Abstract
Many publicly available data repositories and resources have been developed to support protein related information management, data-driven hypothesis generation and biological knowledge discovery. To help researchers quickly find the appropriate protein related informatics resources, we present a comprehensive review (with categorization and description) of major protein bioinformatics databases in this chapter. We also discuss the challenges and opportunities for developing next-generation protein bioinformatics databases and resources to support data integration and data analytics in the Big Data era.
Keywords: Bioinformatics, Database, Protein sequence, Protein structure, Protein family, Protein function, Protein mutation, Protein interaction, Pathway, Proteomics, PTM, Data integration, Data analytics, Big data
1. Introduction
Use of high-throughput technologies to study molecular biology systems in the past decades has revolutionized biological and biomedical research, allowing researchers to systematically study the genomes of organisms (Genomics) [1], the set of RNA molecules (Transcriptomics) [2], and the set of proteins including their structures and functions (Proteomics) [3]. Since proteins occupy a middle ground molecularly between gene and transcript and many higher levels of molecular and cellular structure and organization, and most physiological and pathological processes are manifested at the protein level, biological and biomedical scientists are increasingly interested in applying high-throughput proteomics techniques to achieve a better understanding of basic molecular biology and disease processes [4, 5].
The richness of proteomics data allows researchers to ask complex biological questions and gain new scientific insights. To support data-driven hypothesis generation and biological knowledge discovery, many protein-related bioinformatics databases, query facilities, and data analysis software tools have been developed (http://www.oxfordjournals.org/our_journals/nar/database/cap/) to organize and provide biological annotations for proteins to support sequence, structural, functional and evolutionary analyses in the context of pathway, network and systems biology. With the recent extraordinary advances in genome sciences and Next-Generation Sequencing (NGS) technologies [6] that have uncovered rich genomic information in a huge number of organisms, new protein bioinformatics databases are also being introduced and many existing databases have been enhanced. As more and more genomes are sequenced, the protein sequences archived in databases have increased dramatically in recent years (see Figure 1 for an example). This poses new challenges for computational biologists in building new infrastructure to support protein science research in the age of Big Data.
Figure 1. The total number of protein sequences in UniProtKB.
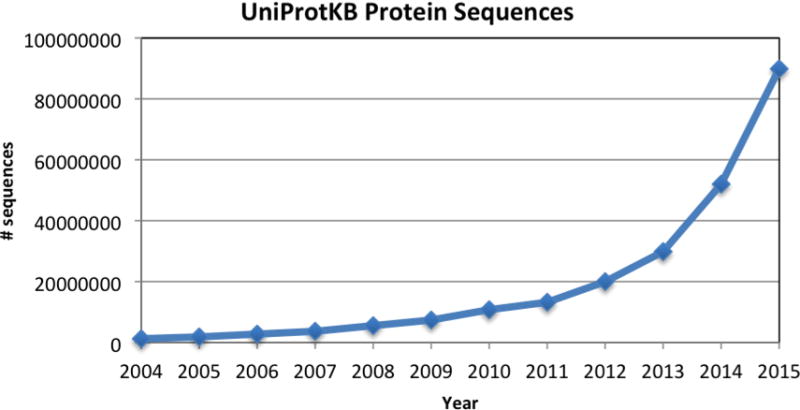
The diagram shows that as the result of the rapid development of genome sequencing projects, protein sequences archived in UniProtKB have increased dramatically in recent years.
We present a summary review (with categorization and description) of protein bioinformatics databases and resources in Table 1. The databases and categories presented in Table 1 are selected from the databases listed in the Nucleic Acids Research (NAR) database issues and database collection, as well as the databases cross-referenced in the UniProtKB. The reason we choose them is because they: 1) are protein related and well grouped; 2) are well-documented with papers and websites; 3) have been peer reviewed or/and selected by the UniProt consortium for UniProtKB database cross-references; and 4) are supposed to be well maintained.
Table 1.
Overview of Protein Bioinformatics Databases
| Category | DB Short Name | DB Name | URLs | Ref. |
|---|---|---|---|---|
| Sequence databases | CCDS | The Consensus CDS protein set database | https://www.ncbi.nlm.nih.gov/CCDS/CcdsBrowse.cgi | [9] |
| DDBJ | DNA Data Bank of Japan | http://www.ddbj.nig.ac.jp/ | [10] | |
| ENA | European Nucleotide Archive | http://www.ebi.ac.uk/ena | [11] | |
| GenBank | GenBank nucleotide sequence database | https://www.ncbi.nlm.nih.gov/genbank/ | [12] | |
| RefSeq* | NCBI Reference Sequence Database | https://www.ncbi.nlm.nih.gov/refseq/ | [13] | |
| UniGene | Database of computationally identifies transcripts from the same locus | https://www.ncbi.nlm.nih.gov/unigene/ | [12] | |
| UniProtKB* | Universal Protein Resource (UniProt) | http://www.uniprot.org/ | [14] | |
| 2D gel databases | COMPLUYEAST-2DPAGE | 2-DE database at Universidad Complutense de Madrid, Spain | http://compluyeast2dpage.dacya.ucm.es/ | [15] |
| REPRODUCTION-2DPAGE | 2-DE database at Nanjing Medical University, China | http://reprod.njmu.edu.cn/cgi-bin/2d/2d.cgi | [16] | |
| SWISS-2DPAGE | 2-DE database at Swiss Institute of Bioinformatics, Switzerland | http://world-2dpage.expasy.org/swiss-2dpage/ | [17] | |
| World-2DPAGE* | The World-2DPAGE database | http://world-2dpage.expasy.org/repository/ | [18] | |
| 3D structure databases | DisProt | Database of Protein Disorder | http://www.disprot.org/ | [19] |
| MobiDB | Database of intrinsically disordered and mobile proteins | http://mobidb.bio.unipd.it/ | [20] | |
| ModBase | Database of Comparative Protein Structure Models | http://modbase.compbio.ucsf.edu/modbase-cgi/index.cgi | [21] | |
| PDBe* | Protein Data Bank at Europe | http://www.ebi.ac.uk/pdbe/ | [22] | |
| PDBj* | Protein Data Bank at Japan | http://pdbj.org/ | [23] | |
| PDBsum | Pictorial database of 3D structures in the Protein Data Bank | http://www.ebi.ac.uk/pdbsum/ | [24] | |
| ProteinModelPortal | Protein Model Portal of the PSI-Nature Structural Biology Knowledgebase | http://www.proteinmodelportal.org/ | [25] | |
| RCSB-PDB* | Protein Data Bank at RCSB | http://www.pdb.org/ | [26] | |
| SMR | Database of annotated 3D protein structure models | http://swissmodel.expasy.org/repository/ | [27] | |
| Chemistry databases | BindingDB | The Binding Datbase | http://www.bindingdb.org/ | [28] |
| ChEMBL* | Database of bioactive drug-like small molecules | https://www.ebi.ac.uk/chembldb | [29] | |
| DrugBank | Drug and Drug Target Databse | http://www.drugbank.ca/ | [30] | |
| Enzyme and pathway databases | MetaCyc/BioCyc* | MetaCyc Database of Metabolic Pathways, BioCyc Collection of Pathway/Genome Databases | http://www.biocyc.org/ | [31] |
| BRENDA* | BRaunschweig ENzyme DAtabase | http://www.brenda-enzymes.org | [32] | |
| ENZYME | Enzyme nomenclature database | http://enzyme.expasy.org/ | [33] | |
| Reactome* | A knowledgebase of biological pathways and processes | http://www.reactome.org/ | [34] | |
| SABIO-RK | SABIO-RK: Biochemical Reaction Kinetics Database | http://sabiork.h-its.org/ | [35] | |
| SignaLink | A signaling pathway resource with multi-layered regulatory networks | http://signalink.org/ | [36] | |
| UniPathway | UniPathway: a resource for the exploration of metabolic pathways | http://www.unipathway.org | [37] | |
| Family and domain databases | Gene3D | Structural and Functional Annotation of Protein Families | http://gene3d.biochem.ucl.ac.uk/Gene3D/ | [38] |
| HAMAP | High-quality Automated and Manual Annotation of Proteins | http://hamap.expasy.org/ | [39] | |
| InterPro* | Integrated resource of protein families, domains and functional sites | http://www.ebi.ac.uk/interpro/ | [40] | |
| PANTHER | The PANTHER Classification System | http://www.pantherdb.org/ | [41] | |
| Pfam* | The Pfam protein families database | http://pfam.xfam.org/ | [42] | |
| PIRSF* | A whole-protein classification database | http://pir.georgetown.edu/pirwww/dbinfo/pirsf.shtml | [43] | |
| PRINTS | Protein Motif fingerprint database | http://www.bioinf.manchester.ac.uk/dbbrowser/PRINTS/ | [44] | |
| ProDom | Protein domain families database | http://prodom.prabi.fr/prodom/current/html/home.php | [45] | |
| PROSITE* | Database of protein domains, families and functional sites | http://prosite.expasy.org/ | [46] | |
| ProtoNet | Automatic hierarchical classification of proteins | http://www.protonet.cs.huji.ac.il/ | [47] | |
| SMART | Simple Modular Architecture Research Tool | http://smart.embl.de/ | [48] | |
| SUPFAM | Superfamily database of structural and functional annotation | http://supfam.org | [49] | |
| TIGRFAMs | TIGRFAMs protein family database | http://www.jcvi.org/cgi-bin/tigrfams/index.cgi | [50] | |
| Gene expression databases | Bgee | Database for Gene Expression Evolution | http://bgee.unil.ch | [51] |
| CleanEx | Database of gene expression profiles | http://cleanex.vital-it.ch/ | [52] | |
| Genevisible | Search portal to normalized and curated expression data from Genevestigator | http://genevisible.com/search | [53] | |
| ExpressionAtlas* | Database of Differential and Baseline Expression | http://www.ebi.ac.uk/gxa/home | [54] | |
| Genome annotation databases | Ensembl* | Ensembl Eukaryotic genome annotation database | http://www.ensembl.org/ | [55] |
| EnsemblBacteria | Ensembl Bacteria genome annotation database | http://bacteria.ensembl.org/ | [56] | |
| EnsemblFungi | Ensembl Fungi genome annotation database | http://fungi.ensembl.org/ | [56] | |
| EnsemblMetazoa | Ensembl Metazoa genome annotation database | http://metazoa.ensembl.org/ | [56] | |
| EnsemblPlants | Ensembl Plants genome annotation database | http://plants.ensembl.org/ | [56] | |
| EnsemblProtists | Ensembl Protists genome annotation database | http://protists.ensembl.org/ | [56] | |
| Entrez Gene* | Database of Genes of Genomes in the Reference Sequence Collection | https://www.ncbi.nlm.nih.gov/gene | [57] | |
| KEGG | Kyoto Encyclopedia of Genes and Genomes | http://www.genome.jp/kegg/ | [58] | |
| PATRIC | Bacterial Bioinformatics Resource Center | http://patricbrc.org/ | [59] | |
| UCSC* | UCSC Genome Bioinformatics | http://genome.ucsc.edu | [60] | |
| VectorBase | Bioinformatics Resource for Invertebrate Vectors of Human Pathogens | http://www.vectorbase.org/ | [61] | |
| WBParaSite | WormBase ParaSite | http://parasite.wormbase.org | [62] | |
| Organism specific databases | ArachnoServer | ArachnoServer: Spider toxin database | http://www.arachnoserver.org | [63] |
| CGD | Candida Genome Database | http://www.candidagenome.org/ | [64] | |
| ConoServer | ConoServer: Cone snail toxin database | http://www.conoserver.org/ | [65] | |
| CTD | Comparative Toxicogenomics Database | http://ctdbase.org/ | [66] | |
| dictyBase | Central resource for Dictyostelid genomics | http://dictybase.org/ | [67] | |
| EchoBASE | EchoBASE – an integrated post-genomic database for E. coli. | http://www.york.ac.uk/res/thomas/ | [68] | |
| EcoGene | Escherichia coli strain K12 genome database | http://www.ecogene.org/ | [69] | |
| euHCVdb | The European Hepatitis C Virus database | https://euhcvdb.ibcp.fr/euHCVdb/ | [70] | |
| EuPathDB | Eukaryotic Pathogen Database Resources | http://eupathdb.org/eupathdb/ | [71] | |
| FlyBase* | A Database of Drosophila Genes & Genomes | http://flybase.org/ | [72] | |
| GenAtlas | A database on genes, functions and related diseases | http://genatlas.medecine.univ-paris5.fr/ | [73] | |
| GeneCards | The Human Gene Database | http://www.genecards.org/ | [74] | |
| GenoList | Integrated Environment for the Analysis of Microbial Genomes | http://genodb.pasteur.fr/cgi-bin/WebObjects/GenoList | [75] | |
| Gramene | A comparative resource for plants | http://www.gramene.org/ | [76] | |
| H-InvDB | H-Invitational Database | http://www.h-invitational.jp/ | [77] | |
| HGNC | HUGO Gene Nomenclature Committee Database | http://www.genenames.org/ | [78] | |
| HPA | The Human Protein Atlas | http://www.proteinatlas.org/ | [79] | |
| HUGE | A Database of Human Unidentified Gene-Encoded Large Proteins | http://www.kazusa.or.jp/huge/ | [80] | |
| LegioList | Legionella pneumophila genome database | http://genolist.pasteur.fr/LegioList/ | [81] | |
| Leproma | Mycobacterium leprae genome database | http://mycobrowser.epfl.ch/leprosy.html | [82] | |
| MaizeGDB | Maize Genetics and genomics Database | http://www.maizegdb.org/ | [83] | |
| MGD* | Mouse Genome Database | http://www.informatics.jax.org/ | [84] | |
| Micado | MICrobial Advanced Database Organization | http://genome.jouy.inra.fr/cgi-bin/micado/index.cgi | [85] | |
| OMIM | Online Mendelian Inheritance in Man | http://www.omim.org/ | [86] | |
| neXtProt* | Exploring the universe of human proteins | http://www.nextprot.org/ | [87] | |
| Orphanet | The portal for rare diseases and orphan drugs | http://www.orpha.net/consor/cgi-bin/home.php?Lng=GB | [88] | |
| PharmGKB | The Pharmacogenomics Knowledgebase | http://www.pharmgkb.org | [89] | |
| PomBase | The scientific resource for fission yeast | http://www.pombase.org/ | [90] | |
| PseudoCAP | The Pseudomonas Genome Database | http://www.pseudomonas.com/ | [91] | |
| RGD | Rat Genome Database | http://rgd.mcw.edu/ | [92] | |
| Rouge | A Database of Rodent Unidentified Gene-Encoded Large Proteins | http://www.kazusa.or.jp/rouge/ | [80] | |
| SGD | Saccharomyces Genome Database | http://www.yeastgenome.org/ | [93] | |
| TAIR | The Arabidopsis Information Resource | http://www.arabidopsis.org/ | [94] | |
| TubercuList | Mycobacterium tuberculosis strain H37Rv genome database | http://tuberculist.epfl.ch | [95] | |
| WormBase | C. elegans and related nematodes Genetics and Genomics Database | http://www.wormbase.org/ | [62] | |
| Xenbase | Xenopus laevis and tropicalis biology and genomics resource | http://www.xenbase.org/ | [96] | |
| ZFIN | The Zebrafish Model Organism Database | http://zfin.org/ | [97] | |
| Phylogenomic databases | eggNOG | Database of orthologous groups and functional annotation | http://eggnog.embl.de/ | [98] |
| HOGENOM | Database of Homologous Genes from Fully Sequenced Organisms | http://pbil.univ-lyon1.fr/databases/hogenom/home.php | [99] | |
| HOVERGEN | Homologous Vertebrate Genes Database | http://pbil.univ-lyon1.fr/databases/hovergen.html | [100] | |
| InParanoid | Eukaryotic Ortholog Groups with inparalogs | http://inparanoid.sbc.su.se/ | [101] | |
| KO | Kyoto Encyclopedia of Genes and Genomes Orthology | http://www.genome.jp/kegg/ | [102] | |
| OMA* | The OMA orthology database | http://omabrowser.org/ | [103] | |
| OrthoDB | Database of Orthologous Groups | http://cegg.unige.ch/orthodb6 | [104] | |
| PhylomeDB | Database for complete catalogs of gene phylogenies (phylomes) | http://phylomedb.org/ | [105] | |
| TreeFam | Database of animal gene trees | http://www.treefam.org | [106] | |
| Polymorphism and mutation databases | BioMuta | Single-nucleotide variation and disease association database | https://hive.biochemistry.gwu.edu/tools/biomuta/ | [107] |
| dbSNP* | Database of Short Genetic Variations | https://www.ncbi.nlm.nih.gov/SNP/ | [12] | |
| DMDM | Domain Mapping of Disease Mutations | http://bioinf.umbc.edu/dmdm/ | [108] | |
| Protein-protein interaction databases | BioGRID | The Biological General Repository for Interaction Datasets | http://thebiogrid.org | [109] |
| DIP | Database of Interacting Proteins | http://dip.doe-mbi.ucla.edu/ | [110] | |
| IntAct* | IntAct Molecular Interaction Database | http://www.ebi.ac.uk/intact/ | [111] | |
| MINT | The Molecular INTeraction database | http://mint.bio.uniroma2.it/mint/ | [112] | |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins | http://string-db.org | [113] | |
| Proteomic databases | MaxQB | The MaxQuant DataBase | http://maxqb.biochem.mpg.de/mxdb/ | [114] |
| PaxDb | Protein Abundance Across Organisms | http://pax-db.org | [115] | |
| PeptideAtlas* | PeptideAtlas | http://www.peptideatlas.org | [116] | |
| PRIDE* | PRoteomics IDEntifications database | http://www.ebi.ac.uk/pride | [117] | |
| ProMEX | Protein Mass spectra EXtraction | http://promex.pph.univie.ac.at/promex/ | [118] | |
| PTM databases | DEPOD* | The Human DEPhOsphorylation Database | http://www.koehnlab.de/depod/index.php | [119] |
| iPTMnet* | Protein post-translational modifications (PTMs) in systems biology context | http://research.bioinformatics.udel.edu/iptmnet/ | [120] | |
| PhosPhAt* | The Arabidopsis Protein Phosphorylation Site Database | http://phosphat.uni-hohenheim.de | [121] | |
| Phospho.ELM* | Database of S/T/Y phosphorylation sites | http://phospho.elm.eu.org | [122] | |
| PhosphoGrid* | Database of experimentally verified in vivo protein phosphorylation sites | http://www.phosphogrid.org | [123] | |
| PhosphoSitePlus* | Phosphorylation site database | http://www.phosphosite.org | [124] | |
| UniCarbKB* | Database of glycomics and glycobiology | http://www.unicarbkb.org/ | [125] | |
| Ontology | GO* | Gene Ontology | http://www.geneontology.org/ | [126] |
| PRO | Protein Ontology | http://pir.georgetown.edu/pro/pro.shtml | [127] | |
| Specialized protein databases | Allergome | Allergome: platform for allergen knowledge | http://www.allergome.org/ | [128] |
| CAZy | Carbohydrate-Active enZYmes Database | http://www.cazy.org/ | [129] | |
| ESTHER | ESTerases and alpha/beta-Hydrolase Enzymes and Relatives database | http://bioweb.ensam.inra.fr/ESTHER/general?what=index | [130] | |
| GPCRDB | Information system for G protein-coupled receptors (GPCRs) | http://www.gpcr.org/7tm/ | [131] | |
| IMGT | The International ImMunoGeneTics information system | http://www.imgt.org/ | [132] | |
| MEROPS* | MEROPS protease database | http://merops.sanger.ac.uk/ | [133] | |
| MoonProt | Moonlighting protein database | http://www.moonlightingproteins.org/ | [134] | |
| mycoCLAP | Characterized Lignocellulose-Active Proteins of Fungal Origin | https://mycoclap.fungalgenomics.ca/mycoCLAP/ | [135] | |
| PeroxiBase | The peroxidases database | http://peroxibase.toulouse.inra.fr/ | [136] | |
| REBASE | The Restriction Enzyme Database | http://rebase.neb.com/rebase/rebase.html | [137] | |
| TCDB | Transporter Classification Database | http://www.tcdb.org/ | [138] | |
| Other [Miscellaneous] databases | ChiTaRS | Database of chimeric transcripts and rna-seq data | http://chitars.bioinfo.cnio.es/ | [139] |
| EvolutionaryTrace | Database of relative evolutionary importance of amino acids within a protein sequence | http://mammoth.bcm.tmc.edu/ETserver.html | [140] | |
| GeneWiki* | Wiki portal for the annotation of gene and protein function | http://en.wikipedia.org/wiki/Portal:Gene_Wiki | [141] | |
| GenomeRNAi | Database of phenotypes from RNA interference screens in Drosophila and Homo sapiens | http://genomernai.dkfz.de/GenomeRNAi/ | [142] | |
| PMAP-CutDB | Proteolytic event database | http://www.proteolysis.org/ | [143] | |
| SOURCE | The Stanford Online Universal Resource for Clones and ESTs | http://smd.princeton.edu/cgi-bin/source/sourceSearch | [144] |
Databases covered in the section 3 of the chapter.
Protein bioinformatics databases can be primarily classified as sequence databases, 2D gel databases, 3D structure databases, chemistry databases, enzyme and pathway databases, family and domain databases, gene expression databases, genome annotation databases, organism specific databases, phylogenomic databases, polymorphism and mutation databases, protein-protein interaction databases, proteomic databases, PTM databases, ontologies, specialized protein databases, and other (miscellaneous) databases. Please visit http://proteininformationresource.org/staff/chenc/MiMB/dbSummary2015.html to access the databases reviewed in this chapter through their corresponding web addresses (URLs). For many of these databases, their identifiers can be mapped to UniProtKB protein AC/IDs [7]. Our coverage of protein bioinformatics databases in this chapter is by no means exhaustive. Our intention is to cover databases that are recent, high quality, publicly available, and are expected to be of interest to more users in the community. It is worth noting that certain databases can be classified into more than one category.
As an update to our previously contributed MiMB series chapter [8], we now focus on databases that are aligned with the content of this book and emphasize the types of data stored and related data access and data analysis supports. For each category of databases listed in Table 1, we select some representatives and describe them briefly in section 2. In section 3, we discuss the challenges and opportunities for developing next-generation protein bioinformatics databases and resources to support data integration and data analytics in Big Data era. We conclude the chapter in section 4.
2. Databases and Resources Highlights
2.1. Sequence Databases
2.1.1. RefSeq
The National Center for Biotechnology Information Reference Sequence (NCBI RefSeq) database [13] provides curated non-redundant sequences of genomic regions, transcripts and proteins for taxonomically diverse organisms including Archaea, Bacteria, Eukaryotes, and Viruses. RefSeq database is derived from the sequence data available in the redundant archival database GenBank [12]. RefSeq sequences include coding regions, conserved domains, variations etc. and enhanced annotations such as publications, names, symbols, aliases, Gene IDs, and database cross-references. The sequences and annotations are generated using a combined approach of collaboration, automated prediction, and manual curation [13]. The RefSeq release 73 on November 6, 2015 includes 54,766,170 proteins, 12,998,293 transcripts and 55,966 organisms. The RefSeq records can be directly accessed from NCBI web sites by search of the Nucleotide or Protein databases, BLAST searches against selected databases and FTP downloads. RefSeq records are also available through indirect links from other NCBI resources such as Gene, Genome, BioProject, dbSNP, ClinVar and Map Viewer etc. In addition, RefSeq supports programmatic access through Entrez Programming Utilities [145].
2.1.2. UniProt
The UniProt Consortium consists of research teams from the European Bioinformatics Institute (EBI), the Swiss Institute of Bioinformatics (SIB) and the Protein Information Resource (PIR). The UniProt Consortium provides a central resource for protein sequences and functional annotations with four core database components to support protein bioinformatics research.
The UniProt Knowledgebase (UniProtKB) is the predominant data store for functional information on protein sequences with rich and accurate annotations (protein name or description, taxonomic information, classification, cross-reference and literature citation) [14]. The UniProtKB consists of two parts: UniProtKB/Swiss-Prot, which contains manually annotated records with information extracted from literature and curator-evaluated computational analysis, and UniProtKB/TrEMBL, which contains computationally analyzed records with automatic annotation and classification. Comparative analysis and query for proteins are supported by UniProtKB extensive cross-references, functional and feature annotations, classification, and literature-based evidence attribution. The 2015_12 release on December 09, 2015 of UniProtKB/Swiss-Prot contains 550,116 sequence entries, comprising 196,219,159 amino acids, and 55,270,679 UniProtKB/TrEMBL sequence entries comprising 18,388,518,872 amino acids.
The UniProt Archive (UniParc) [146] is a comprehensive and non-redundant archival protein sequence database from all major publicly accessible resources. UniParc contains protein sequences and cross-references to their source databases. UniParc stores each unique protein sequence with a stable and unique identifier and tracks sequence changes in its source databases.
The UniProt Reference Clusters (UniRef) [147] are clustered sets of sequences from the UniProt Knowledgebase (including isoforms) and selected UniParc records. UniRef merges sequences and sub-fragments with 100% (UniRef100), ≥ 90% (UniRef90), or ≥ 50% (UniRef50) identity and 80% overlap with the longest sequences in the cluster (seed) into a single UniRef entry and select the highest ranked protein sequences as the cluster representatives.
The UniProt Proteomes [14] provides sets of proteins that are considered to be expressed by organisms whose genomes have been completely sequenced. A UniProt proteome consists of all UniProtKB/Swiss-Prot entries plus those UniProtKB/TrEMBL entries mapped to Ensembl Genomes for that proteome. Some well-studied model organisms and other organisms of interest to biomedical research and phylogeny have been manually and computationally [148] selected as reference proteomes.
The UniProt web site (http://www.uniprot.org) is the primary access point to its data and documentation. The site provides batch retrieval using UniProt identifiers; BLAST-based sequence similarity search; Clustal Omega based sequence alignment; and Database identifier mapping [7]. The UniProt FTP download site provides batch download of protein sequence data in various formats, including flat file TEXT, XML, RDF and FASTA. Programmatic access to data and search result is supported via RESTful web services. For more details about UniProt databases, we refer the readers to chapter 2 of this book.
2.2. 2D Gel Databases: World-2DPAGE
The World-2DPAGE Constellation [18] is an effort of the Swiss Institute of Bioinformatics to promote and publish two-dimensional gel electrophoresis proteomics data online through the ExPASy proteomics server. The World-2DPAGE Constellation consists of three components:
World-2DPAGE List (http://world-2dpage.expasy.org/list/) contains references to known federated 2-D PAGE databases, as well as to 2-D PAGE related servers and services.
World-2DPAGE Portal (http://world-2dpage.expasy.org/portal/) is a dynamic portal that serves as a single interface to query simultaneously worldwide gel-based proteomics databases that are built using the Make2D-DB package [149].
World-2DPAGE Repository (http://world-2dpage.expasy.org/repository/) is a public repository for gel-based proteomics data with protein identifications published in the literature. Mass-spectrometry based proteomics data from related studies can also be submitted to the PRIDE database [117] so that interested readers can explore the data in the views of 2D-gel and/or MS.
The World-2DPAGE Constellation also provides a set of tools:
Make2D-DB package (ver. 3.10.2) is open source packages that can be used to build a user’s own 2-D PAGE web site, access and integrate federated 2D-PAGE databases, portals or data repositories.
Melanie Viewer (ver. 7.0) is a free viewer that can be used to visualize gels and related data obtained through the use of the full version of Melanie 2D electrophoresis gel analysis software.
MIAPEGelDB can be used to produce MIAPE-compliant gel experiments documents.
2.3. 3D Structure Databases: wwPDB
The worldwide PDB (wwPDB, http://www.wwpdb.org) [150] was established in 2003 as an international collaboration to maintain a single and publicly available Protein Data Bank Archive (PDB Archive) of macro-molecular structural data. The wwPDB member includes Protein Data Bank in Europe (PDBe) [22], Protein Data Bank Japan (PDBj) [23], Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) [26], and Biological Magnetic Resonance Bank (BMRB) [151]. The “PDB Archive” is a collection of flat files in three different formats: the legacy PDB format; the PDBx/mmCIF (http://deposit.pdb.org/mmcif/) format; and the Protein Data Bank Markup Language (PDBML) [152] format. Each member site serves as a deposition, data processing and distribution site for the PDB Archive, and each provides its own view of the primary data and a variety of tools and resources. As of December 1, 2015, there are 113,971 biological macromolecular structures in the wwPDB database including 37,049 distinct protein sequences, 30,099 structures of human sequences, 8,096 Nucleic Acid containing structures.
2.4. Chemistry Databases: ChEMBL
ChEMBL [29] is a large-scale bioactivity database containing binding, functional, in vivo absorption, distribution, metabolism, excretion, and toxicity (ADMET) information about drug-like bioactive compounds. ChEMBL data are manually curated from the published literature together with data drawn from other databases. ChEMBL are standardized for using in many types of chemical biology and drug-discovery research problems. ChEMBL database can be accessed from a web-based interface where a variety of search and browsing functionality are provided. ChEMBL data is freely available from their FTP site in the formats of Oracle, MySQL, PostgreSQL, structure-data file (SDF), FASTA and RDF. Programmatic access is also supported by a set of RESTful web services. The ChEMBL release 20 (prepared on Jan 14, 2015) contains 1,715,135 compound records, 1,463,270 compounds (of which 1,456,020 have mol files), 13,520,737 activities, 1,148,942 assays, 10,774 targets, and 59,610 documents.
2.5. Enzyme and Pathway Databases
2.5.1. MetaCyc and BioCyc
MetaCyc is a reference database of non-redundant, experimentally elucidated metabolic pathways and enzymes curated from the scientific literature [31]. MetaCyc stores pathways, compounds, proteins, protein complexes and genes associated with these pathways with extensive links to protein sequence databases, nucleic acid sequence databases, protein structure databases and literature. MetaCyc can also be used as a reference database to predict the metabolic network in sequenced genomes by Pathway Tools software [153] using machine-learning methods [154]. The 2015 release of MetaCyc includes 2,411 metabolic pathways, 13,074 reactions, 10,789 enzymes, 10,928 genes, 12,792 chemical compounds, 2,740 organisms, and 47,838 citations.
BioCyc is a collection of Pathway/Genome Databases (PGDBs) [31]. Each BioCyc PGDB contains the metabolic network of one organism predicted by the Pathway Tool software using MetaCyc as a reference database. The BioCyc databases are organized into three tiers: Tier 1 databases are those that have received at least one person-year of literature-based curation. Tier 2 and Tier 3 databases are computationally predicted metabolic pathways. Web-based query, browsing, visualization and comparative analysis tools are also provided from MetaCyc and BioCyc web sites. A collection of data files in different formats is provided for download. BioCyc also provides RESTful web services, MySQL server and Perl, Java and Lisp APIs access to its data. The 2015 release of BioCyc includes 7,667 Pathway/Genome Databases.
2.5.2. BRENDA
BRENDA (BRaunschweig ENzyme DAtabase) [32] is an information system for functional and molecular properties of enzymes and enzyme-ligands obtained by manual extraction from literature, text and data mining, data integration and computational predictions. BRENDA stores enzyme data in textual, single numeric, numeric range, and graphic formats. The content of BRENDA is based on the IUBMB (International Union of Biochemistry and Molecular Biology) enzyme classification system. BRENDA includes the following databases generated by text mining approach.
KENDA contains kinetic values and kinetic expressions mined from PubMed abstracts.
DRENDA contains disease-related enzyme information (causal interaction, therapeutic application, diagnostic usage, and ongoing research) mined from PubMed abstracts using MeSH terms.
FRENDA contains references found in PubMed abstracts that have the enzyme name and organism combination.
AMENDA is a subset of FRENDA providing organism-specific information on the enzyme sources and the subcellular localization.
The user can access the data and information in BRENDA by searching (Quick Search, Advanced Search, Full text Search, Substructure Search, and Sequence Search) and browsing (TaxTree Explorer, EC Explorer, Ontology Explorer, and Genome Explorer). The search results can be downloaded as CSV file. The BRENDA release 2015.2 in July 2015 contains 6,759 enzymes.
2.5.3. Reactome
Reactome [34] is an open source, expert-curated and peer-reviewed database of biological reactions and pathways with cross-references to major molecular databases. Reactome provides the visual representation of classical intermediary metabolism, signaling, innate and acquired immune function, transcriptional regulation, apoptosis and disease process etc. Reactome website supports the navigation of pathway knowledge and pathway-based analysis and visualization of experimental or computational data. Interaction, reaction and pathway data are downloadable as flat file, MySQL, BioPAX, SBML and PSI-MITAB files. They are also accessible through RESTful web services. Software tools such as Pathway Browser, Analyze Data, Species Comparison, Reactome FI Network are provided to support data mining and analysis of large-scale data sets. The Reactome release 54 in September 2015 contains 101,670 proteins, 74,357 complexes, 68,659 reactions, and 20,261 pathways.
2.6. Family and Domain Databases
2.6.1. InterPro
InterPro [40] is an integrated resource of predictive models or ‘signatures’ representing protein domains, families, regions, repeats and sites from major protein signature databases including CATH-Gene3D [38], HAMAP [37], PANTHER [41], Pfam [42], PIRSF [43], PRINTS [44], ProDom [45], PROSITE [46], SMART [48], SUPERFAMILY [49] and TIGRFAMs [50]. Each entry in the InterPro database is annotated with a descriptive abstract name and cross-references to the original data sources, as well as to specialized functional databases. The search by sequence or domain architecture is provided by InterPro web site. The InterPro signatures in XML format are available via anonymous FTP download. InterPro also provides a software package InterProScan [155] that can be used locally to scan protein sequences against InterPro’s signatures. Programmatic access to InterProScan is possible via RESTful and SOAP web service APIs. The InterPro BioMart [156] allows users to retrieve InterPro data from a query-optimized data warehouse that is synchronized with the main InterPro database, and to build simple or complex queries and control the query results through a unified interface. The InterPro release 54.0 on October 15, 2015 includes 28,462 entries containing signatures of 19,110 families, 8,191 domains, 284 repeats, 115 active sites, 74 binding sites, 672 conserved sites and 16 PTMs.
2.6.2. Pfam
Pfam is a database of protein families represented as multiple sequence alignments and Hidden Markov Models (HMMs) [42]. Pfam entries can be classified as Family (related protein regions), Domain (protein structural unit), Repeat (multiple short protein structural units), Motifs (short protein structural unit outside global domains). Related Pfam entries are grouped into clans based on sequence, structure or profile-HMM similarity. The Pfam database web site provides search interface for querying by sequence, keyword, domain architecture, taxonomy, and browse interfaces for analyzing protein sequences for Pfam matches and viewing Pfam annotations in domain architectures, sequence alignments, interactions, species and protein structures in PDB [26]. The Pfam data can be downloaded from its FTP site or programmatically accessed through RESTful web service APIs. The Pfam release 28.0 in May 2015 contains 16,230 families.
2.6.3. PIRSF
The PIRSF classification system [43] provides comprehensive and non-overlapping clustering of UniProtKB [14] sequences into a hierarchical order to reflect their evolutionary relationships based on whole proteins rather than on the component domains. The PIRSF system classifies the protein sequences into families, whose members are both homologous (evolved from a common ancestor) and homeomorphic (sharing full-length sequence similarity and a common domain architecture) [43]. The PIRSF family classification results are expert-curated based on literature review and integrative sequence and functional analysis. The classification report shows the information on PIRSF members and general statistics, family and function/structure relationships, database cross-references and graphical display of domain and motif architecture of seed members or all members. The web-based PIRSF system has been demonstrated as a useful tool for studying the function and evolution of protein families [43]. It provides batch retrieval of entries from the PIRSF database. The PIRSF scan allows searching a query sequence against the set of fully curated PIRSF families with benchmarked Hidden Markov models. The PIRSF membership hierarchy data is also available for FTP download. The current release of PIRSF contains 11,800 families, which cover 5,407,000 UniProtKB protein sequences.
2.6.4. PROSITE
PROSITE [46] is a database of documentation entries describing protein domains, families and functional sites as well as associated patterns and profiles to identify them. The entries are derived from multiple alignments of homologous sequences and have the advantage of identifying distant relationships between sequences. PROSITE includes a collection of ProRules based on profiles and patterns of functionally and/or structurally critical amino acids that can be used to increase PROSITE’s discriminatory power [46]. The PROSITE web site provides keyword-based search and allows browsing by documentation entry, ProRule description, taxonomic scope and number of positive hits. The software tool ScanProsite [157] supports three options for users to scan proteins for matches to PROSITE motifs or their own sequence patterns: 1) scan protein sequence against the PROSITE motifs; 2) scan motifs against a protein sequence database; 3) submit protein sequences and motifs and scan them against each other. The PROSITE documentation entries and related tools can be downloaded from its FTP site. The PROSITE release 20.120 on November 4, 2015 contains 1,742 documentation entries, 1,309 patterns, 1,139 profiles and 1,138 ProRules.
2.7. Gene Expression Databases: Expression Atlas
The Expression Atlas database [54] provides gene, protein and splice variant expression patterns in different cell types, organism parts, biological and experimental conditions. The high quality Microarray and RNA-Seq data imported from ArrayExpress [158] and Gene Expression Omnibus [12] were manually curated, annotated and processed using standardized analysis methods to detect the expression patterns under the original experimental conditions. Expression Atlas consists of two components: Baseline Atlas and Differential Atlas. The Baseline Atlas is about genes and their expression pattern under the “normal” conditions using only RNA-Seq data. The Differential Atlas is about genes that are up- or down- regulated in differential biological or experimental conditions using both Microarray and RNA-Seq data. Expression Atlas web interface supports query both the Baseline Atlas and Differential Atlas by gene, protein and splice variant. The search for sample attributes and experimental conditions are also supported. All Expression Atlas analysis results can be downloaded from their FTP site. The differential expression data and meta-data can be used in R Bioconductor (https://www.bioconductor.org/) package. The APIs to programmatically access Expression Atlas is under development. The October 29, 2015 release of Expression Atlas contains 2,373 datasets (93,057 assays).
2.8. Genome Annotation Databases
2.8.1. Ensembl
Ensembl is a genome annotation database that provides up-to-date annotations for chordates and model organism genomes [55]. Additional metazoan genomes are available from EnsemblMetazoa [56], Plant and fungal genomes are available from EnsemblPlants [56] and EnsemblFungi [56], Unicellular eukaryotic and prokaryotic genomes are available from EnsemblProtists [56] and EnsemblBacteria [56]. Ensembl supports variety of access routes to their data. Small data set can be exported from online search results. Large dataset or complex analyses can be accessed from MySQL server, Perl and RESTful APIs. Complex cross databases queries are supported by BioMart data mining tool [156]. The whole database can be downloaded from FTP site in FASTA, EMBL, GenBank, GVF, VCF, VEP, GFF formats or through MySQL dumps. In addition, Ensembl also provides a set of data processing software tools. For example, Variant Effect Predictor, BLAST/BLAT, Assembly converter, ID History converter etc. The Ensembl release v83 in September 2015 contains 69 species with annotations for gene and transcript, gene sequence evolution, genome evolution, sequence and structural variants and regulatory elements.
2.8.2. Entrez Gene
Entrez Gene [57] is a NCBI gene-specific database that provides GeneIDs (unique integer identifiers) for genomes that have been completely sequenced. The data in Entrez Gene database (nomenclature, map location, gene products and attributes, markers, phenotypes, citations, sequences, variations, maps, expression, homologs, protein domains etc.) are results of manual curation and automated computational analysis of data from RefSeq [13] and many other NCBI databases [12]. The data in Entrez Gene database can be accessed in several ways: 1) query Entrez from the NCBI home page and display the results in Gene, 2) enter a query in any Entrez query bar and restrict the database search to Gene, 3) cross links from other NCBI resources such as GenBank, BLAST, RefSeq, Map Viewer. Entrez Gene data can be downloaded from NCBI FTP site and accessed by Entrez Programming Utilities [145]. The Entrez Gene release on December 4, 2015 includes 13,778 taxa and 12,841,400 genes.
2.8.3. UCSC
UCSC Genome Browser database [60] contains large collection of genome assemblies and annotations for vertebrate and selected model organisms. The major sources of genome annotations include RefSeq, GENCODE, Ensembl, GenBank, ENCODE, RepeatMasker, dbSNP, the 1000 Genome project and other resources. In addition to Genome Browser, the UCSC bioinformatics group also provides web-based and command-line based tools to facilitate the use of genome annotations data. For example, BLAT can be used to quickly find sequences of 95% and greater similarity and 25 bases or more in length. The Table Browser can retrieve the data associated with a track in Genome Browser and calculate intersections between tracks. The Variant Annotation Integrator can associate UCSC Genome Browser annotations with the user-uploaded variants. The Gene Sorter can be used to show expression, homology and other information on groups of genes. User data can be viewed together with UCSC annotations via ‘custom track’, ‘track data hubs’, ‘assembly hub’ and ‘Genome Browser in a Box (GBiB)’ [159]. Genome data and source codes are downloadable. UCSC Genome Bioinformatics group also provides public MySQL server access. Currently (December 11, 2015), there are 95 genomes in UCSC Genome Browser database.
2.9. Organism Specific Databases
2.9.1. FlyBase
FlyBase [72] is a database of Drosophila melanogaster related genetic and genomic information. The sequence and annotation data for Drosophila melanogaster genome assembly can be downloaded from FlyBase FTP site in multiple formats (GFF3, FASTA, GTF, Chado XML, and Chado PostgreSQL dump). FlyBase uses generic genome browser 2 (GBrowse 2) to display the genome annotations and genome-aligned evidence on the reference genome assembly. FlyBase database can be searched for genes, alleles, aberrations and other genetic objects, phenotypes, sequences, stocks, images and movies, controlled terms. FlyBase provides a standalone BLAST server for 50 different arthropod genomes and supports query results analysis such as hit list refinement and batch download. The latest FlyBase is FB2015_05 released on November 20, 2015 that consists of 212,991 references, 141,104 stocks and 1,258 images.
2.9.2. MGD
The Mouse Genome Database (MGD) [84] is a database of integrated genomic, genetic and biological data on the laboratory mouse that is a model for translational research. MGD integrates mouse genome annotations from NCBI, Ensembl and Havana into a single non-redundant resource. MGD is the authoritative source for the unified catalog of mouse genome features, Gene Ontology (GO) annotations (functional associations) of mouse protein-coding genes, and mouse phenotype annotations. The Human-Mouse: Disease Connection (http://www.diseasemodel.org) is a translational research tool that provides simultaneous access to human-mouse genomic, phenotypic and genetic disease information. MGD uses a powerful new genome browser called JBrowse [160] to integrate mouse gene and protein annotations with large-scale sequence data. In addition to online search tools for genes, genome features and maps, phenotypes, alleles and disease models, gene expression, GO functional annotations, strains, SNPs and polymorphisms, sequences, references, vocabularies, MGD also provides bulk data download as FTP reports and batch query tool and programmatic access by Web services and BioMart [156]. MGD is updated on a weekly basis.
2.9.3. neXtProt
neXtProt [87] is a new protein-centric knowledge platform serves as a central hub for all knowledge about human proteins. neXtProt integrates high-quality and manually curated UniProt/Swiss-Prot entries with large amount of additional human protein related information from other resources such as Human Protein Atlas [79], ArrayExpress [158], UniGene [12], PeptideAtlas [116], Gene Ontology Annotation [126], Ensembl [55], dbSNP [12] etc. Ontologies and controlled vocabularies (CVs) are extensively used in neXtProt to support consistent annotation and data retrieval. neXtProt’s Google-like search interface supports free text search and complex queries with results displayed as lists or short summaries. neXtProt provides export functionality for protein entries in TEXT, Excel, FASTA and XML formats and bulk download from the FTP site. neXtProt release on September 1, 2015 contains 20,066 protein entries, 153,556 controlled vocabularies and 465,706 publications.
2.10. Phylogenomic Databases: OMA
The Orthologous Matrix (OMA) [103] is a method and associated database that infers evolutionary relationships amongst complete proteomes. OMA’s inference algorithm includes three steps: 1) infer homologous sequences (sequences of common ancestry); 2) infer orthologous pairs (subsets of homologs related by speciation events); 3) cluster orthologs into: (i) OMA groups (cliques of orthologous pairs) and (ii) HOGs (groups of genes descended from a common ancestral gene in a given taxonomic range). OMA can be accessed through the OMA browser and programmatic interfaces. OMA genomes including all-against-all computations can be downloaded with OMA stand-alone program to do orthology prediction using user’s custom data. The OMA release in September 2015 contains 1,970 species, 1,001,242 OMA groups, and 10,129,468 proteins.
2.11. Polymorphism and Mutation Databases: dbSNP
The NCBI dbSNP database [12] is a database for short genetic variations from variety of organisms. dbSNP catalogs single nucleotide variations, short nucleotide insertions and deletions, short tandem repeats and microsatellites. dbSNP homepage provides search interface for querying variations by simple term or complex queries. The details of matched variation record is displayed as the Reference SNP Cluster Report that contains summary of the allele, mapping information in Human Genome Variation Society (HGVS) nomenclature, gene-centric view, map table with chromosomal coordinates, variation view, and link to the 1000 Genomes Browser. dbSNP integrates disease-related variations collected by OMIM [86]. dbSNP variation data are accessible through links from other NCBI databases. dbSNP data can also be downloaded from a FTP site and accessed by EUtils API (https://www.ncbi.nlm.nih.gov/books/NBK25500/). dbSNP build 146 on November 24, 2015 for Homo sapiens contains 150,482,731 RefSNP Clusters, among them 100,135,281 are validated.
2.12. Protein-protein Interaction Databases: IntAct
IntAct [111] is an open source database and toolkit for the storage, presentation and analysis of rich curated molecular interaction data in community accepted standard formats. IntAct provides relevant experimental details of protein interactions curated from literatures or directly deposited. All the entries in the database are fully compliant with the IMEx [162] guidelines and MIMIx [163] standard. IntAct web site provides multiple search functionalities: 1) search by anything that might be related to interactions, for example, gene name, identifiers, GO term, publication, and experimental method etc.; 2) search on four ontologies: Gene Ontology [126], InterPro [40], PSI-MI [164], ChEBI [165]; 3) draw all or part of a chemical structure and search for chemical compounds. IntAct data is released monthly and available as FTP download. IntAct release 194 on December 2, 2015 consists of 577,297 binary interactions from 13,952 curated publications and 1,378 biological complexes.
2.13. Proteomics Databases
2.13.1. PeptideAtlas
PeptideAtlas [116] provides an approach and framework to archive proteomic data that enables the data exchange and integration with genomic data. PeptideAtlas statistically validates peptides identified by high-throughput tandem mass spectrometry (MS/MS) experiments and maps peptide sequences to eukaryotic genomes. PeptideAtlas uses a uniform statistical validation process to ensure consistent and high-quality peptide and protein identifications. The raw data used to build PeptideAtlas includes raw MS/MS files, MS/MS files in mzXML [166] format, SEQUEST [167] search results. The user can also download PeptideProphet [168] results and ProteinProphet [169] outputs. The PeptideAtlas builds are available for download or browse via PeptideAtlas web interface. As of December 7, 2015, there are total 72 builds covering 19 organisms.
2.13.2. PRIDE
The PRoteomics IDEntifications database (PRIDE) [117] is a repository for mass-spectrometry based proteomics data including identifications of proteins, peptides and post-translational modifications that have been described in the scientific literature, together with supporting mass spectra and related technical and biological metadata. PRIDE supports tandem MS (MS/MS) and Peptide Fingerprinting datasets with search/analysis workflows originally analyzed by the submitters. PIRDE provides several services such as the Protein Identifier Cross-Reference (PICR) [170], the Ontology Lookup Service (OLS) [171] and Database on Demand [172]. The data in PRIDE database can be accessed in different ways: 1) The PRIDE web interface can be used to explore all public datasets currently available in the repository; 2) Batch data retrieval and integration with other databases can be achieved by PRIDE BioMart [156]; 3) PRIDE public experiments data in mzData (http://www.psidev.info/mzdata) and PRIDE XML formats can be downloaded via FTP, Aspera, and HTTP; 4) A set of RESTful web services can be used to get programmatic access to data in the PRIDE repository. PRIDE supports submissions of protein and peptide identification/quantification data with the accompanying mass spectral evidence by following the ProteomeXchange (PX) consortium [173] guidelines. PRIDE also provides a set of software tools: PRIDE Converter 2 for converting common mass spectrometry data formats into PRIDE XML for data submission, and PRIDE Inspector for visualizing and analyzing MS dataset, such as: mzML [174], mzIdentML (http://www.psidev.info/mzidentml), and PRIDE XML. As of December 8, 2015, PRIDE repository includes 3,774 projects and 55,873 assays.
2.14. PTM Databases
2.14.1. DEPOD
The human DEPhOsphorylation Database (DEPOD) [119] is a comprehensive, high quality, manually curated database for human phosphatases, their experimentally verified protein and non-protein substrates, dephosphorylation sites, involved pathways with cross-references to kinases and small molecule modulators. The human phosphatase substrate information is integrated from a variety of sources including ‘dephosphorylation’ post-translational modification data in Human Protein Reference Database [175], ‘dephosphorylation’ interaction data from PSICQUIC service [176], substrate information from UniProt annotation [14], and scientific literature from PubMed and Google. DEPOD database can be browsed by Human Phosphatases, Protein Substrates, Non-Protein Substrates, Pathway Mapping, and Dephosphorlation Network. DEPOD also allows direct deposit substrate candidates for human active phosphatases. The Human active phosphatase data can be downloaded in XSLX format. The Human phosphatase-substrate interaction and dephosphorylation sites data are available for download in PSI-MI Tab 2.5 format. In addition, KEGG [58], NCI Nature PID and Reactome [34] pathways mapped on phosphatases and substrates are available in TXT format. The latest release of DEPOD on August 15, 2015 contains 228 human active and 11 inactive phosphatases (194 phosphatases have substrate), 298 protein substrates, 89 non-protein substrates, 1,096 dephosphorylation interactions, 213 KEGG pathways, 206 NCI Nature PID pathways and 560 Reactome pathways.
2.14.2. iPTMnet
iPTMnet [120] is an integrated resource for protein post-translational modification network discovery which combines text mining, data mining, and ontological representation to capture rich PTM information, including PTM enzyme-substrate relationships, PTM-specific protein-protein interactions (PPIs) and PTM conservation across species to support PTM analysis in the context of systems biology. It employs RLIMS-P [177] and eFIP [178] text mining tools developed by PIR group for full-scale mining of PubMed abstracts to identify PTM information (kinase, substrate, and site) and phosphorylation-dependent PPI. Experimentally observed PTMs, including high-throughput proteomic data from curated PTM databases, are incorporated. Proteins and PTM protein forms (proteoforms) are organized using the Protein Ontology (PRO) [127], enabling representation and annotation of forms modified on combinations of PTM sites and orthologous relationships between forms. iPTMnet thus serves as an integrated resource that connects between knowledge about biologically relevant modified proteins from disparate sources. Covering seven major PTM types (phosphorylation, acetylation, ubiquitination, methylation, glycosylation, sumoylation and myristoylation), the current iPTMnet database contains more than 250,000 PTM sites in more than 45,000 modified proteins, along with more than 1,000 PTM enzymes for human, mouse, rat, yeast, Arabidopsis and several other organisms. The web portal supports online search and visual analysis for scientific queries. For more details about iPTMnet database, we refer the readers to chapter 21 of this book.
2.14.3. PhosPhAt
The Arabidopsis Protein Phosphorylation Site Database (PhosPhAt) [121] catalogs published information on large-scale Mass Spectrometry experiments identified phosphorylation sites in Arabidopsis. It contains information about the peptides, their annotated biological functions, and experimental and analytical contexts. In addition, PhosPhAt provides a plant specific phosphorylation site predictor trained using Serine, Threonine and Tryosine phosphorylation (pSer, pThr, pTyr) experimental data. The user can access the pre-computed prediction using Arabidopsis gene identifiers or do “on-the-fly” prediction of phosphorylation of user submitted protein sequences. Both the experimentally determined phosphorylation sites and high confidence predicted sites are available for download. As of December 8, 2015, PhosPhAt includes 9,159 experimental phosphoproteins with 19,100 unique tryptic phosphopeptides, and 31,916 predicted proteins with 2,176,360 predicted phosphosites.
2.14.4. Phospho.ELM
Phospho.ELM [122] is a manually curated database of experimentally verified eukaryotic phosphorylation sites. Each entry in Phospho.ELM database is manually annotated with information about the phosphorylated proteins, the positions of known phosphorylations, the kinases responsible for phosphorylation, and literature citations. Additional information such as structure, interaction partners, sub-cellular compartment, and tissue specificities are also provided whenever they are available. Phospho.ELM data can be searched from its web interface. The data sets are also available for download upon request. PhosphoBlast server can be used to search protein (UniProt ID/AC or amino acid sequence) against the curated dataset of phosphorylated peptides. Phospho.ELM (v9.0, September 2010) contains 8,718 substrate proteins covering 3,370 tyrosine, 31,754 serine and 7,449 threonine instances.
2.14.5. PhosphoGrid
PhosphoGrid [123] is a database of experimentally verified in vivo protein phosphorylation sites of Saccharomyces cerevisiae curated from literatures. Both high-throughput MS phospho-proteomics studies and focused low-throughput analyses of individual proteins or complexes are integrated into PhosphoGrid. Each in vivo phosphorylation site is annotated by a hierarchy of experimental evidence codes, experimentally defined protein kinases and/or phosphatases, specific condition(s) under which the phosphorylation event occurs and the effect(s) of phosphorylation on protein function. The user can search PhosphoGrid web-based interface for any substrate, protein kinase or phosphatase. Each record is cross-referenced with BioGRID [109], Saccharomyces Genome Database (SGD) [93], NCBI protein database [12] and its original PubMed articles. Latest release of PhosphoGrid contains 20,177 phosphorylation sites, 3,011 kinases, 266 phosphatases, and 563 publications.
2.14.6. PhosphoSitePlus
PhosphoSitePlus (PSP) [124] is a curated and highly interactive systems biology knowledgebase for studying experimentally observed mammalian post-translational modifications (PTMs) in the regulation of biological process. PSP provides a comprehensive coverage of protein phosphorylation, acetylation, methylation, ubiquitination, and O-glycosylation. PSP includes structural and functional information about the topology, biological function and regulatory significance of modification sites integrated from both low- and high-throughput (LTP and HTP) data. The Homepage of PSP includes “Simple Search” that allows query of all known phosphorylation sites in a specific protein and “Advanced Search” that allows search by Protein, Sequence or Reference. PSP also supports retrieval of a list of modified sites that possess certain specified attributes and browsing curated MS/MS records by disease type, cell line and tissue. Multiple types of datasets and tools are available for download such as PTMVar dataset, Modification site datasets, Regulatory sites, Disease-associated sites, Kinase-substrate dataset, Cytoscape plugin etc. The latest release of PSP (accessed on December 9, 2015) contains 52,872 proteins, 21,619 Low throughput (LTP) sites, 456,434 High throughput (HTP) MS sites, 2,130,888 MS peptides and 19,704 curator-reviewed papers. For more details about PhosphoSitePlus database, we refer the readers to chapter 8 of this book.
2.14.7. UniCarbKB
UniCarbKB [125] is a curated knowledgebase for glycomics and glycobiology research. UniCarbKB provides comprehensive information about the structures, pathways and networks involved in glycosylation and glycol-mediated processes. UniCarbKB integrates GlycoSuiteDB [179] and EUROCarbDB [180] to provide a unified portal to support glycol-bioinformatics research and knowledge dissemination. The content of UniCarbKB is mainly eukaryotic glycoproteins curated from GlycoSuiteDB and a selected few datasets from EUROCarbDB. The data in GlycosuiteDB, EUROCarbDB and GlycoBase [181] can be queried by Taxonomy, Tissue, Protein Name, Protein Accession and Composition. Glycan structures can be searched using carbohydrate sequences in GlycoCT format. The user can browse the curated collection of proteins or search them by names. Glycan Builder provides a GUI interface for building and displaying glycan structures. GlycoDigest is a tool that simulates exoglycosidase digestion, based on controlled rules acquired from expert knowledge and experimental evidence available in GlycoBase. The latest release of UniCarbKB (accessed on December 9, 2015) contains 899 Glycoproteins, 3,238 GlycoSuite structures, 520 UniCarb-DB MS/MS datasets and 909 publications.
2.15. Ontology Databases: Gene Ontology (GO)
The Gene Ontology (GO) [126] is a bioinformatics effort to create the consistent computational representation of gene functions at the molecular, cellular and tissue system levels across all organisms. GO provides controlled vocabulary of terms (ontologies) to describe gene products in terms of the biological processes, cellular components and associated molecular functions. The user of GO terms enables the uniform query and association across many biological databases. From GO web site, the user can search for GO terms, annotations to gene products, and metadata across multiple species and perform GO enrichment analysis. GO web site provides the download of the gene association files (Annotation), Gene Ontology (Ontology) and mappings of GO terms to those in a number of external vocabularies (Mapping). The Gene Ontology as of December 8, 2015 consists of 29,033 biological process terms, 4,039 cellular component terms, and 10,920 molecular function terms.
2.16. Specialized Protein Databases: MEROPS
MEROPS [133] is an integrated database of information about peptidases (also termed proteases, proteinases and proteolytic enzymes) and the proteins that inhibit them. A homologous set of peptidases and protein inhibitors are grouped into peptidase and inhibitor species. Protein species are grouped into family that contains statistically significant similarities in amino acid sequence. Families are grouped into clans that contain related structures. Both family (sub-family) and clan can be browsed by index page with links to their summary page. Each peptidase has a summary page that can be browsed by Name, Identifier, Gene name, Organism and Substrates. The peptidase summary page includes information of Gene Structure, Alignment, Tree, Sequences and their features, Distribution, Structure, Literature, Human EST, Mouse EST, Substrates, Inhibitors and Pharma. The MEROPS database can be searched for a peptidases or inhibitor, peptidases or inhibitor genes, structures of peptidases or inhibitors. User can also search via specificity, organism and citation. MEROPS supports searching peptidase and protein inhibitor sequences with a protein or nucleotide query sequence by WU-BLAST. MEROPS also provides batch substrate cleavage analysis. MEROPS allows online submission of protein cleavage site, however login is required for data download.
2.17. Other (Miscellaneous) Databases: Gene Wiki
Gene Wiki [141] is a collection of community written Wikipedia articles about human genes in NCBI Gene database [57]. Gene Wiki starts with a set of seed stub Wikipedia articles, populated and expanded by community contributors with focus on the functions and disease relevance of gene and corresponding protein. Gene Wiki has an automated system to keep the article structures in sync with the data from trusted primary databases and use WikiTrust [161] reputation system to assess and display the trustworthiness of authors and their contributions. Gene Wiki has over 10,000 distinct gene pages, spanning 2.07 million words and 82 megabytes of data.
3. Challenges and Opportunities
Although a large number of protein bioinformatics databases and resources have been developed to catalog and store different information about proteins, there are challenges and opportunities to develop Next-Generation databases and resources to facilitate data integration, data-driven hypothesis generation, and biological knowledge discovery. Recent rapid developments of high-throughput sequencing technologies bring the molecular biology researchers to the age of Big Data, where the research paradigm has shifted from hypothesis-driven to data-driven. Big Data opens new avenues to study molecular biology as well as brings new challenges for computational biologist to explore ways to efficiently manage and analyze data, and eventually turn data into usable and actionable knowledge. Next, we will review and discuss some recent technology developments that can help addressing some of the challenges.
3.1. Characteristics of Big Data
Big data is a broad term for data sets so large or complex that traditional data processing applications are inadequate (Wikipedia, http://www.wikipedia.org/). In a more articulated definition, Big Data has the following characteristics.
Volume The size of data is definitely an important aspect of Big Data. Large volume of data demands scalable storage solution and distributed information processing and retrieval.
Variety The types of data determine how the data would be analyzed. The heterogeneity of data requires non-trivial analysis methods.
Velocity The speed the data were generated and processed challenges novel real-time data analytics.
Variability The inconsistency of data calls for effective data management and handling.
Veracity The accuracy of data analysis depends on high quality data and data capture methodology.
3.2. Data Storage and Management
The first challenge computational biologists have to face is the efficient storage and management of large volume of data. In addition to better hardware support, massive parallel storage systems (distributed file systems, cluster file systems, and parallel file systems) have been explored. Examples include Lustre [182] and Hadoop Distributed File System (HDFS) [183]. On top of that we need framework for user specific solutions where several tools have been developed. Apache Hive [184] is a distributed data warehouse framework for analyzing data stored in HDFS and compatible systems using SQL-like language called HiveQL. Apache Pig [185] further simplifies the complex data analysis using simpler scripting language targeting domain experts. Traditional relational database management systems often have difficulty handling Big Data. Because they lack horizontal scalability, require hard consistency and become very complex when dealing with large volume of heterogeneous data. Non-relational databases (NoSQL) are alternative to Big Data storage and management because they focus on the scalability and flexibility. The popular NoSQL database management systems include key-value stores, columnar databases, graph databases and document-oriented databases.
3.3. Data Analytics
Data storage and management is only one side of the same coin. In the field of biomedical research and healthcare system, the purpose of high-throughput omics studies is to turn biomedical data into knowledge. In order to accomplish the goal of personalized medicine and better treatments, we need scalable computational facilities and efficient data analytics framework. Compared to traditional HPC cluster computing, cloud computing emerges as economical solution to large-scale data analysis. By hosting large volume high-throughput data in the cloud, bioinformatic analysis is now changing the way the analysis is done. Instead of moving data to the analysis code, code is now moving to the data. In addition, novel and efficient machine learning and data mining algorithms and computational framework are also essential to the success of turning data into knowledge. Apache Spark [186] is a recently developed fast and general computing engine for large-scale lightning-fast in-memory clustering computing. It supports a rich set of higher-level tools including Spark SQL for SQL and structured data processing, MLlib for machine learning, GraphX for graph processing, and Spark Streaming for scalable streaming applications.
3.4. Data Integration
The most challenging task in Big Data research is to deal with the heterogeneity, diversity and complexity of the data and to find a better way to integrate them. In addition to explore the flexibility of NoSQL technology, another promising area is to apply the ontologies and Semantic Web technology. As a formal, explicit specification of a shared conceptualization of a domain of interest, ontology plays a perfect role in addressing the issues of heterogeneity in data sources. Rapid development and adoption of ontologies have enabled the research community to annotate and integrate biological and biomedical data using standardized ontologies, and automate the discovery and composing bioinformatics web services and workflows. Linked Data technology provides a method for publishing structured data on the web and making them interconnected. The successful Linked data projects in the field of bioinformatics include Bio2RDF [187] and EBI RDF platform [188]. They use Semantic Web technologies to build and provide the largest network of Linked data for the Life Sciences by defining a set of simple conventions to create RDF(s) compatible Linked Data from a diverse set of heterogeneously formatted sources obtained from multiple data providers. The challenge for data integration using Linked Data is to develop applications that can consume such data, extract meaningful biological knowledge and present them in a user-friendly fashion.
3.5. User Interfaces
With the pervasiveness of mobile devices (Tablets and Phones), responsive web design that makes the web page look good on all devices become more and more important. Next-generation protein bioinformatics databases should provide user an optimal viewing and interaction experience across a wide range of devices using technology such as Bootstrap [189], JQuery [190], and Dojo Toolkit [191] etc. The need for speed, particularly for web-based applications has also driven the development of NoSQL technology and high performance index and search platform such as Lucene/Solr [192] for fast information retrieval.
4. Conclusions
In this chapter, we presented a comprehensive review (with categorization and description) of major protein bioinformatics databases. We also reviewed and discussed the recent technology improvements that can help addressing some of the challenges in building Next-Generation protein bioinformatics databases and resources in Big Data era.
Acknowledgments
This work was supported by grants from the National Institutes of Health: U41HG007822 and Delaware-INBRE (P20GM103446).
References
- 1.Ridley M. Genome. Harper Perennial; New York: 2006. [Google Scholar]
- 2.Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE, Jr, Hieter P, Vogelstein B, Kinzler KW. Characterization of the yeast transcriptome. Cell. 1997;2:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 3.Anderson NL, Anderson NG. Proteome and proteomics: new technologies, new concepts, and new words. Electrophoresis. 1998;11:1853–1861. doi: 10.1002/elps.1150191103. [DOI] [PubMed] [Google Scholar]
- 4.Hye A, Lynham S, Thambisetty M, Causevic M, Campbell J, Byers HL, Hooper C, Rijsdijk F, Tabrizi SJ, Banner S, Shaw CE, Foy C, Poppe M, Archer N, Hamilton G, Powell J, Brown RG, Sham P, Ward M, Lovestone S. Proteome-based plasma biomarkers for Alzheimer’s disease. Brain. 2006;11:3042–3050. doi: 10.1093/brain/awl279. [DOI] [PubMed] [Google Scholar]
- 5.Decramer S, Wittke S, Mischak H, Zürbig P, Walden M, Bouissou F, Bascands JL, Schanstra JP. Predicting the clinical outcome of congenital unilateral ureteropelvic junction obstruction in newborn by urinary proteome analysis. Nat Med. 2006;4:398–400. doi: 10.1038/nm1384. [DOI] [PubMed] [Google Scholar]
- 6.Metzker M. Sequencing technologies – the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 7.Huang H, McGarvey PB, Suzek BE, Mazumder R, Zhang J, Chen Y, Wu CH. A comprehensive protein-centric ID mapping service for molecular data integration. Bioinformatics. 2011;27:1190–1191. doi: 10.1093/bioinformatics/btr101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C, Huang H, Wu CH. Protein bioinformatics databases and resources. Methods Mol Biol. 2011;694:3–24. doi: 10.1007/978-1-60761-977-2_1. [DOI] [PubMed] [Google Scholar]
- 9.Farrell CM, O’Leary NA, Harte RA, Loveland JE, Wilming LG, Wallin C, Diekhans M, Barrell D, Searle SM, Aken B, Hiatt SM, Frankish A, Suner MM, Rajput B, Steward CA, Brown GR, Bennett R, Murphy M, Wu W, Kay MP, Hart J, Rajan J, Weber J, Snow C, Riddick LD, Hunt T, Webb D, Thomas M, Tamez P, Rangwala SH, McGarvey KM, Pujar S, Shkeda A, Mudge JM, Gonzalez JM, Gilbert JG, Trevanion SJ, Baertsch R, Harrow JL, Hubbard T, Ostell JM, Haussler D, Pruitt KD. Current status and new features of the Consensus Coding Sequence database. Nucleic Acids Res. 2014;42:D865–D872. doi: 10.1093/nar/gkt1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kodama Y, Mashima J, Kosuge T, Katayama T, Fujisawa T, Kaminuma E, Ogasawara O, Okubo K, Takagi T, Nakamura Y. The DDBJ Japanese Genotype-phenotype Archive for genetic and phenotypic human data. Nucleic Acids Res. 2015;43:D18–D22. doi: 10.1093/nar/gku1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulikova T, Akhtar R, Aldebert P, Althorpe N, Andersson M, Baldwin A, Bates K, Bhattacharyya S, Bower L, Browne P, Castro M, Cochrane G, Duggan K, Eberhardt R, Faruque N, Hoad G, Kanz C, Lee C, Leinonen R, Lin Q, Lombard V, Lopez R, Lorenc D, McWilliam H, Mukherjee G, Nardone F, Pastor MP, Plaister S, Sobhany S, Stoehr P, Vaughan R, Wu D, Zhu W, Apweiler R. EMBL Nucleotide Sequence Database in 2006. Nucleic Acids Res. 2007;35:D16–D20. doi: 10.1093/nar/gkl913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agarwala R, Barrett T, Beck J, Benson DA, Bollin C, Bolton E, Bourexis D, Brister J, Bryant SH, Canese K, Clark K, DiCuccio M, Dondoshansky I, Federhen S, Feolo M, Funk K, Geer LY, Gorelenkov V, Hoeppner M, Holmes B, Johnson M, Khotomlianski V, Kimchi A, Kimelman M, Kitts P, Klimke W, Krasnov S, Kuznetsov A, Landrum MJ, Landsman D, Lee JM, Lipman DJ, Lu Z, Madden TL, Madej T, Marchler-Bauer A, Karsch-Mizrachi I, Murphy T, Orris R, Ostell J, O’Sullivan C, Panchenko A, Phan L, Preuss D, Pruitt KD, Rubinstein W, Sayers EW, Schneider V, Schuler GD, Sherry ST, Sirotkin K, Siyan K, Slotta D, Soboleva A, Soussov V, Starchenko G, Tatusova TA, Trawick BW, Vakatov D, Wang Y, Ward M, Wilbur W, Yaschenko E, Zbicz K. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;43:D6–D17. doi: 10.1093/nar/gku1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2006;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitarch A, Sánchez M, Nombela C, Gil C. Analysis of the Candida albicans proteome. II Protein information technology on the Net (update 2002) J Chromatogr B Analyt Technol Biomed Life Sci. 2003;787:129–148. doi: 10.1016/s1570-0232(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 16.Zhou T, Zhou ZM, Guo XJ. Bioinformatics for spermatogenesis: annotation of male reproduction based on proteomics. Asian J Androl. 2013;15:594–602. doi: 10.1038/aja.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoogland C, Mostaguir K, Sanchez JC, Hochstrasser DF, Appel RD. SWISS-2DPAGE, ten years later. Proteomics. 2004;4:2352–2356. doi: 10.1002/pmic.200300830. [DOI] [PubMed] [Google Scholar]
- 18.Hoogland C, Mostaguir K, Appel RD, Lisacek F. The World-2DPAGE Constellation to promote and publish gel-based proteomics data through the ExPASy server. J Proteomics. 2008;71:245–248. doi: 10.1016/j.jprot.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. DisProt: the Database of Disordered Proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potenza E, Di Domenico T, Walsh I, Tosatto SC. MobiDB 2.0: an improved database of intrinsically disordered and mobile proteins. Nucleic Acids Res. 2014;43:D315–D320. doi: 10.1093/nar/gku982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pieper U, Webb BM, Dong GQ, Schneidman-Duhovny D, Fan H, Kim SJ, Khuri N, Spill YG, Weinkam P, Hammel M, Tainer JA, Nilges M, Sali A. ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2014;42:D336–D346. doi: 10.1093/nar/gkt1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velankar S, van Ginkel G, Alhroub Y, Battle GM, Berrisford JM, Conroy MJ, Dana JM, Gore SP, Gutmanas A, Haslam P, Hendrickx PM, Lagerstedt I, Mir S, Fernandez Montecelo MA, Mukhopadhyay A, Oldfield TJ, Patwardhan A, Sanz-García E, Sen S, Slowley RA, Wainwright ME, Deshpande MS, Iudin A, Sahni G, Salavert TJ, Hirshberg M, Mak L, Nadzirin N, Armstrong DR, Clark AR, Smart OS, Korir PK, Kleywegt GJ. PDBe: improved accessibility of macromolecular structure data from PDB and EMDB. Nucleic Acids Res. 2015;44:D385–D395. doi: 10.1093/nar/gkv1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinjo AR, Suzuki H, Yamashita R, Ikegawa Y, Kudou T, Igarashi R, Kengaku Y, Cho H, Standley DM, Nakagawa A, Nakamura H. Protein Data Bank Japan (PDBj): maintaining a structural data archive and resource description framework format. Nucleic Acids Res. 2012;40:D453–D460. doi: 10.1093/nar/gkr811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Beer TA, Berka K, Thornton JM, Laskowski RA. PDBsum additions. Nucleic Acids Res. 2014;42:D292–D296. doi: 10.1093/nar/gkt940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haas J, Roth S, Arnold K, Kiefer F, Schmidt T, Bordoli L, Schwede T. The Protein Model Portal-a comprehensive resource for protein structure and model information. Database. 2013 doi: 10.1093/database/bat031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bento AP, Gaulton A, Hersey A, Bellis LJ, Chambers J, Davies M, Krüger FA, Light Y, Mak L, McGlinchey S, Nowotka M, Papadatos G, Santos R, Overington JP. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014;42:D1083–D1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, Tang A, Gabriel G, Ly C, Adamjee S, Dame ZT, Han B, Zhou Y, Wishart DS. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Weerasinghe D, Zhang P, Karp PD. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014;42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang A, Schomburg I, Placzek S, Jeske L, Ulbrich M, Xiao M, Sensen CW, Schomburg D. BRENDA in 2015: exciting developments in its 25th year of existence. Nucleic Acids Res. 2015;43:D439–D446. doi: 10.1093/nar/gku1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, Jassal B, Jupe S, Matthews L, May B, Palatnik S, Rothfels K, Shamovsky V, Song H, Williams M, Birney E, Hermjakob H, Stein L, D’Eustachio P. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wittig U, Kania R, Golebiewski M, Rey M, Shi L, Jong L, Algaa E, Weidemann A, Sauer-Danzwith H, Mir S, Krebs O, Bittkowski M, Wetsch E, Rojas I, Müller W. SABIO-RK – database for biochemical reaction kinetics. Nucleic Acids Res. 2012;40:D790–D796. doi: 10.1093/nar/gkr1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fazekas D, Koltai M, Türei D, Módos D, Pálfy M, Dúl Z, Zsákai L, Szalay-Bekő M, Lenti K, Farkas IJ, Vellai T, Csermely P, Korcsmáros T. SignaLink 2 – a signaling pathway resource with multi-layered regulatory networks. BMC Syst Biol. 2013;7:7. doi: 10.1186/1752-0509-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgat A, Coissac E, Coudert E, Axelsen KB, Keller G, Bairoch A, Bridge A, Bougueleret L, Xenarios I, Viari A. UniPathway: a resource for the exploration and annotation of metabolic pathways. Nucleic Acids Res. 2012;40:D761–D769. doi: 10.1093/nar/gkr1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeats C, Maibaum M, Marsden R, Dibley M, Lee D, Addou S, Orengo CA. Gene3D: modelling protein structure, function and evolution. Nucleic Acids Res. 2006;34:D281–D284. doi: 10.1093/nar/gkj057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedruzzi I, Rivoire C, Auchincloss AH, Coudert E, Keller G, de Castro E, Baratin D, Cuche BA, Bougueleret L, Poux S, Redaschi N, Xenarios I, Bridge A. HAMAP in 2015: updates to the protein family classification and annotation system. Nucleic Acids Res. 2015;43:D1064–D1070. doi: 10.1093/nar/gku1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, Sangrador-Vegas A, Scheremetjew M, Rato C, Yong SY, Bateman A, Punta M, Attwood TK, Sigrist CJ, Redaschi N, Rivoire C, Xenarios I, Kahn D, Guyot D, Bork P, Letunic I, Gough J, Oates M, Haft D, Huang H, Natale DA, Wu CH, Orengo C, Sillitoe I, Mi H, Thomas PD, Finn RD. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. The Pfam protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu CH, Nikolskaya A, Huang H, Yeh LS, Natale DA, Vinayaka CR, Hu ZZ, Mazumder R, Kumar S, Kourtesis P, Ledley RS, Suzek BE, Arminski L, Chen Y, Zhang J, Cardenas JL, Chung S, Castro-Alvear J, Dinkov G, Barker WC. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res. 2004;32:D112–D114. doi: 10.1093/nar/gkh097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Attwood TK, Bradley P, Flower DR, Gaulton A, Maudling N, Mitchell A, Moulton G, Nordle A, Paine K, Taylor P, Uddin A, Zygouri C. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31:400–402. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Servant F, Bru C, Carrère S, Courcelle E, Gouzy J, Peyruc D, Kahn D. ProDom: Automated clustering of homologous domains. Brief Bioinform. 2002;3:246–251. doi: 10.1093/bib/3.3.246. [DOI] [PubMed] [Google Scholar]
- 46.Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rappoport N, Karsenty S, Stern A, Linial N, Linial M. ProtoNet 6.0: organizing 10 million protein sequences in a compact hierarchical family tree. Nucleic Acids Res. 2011;40:D313–D320. doi: 10.1093/nar/gkr1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J. SUPERFAMILY – Comparative Genomics, Datamining and Sophisticated Visualisation. Nucleic Acids Res. 2009;37:D380–D386. doi: 10.1093/nar/gkn762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, Nelson WC, Richter AR, White O. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bastian F, Parmentier G, Roux J, Moretti S, Laudet V, Robinson-Rechavi M. Bgee: Integrating and Comparing Heterogeneous Transcriptome Data Among Species. Lect Notes Comput Sci. 2008;5109:124–131. [Google Scholar]
- 52.Praz V, Jagannathan V, Bucher P. CleanEx: a database of heterogeneous gene expression data based on a consistent gene nomenclature. Nucleic Acids Res. 2004;32:D542–D547. doi: 10.1093/nar/gkh107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grennan AK. Genevestigator. Facilitating web-based gene-expression analysis. Plant Physiol. 2006;141:1164–1166. doi: 10.1104/pp.104.900198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petryszak R, Burdett T, Fiorelli B, Fonseca NA, Gonzalez-Porta M, Hastings E, Huber W, Jupp S, Keays M, Kryvych N, McMurry J, Marioni JC, Malone J, Megy K, Rustici G, Tang AY, Taubert J, Williams E, Mannion O, Parkinson HE, Brazma A. Expression Atlas update-a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42:D926–D932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kähäri AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SM, Spudich G, Trevanion SJ, Yates A, Zerbino DR, Flicek P. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kähäri A, Kinsella RJ, Kulesha E, Maheswari U, Megy K, Nuhn M, Proctor G, Staines D, Valentin F, Vilella AJ, Yates A. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;38:D563–D569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJ, Yoo HS, Zhang C, Zhang Y, Sobral BW. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hickey G, Hinrichs AS, Hubley R, Karolchik D, Learned K, Lee BT, Li CH, Miga KH, Nguyen N, Paten B, Raney BJ, Smit AF, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, Emmert D, Hammond M, Hill CA, Kennedy RC, Lobo NF, MacCallum MR, Madey G, Megy K, Redmond S, Russo S, Severson DW, Stinson EO, Topalis P, Zdobnov EM, Birney E, Gelbart WM, Kafatos FC, Louis C, Collins FH. VectorBase: a home for invertebrate vectors of human pathogens. Nucleic Acids Res. 2007;35:D503–D505. doi: 10.1093/nar/gkl960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harris TW, Baran J, Bieri T, Cabunoc A, Chan J, Chen WJ, Davis P, Done J, Grove C, Howe K, Kishore R, Lee R, Li Y, Muller HM, Nakamura C, Ozersky P, Paulini M, Raciti D, Schindelman G, Tuli MA, Van Auken K, Wang D, Wang X, Williams G, Wong JD, Yook K, Schedl T, Hodgkin J, Berriman M, Kersey P, Spieth J, Stein L, Sternberg PW. WormBase 2014: new views of curated biology. Nucleic Acids Res. 2014;42:D789–D793. doi: 10.1093/nar/gkt1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herzig V, Wood DL, Newell F, Chaumeil PA, Kaas Q, Binford GJ, Nicholson GM, Gorse D, King GF. ArachnoServer 2.0, an updated online resource for spider toxin sequences and structures. Nucleic Acids Res. 2011;39:D653–D657. doi: 10.1093/nar/gkq1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Inglis DO, Arnaud MB, Binkley J, Shah P, Skrzypek MS, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G. The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res. 2012;40:D667–D674. doi: 10.1093/nar/gkr945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaas Q, Yu R, Jin AH, Dutertre S, Craik DJ. ConoServer: updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2012;40:D325–D330. doi: 10.1093/nar/gkr886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davis AP, Grondin CJ, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL, Wiegers TC, Mattingly CJ. The Comparative Toxicogenomics Database’s 10th year anniversary: update 2015. Nucleic Acids Res. 2015;43:D914–D920. doi: 10.1093/nar/gku935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Basu S, Fey P, Pandit Y, Dodson RJ, Kibbe WA, Chisholm RL. DictyBase 2013: integrating multiple Dictyostelid species. Nucleic Acids Res. 2013;41:D676–D683. doi: 10.1093/nar/gks1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Misra RV, Horler RS, Reindl W, Goryanin II, Thomas GH. EchoBASE: an integrated post-genomic database for Escherichia coli. Nucleic Acids Res. 2005;33:D329–D333. doi: 10.1093/nar/gki028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou J, Rudd KE. EcoGene 3.0. Nucleic Acids Res. 2013;41:D613–D624. doi: 10.1093/nar/gks1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Combet C, Garnier N, Charavay C, Grando D, Crisan D, Lopez J, Dehne-Garcia A, Geourjon C, Bettler E, Hulo C, Mercier PL, Bartenschlager R, Diepolder H, Moradpour D, Pawlotsky JM, Rice CM, Trepo C, Penin F, Deléage G. euHCVdb: the European hepatitis C virus database. Nucleic Acids Res. 2007;35:D363–D366. doi: 10.1093/nar/gkl970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aurrecoechea C, Brestelli J, Brunk BP, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer ET, Li W, Miller JA, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Srinivasamoorthy G, Stoeckert CJ, Jr, Thibodeau R, Treatman C, Wang H. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 2010;38:D415–D419. doi: 10.1093/nar/gkp941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.dos Santos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, Emmert DB, Gelbart WM, FlyBase Consortium FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015;43:D690–D697. doi: 10.1093/nar/gku1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frézal J. Genatlas database, genes and development defects. C R Acad Sci III. 1998;321:805–817. doi: 10.1016/s0764-4469(99)80021-3. [DOI] [PubMed] [Google Scholar]
- 74.Safran M, Solomon I, Shmueli O, Lapidot M, Shen-Orr S, Adato A, Ben-Dor U, Esterman N, Rosen N, Peter I, Olender T, Chalifa-Caspi V, Lancet D. GeneCards 2002: towards a complete, object-oriented, human gene compendium. Bioinformatics. 2002;18:1542–1543. doi: 10.1093/bioinformatics/18.11.1542. [DOI] [PubMed] [Google Scholar]
- 75.Lechat P, Hummel L, Rousseau S, Moszer I. GenoList: an integrated environment for comparative analysis of microbial genomes. Nucleic Acids Res. 2008;36:D469–D474. doi: 10.1093/nar/gkm1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Monaco MK, Stein J, Naithani S, Wei S, Dharmawardhana P, Kumari S, Amarasinghe V, Youens-Clark K, Thomason J, Preece J, Pasternak S, Olson A, Jiao Y, Lu Z, Bolser D, Kerhornou A, Staines D, Walts B, Wu G, D’Eustachio P, Haw R, Croft D, Kersey PJ, Stein L, Jaiswal P, Ware D. Gramene 2013: comparative plant genomics resources. Nucleic Acids Res. 2014;42:D1193–D1199. doi: 10.1093/nar/gkt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamasaki C, Murakami K, Takeda J, Sato Y, Noda A, Sakate R, Habara T, Nakaoka H, Todokoro F, Matsuya A, Imanishi T, Gojobori T. H-InvDB in 2009: extended database and data mining resources for human genes and transcripts. Nucleic Acids Res. 2009;38:D626–D632. doi: 10.1093/nar/gkp1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gray KA, Daugherty LC, Gordon SM, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2013. Nucleic Acids Res. 2013;41:D545–D552. doi: 10.1093/nar/gks1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uhlén M, Björling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, Berglund L, Bergström K, Brumer H, Cerjan D, Ekström M, Elobeid A, Eriksson C, Fagerberg L, Falk R, Fall J, Forsberg M, Björklund MG, Gumbel K, Halimi A, Hallin I, Hamsten C, Hansson M, Hedhammar M, Hercules G, Kampf C, Larsson K, Lindskog M, Lodewyckx W, Lund J, Lundeberg J, Magnusson K, Malm E, Nilsson P, Odling J, Oksvold P, Olsson I, Oster E, Ottosson J, Paavilainen L, Persson A, Rimini R, Rockberg J, Runeson M, Sivertsson A, Sköllermo A, Steen J, Stenvall M, Sterky F, Strömberg S, Sundberg M, Tegel H, Tourle S, Wahlund E, Waldén A, Wan J, Wernérus H, Westberg J, Wester K, Wrethagen U, Xu LL, Hober S, Pontén F. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 80.Kikuno R, Nagase T, Nakayama M, Koga H, Okazaki N, Nakajima D, Ohara O. HUGE: a database for human KIAA proteins, a 2004 update integrating HUGEppi and ROUGE. Nucleic Acids Res. 2004;32:D502–D504. doi: 10.1093/nar/gkh035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moszer I, Glaser P, Danchin A. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology. 1995;141:261–268. doi: 10.1099/13500872-141-2-261. [DOI] [PubMed] [Google Scholar]
- 82.Kapopoulou A, Lew JM, Cole ST. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb) 2011;91:8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 83.Andorf CM, Cannon EK, Portwood JL, Gardiner JM, Harper LC, Schaeffer ML, Braun BL, Campbell DA, Vinnakota AG, Sribalusu VV, Huerta M, Cho KT, Wimalanathan K, Richter JD, Mauch ED, Rao BS, Birkett SM, Richter JD, Sen TZ, Lawrence CJ. MaizeGDB 2015: New tools, data, and interface for the maize model organism database. Nucleic Acids Res. 2015;44:D1195–D1201. doi: 10.1093/nar/gkv1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE, The Mouse Genome Database Group The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43:D726–D736. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Biaudet V, Samson F, Bessières P. Micado-a network-oriented database for microbial genomes. Comput Appl Biosci. 1997;13:431–438. doi: 10.1093/bioinformatics/13.4.431. [DOI] [PubMed] [Google Scholar]
- 86.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gaudet P, Argoud-Puy G, Cusin I, Duek P, Evalet O, Gateau A, Gleizes A, Pereira M, Zahn-Zabal M, Zwahlen C, Bairoch A, Lane L. neXtProt: organizing protein knowledge in the context of human proteome projects. J Proteome Res. 2013;12:293–298. doi: 10.1021/pr300830v. [DOI] [PubMed] [Google Scholar]
- 88.Aymé S, Schmidtke J. Networking for rare diseases: a necessity for Europe. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2007;50:1477–1483. doi: 10.1007/s00103-007-0381-9. [DOI] [PubMed] [Google Scholar]
- 89.Thorn CF, Klein TE, Altman RB. PharmGKB: the pharmacogenetics and pharmacogenomics knowledge base. Methods Mol Biol. 2005;311:179–191. doi: 10.1385/1-59259-957-5:179. [DOI] [PubMed] [Google Scholar]
- 90.Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bähler J, Kersey PJ, Oliver SG. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 2012;40:D695–D699. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Winsor GL, Lo R, Ho Sui SJ, Ung KS, Huang S, Cheng D, Ching WK, Hancock RE, Brinkman FS. Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res. 2005;33:D338–D343. doi: 10.1093/nar/gki047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shimoyama M, De Pons J, Hayman GT, Laulederkind SJ, Liu W, Nigam R, Petri V, Smith JR, Tutaj M, Wang SJ, Worthey E, Dwinell M, Jacob H. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 2015;28:D743–D750. doi: 10.1093/nar/gku1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, Karthikeyan AS, Lee CH, Nelson WD, Ploetz L, Singh S, Wensel A, Huala E. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList – 10 years after. Tuberculosis (Edinb) 2011;1:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 96.Bowes JB, Snyder KA, Segerdell E, Gibb R, Jarabek C, Noumen E, Pollet N, Vize PD. Xenbase: a Xenopus biology and genomics resource. Nucleic Acids Res. 2008;36:D761–D767. doi: 10.1093/nar/gkm826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Howe DG, Bradford YM, Conlin T, Eagle AE, Fashena D, Frazer K, Knight J, Mani P, Martin R, Moxon SA, Paddock H, Pich C, Ramachandran S, Ruef BJ, Ruzicka L, Schaper K, Shao X, Singer A, Sprunger B, Van Slyke CE, Westerfield M. ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Res. 2013;41:D854–D860. doi: 10.1093/nar/gks938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J, Gabaldón T, Rattei T, Creevey C, Kuhn M, Jensen LJ, von Mering C, Bork P. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014;42:D231–D239. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Perrière G, Duret L, Gouy M. HOBACGEN: database system for comparative genomics in bacteria. Genome Res. 2000;10:379–385. doi: 10.1101/gr.10.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duret L, Mouchiroud D, Gouy M. HOVERGEN: a database of homologous vertebrate genes. Nucleic Acids Res. 1994;22:2360–2365. doi: 10.1093/nar/22.12.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sonnhammer EL, Östlund G. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015;43:D234–D239. doi: 10.1093/nar/gku1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Altenhoff AM, Škunca N, Glover N, Train CM, Sueki A, Piližota I, Gori K, Tomiczek B, Müller S, Redestig H, Gonnet GH, Dessimoz C. The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res. 2015;43:D240–D249. doi: 10.1093/nar/gku1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva EV. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 2013;41:D358–D365. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huerta-Cepas J, Capella-Gutiérrez S, Pryszcz LP, Marcet-Houben M, Gabaldón T. PhylomeDB v4: zooming into the plurality of evolutionary histories of a genome. Nucleic Acids Res. 2014;42:D897–D902. doi: 10.1093/nar/gkt1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ruan J, Li H, Chen Z, Coghlan A, Coin LJ, Guo Y, Hériché JK, Hu Y, Kristiansen K, Li R, Liu T, Moses A, Qin J, Vang S, Vilella AJ, Ureta-Vidal A, Bolund L, Wang J, Durbin R. TreeFam: 2008 Update. Nucleic Acids Res. 2008;36:D735–D740. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu TJ, Shamsaddini A, Pan Y, Smith K, Crichton DJ, Simonyan V, Mazumder R. A framework for organizing cancer-related variations from existing databases, publications and NGS data using a High-performance Integrated Virtual Environment (HIVE) Database. 2014 doi: 10.1093/database/bau022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Peterson TA, Adadey A, Santana-Cruz I, Sun Y, Winder A, Kann MG. DMDM: Domain Mapping of Disease Mutations. Bioinformatics. 2010;26:2458–2459. doi: 10.1093/bioinformatics/btq447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A, Kolas N, O’Donnell L, Reguly T, Nixon J, Ramage L, Winter A, Sellam A, Chang C, Hirschman J, Theesfeld C, Rust J, Livstone MS, Dolinski K, Tyers M. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, Campbell NH, Chavali G, Chen C, del-Toro N, Duesbury M, Dumousseau M, Galeota E, Hinz U, Iannuccelli M, Jagannathan S, Jimenez R, Khadake J, Lagreid A, Licata L, Lovering RC, Meldal B, Melidoni AN, Milagros M, Peluso D, Perfetto L, Porras P, Raghunath A, Ricard-Blum S, Roechert B, Stutz A, Tognolli M, van Roey K, Cesareni G, Hermjakob H. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schaab C, Geiger T, Stoehr G, Cox J, Mann M. Analysis of high accuracy, quantitative proteomics data in the MaxQB database. Mol Cell Proteomics. 2012;11:M111.014068. doi: 10.1074/mcp.M111.014068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, von Mering C. Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics. 2015;15:3163–3168. doi: 10.1002/pmic.201400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655–D658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O’Kelly G, Schoenegger A, Ovelleiro D, Perez-Riverol Y, Reisinger F, Rios D, Wang R, Hermjakob H. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wienkoop S, Staudinger C, Hoehenwarter W, Weckwerth W, Egelhofer V. ProMEX – a mass spectral reference database for plant proteomics. Front Plant Sci. 2012;3:125. doi: 10.3389/fpls.2012.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Duan G, Li X, Köhn M. The human DEPhOsphorylation database DEPOD: a 2015 update. Nucleic Acids Res. 2015;43:D531–D535. doi: 10.1093/nar/gku1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ross KE, Arighi CN, Ren J, Huang H, Wu CH. Construction of protein phosphorylation networks by data mining, text mining and ontology integration: analysis of the spindle checkpoint. Database. 2013 doi: 10.1093/database/bat038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Durek P, Schmidt R, Heazlewood JL, Jones A, Maclean D, Nagel A, Kersten B, Schulze WX. PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 2010;38:D828–D834. doi: 10.1093/nar/gkp810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dinkel H, Chica C, Via A, Gould CM, Jensen LJ, Gibson TJ, Diella F. Phospho.ELM: a database of phosphorylation sites-update 2011. Nucleic Acids Res. 2011;39:D261–D27. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sadowski I, Breitkreutz BJ, Stark C, Su TC, Dahabieh M, Raithatha S, Bernhard W, Oughtred R, Dolinski K, Barreto K, Tyers M. The PhosphoGRID Saccharomyces cerevisiae protein phosphorylation site database: version 2.0 update. Database. 2013 doi: 10.1093/database/bat026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2014;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Campbell MP, Peterson R, Mariethoz J, Gasteiger E, Akune Y, Aoki-Kinoshita KF, Lisacek F, Packer NH. UniCarbKB: building a knowledge platform for glycoproteomics. Nucleic Acids Res. 2014;42:D215–D221. doi: 10.1093/nar/gkt1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.The Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucl Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Natale DA, Arighi CN, Blake JA, Bult CJ, Christie KR, Cowart J, D’Eustachio P, Diehl AD, Drabkin HJ, Helfer O, Huang H, Masci AM, Ren J, Roberts NV, Ross KE, Ruttenberg A, Shamovsky V, Smith B, Yerramalla MS, Zhang J, AlJanahi A, Çelen I, Gan C, Lv M, Schuster-Lezell E, Wu CH. Protein Ontology: a controlled structured network of protein entities. Nucleic Acids Res. 2014;42:D415–D421. doi: 10.1093/nar/gkt1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mari A, Rasi C, Palazzo P, Scala E. Allergen databases: current status and perspectives. Curr Allergy Asthma Rep. 2009;9:376–983. doi: 10.1007/s11882-009-0055-9. [DOI] [PubMed] [Google Scholar]
- 129.Lombard V, Golaconda RH, Drula E, Coutinho PM, Henrissat B. The Carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lenfant N, Hotelier T, Velluet E, Bourne Y, Marchot P, Chatonnet A. ESTHER, the database of the alpha/beta-hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res. 2013;41:D423–D429. doi: 10.1093/nar/gks1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Isberg V, Vroling B, van der Kant R, Li K, Vriend G, Gloriam D. GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res. 2014;42:D422–D425. doi: 10.1093/nar/gkt1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Giudicelli V, Duroux P, Ginestoux C, Folch G, Jabado-Michaloud J, Chaume D, Lefranc MP. IMGT/LIGM-DB, the IMGT comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res. 2006;34:D781–D784. doi: 10.1093/nar/gkj088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42:D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jeffery CJ. Moonlighting proteins. Trends Biochem Sci. 1999;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- 135.Murphy C, Powlowski J, Wu M, Butler G, Tsang A. Curation of characterized glycoside hydrolases of fungal origin. Database. 2011 doi: 10.1093/database/bar020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fawal N, Li Q, Savelli B, Brette M, Passaia G, Fabre M, Mathé C, Dunand C. PeroxiBase: a database for large-scale evolutionary analysis of peroxidases. Nucleic Acids Res. 2013;41:D441–D414. doi: 10.1093/nar/gks1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE-a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015;43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Saier MH, Reddy VS, Tamang DG, Vastermark A. The transporter classification database. Nucleic Acids Res. 2014;42:D251–D258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Frenkel-Morgenstern M, Gorohovski A, Lacroix V, Rogers M, Ibanez K, Boullosa C, Andres Leon E, Ben-Hur A, Valencia A. ChiTaRS: a database of human, mouse and fruit fly chimeric transcripts and RNA-sequencing data. Nucleic Acids Res. 2013;41:D142–D151. doi: 10.1093/nar/gks1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mihalek I, Res I, Lichtarge O. A Family of Evolution-Entropy Hybrid Methods for Ranking of Protein Residues by Importance. J Mol Bio. 2004;336:1265–1282. doi: 10.1016/j.jmb.2003.12.078. [DOI] [PubMed] [Google Scholar]
- 141.Good BM, Clarke EL, de Alfaro L, Su AI. The Gene Wiki in 2011: community intelligence applied to human gene annotation. Nucleic Acids Res. 2012;40:D1255–D1261. doi: 10.1093/nar/gkr925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schmidt EE, Pelz O, Buhlmann S, Kerr G, Horn T, Boutros M. GenomeRNAi: a database for cell-based and in vivo RNAi phenotypes, 2013 update. Nucleic Acids Res. 2013;41:D1021–D1026. doi: 10.1093/nar/gks1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Igarashi Y, Heureux E, Doctor KS, Talwar P, Gramatikova S, Gramatikoff K, Zhang Y, Blinov M, Ibragimova SS, Boyd S, Ratnikov B, Cieplak P, Godzik A, Smith JW, Osterman AL, Eroshkin AM. PMAP: databases for analyzing proteolytic events and pathways. Nucleic Acids Res. 2009;37:D611–D618. doi: 10.1093/nar/gkn683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Diehn M, Sherlock G, Binkley G, Jin H, Matese JC, Hernandez-Boussard T, Rees CA, Cherry JM, Botstein D, Brown PO, Alizadeh AA. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 2003;31:219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Entrez Programming Utilities Help [Internet] Bethesda (MD): National Center for Biotechnology Information (US); 2010. https://www.ncbi.nlm.nih.gov/books/NBK25501/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Leinonen R, Diez FG, Binns D, Fleischmann W, Lopez R, Apweiler R. UniProt archive. Bioinformatics. 2004;20:3236–3237. doi: 10.1093/bioinformatics/bth191. [DOI] [PubMed] [Google Scholar]
- 147.Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH, UniProt Consortium UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen C, Natale DA, Finn RD, Huang H, Zhang J, Wu CH, Mazumder R. Representative proteomes: a stable, scalable and unbiased proteome set for sequence analysis and functional annotation. PLoS One. 2011;6:e18910. doi: 10.1371/journal.pone.0018910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mostaguir K, Hoogland C, Binz PA, Appel RD. The Make 2D-DB II package: conversion of federated two-dimensional gel electrophoresis databases into a relational format and interconnection of distributed databases. Proteomics. 2003;3:1441–1444. doi: 10.1002/pmic.200300483. [DOI] [PubMed] [Google Scholar]
- 150.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 151.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent WR, Yao H, Markley JL. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Westbrook J, Ito N, Nakamura H, Henrick K, Berman HM. PDBML: the representation of archival macromolecular structure data in XML. Bioinformatics. 2005;21:988–992. doi: 10.1093/bioinformatics/bti082. [DOI] [PubMed] [Google Scholar]
- 153.Karp PD, Paley SM, Krummenacker M, Latendresse M, Dale JM, Lee TJ, Kaipa P, Gilham F, Spaulding A, Popescu L, Altman T, Paulsen I, Keseler IM, Caspi R. Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinform. 2010;11:40–79. doi: 10.1093/bib/bbp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dale JM, Popescu L, Karp PD. Machine learning methods for metabolic pathway prediction. BMC Bioinformatics. 2010;11:15. doi: 10.1186/1471-2105-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, Arnaiz O, Awedh MH, Baldock R, Barbiera G, Bardou P, Beck T, Blake A, Bonierbale M, Brookes AJ, Bucci G, Buetti I, Burge S, Cabau C, Carlson JW, Chelala C, Chrysostomou C, Cittaro D, Collin O, Cordova R, Cutts RJ, Dassi E, Di Genova A, Djari A, Esposito A, Estrella H, Eyras E, Fernandez-Banet J, Forbes S, Free RC, Fujisawa T, Gadaleta E, Garcia-Manteiga JM, Goodstein D, Gray K, Guerra-Assunção JA, Haggarty B, Han DJ, Han BW, Harris T, Harshbarger J, Hastings RK, Hayes RD, Hoede C, Hu S, Hu ZL, Hutchins L, Kan Z, Kawaji H, Keliet A, Kerhornou A, Kim S, Kinsella R, Klopp C, Kong L, Lawson D, Lazarevic D, Lee JH, Letellier T, Li CY, Lio P, Liu CJ, Luo J, Maass A, Mariette J, Maurel T, Merella S, Mohamed AM, Moreews F, Nabihoudine I, Ndegwa N, Noirot C, Perez-Llamas C, Primig M, Quattrone A, Quesneville H, Rambaldi D, Reecy J, Riba M, Rosanoff S, Saddiq AA, Salas E, Sallou O, Shepherd R, Simon R, Sperling L, Spooner W, Staines DM, Steinbach D, Stone K, Stupka E, Teague JW, Dayem Ullah AZ, Wang J, Ware D, Wong-Erasmus M, Youens-Clark K, Zadissa A, Zhang SJ, Kasprzyk A. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015;43:W589–W598. doi: 10.1093/nar/gkv350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.De Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kolesnikov N, Hastings E, Keays M, Melnichuk O, Tang YA, Williams E, Dylag M, Kurbatova N, Brandizi M, Burdett T, Megy K, Pilicheva E, Rustici G, Tikhonov A, Parkinson H, Petryszak R, Sarkans U, Brazma A. ArrayExpress update-simplifying data submissions. Nucleic Acids Res. 2015;43:D1113–D1116. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Haeussler M, Raney BJ, Hinrichs AS, Clawson H, Zweig AS, Karolchik D, Casper J, Speir ML, Haussler D, Kent WJ. Navigating protected genomics data with UCSC Genome Browser in a Box. Bioinformatics. 2015;31:764–766. doi: 10.1093/bioinformatics/btu712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: a next-generation genome browser. Genome Res. 2009;19:630–638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Adler BT, de Alfaro L, Kulshreshtha A, Pye I. Reputation Systems for Open Collaboration. Commun ACM. 2011;54:81–87. doi: 10.1145/1978542.1978560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Orchard S, Kerrien S, Abbani S, Aranda B, Bhate J, Bidwell S, Bridge A, Briganti L, Brinkman FS, Cesareni G, Chatr-aryamontri A, Chautard E, Chen C, Dumousseau M, Goll J, Hancock RE, Hannick LI, Jurisica I, Khadake J, Lynn DJ, Mahadevan U, Perfetto L, Raghunath A, Ricard-Blum S, Roechert B, Salwinski L, Stümpflen V, Tyers M, Uetz P, Xenarios I, Hermjakob H. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat Methods. 2012;9:345–350. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stümpflen V, Ceol A, Chatr-aryamontri A, Armstrong J, Woollard P, Salama JJ, Moore S, Wojcik J, Bader GD, Vidal M, Cusick ME, Gerstein M, Gavin AC, Superti-Furga G, Greenblatt J, Bader J, Uetz P, Tyers M, Legrain P, Fields S, Mulder N, Gilson M, Niepmann M, Burgoon L, De Las Rivas J, Prieto C, Perreau VM, Hogue C, Mewes HW, Apweiler R, Xenarios I, Eisenberg D, Cesareni G, Hermjakob H. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 164.Hermjakob H. The HUPO Proteomics Standards Initiative – overcoming the fragmentation of proteomics data. Proteomics. 2006;6:34–38. doi: 10.1002/pmic.200600537. [DOI] [PubMed] [Google Scholar]
- 165.Hastings J, de Matos P, Dekker A, Ennis M, Harsha B, Kale N, Muthukrishnan V, Owen G, Turner S, Williams M, Steinbeck C. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res. 2013;41:D456–D463. doi: 10.1093/nar/gks1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pedrioli PG, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, Pratt B, Nilsson E, Angeletti RH, Apweiler R, Cheung K, Costello CE, Hermjakob H, Huang S, Julian RK, Kapp E, McComb ME, Oliver SG, Omenn G, Paton NW, Simpson R, Smith R, Taylor CF, Zhu W, Aebersold R. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 167.Eng JK, McCormack AL, Yates JR. An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 168.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical Statistical Model To Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 169.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 170.Wein SP, Cote RG, Dumousseau M, Reisinger F, Hermjakob H, Vizcaino JA. Improvements in the protein identifier cross-reference service. Nucleic Acids Res. 2012;40:W276–W280. doi: 10.1093/nar/gks338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cote R, Reisinger F, Martens L, Barsnes H, Vizcaino JA, Hermjakob H. The Ontology Lookup Service: bigger and better. Nucleic Acids Res. 2010;38:W155–W160. doi: 10.1093/nar/gkq331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Reisinger F, Martens L. Database on demand – an online tool for the custom generation of FASTA formatted sequence databases. Proteomics. 2009;9:4421–4424. doi: 10.1002/pmic.200900254. [DOI] [PubMed] [Google Scholar]
- 173.Hermjakob H, Apweiler R. The Proteomics Identifications Database (PRIDE) and the ProteomExchange Consortium: making proteomics data accessible. Expert Rev Proteomics. 2006;3:1–3. doi: 10.1586/14789450.3.1.1. [DOI] [PubMed] [Google Scholar]
- 174.Pedrioli PGA, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, Pratt B, Nilsson E, Angeletti R, Apweiler R, Cheung K, Costello CE, Hermjakob H, Huang S, Julian RK, Jr, Kapp E, McComb ME, Oliver SG, Omenn G, Paton NW, Simpson R, Smith R, Taylor CF, Zhu W, Aebersold R. A Common Open Representation of Mass Spectrometry Data and its Application in a Proteomics Research Environment. Nat Biotechnol. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 175.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Human Protein Reference Database-2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Aranda B, Blankenburg H, Kerrien S, Brinkman FS, Ceol A, Chautard E, Dana JM, De Las Rivas J, Dumousseau M, Galeota E, Gaulton A, Goll J, Hancock RE, Isserlin R, Jimenez RC, Kerssemakers J, Khadake J, Lynn DJ, Michaut M, O’Kelly G, Ono K, Orchard S, Prieto C, Razick S, Rigina O, Salwinski L, Simonovic M, Velankar S, Winter A, Wu G, Bader GD, Cesareni G, Donaldson IM, Eisenberg D, Kleywegt GJ, Overington J, Ricard-Blum S, Tyers M, Albrecht M, Hermjakob H. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nat Methods. 2011;8:528–529. doi: 10.1038/nmeth.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Torii M, Arighi CN, Li G, Wang Q, Wu CH, Vijay-Shanker K. RLIMS-P 2.0: A Generalizable Rule-Based Information Extraction System for Literature Mining of Protein Phosphorylation Information. IEEE/ACM Trans Comput Biol Bioinform. 2015;12:17–29. doi: 10.1109/TCBB.2014.2372765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tudor CO, Ross KE, Li G, Vijay-Shanker K, Wu CH, Arighi CN. Construction of phosphorylation interaction networks by text mining of full-length articles using the eFIP system. Database. 2015 doi: 10.1093/database/bav020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Cooper CA, Harrison MJ, Wilkins MR, Packer NH. GlycoSuiteDB: a new curated relational database of glycoprotein glycan structures and their biological sources. Nucleic Acids Res. 2001;29:332–335. doi: 10.1093/nar/29.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.von der Lieth CW, Freire AA, Blank D, Campbell MP, Ceroni A, Damerell DR, Dell A, Dwek RA, Ernst B, Fogh R, Frank M, Geyer H, Geyer R, Harrison MJ, Henrick K, Herget S, Hull WE, Ionides J, Joshi HJ, Kamerling JP, Leeflang BR, Lütteke T, Lundborg M, Maass K, Merry A, Ranzinger R, Rosen J, Royle L, Rudd PM, Schloissnig S, Stenutz R, Vranken WF, Widmalm G, Haslam SM. EUROCarbDB: An open-access platform for glycoinformatics. Glycobiology. 2011;21:493–502. doi: 10.1093/glycob/cwq188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Campbell MP, Royle L, Radcliffe CM, Dwek RA, Rudd PM. GlycoBase and autoGU: tools for HPLC-based glycan analysis. Bioinformatics. 2008;24:1214–1216. doi: 10.1093/bioinformatics/btn090. [DOI] [PubMed] [Google Scholar]
- 182.The OpenSFS and Lustre Community Portal. http://lustre.opensfs.org.
- 183.The Apache Hadoop Project. http://hadoop.apache.org.
- 184.The Apache Hive data warehouse software. http://hive.apache.org.
- 185.The Apache Pig platform. http://pig.apache.org.
- 186.The Apache Spark. http://spark.apache.org.
- 187.Belleau F, Nolin MA, Tourigny N, Rigault P, Morissette J. Bio2RDF: Towards a mashup to build bioinformatics knowledge systems. J Biomed Inform. 2008;41:706–716. doi: 10.1016/j.jbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 188.Jupp S, Malone J, Bolleman J, Brandizi M, Davies M, Garcia L, Gaulton A, Gehant S, Laibe C, Redaschi N, Wimalaratne SM, Martin M, Le Novère N, Parkinson H, Birney E, Jenkinson AM. The EBI RDF platform: linked open data for the life sciences. Bioinformatics. 2014;30:1338–1339. doi: 10.1093/bioinformatics/btt765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bootstrap. http://www.getbootstrap.com.
- 190.JQuery. https://www.jquery.com.
- 191.Dojo Toolkit. https://dojotoolkit.org.
- 192.The Apache Lucene. http://lucene.apache.org.


