Figure 2. Intermolecular salt bridges between the N-terminus of SLN and M4S4 of SERCA.
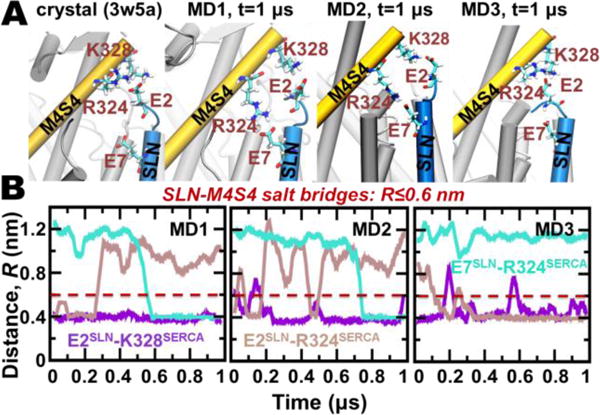
A) Salt bridges between SLN residues E2/E7 and SERCA residues R324/K328 are identified in crystal structure 3w5a6 (left) and at the end of each 1-μs MDS trajectories (right). SERCA and SLN are shown as cartoons, and residues participating in salt-bridge interactions are shown as sticks. (B). MDS distance evolution is shown between E2SLN‒K328SERCA, E2SLN‒R324SERCA, and E7SLN‒R324SERCA. Distances between E‒K and E‒R residue pairs were calculated between Cδ‒Cζ and Cδ‒Nζ atoms, respectively. The cutoff distance for salt bridge interaction using these distance definitions is 0.6 nm.30
