Abstract
Background
Molecular technologies have allowed laboratories to detect and establish the profiles of human cancers by identifying a variety of somatic variants. In order to improve personalized patient care, we have established a next-generation sequencing (NGS) test to screen for somatic variants in primary or advanced cancers. In this study, we describe the laboratory quality management program for NGS testing, and also provide an overview of the somatic variants identified in over 1000 patient samples as well as their implications in clinical practice.
Methods
Over the past one-and-a-half years, our laboratory received a total of 1028 formalin-fixed, paraffin-embedded (FFPE) tumor tissues, which consisted of non-small-cell lung carcinomas (NSCLCs), colon adenocarcinomas, glioma/glioblastomas, melanomas, breast carcinomas, and other tumor types. During this time period, we implemented a series of quality control (QC) checks that included (1) pre-DNA extraction, (2) DNA quantification, (3) DNA quality, (4) library quantification, (5) post-emulsification PCR, and (6) post-sequencing metrics. At least 10 ng of genomic DNA (gDNA) were used to prepare barcoded libraries using the AmpliSeq CHPv2. Samples were multiplexed and sequenced on Ion Torrent 318 chips using the Ion PGM System. Variants were identified using the Variant Caller Plugin, and annotation and functional predictions were performed using the Golden Helix SVS.
Results
A total of 1005 samples passed QC1–3, and following additional library preparation QC checkpoints, 877 samples were sequenced. Samples were classified into two categories: wild-type (127) and positive for somatic variants (750). Somatic variants were classified into clinically actionable (60%) and non-actionable (40%).
Conclusions
The use of NGS in routine clinical laboratory practice allowed for the detection of tumor profiles that are essential for the selection of targeted therapies and identification of applicable clinical trials, contributing to the improvement of personalized patient care in oncology.
Keywords: massively parallel sequencing, next-generation sequencing, solid tumor, somatic variant
Introduction
Somatic variant analysis of human cancers has become a critical step in the selection of appropriate therapies for patient management. Using a variety of molecular tools, clinical laboratories are able to profile human cancers for numerous clinically actionable variants. Initially, most of these analyses were performed by fluorescence in situ hybridization (FISH), polymerase chain reaction (PCR), real-time PCR, Sanger sequencing, and microarrays. For many years, Sanger sequencing and allele-specific real-time PCR represented the traditional methods used to detect somatic variants in genes with known clinical significance, such as KRAS, BRAF, and EGFR. Both technologies are limited by the types and number of variants they are able to detect. While allele-specific real-time PCR is more sensitive than Sanger sequencing, both are still more time-consuming and less cost effective when compared to massively parallel or next-generation sequencing (NGS) [1, 2].
NGS has allowed for the analysis of numerous variants and genes in multiple patient samples simultaneously. Its ability to assess multiple genetic targets concurrently (including point mutations, small INDELs, copy number alterations, and rearrangements or gene fusions) generates a mutation profile of the individual tumor that helps in selecting the most efficacious management strategy. Accurate assessment of human cancer, which correlates with best patient outcomes, needs to be performed in such a way as to provide the most clinically actionable information to the provider. Our understanding of the complexity of human cancer, including driver, passenger, sensitizing, and resistance mutations, has become the target of many novel therapies that now require profiling as a companion diagnostic.
In the past few years, the number of laboratories offering NGS testing has grown considerably because of the lower cost, high throughput, and significant impact on patient care. Since 2013, a number of guidelines have been put in place to ensure the quality of laboratory-developed NGS assays [3, 4]. However, there still exists significant room for interpretation of these requirements in individual laboratories. Our laboratory has been performing NGS as part of our clinical molecular diagnostics service for approximately 2 years. In this study, we describe our laboratory’s implementation of a quality management program for NGS testing, and also provide an overview of the somatic variants identified in over 1000 patient samples as well as their implications in clinical practice.
Materials and methods
Samples
One thousand and twenty-eight formalin-fixed, paraffin-embedded (FFPE) tumor tissues were received at the Dartmouth Hitchcock Medical Center (DHMC) for somatic mutation screening between 2013 and 2015 as part of our clinical molecular diagnostic service. FFPE samples consisted of 408 non-small-cell lung carcinomas (NSCLCs), 304 colon adenocarcinomas, 120 gliomas/glioblastomas, 95 melanomas, 27 breast carcinomas, and 74 samples consisting of other tumor types.
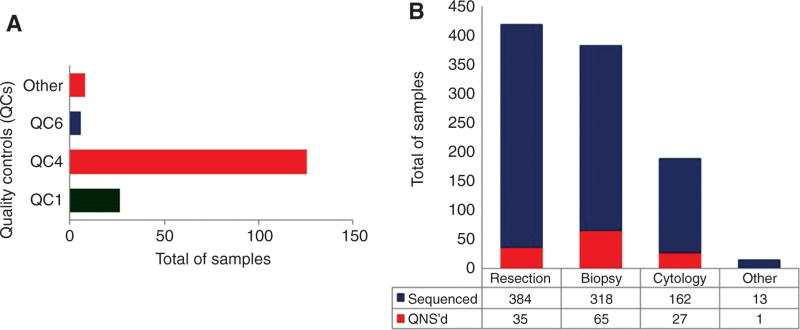
The FFPE sample types received at DHMC included resections (421), biopsies (397), cytology (195) (FNA, pleural fluid, and bronchial alveolar lavage), and others (15 consult samples). The resection samples were composed mainly of colon adenocarcinoma samples (38%) and glioma/glioblastomas (26%). Among the biopsy samples, the majority were NSCLC (41%), followed by colon adenocarcinoma (35%). Approximately 90% of the cytology samples were NSCLC (Table 1).
Table 1.
Total of sample types (resection, biopsy, and cytology) among each tumor type (NSCLC, colon adenocarcinoma, glioma/glioblastoma, melanoma, and breast carcinomas).
| Sample type | Total | ||||
|---|---|---|---|---|---|
|
|
|||||
| Resection | Biopsy | Cytology | Othersa | ||
| Tumor type | |||||
| NSCLC | 71 | 161 | 169 | 7 | 408 |
| Colon adenocarcinoma | 158 | 137 | 6 | 3 | 304 |
| Glioma/glioblastoma | 108 | 12 | 0 | 0 | 120 |
| Melanoma | 49 | 35 | 6 | 5 | 95 |
| Breast carcinoma | 9 | 14 | 4 | 0 | 27 |
| Others | 26 | 38 | 10 | 0 | 74 |
| Total | 421 | 397 | 195 | 15 | 1028 |
Consult samples.
Quality management (QM) program
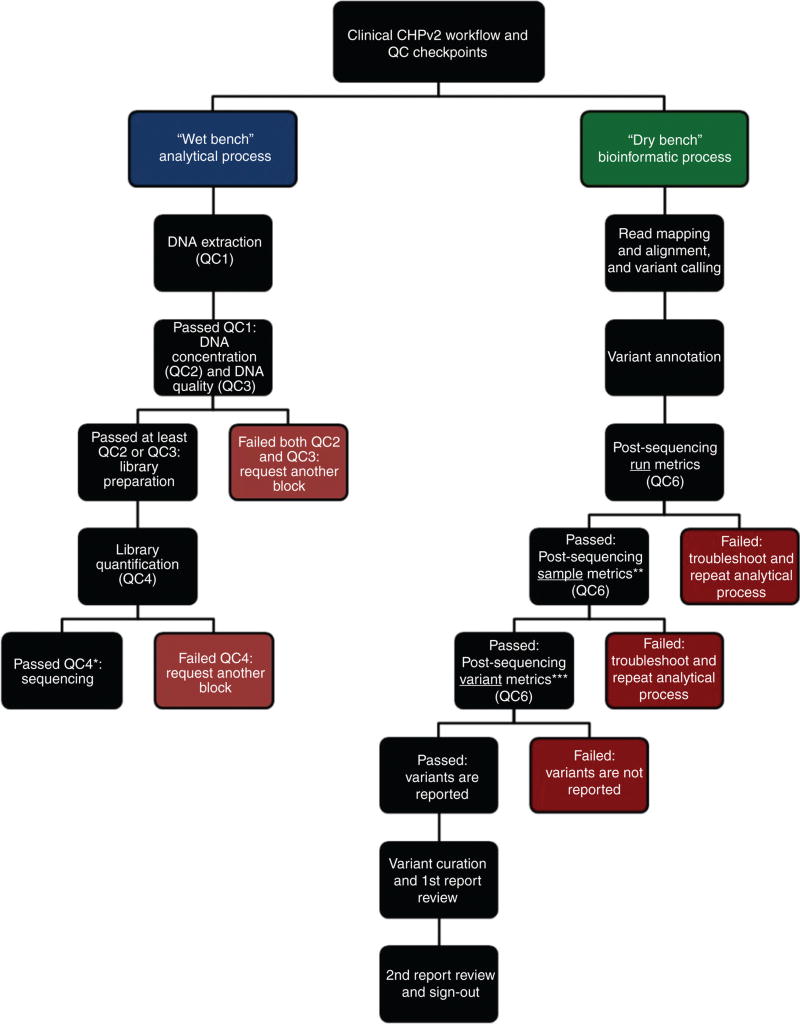
NGS can be divided into two processes: (1) the “wet bench” or analytic process and (2) the “dry bench” or bioinformatics process. The “wet bench” is composed of the following processes: DNA extraction, verification of DNA quantity and DNA quality, library preparation, library quantification, library pooling, and sequencing. The “dry bench” is composed of a number of processes designed to convert raw sequencing reads to reportable variants. These processes include read mapping and alignment, variant calling, variant annotation, variant curation, and final clinical report. These extensive wet- and dry-bench NGS processes are uniquely complex amongst traditional assays within the clinical laboratory. Due to these complexities, these assays require an exceptional level of quality control (QC) to ensure the accuracy and reproducibility of reportable results. Standard QM programs already in place within many clinical laboratories will generally require additional policies and procedures to accommodate the implementation of NGS testing.
With these considerations in mind, we have implemented a comprehensive QM program that monitors these processes on a periodic basis, including biannual CAP proficiency testing for NGS testing and for multiple genes within the panel (BRAF, KRAS, EGFR, KIT, IDH1, and IDH2 genes), confirmatory testing using a SNaPshot assay for specific tumor types or specific variants within selected genes, and biannual sample exchange with an outside laboratory, as well as performance of routine QC.
Throughout the analytical and bioinformatics workflows we have established six QCs: (QC1) pre-DNA extraction: samples must have tumor content ≥10% (limit of detection established during validation) [5] and sufficient tissue on eight unstained slides (5 µm each), (QC2) DNA concentration must be ≥1.7 ng/µL, (QC3) DNA quality: samples must have a Q129/Q41 ratio ≥0.4 using the KAPA hgDNA Quantification and QC Kit (KAPA Biosystems), (QC4) library quantification: libraries must have ≥100 pM, (QC5) post-emulsification PCR: the percent of templated ISPs (Ion Sphere™ particles) must be between 10% and 30%, and (QC6) post-sequencing metrics were established at the run, sample, and variant levels. For each run, the following sequencing metrics were verified: chip loading (>70%), usable sequences (>55%), polyclonality (<35%), and low quality reads (<20%). For each individual sample, the metrics assessed were: on-target reads (>90%), coverage uniformity (>90%), and ≥95% amplicons with 500× coverage (in order to avoid amplicon drop-outs and false negatives). And finally, for each variant, the metrics assessed were: coverage ≥500×, allelic frequency of ≥5%, and strand bias of approximately 0.40–0.59. Also, multiple alignment metrics were evaluated for overall quality, which included percent of aligned bases (>85%), percent of aligned reads (>98%), and average (1×) accuracy across each individual base position in read (>97%).
In addition to the QCs implemented, we have included a FFPE QC cell line (EGFR ΔE746-A750 50% FFPE Reference Standard) (Horizon Diagnostics) that is taken through the entire clinical NGS workflow. This QC material is used to detect deficiencies that may occur before, during, or after the analytic process. Two such issues are changes in reagent lot numbers, and instrument or software upgrades. The QC material must pass all six QCs established throughout the analytical wet-bench and bioinformatics workflows. In QC4 (library quantification), the QC material must have a quantification value >100 pM. If the QC material fails QC4, none of the samples are sequenced, and run failure documentation is generated. After troubleshooting, another library preparation must be started. In QC6 (post-sequencing run, sample, and variant metrics), the QC material must have the expected sample and variant metrics. Also, if the variants present in the QC are absent or have different allelic frequencies than expected (EGFR c.2235_2249del15, p.E746_A750del: 50%; PIK3CA c.3140A>G, p.H1047R: 50%; BRAF c.1799T>A, p.V600E: 66%), the QC has failed the QC6 variant metrics. If the QC material fails either component of the QC6 checkpoint, none of the samples are analyzed, and run failure documentation is generated. Following troubleshooting, another library preparation must be started.
DNA extraction
Hematoxylin and eosin (H&E) stained slides from each sample were reviewed by an attending pathologist who determined the tumor area and the percentage of tumor cells present in the tissue section. Samples that passed QC1 were macrodissected, and genomic DNA (gDNA) was obtained using the Gentra Puregene Kit (Qiagen, Valencia, CA, USA). Samples were quantified using the PicoGreen Quant-iT™ PicoGreen® dsDNA Assay Kit (Invitrogen, Paisley, UK). DNA quality was assessed using the KAPA hgDNA Quantification and QC Kit (KAPA Biosystems, Wilmington, MA, USA). Samples must have DNA concentrations higher than 1.7 ng/µL (QC2) and a Q129/Q41 ratio ≥0.4 (QC3). Samples that failed both QC2 and QC3 were identified as quantity not sufficient (QNS) for testing before proceeding to library preparation.
Next-generation sequencing, data analysis, and reporting
Approximately 10 ng of gDNA from each sample was used to create libraries using the Ion AmpliSeq™ Cancer Hotspot Panel v2 (CHPv2) (ThermoFisher). The CHPv2 consists of 207 amplicons covering 50 hotspot regions of oncogenes and tumor suppressor genes. Libraries were barcoded using Ion Xpress™ barcode adapters (ThermoFisher) and then combined to a final concentration of 100 pM using the Ion Library Quantitation Kit (ThermoFisher). Samples with concentrations below 100 pM failed QC4 and, for this reason, were identified as QNS. Emulsion PCR was performed using the Ion Torrent One-Touch™ 2 System. Library pools are required to obtain a template percentage between 10–30 (QC5) and values outside of that range, were repeated with adjusted library input. A maximum of 10 samples per Ion 318™ Chip v2 were sequenced using the Ion PGM™ System (ThermoFisher).
For data analysis, Torrent Suite software (v4.0.2) was used for read mapping, alignment of sequences to human genome version 19 (hg19), variant calling, and coverage analysis. Golden Helix SNP & Variation Suite (SVS) software (v8.2.1) was used for variant annotation and prediction of functional significance. Reported variants were also manually assessed using the Broad Institute’s Integrated Genomics Viewer (IGV v2.3). Runs that did not pass post-sequencing metrics (QC6) underwent a troubleshooting process and were ultimately repeated; for samples that did not pass QC6, an alternative tissue block was requested for testing, and variants that did not pass QC6 were not reported as per assay validation [5]. Finally variants that passed post-sequencing metrics were curated and reported.
For clinical reporting, variants were characterized as clinically actionable or non-actionable according to the NCCN Clinical Practice Guidelines in Oncology and My Cancer Genome: Genetically Informed Cancer Medicine. Curation of the variants was performed using the following databases: NCCN (www.nccn.org), My Cancer Genome (www.mycancergenome.org), COSMIC: Catalogue of Somatic Mutations in Cancer (cancer.sanger.ac.uk/), ClinVar National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/clinvar/), and dbSNP National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/SNP/). For the curation of rare variants, literature searches using PubMed (www.ncbi.nlm.nih.gov/pubmed) were used to verify the importance of the variants. All reports were reviewed by a laboratory computational science staff member before being reviewed and curated by a genomic analyst. Somatic mutation reports were then signed out by an attending pathologist. All identified variants for each sample were stored in a laboratory-developed clinical knowledge base along with all references and curation information.
Figure 1 outlines the routine clinical workflow for our NGS pipeline, including the pre- and post-analytical QC checkpoints.
Figure 1.
Clinical workflow for the NGS CHPv2 workflow including the pre and post-analytical QC checkpoints.
*If the quality control material fails this step, none of the samples are sequenced, and run failure documentation is generated. After troubleshooting, another library preparation must be started. **If the quality control material fails this step, none of the samples are analyzed, and run failure documentation is generated. After troubleshooting, another library preparation must be started. ***If the variants present in the quality control are absent or have different allelic frequencies than expected, none of the samples are analyzed, and run failure documentation is generated. After troubleshooting, another library preparation must be started.
Results
Sample concentration and sample quality
Twenty-three samples failed QC1, which were identified as QNS as a result of having either tumor content below the limit of detection (LOD) (10%), or insufficient tissue on eight unstained slides before DNA extraction (Figure 2A). A total of 1005 FFPE samples were used for DNA extraction. While most of the samples had DNA concentrations between 1.7 and 50 ng/µL (532), 313 samples had DNA concentrations >50 ng/µL, and 103 had DNA concentrations below 1.7 ng/µL. Approximately 82% of the samples with low DNA concentrations (<1.7 ng/µL) were the smaller biopsy and cytology samples, and approximately 75% of the samples with highest DNA concentrations (≥50 ng/µL) were resection samples.
Figure 2.
Total number of samples that failed quality controls (QCs) established throughout the CHPv2 workflow, and total number of samples that were sequenced.
(A) Represents all samples that were QNS before or after sequencing. In green are the samples that did not pass QC1, and consequently did not have their DNA extracted (they are not present in Figure 2B). In blue and red are the samples that had their DNA extracted, and library preparation was performed. Since DNA quality was recently implemented, none of the samples that failed both DNA concentration and DNA quality were QNS. However, after extensive analysis, this is part of our current workflow. (B) Represents only samples that passed QC1. In blue are the samples that passed QC4 and they were sequenced. Only two of them failed QC6. In red are the samples that failed QC4, and were QNS.
Of the 243 samples evaluated for DNA quality, 204 had a Q129/Q41 ratio score higher than 0.4, and approximately 92.6% (189) also passed QC2. Of the 39 with a Q129/Q41 ratio below 0.40, 24 passed QC2. However, 11 failed QC4. A total of 12 samples failed QC2, QC3, and QC4, respectively.
Next-generation sequencing
Library preparation was performed for 1005 samples. However, 126 (12.5%) samples failed QC4 (Figure 2A). Ninety-two (75%) were either biopsy (65) or cytology (27) samples (Figure 2B).
Among the 879 sequenced samples, there were 339 NSCLCs, 280 colon adenocarcinomas, 104 glioma/glioblastomas, 79 melanomas, 23 breast carcinomas, as well as 54 other tumor types. Only two samples did not pass QC6 (Figure 2A), which is a strong indication that the implementation of a pre-sequencing QC program is extremely effective in preventing the sequencing of poor-quality samples.
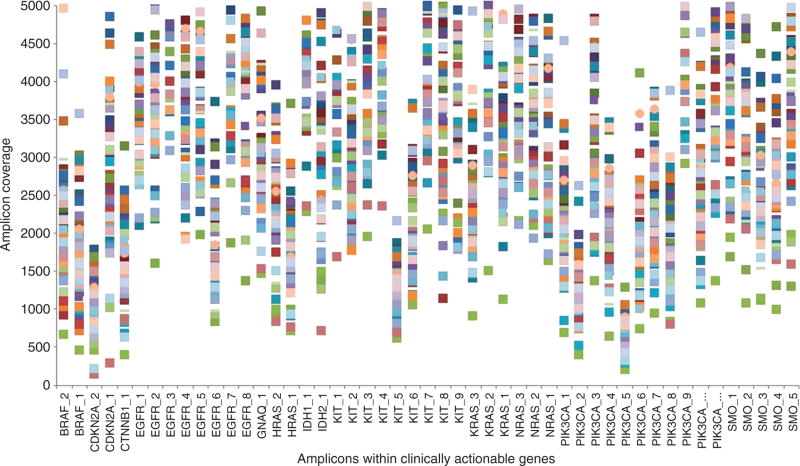
In order to show the performance of the NGS workflow resulting from the implementation of a pre-sequencing QC program, a random set of 200 samples was analyzed demonstrating that the QC6 metrics were consistently higher than the minimum established threshold. For instance, greater than 97% of the 207 panel amplicons were consistently covered at >500×. In addition, when only considering the 49 amplicons covering the 13 clinically indicated genes (excluding those indicated for breast cancer), more than 98.5% of the amplicons were consistently covered at >500× (Figure 3).
Figure 3.
Amplicon coverage within clinically actionable genes.
A total of 127 (14.5%) samples were wild-type (or negative) for the hotspot mutations present in the CHPv2 panel, and 750 (85.5%) samples were positive for variants within hotspot regions of genes in this panel. Almost 40% of the samples had one molecular variant and 60% had more than two variants. Patients that were positive for variants in clinically actionable genes (449) also showed either a single variant (24.9%) or more than one variant (75.1%) involving other genes in the panel.
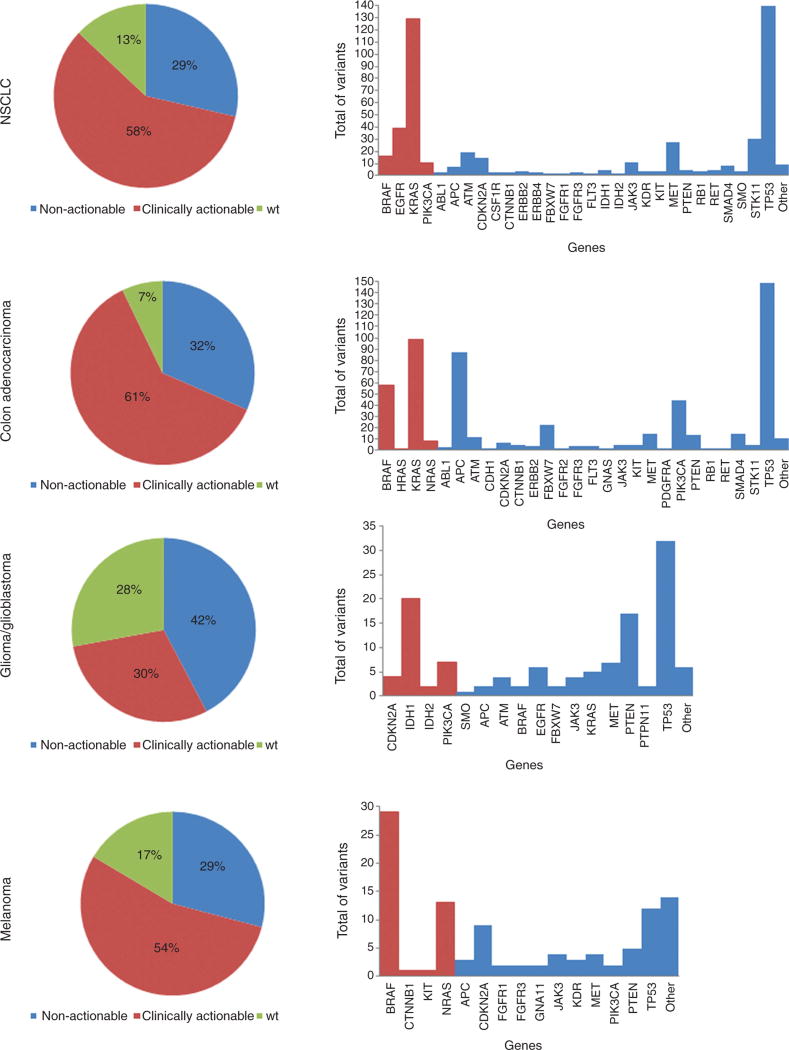
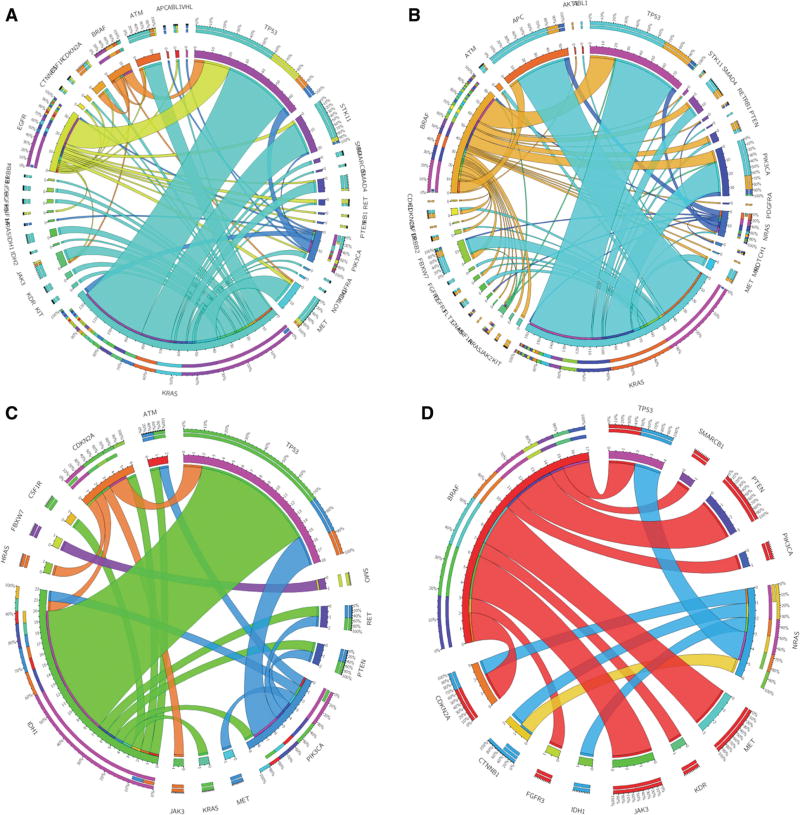
The mutation distributions identified in these patient samples are shown by individual tumor type (NSCLC, colon adenocarcinoma, glioma/glioblastoma, and melanoma) (Figure 4). The frequency of variants in clinically actionable genes with at least one variant in other genes is shown in Figure 5.
Figure 4.
Variants detected in clinically actionable and non-actionable genes in each tumor type.
The color “red” represents the clinically actionable genes, and the color “blue”, the non-actionable genes. The pie chart graphs represent the classification of the samples into one of the three groups: absence of mutations (wild-type: wt), presence of only clinically actionable mutation(s), and presence of only non-actionable mutation(s). The bar graphs represent a profile of all mutated genes identified in all samples. In the non-actionable category, genes with only one mutation detected were grouped into “other”.
Figure 5.
Circos plots for each tumor type showing the frequency of patients with mutations in both clinically actionable and non-actionable genes.
In the NSCLC cases, 58% of the patients showed variants in BRAF, EGFR, KRAS, or PIK3CA, which were classified as clinically actionable genes (Figure 4). The most frequently identified variants were point mutations in codons 12, 13, and 61 of the KRAS gene, followed by point mutations and INDELs in the EGFR gene; point mutations in the BRAF (V600E) and PIK3CA genes were also commonly identified. Point mutations and INDELs were also observed in other genes including TP53, STK11, MET, ATM, CDKN2A, JAK3, and others (Figure 4). A total of 122 patients had only one variant, of which 57 were in clinically actionable genes and 65 in other genes. Among the samples that had more than one variant, 81.5% had at least one variant in a clinically actionable gene (Figure 4A). Patients with a KRAS mutation also showed variants in TP53 (43%), STK11 (18%), ATM (12%), MET (14%), and other genes. Mutations in the TP53 gene were also observed in patients with EGFR (66%), BRAF (50%), and PIK3CA (25%) mutations.
Among patients diagnosed with colon adenocarcinoma, 57% had variants in clinically actionable genes (BRAF, KRAS, HRAS, and NRAS) (Figure 4). Mutations in codon 12 of the KRAS gene, followed by the V600E mutation in the BRAF gene were the most frequently identified variants. TP53, APC, PIK3CA, FBXW7, MET, SMAD4, PTEN, and ATM were the most commonly mutated other genes (Figure 4). Out of 58 patients with only one mutation, 18 showed variants in clinically actionable genes. Figure 5B shows that samples with KRAS mutations also had variants in APC (47%), TP53 (59%), PIK3CA (24%), and other genes. Patients with BRAF variants presented similar profile (TP53: 44%, PIK3CA: 20%, APC: 13%, and other gene mutations).
IDH1, IDH2, CDKN2A, PIK3CA, and SMO constituted our clinically actionable gene list for the glioma/glioblastoma category. Thirty-one patients had variants in clinically actionable genes; however, 12 of them had a single gene variant (Figure 4). IDH2 mutations (R172K) were detected in two patients. IDH1 (R132H) and PIK3CA (H1047, E545K) were the two most frequently mutated genes. Among other genes, TP53 and PTEN were the most commonly mutated (Figure 4). TP53 was mutated in 60%, 43%, and 50% of the patients with IDH1, PIK3CA, and CDKN2A mutations, respectively (Figure 5C).
Forty-three patients diagnosed with melanoma were identified with variants in the following clinically actionable genes: BRAF, CTNNB1, KIT, and NRAS (Figure 4). Almost 55% of them had a single variant in either BRAF (V600E, D594N), KIT (L576P), or NRAS (G12C and codon 61) genes. Mutations in other genes were identified in 23 patients, and the most frequently mutated genes were TP53 and CDKN2A (Figure 5D). Some patients with BRAF mutations also showed concurrent variants in PTEN (21%), PIK3CA (7%), MET (21%), JAK3 (21%), and CDKN2A (14%) genes.
Discussion
In the past decade, molecular tests have been intensively used in peripheral blood, fresh frozen, and FFPE tissues to identify somatic variants involved in carcinogenesis. These variants can contribute to better prognosis, prediction, and potential selection of targeted therapies for patients. Recently, many studies have shown that FFPE samples are a reliable source for molecular oncology testing [6–8]. In this study, we demonstrated that with a well-established and validated analytical workflow, such as pre-DNA extraction (verification of tumor content and amount of tissue on each unstained slide) and post-DNA extraction (verification of DNA concentration and DNA quality), FFPE tissues from resection, biopsy or cytology samples can be routinely used for NGS in a clinical molecular diagnostic laboratory. In our laboratory, more than 50% of the specimens received represent either small biopsies or cytology samples, which are considered a standard diagnostic specimen for some solid tumors (most notably lung specimens). Despite the often poor DNA quality and limited DNA yield, cytology samples were shown to be usable for NGS to detect somatic variants with better sensitivity than traditional sequencing methods.
NGS has rapidly evolved as part of the routine clinical laboratory due to its unique ability to assess multiple patients and multiple genetic targets simultaneously (including point mutations, small INDELs, copy number alteration, and rearrangements or gene fusions). It is a critical technology for generation of accurate mutation profiles of the individual tumor that can be used to identify the most effective available therapy, as well as determine the eligibility for evolving clinical trials. However, one of its limitations is in data analysis, which includes variant annotation and curation [9]. A comprehensive reference database containing somatic variants, their clinical annotation and relevance, classification (clinically actionable or non-actionable), and applicable clinical trials would greatly help in standardized and routine curation and reporting of detected variants. Some of the most clinically relevant databases available online include ClinVar, My Cancer Genome, COSMIC, Massachusetts General Hospital Cancer Center, and ClinicalTrials.gov. To address inter-database discrepancies involving variant classification into one of three major categories (“pathogenic or likely pathogenic”, “uncertain significance”, and “likely benign or benign”), some databases utilize a notification system that can alert them to the need to reevaluate their records of gene-disease relevance [9]. The most recent database, ClinGen (Clinical Genome Resource), is developing interconnected community resources to improve the understanding of genomic variation and improve its use in clinical care [10].
In our study, 127 patients were negative and 750 patients were positive for somatic varaints in the 50 genes present in this panel. The identification of wild-type patients for mutations within the panel is also important to guide treatment decisions. For example, clinicians can direct them to broader molecular testing capable of detecting copy number alterations, gene fusions, large INDELs, or to a non-genotyping clinical trial (such as immunotherapy) [11]. Additionally, eligibility for some clinical trials is based on absence of variant within certain genomic targets.
We categorized the somatic variants in this hotspot panel as either clinically actionable or other gene variant according to the NCCN Clinical Practice Guidelines in Oncology and My Cancer Genome. It is important to track the other gene variants in a clinical knowledge base in the event new information arises which supports their clinical actionability. Approximately 60% of the patients in this study had an actionable variant, meaning that these patients had a variant that was targetable by an approved drug according to their tumor type (on-label). For example, BRAF (V600E) mutations confer sensitivity to BRAF inhibitors and have been found in 50% of advanced melanoma patients and 10% of colorectal carcinomas. Additionally, KRAS mutant NSCLC patients are resistant to anti-EGFR therapy, and mutations in the EGFR gene can confer either sensitivity (exon 19 deletions and L858R) or resistance (T790M and small insertions or deletions in exon 20) to patients diagnosed with NSCLC [2]. However, recent studies have shown that patients with G12V or G12C mutations in the KRAS gene have increased sensitivity to MEK inhibitors compared with tumors carrying other KRAS mutations [12]. While these variants are considered actionable, not all drugs are approved for use in different tumor types (off-label use). For this reason, access to these drugs may be limited by the payor.
The remaining 40% of patients in this study had variants in other genes, meaning that while there is no approved therapy for mutations in that particular gene, there may either be a clinical trial available for the detected variant, or the clinical significance of that variant is still unknown. Among the patients with these other variants, only a few of them presented similar molecular profiles [13]. The presence or absence of somatic variants associated with the tumor type often represents the main criteria to determine clinical trial eligibility. In addition, previous treatment, restriction to use of off-label drugs, accuracy and quality of the molecular test, and proximity to trial sites may limit patient access to novel targeted therapies.
The introduction of NGS into routine clinical use has required healthcare providers to recognize the significance of common molecular biomarkers and their therapeutic implications. In order to provide interpretive support for non-actionable variants in patients with advanced malignancies, numerous institutions have implemented Molecular Tumor Boards [14, 15]. With the collaboration of a multidisciplinary team including oncologists, clinical laboratory scientists, pathologists, clinical geneticists, basic and translational science researchers, and bioinformatics and pathway analysis specialists, Molecular Tumor Boards can begin to decipher genetic profiles and guide treatment decisions that improve patient care.
With the recent advances in understanding tumor heterogeneity, establishing a molecular tumor profile has become an essential component of determining patient prognosis and making informed treatment decisions. Tumor heterogeneity can occur at the genetic, epigenetic, transcriptomic, and proteomic levels [16–18]. There are many possible types of genetic heterogeneity that providers need to be aware of in human cancer, including interpatient tumor heterogeneity, intratumor heterogeneity, intermetastatic heterogeneity and intrametastatic heterogeneity [19], all of which can be attributed to genomic instabilities. These may be responsible for small (point mutations, and INDELs) and large (structural and numerical chromosome changes) scale alterations that occur early or later in tumor evolution, following branched evolution [16, 20]. According to Swanton [20], early-stage tumors are composed of multiple clones, while late-stage tumors are composed of both multiple clones and sub-clones. These sub-clones can coexist either within the same tumor or intermixed within the same tissue biopsy specimen. For this reason, biologically complex tumor heterogeneity is associated with disease progression, and clinically, it is also associated with drug resistance and poor prognosis. Broader gene panels may identify tumor alterations that can predict either sensitivity or resistance to targeted therapies. In some cases, studies have indicated that patients may benefit from a combination of drugs that can simultaneously target multiple tumor subclonal cell populations [18, 21]. As demonstrated in Figures 4 and 5, routine sequencing of advanced tumors using the CHPv2 has allowed us to characterize each patient’s complex tumor profile. The detection of sensitizing and resistance variants contributed both to the selection of the most effective therapeutic agent and eligibility for clinical trials when available, ultimately leading to a more personalized cancer treatment.
Given the complexity of the biology of tumor cells and the growing datasets evolving from large numbers of NGS studies, the data still raises more questions than answers. While heterogeneity may play a large role in clinical outcomes, we do not understand the confounding effects of multiple variants in the same tumor. Some variants may increase drug sensitivity while others may result in decreased sensitivity and/or resistance. Resistance may be immediate or latent and emerge as a new and more aggressive clone as a relapse or metastatic disease. While response rates for targeted therapies are often better than traditional cytotoxic chemotherapy with less adverse reactions, they are currently still below what one would expect [22]. The use of patient-derived xenograft models to assess tumor response rates to therapeutics is one solution that is being fully explored by many investigators [23]. There is still a need for better therapeutic modalities of all types and better monitoring of disease.
In the era of precision medicine, it is crucial for clinical laboratories to offer comprehensive somatic mutation testing that can be used to create molecular profiles for individual patient tumors. These profiles are essential in guiding clinicians in the selection of targeted therapies and identification of applicable clinical trials. It is extremely important to implement a quality management program to ensure the accuracy and reproducibility of reportable results. As a CLIA-certified and CAP-accredited laboratory, we fully validated a targeted NGS panel and implemented an extensive QM program that allows us to routinely generate these molecular profiles that have become such an essential component of personalized cancer treatment.
Footnotes
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
Research funding: None declared.
Employment or leadership: None declared.
Honorarium: None declared.
Competing interests: The funding organization(s) played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the report for publication.
Contributor Information
Francine B. de Abreu, Department of Pathology, Dartmouth Hitchcock Medical Center and Norris Cotton Cancer Center, Lebanon, NH, USA
Jason D. Peterson, Department of Pathology, Dartmouth Hitchcock Medical Center and Norris Cotton Cancer Center, Lebanon, NH, USA
Christopher I. Amos, Center for Genomic Medicine and Department of Community and Family Medicine, Giesel School of Medicine at Dartmouth, Hanover, NH, USA
Wendy A. Wells, Department of Pathology, Dartmouth Hitchcock Medical Center and Norris Cotton Cancer Center, Lebanon, NH, USA
References
- 1.Ross JS, Cronin M. Whole cancer genome sequencing by next-generation methods. Am J Clin Pathol. 2011;136:527–39. doi: 10.1309/AJCPR1SVT1VHUGXW. [DOI] [PubMed] [Google Scholar]
- 2.Yu B, O’Toole SA, Trent RJ. Somatic DNA mutation analysis in targeted therapy of solid tumours. Transl Pediatr. 2015;4:125–38. doi: 10.3978/j.issn.2224-4336.2015.04.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–47. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aziz N, Zhao Q, Bry L, Driscoll DK, Funke B, Gibson JS, et al. College of American pathologists’ laboratory standards for next-generation sequencing clinical tests. Arch Pathol Lab Med. 2015;139:481–93. doi: 10.5858/arpa.2014-0250-CP. [DOI] [PubMed] [Google Scholar]
- 5.Tsongalis GJ, Peterson JD, de Abreu FB, Tunkey CD, Gallagher TL, Strausbaugh LD, et al. Routine use of the ion torrent AmpliSeq™ Cancer Hotspot Panel for identification of clinically actionable somatic mutations. Clin Chem Lab Med. 2013;13:1–8. doi: 10.1515/cclm-2013-0883. [DOI] [PubMed] [Google Scholar]
- 6.Kanagal-Shamanna R, Portier BP, Singh RR, Routbort MJ, Aldape KD, Handal BA, et al. Next-generation sequencing-based multi-gene mutation profiling of solid tumors using fine needle aspiration samples: promises and challenges for routine clinical diagnostics. Mod Pathol. 2014;27:314–27. doi: 10.1038/modpathol.2013.122. [DOI] [PubMed] [Google Scholar]
- 7.Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682–8. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gailey MP, Stence AA, Jensen CS, Ma D. Multiplatform comparison of molecular oncology tests performed on cytology specimens and formalin-fixed, paraffin-embedded tissue. Cancer Cytopathol. 2015;123:30–9. doi: 10.1002/cncy.21476. [DOI] [PubMed] [Google Scholar]
- 9.Dumur CI. Available resources and challenges for the clinical annotation of somatic variations. Cancer Cytopathol. 2014;122:730–6. doi: 10.1002/cncy.21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, et al. ClinGen – the clinical genome resource. N Engl J Med. 2015;372:2235–42. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson DB, Dahlman KH, Knol J, Gilbert J, Puzanov I, Means-Powell J, et al. Enabling a genetically informed approach to cancer medicine: a retrospective evaluation of the impact of comprehensive tumor profiling using a targeted next-generation sequencing panel. Oncologist. 2014;19:616–22. doi: 10.1634/theoncologist.2014-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janne PA, Smith I, McWalter G, Mann J, Doughrty B, Walker J, et al. Impact of KRAS codon subtypes from a randomised phase II trial of selumetinib plus docetaxel in KRAS mutant advanced non-small-cell lung cancer. Br J Cancer. 2015;113:199–203. doi: 10.1038/bjc.2015.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwaederle M, Daniels GA, Piccioni DE, Fanta PT, Schwab RB, Shimabukuro KA, et al. On the road to precision cancer medicine: analysis of genomic biomarker actionability in 439 patients. Mol Cancer Ther. 2015;14:1488–94. doi: 10.1158/1535-7163.MCT-14-1061. [DOI] [PubMed] [Google Scholar]
- 14.Schwaederle M, Parker BA, Schwab RB, Fanta PT, Boles SG, Daniels GA, et al. Molecular tumor board: the University of California San Diego moores cancer center experience. Oncologist. 2014;19:631–6. doi: 10.1634/theoncologist.2013-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tafe LJ, Gorlov I, de Abreu FB, Lefferts JA, Liu X, Pettus JR, et al. Implementation of a Molecular tumor board: the impact on treatment decisions for the first 35 patients evaluated at Dartmouth-Hitchcock Medical Center. Oncologist. 2015;20:1011–8. doi: 10.1634/theoncologist.2015-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burrel RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–45. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 17.Burrel RA, Swanton C. Tumor heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol. 2014;8:1095–111. doi: 10.1016/j.molonc.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pribluda A, de la Cruz CC, Jackson EL. Intratumoral heterogeneity: from diversity comes resistance. Clin Cancer Res. 2015;21:2916–22. doi: 10.1158/1078-0432.CCR-14-1213. [DOI] [PubMed] [Google Scholar]
- 19.Jamal-Hanjani M, Quezada SA, Larkin J, Swanton C. Translational implication of tumor heterogeneity. Clin Cancer Res. 2015;21:1258–66. doi: 10.1158/1078-0432.CCR-14-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swanton C. Cancer evolution: the final frontier of precision medicine? Ann Oncol. 2014;25:549–51. doi: 10.1093/annonc/mdu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janku F. Tumor heterogeneity in the clinic: is it a real problem? Ther Adv Med Oncol. 2014;6:43–51. doi: 10.1177/1758834013517414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jorgensen JT. Clinical application of companion diagnostics. Trends Mol Med. 2015;21:405–7. doi: 10.1016/j.molmed.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Stewart EL, Mascaux C, Pham NA, Sakashita S, Sykes J, Kim L, et al. Clinical utility of patient derived xenografts to determine biomarkers of prognosis and map resistance pathways in EGFR-mutant lung adenocarcinoma. J Clin Oncol. 2015;33:2472–80. doi: 10.1200/JCO.2014.60.1492. [DOI] [PubMed] [Google Scholar]