Abstract
Viral infections have been proposed to elicit pathological processes leading to the initiation of T helper 1 (TH1) immunity against dietary gluten and celiac disease (CeD). To test this hypothesis and gain insights into mechanisms underlying virus-induced loss of tolerance to dietary antigens, we developed a viral infection model that makes use of two reovirus strains that infect the intestine but differ in their immunopathological outcomes. Reovirus is an avirulent pathogen that elicits protective immunity, but we discovered that it can nonetheless disrupt intestinal immune homeostasis at inductive and effector sites of oral tolerance by suppressing peripheral regulatory T cell (pTreg) conversion and promoting TH1 immunity to dietary antigen. Initiation of TH1 immunity to dietary antigen was dependent on interferon regulatory factor 1 and dissociated from suppression of pTreg conversion, which was mediated by type-1 interferon. Last, our study in humans supports a role for infection with reovirus, a seemingly innocuous virus, in triggering the development of CeD.
Celiac disease (CeD) is a complex immune disorder with an autoimmune component in which genetically susceptible individuals expressing the human leukocyte antigen (HLA) DQ2 or DQ8 molecules display an inflammatory T helper 1 (TH1) immune response against dietary gluten present in wheat (1–3). The HLA-DQ2– or HLA-DQ8–restricted TH1 response against gluten is central to CeD pathogenesis and thought to precede development of villous atrophy (4). However, epidemiological and immunological observations support a role for additional genetic and environmental factors in CeD pathogenesis. Similar levels of wheat consumption and expression of CeD-predisposing HLA molecules can be accompanied by striking differences in CeD prevalence (1). A remarkable example supporting a role for environmental factors is the high frequency of CeD in Finnish Karelia (>2%), which contrasts with the low incidence of CeD in the adjacent Russian republic of Karelia (0.2%), two neighboring regions harboring genetically similar populations. Furthermore, the incomplete digestion of gluten by intestinal enzymes (2, 5) explains why gluten would be conducive to inducing intestinal T cell responses. However, it does not explain why CeD patients develop a gluten-specific TH1 response instead of a regulatory immune response, the default intestinal immune reaction to orally ingested protein, characterized by the induction of peripheral regulatory T cells (pTregs) expressing the transcription factor forkhead box P3 (Foxp3) (6).
Viral infection experimental model using genetically engineered reoviruses
Despite epidemiological evidence of associations between viral infections and the initiation of CeD (1), experimental evidence is lacking. Previous studies have implicated adenovirus, enteroviruses, hepatitis C virus, and rotavirus as triggers of CeD (7, 8). However, little is known about the mechanisms by which viruses evoke the disease. Viruses in the family Reoviridae are segmented, double-stranded RNA (dsRNA) viruses that infect humans frequently throughout their lifetime (9). Mammalian Orthoreovirus (reovirus) strains isolated from humans can infect mice via the oral route and activate innate immune pathways similar to the related rotavirus (10, 11). Two human reovirus isolates, type 1 Lang (T1L) and type 3 Dearing (T3D), differ in replication biology, apoptosis induction, innate immune response activation, cellular tropism, and pathogenesis (11). Furthermore, T1L infects the intestine and perturbs intestinal immune homeostasis (11, 12), whereas T3D is incapable of infecting the intestine (11). On the basis of the fundamental differences between T1L and T3D, we hypothesized that engineering a T3D reassortant virus capable of intestinal infection would yield two viruses with potentially different effects on tolerance to dietary antigen. Therefore, we recovered a T3D reassortant virus called T3D-RV by introducing the S1 and L2 gene segments of T1L into a T3D genetic background, thus allowing the virus to infect the intestine (fig. S1A) (13). Such reassortants arise naturally (10, 11) and can be readily recovered in the laboratory by using reverse genetics (11). We first established that the two viruses are similar in their capacity to replicate (fig. S1B) and infect the intestine (fig. S1, C and D). Furthermore, both viruses are cleared (fig. S1E) without inducing intestinal damage (fig. S1F). Although both viruses induced high antireovirus antibody titers, antibody levels observed after T1L infection were significantly higher than those after T3D-RV infection (fig. S1G). However, comparison of the host T cell response in sham- and virus-infected mice revealed that T1L and T3D-RV induced similar TH1 responses in Peyer’s patches (PP) (fig. S1H), the site at which protective immunity to reovirus is induced (12).
Reovirus T1L infection promotes inflammatory immunity to dietary antigen
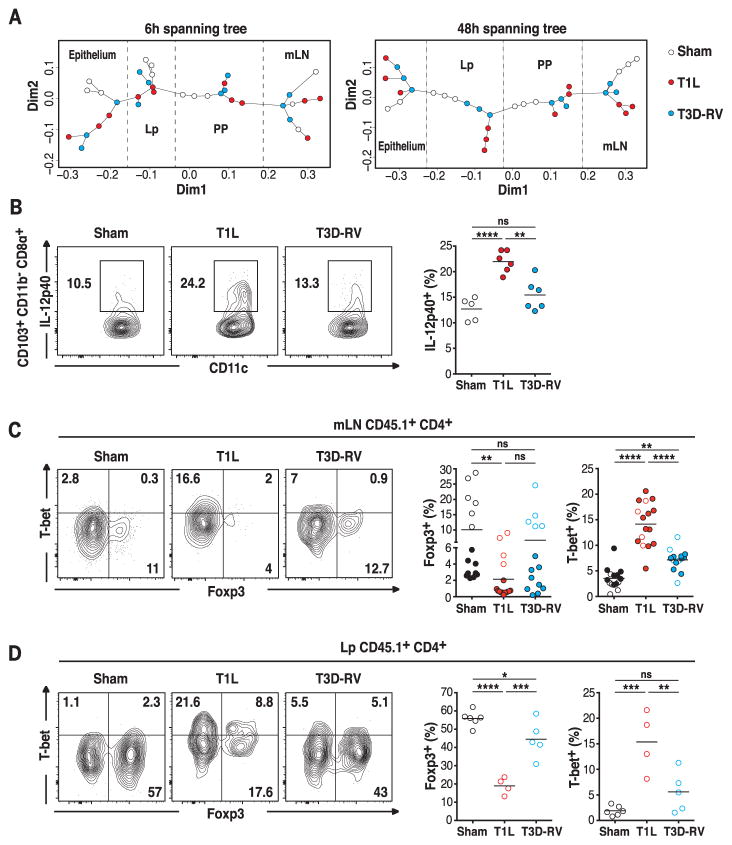
Having established that the two strains infect and induce protective immunity in PP, we next investigated whether they affect immune responses to dietary antigens at inductive and effector sites of oral tolerance—mesenteric lymph nodes (mLN) and lamina propria (Lp) (14, 15), respectively. We took an unbiased approach, using transcriptional profiling to compare and contrast the dynamics of the virus-host interaction across multiple sites of the gut. The application of an unsupervised approach incorporating minimum spanning trees (MSTs) (16, 17) and multidimensional scaling (MDS) ordination (Fig. 1A) revealed transcriptional profile clusters that were strongly driven by differences in location (epithelium, Lp, PP, and mLN) and were influenced by reovirus infection in a location- and time-dependent manner, which is consistent with our understanding of mucosal immunity and intestinal structure and function. This strong location effect was directly evident and predominantly captured by dimension 1 in the MDS scaling operation, which together with dimension 2 also captured the virus-dependent differences. At the early time point (6 hours), both viruses have similar effects primarily on PP and the epithelial compartment (primary sites of infection) (12) and no effect on mLN (Fig. 1A). In contrast, at 48 hours, the viruses altered the transcriptional profile of mLN and Lp (inductive and effector sites of immune responses to dietary antigens) (14, 15), and response differences between the two viruses emerged (Fig. 1A). We therefore followed up this observation with in-depth factorial design analyses (FDAs) (fig. S2) to identify host genes that were differentially expressed in response to the two viruses, characterizing their expression in a time- and location-dependent manner. In both Lp and mLN at 48 hours, we observed a strong enrichment of immune, defense, and antiviral response pathways, including type-1 interferon (IFN) signaling, among genes that were differentially expressed after T1L or T3D-RV infection (fig. S3). Additionally, in mLN, the differential response to the viruses was also characterized by an overrepresentation of cytokine and mitogen-activated protein (MAP) kinase signaling pathways, as well as pathways associated with nutrient response, stress, and vesicle-mediated transport (fig. S3). The finding that T1L induced more extensive transcriptional changes (in the number and level of genes induced) in mLN and Lp at 48 hours compared with that of T3D-RV (Fig. 1A and figs. S2 to S4) raised the possibility that T1L might alter the response to dietary antigen.
Fig. 1. T1L blocks the differentiation of pTregs and promotes TH1 immunity to dietary antigen at inductive and effector sites of the gut.
(A) For each time point, WT mice were inoculated perorally with 1010 plaque forming units (PFU) of T1L (n = 3 mice; red circles), 1010 PFU of T3D-RV (n = 3 mice; blue circles), or phosphate-buffered saline (PBS) (sham, n = 3 mice; open circles) and euthanized 6 or 48 hours after inoculation. RNA of mLN, PP, epithelium, and Lp were isolated and analyzed by means of microarray. MST is represented on multidimensional scaling ordination. The MST traces a path of minimum weight through each vertex or node that represents the profile of differentially expressed genes for each sample state shown. The lengths of edges (or connecting paths) indicate the level of dissimilarity between samples. Each sample state and the distances between them are represented in two-dimensional space. The coordinates of each sample along each dimension are indicated by the two axes. (B) Mice were inoculated perorally with 108 PFU of T1L (n = 6 mice), 108 PFU of T3D-RV (n = 6 mice), or PBS (sham, n = 5 mice) for 2 days. The expression of IL-12p40 on gated MHC-II+ CD11c+ CD103+ CD11b− CD8α+ mLN DCs was evaluated by means of flow cytometry. Representative dot plots and percentages of IL-12p40 in the mLN are shown in the CD103+ CD11b− CD8α+ DC subset. (C and D) OT-II+ CD45.1+ CD4+ T cells were transferred into WT CD45.2+ mice. One day after transfer, mice were inoculated perorally with 1010 PFU of T1L (n = 4 to 16 mice), 1010 PFU of T3D-RV (n = 5 to 14 mice), or PBS (sham, n = 6 to 15 mice) and fed 1.5% OVA in the drinking water (filled circles) or an OVA-containing diet (open circles) for 6 days. The intracellular expression of Foxp3 and T-bet in transferred OT-II+ CD45.1+ CD4+ T cells in the mLN and in the Lp was evaluated by means of flow cytometry. Representative dot plots and percentages of Foxp3+ T-bet− and T-bet+ Foxp3− cells are shown in the mLN (C) and in Lp (D), respectively. [(B) to (D)] Graphs depict at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; one-way analysis of variance (ANOVA)/Tukey’s multiple comparison.
To test this hypothesis, we first determined the effect of in vivo reovirus infection on mLN dendritic cells (DCs). The CD103+ CD11b− DC subset has the highest tolerogenic potential (18, 19) but also drives TH1 responses to intestinal infections (20–22). T1L induced more IL-12p40 than did T3D-RV in CD103+ CD11b− CD8α+ DCs (Fig. 1B), which was the subset that exhibited the highest level of ovalbumin (OVA) uptake after oral administration, irrespective of the virus used (fig. S5, A and B). However, both viruses induced similar levels of IL-12p40 in resident CD103− CD11b− CD8α+ DCs (fig. S5C), whereas no up-regulation of IL-12p40 was detectable in the other mLN DC subsets (fig. S5D). Furthermore, T1L up-regulated more substantially the costimulatory molecule CD86 as well as transcripts of interleukin-27 (IL-27), a cytokine produced by antigen-presenting cells (APCs) that promote TH1 immunity (fig. S5, E and F) (23, 24).
Having found that T1L, but not T3D-RV, induces a proinflammatory phenotype in DCs that take up OVA, we next investigated the effect of the two reoviruses on the T cell response to dietary antigen using an in vivo T cell conversion assay (fig. S6A). In accordance with its capacity to alter the tolerogenic phenotype of CD103+ CD11b− DCs, T1L infection significantly inhibited the conversion of OVA-specific OT-II CD4+ T cells into pTregs and instead promoted their differentiation into T-bet+ and IFNγ+ CD4+ T cells in mLN and Lp, respectively (Fig. 1, C and D, and fig. S6, B to D). In contrast, T3D-RV neither markedly blocked the induction of pTregs nor induced TH1 immune responses against dietary OVA (Fig. 1, C and D, and fig. S6, B to D). Although T1L and T3D-RV differed in their capacity to interfere with responses to dietary antigens, they induced similar host TH1 responses to viral infections in mLN and Lp (fig. S7, A to D).
Oral tolerance is defined as the establishment of peripheral immune tolerance through oral administration of antigen and is thought to be dependent on the induction of pTregs (6). As expected, T1L but not T3D-RV prevented induction of peripheral tolerance upon oral administration of OVA (fig. S7, E to G). Collectively, these results suggest that as a consequence of T1L-host interactions, the tolerogenic response to dietary antigens is abrogated and, instead, TH1 immunity to dietary antigens is induced.
Distinct host pathways block induction of pTregs and induce TH1 immunity to dietary antigen
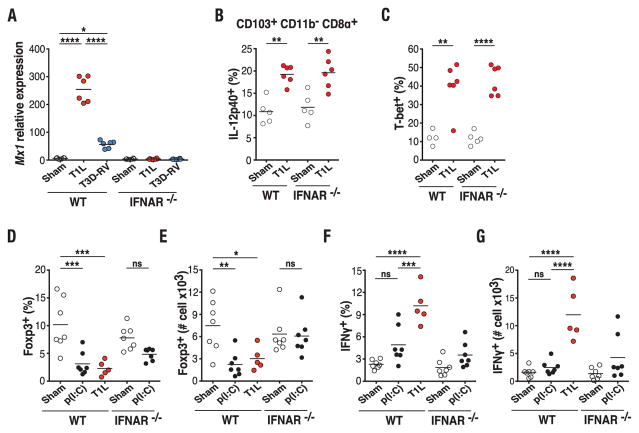
We next sought to determine the mechanistic basis for the differential effect of T1L and T3D-RV infection on the response to dietary antigen. Type-1 IFNs are up-regulated in CeD and have been suggested to explain development of TH1 immunity against dietary gluten (25). RNA sequencing (RNA-seq) analysis of mLN from wild-type (WT) mice and mice in which type-1 IFN receptor has been knocked out (IFNAR−/−) 48 hours after T1L and T3D-RV infection revealed that among the 285 genes found to be differentially expressed between T1L- and T3D-RV–infected mice, 200 genes were regulated in a type-1 IFN-dependent manner (fig. S8), 58 were partially type-1 IFN–dependent, and 27 were type-1 IFN–independent, as assessed with FDA to examine differences in expression response. Further analysis in mLN confirmed that T1L induced higher levels of canonical type-1 IFN inducible genes such as Mx1 and Isg15 than did T3D-RV (Fig. 2A and fig. S9A). This result was contrary to the in vitro data (fig. S9, B to D) and the reported capacity of T1L but not T3D to interfere with type-1 IFN signaling (26), suggesting that yet-to-be-determined differences in virus-host interactions displayed by T1L and T3D-RV can lead to alternative outcomes in mLN in vivo, and that type-1 IFN may be responsible for initiating TH1 immunity against dietary antigen in T1L-infectedmice. To assess this possibility—but to avoid confounding factors associated with uncontrolled T1L replication in the absence of type-1 IFN signaling (27)—we analyzed DCs and OVA-specific T cell conversion at 48 hours, a time point at which viral titers in the ileum are similar in WT and IFNAR−/− mice (fig. S9E). Surprisingly, although CD86 was not induced in IFNAR−/− CD103+ CD11b− CD8α+ DCs (fig. S9F), up-regulation of IL-12p40 in DCs (Fig. 2B and fig. S9G) and Il27 mRNA in mLN (fig. S9H) occurred after T1L infection, suggesting that type-1 IFNs are not required for acquisition of an inflammatory phenotype by mLN APCs. Consistent with these findings, T1L infection induced comparable T-bet expression in OVA-specific CD4+ T cells in WT and IFNAR−/− mice (Fig. 2C and fig. S9I). At this early time point, neither Foxp3 nor IFN-γ can be detected in OVA-specific T cells in the mLN. To assess the role of type-1 IFN in pTreg conversion, an in vivo OT-II T cell conversion assay was performed in WT and IFNAR−/− mice injected intraperitoneally with the dsRNA analog polyinosinic:polycytidylic acid [poly(I:C)]. As shown in Fig. 2, D and E, and fig. S10, A to B, dsRNA was sufficient to block pTreg conversion in a type-1 IFN–dependent manner. Furthermore, type-1 IFNs blocked pTreg conversion comparable with infection with T1L (fig. S10C). In contrast, poly(I:C) (Fig. 2, F and G, and fig. S10D) and type-1 IFN (fig. S10C) were unable to promote TH1 immunity.
Fig. 2. Type-1 IFN is required for blockade of pTreg conversion but not for induction of TH1 immunity to dietary antigen.
(A) WT and IFNAR−/− mice were inoculated perorally with 108 PFU of T1L (n = 6 mice), 108 PFU of T3D-RV (n = 6 mice), or PBS (sham, n = 6 mice) for 2 days. Mx1 expression in the mLN was analyzed by means of reverse transcription polymerase chain reaction (RT-PCR). (B and C) OT-II+ CD45.1+ CD4+ T cells were transferred into WT CD45.2+ or IFNAR−/− CD45.2+ mice. One day after transfer, mice were inoculated perorally with 108 PFU of T1L (n = 6 mice) or PBS (sham, n = 4 or 5 mice) and fed 1.5% OVA in the drinking water for 2 days. The expression of IL-12p40 on gated MHC-II+ CD11c+ CD103+ CD11b− CD8α+ mLN DCs (B) and T-bet in OT-II+ CD45.1+ CD4+ T cells (C) in the mLN was evaluated by means of flow cytometry. (D to G) OT-II+ CD45.1+ CD4+ T cells were transferred into WT CD45.2+ or IFNAR−/− CD45.2+ mice. One day after transfer, mice were inoculated with PBS (n = 7 mice) or 50 mg of poly(I:C) (n = 7 mice) intraperitoneally or 1010 PFU of T1L (n = 5 mice) perorally and fed an OVA-containing diet for 6 days. The intracellular expression of Foxp3 and IFN-γ in OT-II+ CD45.1+ CD4+ T cells in the mLN was evaluated by means of flow cytometry. Percentages and absolute numbers of Foxp3 [(D) and (E)] and IFN-γ [(F) and (G)] are shown. [(A) to (G)] Graphs depict at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; one-way ANOVA/Tukey’s multiple comparison.
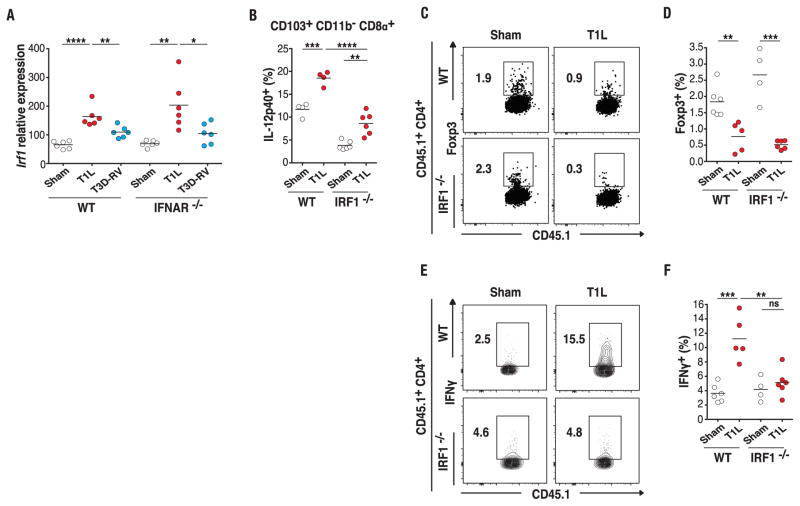
To define the mechanism underlying T1L-induced TH1 immunity to dietary antigen, we interrogated the mLN RNA-seq data from T1L- and T3D-RV–infected WT and IFNAR−/− mice more deeply in order to identify genes that were differentially expressed in a type-1 IFN–independent manner, using FDA (fig. S11A). IL-15, previously shown to prevent pTreg conversion and induce TH1 immunity to dietary antigens (28), was not upregulated after T1L infection in mLN of IFNAR−/− mice (fig. S11B). In contrast, interferon regulatory factor 1 (IRF1)—a transcription factor regulated at the transcriptional level (29) and implicated in multistage regulation of TH1 immune responses (30)—was significantly up-regulated after T1L infection in both WT and IFNAR−/− mice (Fig. 3A and fig. S11A). IRF1 was a particularly intriguing candidate because it is up-regulated in the mucosa of children with CeD (31). To determine whether IRF1 is required in T1L-mediated immunopathology, we analyzed DCs and OT-II T cell conversion in IRF1−/− mice. Viral titers were similar at 48 hours and 6 days after infection in WT and IRF1−/− mice (fig. S11, C and D), enabling us to analyze the response to dietary antigen after oral OVA administration at these two time points. We found that IL-12p40 (Fig. 3B and fig. S11E) and to a lesser extent CD86 (fig. S11F) showed significantly less induction in IRF1−/− mice relative to WT mice. Additionally, induction of Il12b and Il27 mRNA expression was significantly impaired in IRF1−/− mice (fig. S11, G and H). Consistent with preserved type-1 IFN up-regulation after T1L infection in IRF1−/− mice (fig. S11, I and J), absence of IRF1 failed to restore pTreg conversion (Fig. 3, C and D, and fig. S11K). However, whereas type-1 IFN signaling was dispensable at 48 hours (Fig. 2C), IRF1 was required for T1L-mediated TH1 immunity to oral antigen 48 hours (fig. S11, L to N) and 6 days (Fig. 3, E and F, and fig. S11O) after OVA feeding. This result is consistent with the role of IL-12 and IL-27 in TH1 immunity (23, 30, 32) and the abrogation of their up-regulation in IRF1−/− mice (Fig. 3B and fig. S11, G and H). Knowing that the T cell conversion experiments were performed by using OT-II T cells that were IRF1-proficient, we next investigated a role for IRF1 in DCs by performing in vitro T cell conversion assays with mLN DCs isolated 48 hours after T1L infection of WT and IRF1−/− mice. Because IRF1 deficiency skews differentiation of DC subsets (33), we adjusted the relative proportion of the main mLN DC subsets of IRF1−/− mice to approximate that of WT mice (fig. S12, A and B). In keeping with a critical role for IRF1 in DCs, we found that mLN DCs from T1L-infected mice failed to promote TH1 cell differentiation in the absence of IRF1 expression (fig. S12, C and D). Our studies cannot eliminate a potential T cell–intrinsic function of IRF1 in virus-mediated pTreg blockade (34). Furthermore, and in accordance with the defect in IL-12 induction observed in vivo (Fig. 3B and fig. S11G), IL-12 production was impaired in IRF1−/− mLN DCs relative to that in WT mLN DCs (fig. S12E). Last, in support of a direct role for IL-12 in T1L-mediated TH1 immunity to oral antigen, we found that blocking IL-12 in vitro impaired the capacity of T1L-infected WT mLN DCs to convert OT-II T cells into TH1 cells (fig. S12, C and D). These results are in agreement with studies reporting that IRF1 functions in TH1 immunity and IL-12 induction (32, 35). Last, in addition to IL-12, IRF1 is required for IL-27 up-regulation in vivo (fig. S11H), suggesting that it may also participate in the induction of TH1 immunity to dietary antigen upon T1L infection.
Fig. 3. A central role for IRF1 in reovirus-mediated TH1 immunity to dietary antigen.
(A) WT and IFNAR−/− mice were inoculated perorally with 108 PFU of T1L (n = 6 mice), 108 PFU of T3D-RV (n = 6 mice), or PBS (sham, n = 6 mice) for 2 days. Irf1 expression in the mLN was analyzed by means of RT-PCR. (B) OT-II+ CD45.1+ CD4+ T cells were transferred into WT CD45.2+ or IRF1−/− CD45.2+ mice. One day after transfer, mice were inoculated perorally with 108 PFU of T1L (n = 4 to 6 mice) or PBS (sham, n = 4 to 6 mice) and fed 1.5% OVA in the drinking water for 2 days. The expression of IL-12p40 on gated MHC-II+ CD11c+ CD103+ CD11b− CD8α+ mLN DCs was evaluated by means of flow cytometry. (C to F) OT-II+ CD45.1+ CD4+ T cells were transferred into WT CD45.2+ or IRF1−/− CD45.2+ mice. One day after transfer, mice were inoculated perorally with 1010 PFU of T1L (n = 5 or 6 mice) or PBS (sham, n = 4 to 6 mice) and fed 1.5% OVA in the drinking water for 6 days. Intracellular expression of Foxp3 and IFN-γ was evaluated by means of flow cytometry. Representative dot plots (C), percentages of Foxp3 (D), representative dot plots (E), and percentages of IFN-γ (F) are shown in transferred OT-II+ CD4+ T cells in the mLN. [(A) to (F)] Graphs depict two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; one-way ANOVA/Tukey’s multiple comparison.
Taken together, our studies analyzing the effect of T1L on the response to dietary antigen using OVA as a model antigen (fig. S13) suggest that two reovirus strains can induce similar antiviral TH1 responses in PP and yet display distinct immunopathological properties. We show that T1L, but not T3D-RV, promotes an inflammatory phenotype in DCs taking up dietary antigen. In addition, our results indicate that type-1 IFN signaling and IRF1 up-regulation are differentially implicated in blocking pTreg conversion and promoting TH1 immunity to dietary antigen after T1L infection. IRF1 functions in T1L-induced TH1 immunity to dietary antigen by promoting IL-12 and IL-27 in DCs. Although type-1 IFNs are not required (Fig. 3A), they likely contribute to IRF1 up-regulation (36) after reovirus infection in WT mice.
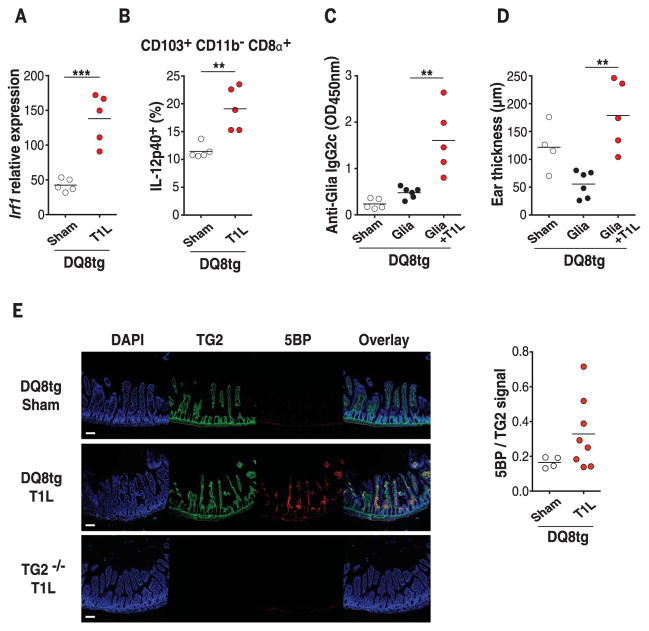
T1L infection breaks oral tolerance to gluten and induces TG2 activation in DQ8tg mice
To determine the relevance of these findings to CeD, we analyzed the effect of T1L infection in transgenic mice expressing the CeD-predisposing HLA molecule DQ8 (DQ8tg mice) (28). First, we confirmed that Irf1 was up-regulated in DQ8tg mice after T1L infection (Fig. 4A). Next, we verified that like OVA-fed WT mice (Fig. 1B and fig. S5B), gluten peptides were preferentially found in CD103+ CD11b− CD8α+ DCs after gavage of sham- or T1L-infected DQ8tg mice (fig. S14, A to C). This DC subset up-regulated IL-12p40 (Fig. 4B and fig. S14D) and CD86 (fig. S14E). The pattern of IL-12p40 expression in the other mLN DC subsets of DQ8tg mice was also similar to that in WT mice (fig. S14F). In addition, Il27 was up-regulated (fig. S14G). In agreement with mLN DCs acquiring a proinflammatory phenotype, T1L infection induced loss of oral tolerance to gluten in DQ8tg mice as assessed by the presence of anti-gliadin IgG2c antibodies (Fig. 4C) and the development of a TH1-delayed hypersensitivity reaction (Fig. 4D and fig. S14H). In addition to intestinal environmental conditions favoring TH1 immunity against dietary antigens, transglutaminase 2 (TG2) activation is thought to promote CeD pathogenesis by increasing the affinity of gluten peptides for HLA-DQ2 and HLA-DQ8 molecules (1–3) through posttranslational modifications. T1L infection induced TG2 activation, as quantified by incorporation of 5-(biotinamido)-pentylamine (5BP), a small-molecule TG2 activity probe, in more than 60% of infected DQ8tg mice (Fig. 4E) without inducing detectable intestinal damage. Thus, in a CeD-relevant mouse model, T1L infection breaks oral tolerance to gluten and promotes TG2 activation, supporting the hypothesis that reovirus infection, despite being clinically silent, can initiate critical events that set the stage for development of CeD.
Fig. 4. Role of reovirus infection in loss of oral tolerance to gluten and TG2 activation.
(A and B) DQ8tg mice were inoculated perorally with 108 PFU of T1L (n = 5 mice) or PBS (sham, n = 5 mice) for 2 days. (A) Levels of Irf1 expression in the mLN were analyzed by means of RT-PCR. (B) The expression of IL-12p40 on gated CD11c+ CD103+ CD11b− CD8α+ mLN DCs was evaluated by means of flow cytometry. (C and D) DQ8tg mice were inoculated perorally with 1010 PFU of T1L at the initiation of an oral tolerance/delayed type hypersensitivity protocol. Mice were fed orally with gliadin (Glia) for 2 days and then immunized subcutaneously with a CFA-Glia emulsion. (C) Levels of Glia-specific IgG2c antibodies in the serum were quantified at day 18 by means of enzyme-linked immunosorbent assay (ELISA). (D) On day 28, mice were challenged subcutaneously with Glia, and the degree of ear swelling was determined 24 hours after challenge. Sham, n = 4 or 5 mice; Glia, n = 6 mice; and Glia + T1L, n = 5 mice. (E) DQ8tg mice were inoculated perorally with 1010 PFU of T1L (n = 8 mice) or PBS (sham, n = 4 mice). TG2−/− mice were inoculated perorally with 1010 PFU of T1L (n = 2 mice) and used as a negative control. Mice were euthanized at 18 hours after infection, and small intestines were collected and frozen in optimal cutting temperature compound. Representative images from stained frozen sections of the proximal small intestines are shown. Scale bars, 100 mm. Staining with 4′,6-diamidino-2-phenylindole (DAPI) is shown in blue, TG2 protein is shown in green, and TG2 enzymatic activity as assessed by means of 5BP cross-linking is shown in red.TG2 enzymatic activity normalized to TG2 protein levels was quantified for each villus. The mean enzymatic activity in the proximal small intestine per mouse is shown. [(A) to (E)] Graph depicts two independent experiments. (A), (B), and (E), **P < 0.01, ***P < 0.001; unpaired t test. (C) and (D), **P < 0.01; one-way ANOVA/Tukey’s multiple comparison.
Evidence for a role of reovirus infection in celiac disease
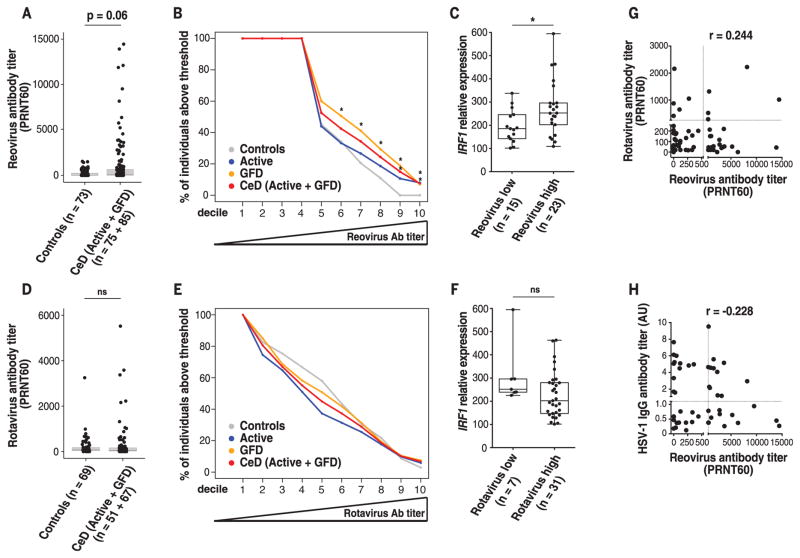
To directly investigate a role for reovirus in CeD, we compared anti-reovirus antibody titers in control individuals with those with active CeD and CeD patients on a gluten-free diet (GFD). CeD patients tended to have higher anti-reovirus antibody titers (P = 0.06) (Fig. 5A) and were significantly overrepresented among patients with very high titers (Fig. 5B). Furthermore, CeD patients on a GFD with high anti-reovirus titers (above the median of our samples) have significantly higher IRF1 levels in the small intestinal mucosa compared with that of individuals with low anti-reovirus titers (Fig. 5C). However, there was no direct correlation between the anti-reovirus antibody titers and the level of IRF1 expression, indicating that there is not a linear relationship between antibody titers and IRF1 levels. Together, these results suggest that the presence of anti-reovirus antibody titers above a certain threshold indicates an antecedent virus-host interaction that caused long-lasting changes in immune homeostasis associated with high IRF1 expression. This hypothesis is in agreement with the concept that viruses may leave a permanent mark on the transcriptional program of the host (37) and is consistent with our observation that mice infected with T1L have higher antibody titers than those of mice infected with T3D-RV (fig. S1G), which does not disrupt tolerance to dietary antigens, unlike T1L. A link between rotavirus infection and development of CeD was suggested in a longitudinal study in children (8). However, our study failed to show such an association (Fig. 5, D to F). Our results do not exclude a role for rotavirus in CeD pathogenesis and could be explained by rotavirus-host immune interactions that differ from those observed with reovirus and hence do not lead to the same immune signatures. Last, high anti-reovirus antibody titers in CeD patients did not correlate with either high anti-rotavirus (Fig. 5G) or high antiherpes simplex virus type 1 (HSV-1) antibody titers (Fig. 5H), indicating that CeD patients displaying high anti-reovirus antibody titers do not mount generally high antibody responses against viruses. Taken together, these results suggest that reovirus infection can trigger the onset of CeD in a subset of CeD patients.
Fig. 5. Role of human reovirus infections in celiac disease pathogenesis.
(A and D) Boxplots showing levels of reovirus antibody titers (A) and rotavirus antibody titers (D) in control (n = 73) and CeD patients (n = 160). (B and E) Percentage of control patients (gray), active CeD patients (blue), CeD patients on a gluten-free diet (GFD) (orange), and active CeD and GFD patients combined (red) that have reovirus (B) or rotavirus (E) antibody titers above an increasingly higher cutoff (left to right). Cutoffs were determined by defining the deciles of the distribution of reovirus (B) or rotavirus (E) antibody titers observed in the patient samples analyzed. GFD and CeD (Active + GFD) patient groups are significantly overrepresented among individuals with reovirus titers above a PRNT60 = 156 (6th decile) and PRNT60 = 1597 (9th decile), respectively. (C and F) IRF1 expression in small intestinal biopsies of GFD patients (n = 38) was analyzed by means of RT-PCR. To avoid any confounding factors associated with increased inflammation, analysis of IRF1 expression was performed in CeD patients on a GFD who, unlike active CeD patients, display normal levels of IFN-γ. Relative expression level of IRF1 in GFD patients with low (left) and high (right) levels of reovirus (C) or rotavirus (F) antibody titers is shown. (C) Reovirus low and reovirus high were respectively defined as individuals with antibody titers below and above the median PRNT60 = 47 [(B), 5th decile]. (F) Rotavirus low and rotavirus high were respectively defined as individuals with antibody titers below and above the median PRNT60 = 53 [(E), 5th decile]. (G and H) Reovirus and rotavirus antibody titers in serum of patients were determined by means of plaque-reduction neutralization assay. Levels of HSV-1 antibody titers were determined by means of ELISA. Correlations between the levels of reovirus antibody titers and rotavirus antibody titers (G) or HSV-1 antibody titers (H) in GFD patients are shown. r, Pearson correlation. (A) to (F), *P < 0.05; Mann-Whitney U test.
Conclusion
This study provides support for the concept that viruses can disrupt intestinal immune homeostasis and initiate loss of oral tolerance and TH1 immunity to dietary antigen. Furthermore, our findngs suggest that an avirulent pathogen, such as reovirus, which is successfully cleared from the infected host, can nonetheless promote immunopathology. In support of this idea, clinically silent norovirus infections increase susceptibility to development of colitis (38). Our data also indicate that two viruses belonging to the same species can have substantially different immunopathological effects. Analysis of the transcriptional profiles induced in response to T1L and T3D-RV suggests that the capacity of a virus to trigger the loss of oral tolerance is associated with its capacity to disrupt immune homeostasis at sites where responses to oral antigens are initiated (fig. S13).
In addition to reovirus, it is probable that other enteric viruses, including viruses belonging to families detected by different immune sensors and involving different signaling pathways, also trigger loss of tolerance to dietary antigen. Expanding the concept of virotypes (37), we propose that viruses eliciting proinflammatory immune responses to dietary antigen alter immune homeostasis and in particular endow DCs with proinflammatory properties at sites where oral tolerance is induced (fig. S13). Identification of other viruses and defining key common features of virus-host interactions leading to the abrogation of oral tolerance will help to design vaccine strategies to prevent CeD and possibly other autoimmune disorders in at-risk populations. On the basis of our findings, even viruses that do not lead to overt clinical pathology could be candidates for such prophylactic intervention.
CeD is a complex disorder that likely requires several environmental perturbations to permanently disrupt tolerance to gluten. Indeed, epidemiological studies longitudinally monitoring genetically at-risk children report transient anti-gluten immune responses before development of fully developed CeD (39, 40) and suggest that anti-gluten antibodies precede anti-TG2 antibodies (41). Furthermore, induction of TH1 immunity to gluten, although required, is insufficient to cause villous atrophy, both in humans and mouse models of CeD (42–44). Our study indicates that although reovirus infections may trigger development of TH1 immunity to gluten as well as activation of TG2, additional events will be required for induction of anti-TG2 antibodies and villous atrophy. Furthermore, nonviral triggers, such as pathogenic members of the microbiota (45, 46), may have disease causing properties similar to reovirus, and the combination of different types of environmental factors likely will eventually lead to formation of a memory pool of TH1 anti-gluten T cells of sufficient magnitude to cause enduring CeD with villous atrophy.
Supplementary Material
Acknowledgments
Library generation and sequencing was performed by the University of Chicago Genomics Facility (http://genomics.bsd.uchicago.edu). Data reported in this paper were deposited into the National Center for Biotechnology Information Gene Expression Omnibus database under the SuperSeries record GSE94604 containing the following SubSeries records: GSE94602 (RNA-seq data) and GSE94517 (Microarray data). Histological slides were generated by the Vanderbilt Translational Pathology Shared Resource supported by National Cancer Institute/National Institutes of Health Cancer Center Support Grant P30 CA068485 and the Vanderbilt Mouse Metabolic Phenotyping Center Grant U24 DK059637. Whole-slide imaging and quantification of immunostaining were performed in the Digital Histology Shared Resource at Vanderbilt University Medical Center (www.mc.vanderbilt.edu/dhsr). We thank P. Savage (University of Chicago) for providing OT-II mice. We thank D. Mucida and D. Esterhazy (The Rockefeller University) for expertise and reagents. We thank B. Xiang for technical assistance. We thank T. Golovkina for helpful suggestions and discussions. This work was supported by the following grants from the NIH: R01DK098435 to B.J., T.S.D., and R.J.X.; R01DK100619 and R01DK067180 to B.J.; R01AI038296 to T.S.D.; R01DK43351 to R.J.X.; F31DK108562 to J.J.B.; R01DK063158 to C.K.; and R01AI113333 and R01DK068181 to H.-C.R. Additional support includes a Cancer Center Support Grant P30CA014599 and Digestive Diseases Research Core Center P30 DK42086 at the University of Chicago to B.J.; an Investigator Start-up Fund, Department of Medicine, MGH to A.N.; an award from the Bettencourt Schueller Foundation to R.B.; a research fellowship from the Dutch Sophia Research Foundation to L.M.M.C.; and the Austrian Science Fund (FWF) J3408-B13 to R.H. Analysis of patient samples in this work was approved by the Institutional Review Board of the University of Chicago (protocol 12623B) and Vanderbilt University (protocol 151358). Last, we thank the CeD patients and their family members as well as the University of Chicago Celiac Disease Center for supporting our research.
Footnotes
REFERENCES AND NOTES
- 1.Abadie V, Sollid LM, Barreiro LB, Jabri B. Annu Rev Immunol. 2011;29:493–525. doi: 10.1146/annurev-immunol-040210-092915. [DOI] [PubMed] [Google Scholar]
- 2.Tjon JM, van Bergen J, Koning F. Immunogenetics. 2010;62:641–651. doi: 10.1007/s00251-010-0465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sollid LM, Jabri B. Nat Rev Immunol. 2013;13:294–302. doi: 10.1038/nri3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nilsen EM, et al. Gastroenterology. 1998;115:551–563. doi: 10.1016/s0016-5085(98)70134-9. [DOI] [PubMed] [Google Scholar]
- 5.Shan L, et al. Science. 2002;297:2275–2279. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- 6.Curotto de Lafaille MA, Lafaille JJ. Immunity. 2009;30:626–635. doi: 10.1016/j.immuni.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Plot L, Amital H. Autoimmun Rev. 2009;8:316–319. doi: 10.1016/j.autrev.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Stene LC, et al. Am J Gastroenterol. 2006;101:2333–2340. doi: 10.1111/j.1572-0241.2006.00741.x. [DOI] [PubMed] [Google Scholar]
- 9.Tai JH, et al. J Infect Dis. 2005;191:1221–1224. doi: 10.1086/428911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angel J, Franco MA, Greenberg HB. Curr Opin Virol. 2012;2:419–425. doi: 10.1016/j.coviro.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dermody TS, Parker JS, Sherry B. Orthoreoviruses. 6. Lippincott Williams & Wilkins; 2013. pp. 1304–1346. [Google Scholar]
- 12.Fleeton MN, et al. J Exp Med. 2004;200:235–245. doi: 10.1084/jem.20041132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodkin DK, Fields BN. J Virol. 1989;63:1188–1193. doi: 10.1128/jvi.63.3.1188-1193.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macpherson AJ, Smith K. J Exp Med. 2006;203:497–500. doi: 10.1084/jem.20060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pabst O, Mowat AM. Mucosal Immunol. 2012;5:232–239. doi: 10.1038/mi.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannistraci CV, Ravasi T, Montevecchi FM, Ideker T, Alessio M. Bioinformatics. 2010;26:i531–i539. doi: 10.1093/bioinformatics/btq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Olman V, Xu D. Bioinformatics. 2002;18:536–545. doi: 10.1093/bioinformatics/18.4.536. [DOI] [PubMed] [Google Scholar]
- 18.Coombes JL, et al. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esterházy D, et al. Nat Immunol. 2016;17:545–555. doi: 10.1038/ni.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luda KM, et al. Immunity. 2016;44:860–874. doi: 10.1016/j.immuni.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Hinterleitner R, Jabri B. Nat Immunol. 2016;17:474–476. doi: 10.1038/ni.3435. [DOI] [PubMed] [Google Scholar]
- 22.Martínez-López M, Iborra S, Conde-Garrosa R, Sancho D. Eur J Immunol. 2015;45:119–129. doi: 10.1002/eji.201444651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida H, Hunter CA. Annu Rev Immunol. 2015;33:417–443. doi: 10.1146/annurev-immunol-032414-112134. [DOI] [PubMed] [Google Scholar]
- 24.van Leeuwen MA, et al. Mucosal Immunol. 2016 doi: 10.1038/mi.2016.76. [DOI] [Google Scholar]
- 25.Di Sabatino A, et al. Gastroenterology. 2007;133:1175–1187. doi: 10.1053/j.gastro.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 26.Irvin SC, et al. J Virol. 2012;86:2302–2311. doi: 10.1128/JVI.06236-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson C, et al. J Exp Med. 2007;204:1349–1358. doi: 10.1084/jem.20061587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DePaolo RW, et al. Nature. 2011;471:220–224. doi: 10.1038/nature09849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura T, Yanai H, Savitsky D, Taniguchi T. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 30.Taki S, et al. Immunity. 1997;6:673–679. doi: 10.1016/s1074-7613(00)80443-4. [DOI] [PubMed] [Google Scholar]
- 31.Salvati VM, et al. Pediatr Res. 2003;54:312–318. doi: 10.1203/01.PDR.0000079184.70237.9C. [DOI] [PubMed] [Google Scholar]
- 32.Maruyama S, et al. J Immunol. 2003;170:997–1001. doi: 10.4049/jimmunol.170.2.997. [DOI] [PubMed] [Google Scholar]
- 33.Gabriele L, et al. J Leukoc Biol. 2006;80:1500–1511. doi: 10.1189/jlb.0406246. [DOI] [PubMed] [Google Scholar]
- 34.Fragale A, et al. J Immunol. 2008;181:1673–1682. doi: 10.4049/jimmunol.181.3.1673. [DOI] [PubMed] [Google Scholar]
- 35.Tada Y, Ho A, Matsuyama T, Mak TW. J Exp Med. 1997;185:231–238. doi: 10.1084/jem.185.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivashkiv LB, Donlin LT. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Virgin HW. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cadwell K, et al. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lionetti E, et al. J Pediatr. 2012;161:908–914. doi: 10.1016/j.jpeds.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Auricchio R, et al. Am J Gastroenterol. 2014;109:913–921. doi: 10.1038/ajg.2014.77. [DOI] [PubMed] [Google Scholar]
- 41.Simell S, et al. Am J Gastroenterol. 2007;102:2026–2035. doi: 10.1111/j.1572-0241.2007.01360.x. [DOI] [PubMed] [Google Scholar]
- 42.Setty M, et al. Gastroenterology. 2015;149:681–691.e10. doi: 10.1053/j.gastro.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mäki M, et al. N Engl J Med. 2003;348:2517–2524. doi: 10.1056/NEJMoa021687. [DOI] [PubMed] [Google Scholar]
- 44.Jabri B, Abadie V. Nat Rev Immunol. 2015;15:771–783. doi: 10.1038/nri3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caminero A, et al. Gastroenterology. 2016;151:670–683. doi: 10.1053/j.gastro.2016.06.041. [DOI] [PubMed] [Google Scholar]
- 46.D’Argenio V, et al. Am J Gastroenterol. 2016;111:879–890. doi: 10.1038/ajg.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.