012), or disruption of physiological ‘clock’ genes (Lakowski & Hekimi, 1996), among others. Lee & Kenyon (2009) and Horikawa et al. (2015) identified the steroid receptor, daf‐12, and p23 cochaperone, daf‐41, as mediators of the influence neurosensory machinery on temperature‐specific longevity. Zhang et al. (2015) defined the critical life stages during which temperature influences longevity and identified a thermosensitive TRP channel as a mediator of this influence. These studies combined with our observed lack of correlation between lifespan extension at 15 and 25 °C in the CHARGE gene set (Fig. 2C) suggest that different sets of processes drive aging in each environmental context. For instance, we found tdo‐2(RNAi) lifespan extension to require daf‐16, but not rsks‐1, at 25 °C, and require rsks‐1, but not daf‐16, at 15 °C (Fig. 5E,G). Caenorhabditis elegans aging studies at 15 °C are relatively rare compared to studies at 20 and 25 °C, and we anticipate that future studies at 15 °C will uncover novel molecular processes capable of influencing aging.
The observed temperature‐specific enrichment for aging genes suggests that the environmental context experienced by aging human blood cells is more analogous to the physiological state of C. elegans at 25 than 15 °C. Similar screens using age‐associated expression data from other tissues will provide insight into whether this temperature specificity is a general or tissue‐specific phenomenon. Alternatively, 25 °C may present a lower hurdle to lifespan extension, with a nearly twofold lower mean lifespan for wild‐type worms than at 15 °C. However, our observation that a similar fraction of RNAi are capable of increasing lifespan at 15 and 25 °C in the Random gene set suggests that this is not the case. A growing body of work has begun to define the genetic landscape underlying the response of aging worms to environmental temperature. To what extent gene–environment interactions can be attributed to specific mechanisms, such as differences in metabolic rate or exposure to environmental stressors, will require further study.
Interaction with established aging pathways prioritizes candidates
In further examining the five genes with the greatest impact of lifespan, we found distinct interactions between each candidate gene and established aging pathways. While interaction data alone are insufficient to definitively identify molecular mechanisms, examining these interactions in the context of published work helps determine which genes are of interest for detailed follow‐up studies:
TSP‐3 and the hypoxic response
TSP‐3 is a C. elegans tetraspanin, a family of transmembrane scaffolding proteins involved in cell adhesion, motility, activation, and proliferation. We observed that tsp‐3(RNAi) lifespan extension is prevented by deletion of either hif‐1 or daf‐16 (Fig. 4B). DAF‐16 is also required for lifespan extension resulting from deletion of hif‐1 at 25 °C (Leiser et al., 2011), suggesting that knockdown of tsp‐3 may extend lifespan by HIF‐1‐mediated activation of DAF‐16. Expression of human TSPAN13 is downregulated both with age (Peters et al., 2015) and in breast cancers (Huang et al., 2005). Human HIF‐1α and HIF‐2α are upregulated in many cancers, promoting a procancer transcriptional program (Semenza, 2010). Age‐dependent loss of TSPAN13 may contribute to cancer risk by elevating HIF‐1‐mediated transcription. Tetraspanins and their link to hypoxic signaling will be an interesting topic for future study in the context of longevity and cancer.
RCAN‐1 as a context‐dependent regulator of calcineurin (RCN)
RCAN‐1 regulates calcineurin, a calcium‐ and calmodulin‐dependent serine/threonine phosphatase that modulates cellular growth, survival, and inflammation. We find that lifespan extension from rcan‐1(RNAi) was eliminated in eat‐2 mutants and suppressed in rsks‐1 mutants (Fig. 4C), suggesting that rcan‐1(RNAi) increases lifespan by inhibiting mTOR signaling in a manner similar to DR. In support of this model, increased autophagy resulting from calcineurin inhibition in rat cardiomyocytes is mediated by mTOR through decreased S6K activity (He et al., 2014). RCAN‐1 is typically studied as an inhibitor of calcineurin (Lee et al., 2003a), implying that increased calcineurin activity should promote longevity. However, reduced calcineurin activity is reported to increase worm lifespan, mediated by increased autophagy (Dwivedi et al., 2009). Studies reporting calcineurin inhibition by RCNs have often been conducted in the context of RCN overexpression, while in the normal range of physiological expression human and yeast RCN can either activate or inhibit calcineurin, depending on context (Hilioti et al., 2004). This suggests that C. elegans at 25 °C may be in a physiological state where RCAN‐1 activates calcineurin, and rcan‐1(RNAi) knockdown extends lifespan by reducing activity through calcineurin and mTOR. Clarifying the complex regulatory relationship between rcan‐1 and calcineurin may open the way to new interventions designed to regulate autophagy by modulating RCN activity in specific physiological contexts.
Kynurenine metabolism offers multiple, distinct aging intervention targets
Among the five genes examined, only kynu‐1(RNAi) extended lifespan at both 15 and 25 °C, increased lifespan in all aging pathway mutants, and delayed pathology in the Alzheimer's and Huntington's disease models. Inhibition of kynu‐1 influenced aging in a manner distinct from inhibition of the previously studied kynurenine pathway gene, tdo‐2. Specifically, RNAi targeting each gene resulted in distinct reproductive patterns, different effects on age‐dependent Q35 aggregation, and distinct interaction with established aging pathways in the context of lifespan.
The structure of the kynurenine pathway suggests that the observed phenotypic differences are mediated through differential influence on kynurenine metabolites. The kynurenine pathway is the primary metabolic destination for TRP. TDO catalyzes the initial enzymatic step, resulting in the conversion of TRP to KYN. KYN is then metabolized through two downstream branches: The NAD branch converts KYN to nicotinamide adenine dinucleotide (NAD) through a series of intermediate enzymatic steps, while the KYNA branch converts KYN to kynurenic acid (KYNA), catalyzed by kynurenine aminotransferases. While tdo‐2 mediates the initial entry of TRP into the pathway, KYNU/kynu‐1 falls downstream of KYN in the NAD branch. Knockdown of tdo‐2 is thought to influence C. elegans aging by increasing TRP concentrations (van der Goot et al., 2012). Similar to tdo‐2(RNAi), adding TRP to the culture media activates DAF‐16 reporters and extends lifespan of wild‐type worms in a manner that is nonadditive with eat‐2 mutation (Edwards et al., 2015).
Worms lacking kynu‐1 do have slightly increased TRP, but substantially less than animals fed tdo‐2(RNAi) (van der Goot et al., 2012), indicating that kynu‐1 aging phenotypes are likely TRP independent. Instead, inhibition of kynu‐1 increases KYN levels and may influence aging by increasing activity through the KYNA branch of the pathway. KYNA inhibits NMDA and α7 nicotinic acetylcholine receptors and can act as an antioxidant, providing potential mechanisms of action. Interventions that increase physiological KYNA levels attenuate neurodegeneration in mouse and fly models of Huntington's disease (Schwarcz et al., 2012). Alternatively, the influence of kynu‐1 on aging may be mediated by increased concentration of 3HK, the metabolite immediately upstream of kynu‐1 in the kynurenine pathway (van der Goot et al., 2012). 3HK has complex pro‐ and antioxidant properties and can also be converted to xanthurenic acid (XA) by KAT enzymes. XA is thought to impair the production, release, and activity of insulin (Oxenkrug, 2013). Increased XA production may explain a partial repression of kynu‐1(RNAi) lifespan extension by deletion of daf‐16 that we observed at 25 °C (Fig. 5E).
Altered kynurenine pathway metabolism is linked to many human diseases of aging, including neurodegeneration (Schwarcz et al., 2012), chronic inflammation (Oxenkrug, 2011), and diabetes (Oxenkrug, 2013). Kynurenine enzymes are being pursued as clinical targets for neurodegenerative and inflammatory disease, with at least two studies reporting that pharmacological inhibitors improve cognition (Pocivavsek et al., 2011; Zwilling et al., 2011). Mechanistic differences between branches of the kynurenine pathway are directly relevant to age‐associated disease in humans, where the dominant pathway branch varies by cell and tissue type (Schwarcz et al., 2012). While kynurenine metabolism is an active area of interest for many types of age‐associated disease, KYNU/kynu‐1 is one of the least studied targets. Our work highlights KYNU as an intriguing target with the potential to produce distinct clinical outcomes compared to more commonly studied kynurenine pathway enzymes.
iglr‐1 interacts with multiple aging pathways
The highly conserved NLRR proteins are involved in organizing neural connectivity, particularly during development. The human gene that led to the selection of iglr‐1, LRRN3, is consistently found to be among the most downregulated with age in human blood (Hong et al., 2008; Harries et al., 2011; Marttila et al., 2013). Lifespan extension from iglr‐1(RNAi) was eliminated in each aging mutant examined except sir‐2.1 (Fig. 4A), indicating a complex interaction with multiple aging pathways. The consistent age‐associated downregulation in humans makes LRRN3/iglr‐1 an interesting candidate, but additional characterization is needed to identify a specific direction for mechanistic follow‐up studies.
Lifespan extension via unc‐36 inhibition is worm specific
UNC‐36 is a voltage‐gated calcium channel involved in muscle calcium signaling. Similar to rcan‐1, RNAi knockdown of unc‐36 failed to extend lifespan in either eat‐2 or rsks‐1 worms (Fig. 4D). The link between unc‐36 and the DR/mTOR pathway is straightforward. Worms with loss‐of‐function mutations in unc‐36 are known to have reduced pharyngeal pumping, limiting food intake (Avery, 1993) and resulting in a genetic mimetic of DR similar to eat‐2 mutation. Mimicking DR via reduced pharyngeal pumping is clearly a worm‐specific phenomenon, placing unc‐36 at a low priority for further study.
Conclusions and future directions
This study highlights the strength of C. elegans as a comparative tool for prioritizing human candidate aging genes and confirms age‐associated gene expression as a rich source for novel genes that play a causative role in aging. The temperature‐dependent enrichment for longevity determinants and the temperature‐specific interactions observed for specific candidate genes emphasize the importance of environmental context in mechanistic studies of C. elegans aging. Our results identify many novel aging genes and specifically promote kynu‐1, tsp‐3, and rcan‐1 as priority candidates for detailed mechanistic follow‐up.
Experimental procedures
Worm culture
We maintained worms on solid nematode growth media (NGM) seeded with UV‐killed Escherichia coli (OP50) bacteria at 20 °C as described (Sutphin & Kaeberlein, 2009). We conducted RNAi feeding, lifespan, healthspan, brood size, paralysis, and aggregate quantification assays according to standard protocols.
Candidate gene selection
We constructed candidate gene sets by selecting the 125 genes with the lowest P‐values in the whole blood expression meta‐analysis from the CHARGE study (Peters et al., 2015) (CHARGE gene set) or by randomly selecting genes from the complete Ensembl human genome (v77, www.ensembl.org) (Random gene set). We used the WORMHOLE ortholog prediction tool (wormhole.jax.org; Sutphin et al. (2016)) to identify C. elegans orthologs for each human candidate and obtained RNAi clones from the Ahringer or Vidal C. elegans RNAi feeding libraries (Table S13, Supporting information).
Lifespan screen
We conducted lifespan screening in two tiers, first measuring the effect of each candidate RNAi on lifespan at 15 and 25 °C. Any RNAi that significantly extended lifespan relative to experiment‐matched EV(RNAi) was validated in at least two additional rounds of lifespan measurement.
Appendix S1: Experimental Procedures provides additional detail for all methods.
Funding
This work was supported by the National Institute on Aging grant AG038070 and the National Cancer Institute Core grant CA034196 to The Jackson Laboratory. MJP and JBJM were supported by the Netherlands Organization for Scientific Research (NWO) VIDI grant 917103521. ADJ purchased worm screening libraries and was supported by National Heart, Lung, and Blood Institute Intramural Funds.
Author contributions
GS, JMM, ADJ, and RK conceived of the study and planned experiments. GS, GB, SS, SB, CC, and TL carried out experiments. All authors analyzed and interpreted data. GS prepared the manuscript. GS, GB, SS, MJP, JBJM, JMM, ADJ, and RK contributed to writing and critically reviewed the manuscript.
Conflict of interest
None declared.
Supporting information
Fig. S1 RNAi knockdown of 5 out of 82 genes in CHARGE gene set increases lifespan at 15 °C.
Fig. S2 Lifespan screen survival curves for the CHARGE gene set.
Fig. S3 Lifespan screen survival curves for the Random gene set.
Fig. S4 Knockout mutants validate RNAi lifespan phenotypes.
Fig. S5 Number of eggs produced per day.
Fig. S6 Impact of candidate RNAi on pathology in C. elegans Alzheimer's (Aβ) and Huntington's (Q35) disease models.
Fig. S7 Lifespan genetic interaction survival curves.
Fig. S8 RNAi targeting kynurenine pathway genes produces expected metabolic response.
Fig. S9 Kynurenine pathway inhibition extends healthspan.
Table S1 125 human genes most significantly differentially expressed in CHARGE aging study.
Table S2 C. elegans orthologs predicted based on CHARGE human candidate genes.
Table S3 C. elegans orthologs predicted based on randomly selected human genes.
Table S4 Summary statistics for C. elegans lifespan screen (CHARGE gene set).
Table S5 Summary statistics for C. elegans lifespan screen (Random gene set).
Table S6 Summary of linear mixed models examining the effect of gene set on lifespan when target genes are knocked down using RNAi in C. elegans.
Table S7 Summary statistics for null mutant lifespan experiments.
Table S8 Summary statistics for brood size experiments.
Table S9 Summary statistics for healthspan experiments.
Table S10 Summary statistics for paralysis experiments in C. elegans Alzheimer's (Aβ) and Huntington's (Q35) disease models.
Table S11 Summary statistics for lifespan genetic interaction experiments.
Table S12 Summary statistics for kynurenine pathway lifespan experiments.
Table S13 C. elegans RNAi feeding library clones used in this study.
Appendix S1 Experimental procedures.
Acknowledgments
We thank Dr. Aric Rogers, Dr. Jarod Rollins, and Santina Snow at the Mount Desert Island Biological Laboratory (MDIBL) for helpful discussion. We further thank Dr. Rogers for use of the WormLab system. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services.
Contributor Information
George L. Sutphin, Email: sutphin@gmail.com.
Ron Korstanje, Email: ron.korstanje@jax.org.
References
- Avery L (1993) The genetics of feeding in Caenorhabditis elegans . Genetics 133, 897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell R, Hubbard A, Chettier R, Chen D, Miller JP, Kapahi P, Tarnopolsky M, Sahasrabuhde S, Melov S, Hughes RE (2009) A human protein interaction network shows conservation of aging processes between human and invertebrate species. PLoS Genet. 5, e1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF, Vander Wende H, Simko M, Klum S, Barfield S, Choi H, Pineda VV, Kaeberlein M (2014) Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans . Nat. Commun. 5, 3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E, Hegi de Castro S, Johnson TE (2004) Isolation of long‐lived mutants in Caenorhabditis elegans using selection for resistance to juglone. Free Radic. Biol. Med. 37, 139–145. [DOI] [PubMed] [Google Scholar]
- Chen D, Pan KZ, Palter JE, Kapahi P (2007) Longevity determined by developmental arrest genes in Caenorhabditis elegans . Aging Cell 6, 525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G (2007) Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 3, e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi M, Song HO, Ahnn J (2009) Autophagy genes mediate the effect of calcineurin on life span in Caenorhabditis elegans . Autophagy 5, 604–607. [DOI] [PubMed] [Google Scholar]
- Edwards C, Canfield J, Copes N, Brito A, Rehan M, Lipps D, Brunquell J, Westerheide SD, Bradshaw PC (2015) Mechanisms of amino acid‐mediated lifespan extension in Caenorhabditis elegans . BMC Genet. 16, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL (1998) Two pleiotropic classes of daf‐2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans . Genetics 150, 129–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goot AT, Zhu W, Vazquez‐Manrique RP, Seinstra RI, Dettmer K, Michels H, Farina F, Krijnen J, Melki R, Buijsman RC, Ruiz Silva M, Thijssen KL, Kema IP, Neri C, Oefner PJ, Nollen EA (2012) Delaying aging and the aging‐associated decline in protein homeostasis by inhibition of tryptophan degradation. Proc. Natl Acad. Sci. USA 109, 14912–14917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS (2005) A systematic RNAi screen for longevity genes in Caenorhabditis elegans . Genes Dev. 19, 1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C (2005) New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 1, 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries LW, Hernandez D, Henley W, Wood AR, Holly AC, Bradley‐Smith RM, Yaghootkar H, Dutta A, Murray A, Frayling TM, Guralnik JM, Bandinelli S, Singleton A, Ferrucci L, Melzer D (2011) Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell 10, 868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Liu X, Lv L, Liang H, Leng B, Zhao D, Zhang Y, Du Z, Chen X, Li S, Lu Y, Shan H (2014) Calcineurin suppresses AMPK‐dependent cytoprotective autophagy in cardiomyocytes under oxidative stress. Cell Death Dis. 5, e997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilioti Z, Gallagher DA, Low‐Nam ST, Ramaswamy P, Gajer P, Kingsbury TJ, Birchwood CJ, Levchenko A, Cunningham KW (2004) GSK‐3 kinases enhance calcineurin signaling by phosphorylation of RCNs. Genes Dev. 18, 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong MG, Myers AJ, Magnusson PK, Prince JA (2008) Transcriptome‐wide assessment of human brain and lymphocyte senescence. PLoS ONE 3, e3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa M, Sural S, Hsu AL, Antebi A (2015) Co‐chaperone p23 regulates Caenorhabditis elegans lifespan in response to temperature. PLoS Genet. 11, e1005023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Groth J, Sossey‐Alaoui K, Hawthorn L, Beall S, Geradts J (2005) Aberrant expression of novel and previously described cell membrane markers in human breast cancer cell lines and tumors. Clin. Cancer Res. 11, 4357–4364. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Pennypacker JK (2015) Aging interventions get human. Oncotarget 6, 590–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sun H (2007) Functional genomic approach to identify novel genes involved in the regulation of oxidative stress resistance and animal lifespan. Aging Cell 6, 489–503. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S (1996) Determination of life‐span in Caenorhabditis elegans by four clock genes. Science 272, 1010–1013. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Kenyon C (2009) Regulation of the longevity response to temperature by thermosensory neurons in Caenorhabditis elegans . Curr. Biol. 19, 715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JI, Dhakal BK, Lee J, Bandyopadhyay J, Jeong SY, Eom SH, Kim DH, Ahnn J (2003a) The Caenorhabditis elegans homologue of Down syndrome critical region 1, RCN‐1, inhibits multiple functions of the phosphatase calcineurin. J. Mol. Biol. 328, 147–156. [DOI] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G (2003b) A systematic RNAi screen identifies a critical role for mitochondria in Caenorhabditis elegans longevity. Nat. Genet. 33, 40–48. [DOI] [PubMed] [Google Scholar]
- Leiser SF, Begun A, Kaeberlein M (2011) HIF‐1 modulates longevity and healthspan in a temperature‐dependent manner. Aging Cell 10, 318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marttila S, Jylhava J, Nevalainen T, Nykter M, Jylha M, Hervonen A, Tserel L, Peterson P, Hurme M (2013) Transcriptional analysis reveals gender‐specific changes in the aging of the human immune system. PLoS ONE 8, e66229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz MJ, Riddle DL (2003) Positive selection of Caenorhabditis elegans mutants with increased stress resistance and longevity. Genetics 163, 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenkrug GF (2011) Interferon‐gamma‐inducible kynurenines/pteridines inflammation cascade: implications for aging and aging‐associated psychiatric and medical disorders. J. Neural. Transm. 118, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenkrug G (2013) Insulin resistance and dysregulation of tryptophan‐kynurenine and kynurenine‐nicotinamide adenine dinucleotide metabolic pathways. Mol. Neurobiol. 48, 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MJ, Joehanes R, Pilling LC, Schurmann C, Conneely KN, Powell J, Reinmaa E, Sutphin GL, Zhernakova A, Schramm K, Wilson YA, Kobes S, Tukiainen T, Consortium NU, Ramos YF, Goring HH, Fornage M, Liu Y, Gharib SA, Stranger BE, De Jager PL, Aviv A, Levy D, Murabito JM, Munson PJ, Huan T, Hofman A, Uitterlinden AG, Rivadeneira F, van Rooij J, Stolk L, Broer L, Verbiest MM, Jhamai M, Arp P, Metspalu A, Tserel L, Milani L, Samani NJ, Peterson P, Kasela S, Codd V, Peters A, Ward‐Caviness CK, Herder C, Waldenberger M, Roden M, Singmann P, Zeilinger S, Illig T, Homuth G, Grabe HJ, Volzke H, Steil L, Kocher T, Murray A, Melzer D, Yaghootkar H, Bandinelli S, Moses EK, Kent JW, Curran JE, Johnson MP, Williams‐Blangero S, Westra HJ, McRae AF, Smith JA, Kardia SL, Hovatta I, Perola M, Ripatti S, Salomaa V, Henders AK, Martin NG, Smith AK, Mehta D, Binder EB, Nylocks KM, Kennedy EM, Klengel T, Ding J, Suchy‐Dicey AM, Enquobahrie DA, Brody J, Rotter JI, Chen YD, Houwing‐Duistermaat J, Kloppenburg M, Slagboom PE, Helmer Q, den Hollander W, Bean S, Raj T, Bakhshi N, Wang QP, Oyston LJ, Psaty BM, Tracy RP, Montgomery GW, Turner ST, Blangero J, Meulenbelt I, Ressler KJ, Yang J, Franke L, Kettunen J, Visscher PM, Neely GG, Korstanje R, Hanson RL, Prokisch H, Ferrucci L, Esko T, Teumer A, van Meurs JB, Johnson AD (2015) The transcriptional landscape of age in human peripheral blood. Nat. Commun. 6, 8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella LN (2014) Natural variants of Caenorhabditis elegans demonstrate defects in both sperm function and oogenesis at elevated temperatures. PLoS ONE 9, e112377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocivavsek A, Wu HQ, Potter MC, Elmer GI, Pellicciari R, Schwarcz R (2011) Fluctuations in endogenous kynurenic acid control hippocampal glutamate and memory. Neuropsychopharmacology 36, 2357–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson AV, Klimczak RR, Thompson DB, Carr CE, Ruvkun G (2007) Identification of Caenorhabditis elegans genes regulating longevity using enhanced RNAi‐sensitive strains. Cold Spring Harb. Symp. Quant. Biol. 72, 489–497. [DOI] [PubMed] [Google Scholar]
- Schwarcz R, Bruno JP, Muchowski PJ, Wu HQ (2012) Kynurenines in the mammalian brain: when physiology meets pathology. Nat. Rev. Neurosci. 13, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL (2010) Defining the role of hypoxia‐inducible factor 1 in cancer biology and therapeutics. Oncogene 29, 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetomi K, Mereiter S, Mori C, Takanami T, Higashitani A (2013) Caenorhabditis elegans ATR checkpoint kinase ATL‐1 influences life span through mitochondrial maintenance. Mitochondrion 13, 729–735. [DOI] [PubMed] [Google Scholar]
- Sutphin GL, Kaeberlein M (2009) Measuring Caenorhabditis elegans life span on solid media. J. Vis. Exp. 27, e1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutphin GL, Korstanje R (2016) Longevity as a complex genetic trait In Handbook of the Biology of Aging, 8th edn (Kaeberlein MR, Martin GM, eds). San Diego, CA: Elsevier Academic Press, pp. 3–54. [Google Scholar]
- Sutphin GL, Bishop E, Yanos ME, Moller RM, Kaeberlein M (2012) Caffeine extends life span, improves healthspan, and delays age‐associated pathology in Caenorhabditis elegans . Longev. Healthspan 1, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutphin GL, Mahoney JM, Sheppard K, Walton DO, Korstanje R (2016) WORMHOLE: novel least diverged ortholog prediction through machine learning. PLoS Comput. Biol. 12, e1005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacutu R, Shore DE, Budovsky A, de Magalhaes JP, Ruvkun G, Fraifeld VE, Curran SP (2012) Prediction of Caenorhabditis elegans longevity genes by human and worm longevity networks. PLoS ONE 7, e48282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura N, Rea SL, Schiavi A, Torgovnick A, Testi R, Johnson TE (2009) p53/CEP‐1 increases or decreases lifespan, depending on level of mitochondrial bioenergetic stress. Aging Cell 8, 380–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, Oakley HD, Carr CE, Sowa JN, Ruvkun G (2014) Gene pathways that delay Caenorhabditis elegans reproductive senescence. PLoS Genet. 10, e1004752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanos ME, Bennett CF, Kaeberlein M (2012) Genome‐wide RNAi longevity screens in Caenorhabditis elegans . Curr. Genomics 13, 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Xiao R, Ronan EA, He Y, Hsu AL, Liu J, Xu XZ (2015) Environmental temperature differentially modulates Caenorhabditis elegans longevity through a thermosensitive TRP channel. Cell Rep. 11, 1414–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwilling D, Huang SY, Sathyasaikumar KV, Notarangelo FM, Guidetti P, Wu HQ, Lee J, Truong J, Andrews‐Zwilling Y, Hsieh EW, Louie JY, Wu T, Scearce‐Levie K, Patrick C, Adame A, Giorgini F, Moussaoui S, Laue G, Rassoulpour A, Flik G, Huang Y, Muchowski JM, Masliah E, Schwarcz R, Muchowski PJ (2011) Kynurenine 3‐monooxygenase inhibition in blood ameliorates neurodegeneration. Cell 145, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 RNAi knockdown of 5 out of 82 genes in CHARGE gene set increases lifespan at 15 °C.
Fig. S2 Lifespan screen survival curves for the CHARGE gene set.
Fig. S3 Lifespan screen survival curves for the Random gene set.
Fig. S4 Knockout mutants validate RNAi lifespan phenotypes.
Fig. S5 Number of eggs produced per day.
Fig. S6 Impact of candidate RNAi on pathology in C. elegans Alzheimer's (Aβ) and Huntington's (Q35) disease models.
Fig. S7 Lifespan genetic interaction survival curves.
Fig. S8 RNAi targeting kynurenine pathway genes produces expected metabolic response.
Fig. S9 Kynurenine pathway inhibition extends healthspan.
Table S1 125 human genes most significantly differentially expressed in CHARGE aging study.
Table S2 C. elegans orthologs predicted based on CHARGE human candidate genes.
Table S3 C. elegans orthologs predicted based on randomly selected human genes.
Table S4 Summary statistics for C. elegans lifespan screen (CHARGE gene set).
Table S5 Summary statistics for C. elegans lifespan screen (Random gene set).
Table S6 Summary of linear mixed models examining the effect of gene set on lifespan when target genes are knocked down using RNAi in C. elegans.
Table S7 Summary statistics for null mutant lifespan experiments.
Table S8 Summary statistics for brood size experiments.
Table S9 Summary statistics for healthspan experiments.
Table S10 Summary statistics for paralysis experiments in C. elegans Alzheimer's (Aβ) and Huntington's (Q35) disease models.
Table S11 Summary statistics for lifespan genetic interaction experiments.
Table S12 Summary statistics for kynurenine pathway lifespan experiments.
Table S13 C. elegans RNAi feeding library clones used in this study.
Appendix S1 Experimental procedures.


