Abstract
Tumor promotion is an early and critical stage during lung adenocarcinoma (ADC). We previously demonstrated that Tlr4 mutant mice were more susceptible to butylated hydroxytoluene (BHT)-induced pulmonary inflammation and tumor promotion in comparison to Tlr4-sufficient mice. Our study objective was to elucidate the underlying differences in Tlr4 mutant mice in innate immune cell populations, their functional responses, and the influence of these cellular differences on ADC progenitor (type II) cells following BHT-treatment. BALB (Tlr4-sufficient) and C.C3-Tlr4Lps-d/J (BALBLpsd; Tlr4 mutant) mice were treated with BHT (promoter) followed by bronchoalveolar lavage (BAL) and flow cytometry processing on the lungs. ELISAs, Club cell enrichment, macrophage function and RNA isolation were also performed. Bone marrow-derived macrophages (BMDM) co-cultured with a type II cell line were used for wound healing assays. Innate immune cells significantly increased in whole lung in BHT treated BALBLpsd mice compared to BALB mice. BHT treated BALBLpsd mice demonstrated enhanced macrophage functionality, increased epithelial wound closure via BMDMs, and increased Club cell number in BALBLpsd mice, all compared to BALB BHT-treated mice. Cytokine/chemokine (Kc, Mcp1) and growth factor (Igf1) levels also significantly differed among the strains and within macrophages, gene expression and cell surface markers collectively demonstrated a more plastic phenotype in BALBLpsd mice. Therefore, these correlative studies suggest that distinct innate immune cell populations are associated with the differences observed in the Tlr4-mutant model. Future studies will investigate the macrophage origins and the utility of the pathways identified herein as indicators of immune system deficiencies and lung tumorigenesis.
Keywords: inflammation, tumor promotion, innate immunity, toll-like receptor 4, macrophage
Introduction
Lung cancer is the most prevalent type of cancer worldwide, comprising 13% of all new cancer cases and 1.59 million deaths in 2012, thus, the public health burden is evident (1). Adenocarcinoma (ADC), a non-small cell lung carcinoma (NSCLC), is now the predominant subtype in most countries (2), is the most common NSCLC subtype among smokers, andis the only lung cancer found in non-smokers (3). Although there are several recent advances in lung cancer detection, such as the use of CT scans (4) and genetic analysis (5), these methods are still largely investigational and not yet used routinely in diagnosis. Unfortunately, for most patients afflicted with ADC, this very heterogeneous disease is lacking available biomarkers and is typically not diagnosed until advanced stages. Therefore, this emphasizes the need to determine early biomarkers and additional pathways to provide targets for more effective therapies for ADC.
Lung cancer development typically involves several stages (initiation, promotion, and progression) (6–9). Promotion is the only reversible stage of carcinogenesis, the most amenable to therapeutic strategies, and often involves inflammation (6–10). These early stages of cancer are difficult to model in humans, but not mice. Thus we employed a mouse model to focus on the pulmonary microenvironment during tumor promotion (6, 7, 9–11). Our laboratory and others have previously demonstrated using a well-established and human relevant two-stage lung tumor initiation/promotion model (low dose tobacco prevalent carcinogen, 3-methylcholanthrene (MCA), initiator / butylated hydroxytoluene (BHT), promoter) that inflammation and lung injury elicited by BHT correlates to tumor promotion (12–15). Specifically, the oxidative metabolites of BHT in the lung have been found to result in inflammation and pneumotoxicities (16). Multiple doses (four to six) of BHT induce inflammation characterized by pulmonary influx of macrophages, lymphocytes, and to a lesser extent, polymorphonuclear leukocytes (PMNs) (13, 15, 17). Specific depletion of either macrophages (sodium hypochlorite) or PMNs (anti-Ly6G antibody) significantly reduces BHT-induced tumor promotion, however depletion of T-cells (CD4 and CD8; anti-CD4 and CD8 antibodies in thymectomized BALB mice) does not impact promotion (13, 15). Hence, there is sufficient evidence to suggest a role for inflammation and further, a specific role for macrophages and PMNs in our model, as well as in human lung cancer and other mouse models (7, 10, 18).
Toll-like receptor 4 (TLR4) is an innate immune receptor responsible for recognition of lipopolysaccharide (LPS) (19) and can both exacerbate and protect against lung insults. For example, TLR4 is involved in lung injury and inflammation in response to ozone (20–22), however, its absence leads to acute lung injury, sepsis, and emphysema (23, 24). Interestingly, chronic occupational exposures (e.g. farm and textile workers) to endotoxin which contains LPS (25), the primary ligand for TLR4, also associate with a significantly decreased risk of developing lung cancer (26, 27). Our previous studies investigated the role of TLR4 in BHT-induced pulmonary inflammation and tumorigenesis (28–30). We demonstrated that Tlr4-deficiency led to significant increases in lung hyperpermeability (indicative of lung injury), inflammation (macrophage and lymphocyte infiltration), primary tumor formation, and decreased gap junctional intercellular communication compared to Tlr4-sufficient control (BALB) mice at both early and late stages of carcinogenesis (please see these figures in previous publications for more details: Fig. 1 (Bauer et al., 2005), http://jnci.oxfordjournals.org/content/97/23/1778.long; Fig 3 (Bauer et al., 2009), http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2785769/figure/F3/; Fig. 3 (Hill et al., 2013), http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4098145/figure/F3/). We also observed differential transcriptional profiles in the whole lung tissue between the Tlr4-mutant and -sufficient mice, such as some immune and/or cytokine/chemokine signaling pathways (eg. Ccr2). Some of these pathways identified supported a role for inflammation driving the ADC progenitor cells (bronchiolar Club cells and alveolar type II cells) to proliferate in the BHT promotion model (29, 31). Thus, prior mouse studies in our laboratory support the association in humans between LPS exposure and lung cancer protection and suggest that individuals with defective innate immune pathways may be more susceptible to lung carcinogenesis.
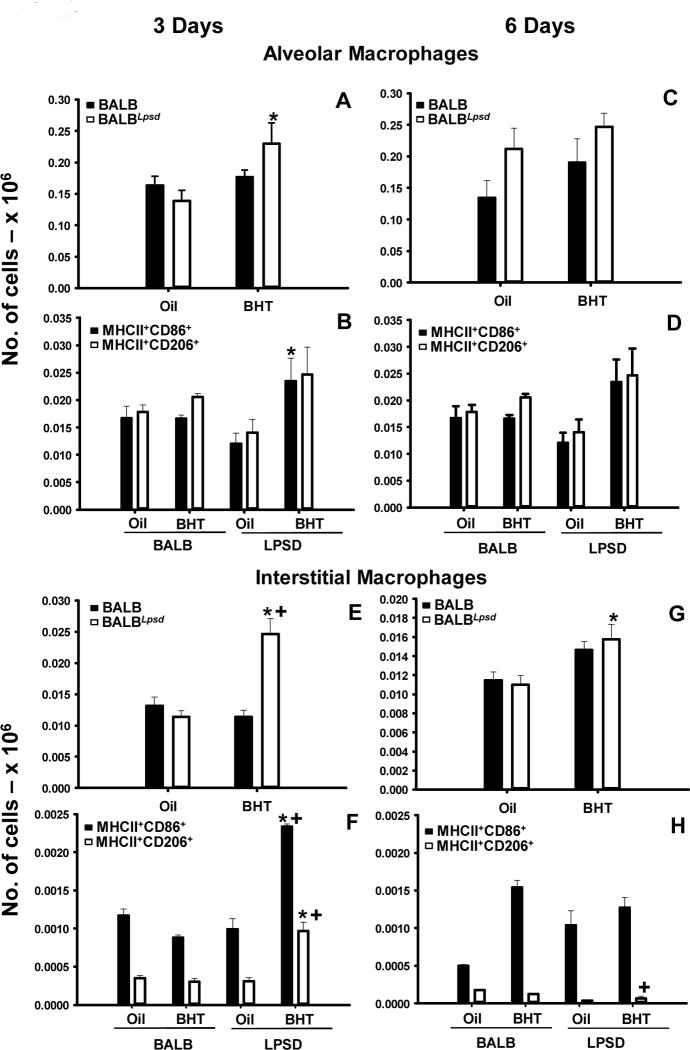
Figure 1. Pulmonary alveolar and interstitial macrophage populations differ between strains 3 days following BHT.
Lung cells were stained with CD11c, CD11b, F4/80, Ly6G, MHCII, CD86 and CD206 for immunophenotyping. A) Alveolar macrophages (AM) -CD11c+CD11b 3 days following BHT or oil; B) Additional markers for alveolar macrophages at the 3 day time point - +Ly6GMHCII+CD86+ or CD206+; C) Alveolar macrophages (AM) -CD11c+CD11b 6 days following BHT or oil ; D) Additional markers for alveolar macrophages at the 6 day time point - +Ly6GMHCII+CD86+ or CD206+. E) Interstitial macrophages (IM) - CD11cCD11b+F4/80+Ly6G 3 days following BHT or oil ; F) Additional markers for interstitial macrophages at the 3 day time point - MHCII+CD86+ or CD206+; G) Interstitial macrophages (IM) - CD11cCD11b+F4/80+Ly6G 6 days following BHT or oil ; H) Additional markers for alveolar macrophages at the 6 day time point - MHCII+CD86+ or CD206+. Mean ± SEM presented; n = 5/group; repeated 2 times. *, P<0.05 compared to oil controls; +, p<0.05 compared to BALB mice.
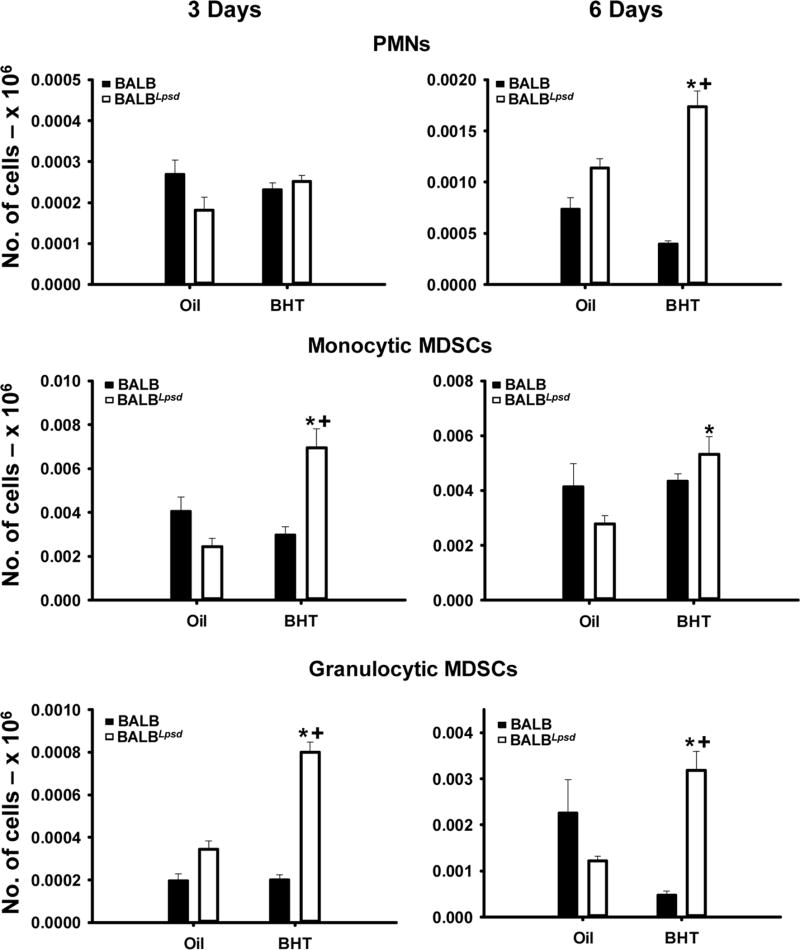
Figure 3. PMNs and both MDSC populations demonstrated differences between BALBLpsd and BALB strains treated with BHT.
A) PMNs (CD11b+F480Ly6G+) 3 (left) and 6 (right) days following BHT. B) monocyte (m)MDSCs (CD11bhiF480+Ly6G) were identified in both strains with BHT treatment or controls 3 (left) and 6 (right) days following BHT. C) granulocytic (g)MDSCs (CD11bhiF480Ly6G+) were identified in both strains with BHT treatment or controls 3 (left) and 6 (right) days following BHT. Mean ± SEM presented; n = 3; repeated 3 times. *, P<0.05 compared to oil controls; +, p<0.05 compared to BALB mice.
Based on the studies described above and the remaining gap in our understanding of the specific types of immune cells involved in this observed protection, we then investigated the pulmonary microenvironment, which during chronic lung disease, is a complex mixture of inflammatory cell types, with many unique functions, such as multiple macrophage populations (e.g. alveolar vs. interstitial), lymphocyte populations (e.g. CD4+ vs. CD8+), dendritic cells (DCs), PMNs, and myeloid-derived suppressor cells (MDSCs) (32–34). Macrophages within different regions of the lung (alveolar vs. interstitial) can also be further phenotyped, traditionally identified by classical activation (ie. M1 macrophages) eliciting more host defense responses compared to alternative activation (ie. M2 macrophages) eliciting more proliferative responses (wound healing) in diseases such as fibrosis and cancer (35–38). However, recent studies demonstrate that macrophage programming is more complex than simply an M1 vs. M2 macrophage, with many macrophage signatures co-existing depending on the microenvironment. In the lung the distribution of macrophage phenotypes is dependent on location (17, 32, 33, 37, 39). We previously observed both bronchoalveolar lavage fluid (BALF) macrophage and lymphocyte populations significantly elevated in Tlr4-mutant mice above that in the Tlr4-sufficient mice in response to BHT, however the distinct types of inflammatory cells were unknown as well as their functions (e.g. phagocytosis vs. efferocytosis) (40). Therefore, to understand the novel phenotypes of these inflammatory cells involved in the observed TLR4 protection, our study objectives were to elucidate the underlying differences in Tlr4 mutant mice in innate immune cell populations and their functional responses. Herein, we immunophenotyped these cells, determined increases in specific cell populations and with some, their corresponding functionality, in the Tlr4-mutant mice in comparison to Tlr4-sufficient mice. In addition, we evaluated transcriptome differences in the BALF macrophages from BALB vs. BALBLpsd as well as investigated numerous inflammatory proteins in the lung that likely influence tumor promotion, such as cytokines and growth factors (e.g. insulin-like growth factor 1 (IGF1)). Supporting a key role of pulmonary macrophages, macrophage progenitor cells and other innate immune cells in driving promotion, BHT treatment significantly increased wound healing in epithelial cells grown in co-culture with bone marrow-derived macrophages (BMDM) from BHT-treated BALBLpsd mice and bronchiolar Club cell number in BALBLpsd mice over that observed in BALB (Tlr4-sufficient) mice. Collectively, these studies are the first to immunophenotype the cells involved in BHT-induced inflammation that is associated with tumor promotion. Further, these novel studies demonstrate variations between immune cell type specific signatures, functionality of the macrophages, and proliferative effects on ADC progenitor cells in mice lacking TLR4.
Materials and Methods
Animals
Five- to seven-week old male C.C3-Tlr4Lps-d/J (BALBLpsd; Tlr4 dominant negative mutant) (41) mice were bred in animal facilities at the University of Colorado, Anschutz Medical Campus. Age-matched male BALB/cJ (BALB; Tlr4 wild-type) mice were purchased from Jackson Laboratories (Bar Harbor, ME). Mice were acclimated 1 wk prior to studies in a pathogen-free facility. Mice were fed irradiated mouse chow (Harlan) and water ad libitum and housed in shoebox cages with humidity and temperature-controlled. All animal use was conducted in facilities accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care and approved by the University of Colorado Denver Institutional Animal Care and Use Committee and follows the Helsinki convention for the use and care of animals.
Experimental Design
Mice were injected intraperitoneally (ip.) with either butylated hydroxytoluene (BHT; Sigma, St. Louis MO) to induce inflammation, as done previously; or corn oil (Mazola) as vehicle control. Injections were administered weekly, for 4 weeks (150 mg/kg for the first dose followed by 200 mg/kg for the following 3 doses) after which the mice were sacrificed at 3 or 6 days following the last injection, based on previously determined time points (28, 29). Mice were euthanized with Fatal Plus (120 mg/kg; MWI, Boise, ID). Cell differentials to determine the number and type of immune cells were performed on each mouse (28, 29).
Immunophenotyping of immune cells in the lung
At sacrifice, lungs were ½ lung bronchoalveolar lavaged (42) with Hanks Balanced Salt Solution (HBSS) with ethylenediaminetetraacetic disodium salt (EDTA) to determine immune cell constituents using cellular differentials in the BAL fluid (BALF), as done previously (29). For flow cytometry, the lavaged right lung was minced and digested with 1 mg/ml collagenase (43) at 37°C for 30 min (44). The digested lung was then continuously agitated using a Pasteur pipet, filtered through a 70 µm filter and washed with HBSS. The cell pellet was re-suspended in RBC lysis buffer, washed, and re-suspended in PBS-BSA. The cell suspension was then counted using the Countess Automated Cell Counter (Life Technologies-Invitrogen,Grand Island NY). To minimize non-specific staining, the cells were incubated with Mouse BD Fc Block (CD16/CD32 clone 2.4G2) (BD Pharmingen, San Jose CA) and rat serum for 20 min prior to primary staining. The cells were stained with the following antibodies: Ly-6G-FITC (clone 1A8) (BD Pharmingen); CD206-PerCP/Cy5.5 (clone C068C2) and CD86-Alexa Fluor 700 (clone PO3) (BioLegend, San Diego, CA); and F4/80-PE-Cy7 (clone BM8), CD11b-eFluor 450 (clone M1/70), CD11c–APC-eFluor 780 (clone N418) and MHC II-APC (clone M5/114.15.2) (eBioscience, San Diego, CA). Siglec-F-PE (clone E50–2440; BD Pharmingen) was also used to confirm alveolar macrophages. After staining, cells were washed twice with PBS-BSA and fixed in 2% paraformaldehyde. Cells were analyzed using the BD LSR II Flow Cytometer (BD Biosciences) at the University of Colorado Flow Cytometry and Cell Sorting Facility (UC Cancer Center).
Phagocytosis and efferocytosis functional assays
BALF was collected, cells pelleted, and counted, and the remaining cell-free BALF removed for further analyses. BALF cells (50,000/well; ∼95% macrophages) were plated in a 96-well plate with macrophage complete media [Dulbecco’s Modified Eagle Medium (DMEM; Life Technologies, Grand Island NY), 50 ml fetal bovine serum (FBS; Atlanta Biological, Atlanta GA), 5 mL L-Glutamine (Life Technologies) and 5 mL Penicillin Streptomycin); incomplete media contains the same components without serum] and incubated at 37°C for 30 mins-1 h to allow macrophages to attach. The media was replaced with fresh media and the plate incubated overnight at 37°C. For the phagocytosis assay, Zymosan bioparticles (Life Technologies) were added to each well (18 hr after plating) with macrophage incomplete media and incubated at 37°C for 1 hr. The macrophages were washed 3 times with PBS followed by staining with 0.4% trypan blue for 1 min. The cells were washed 4 times with PBS, re-suspended in macrophage complete media then stained with Hoechst’s Dye (Life Technologies) for 7 min. Macrophages were washed 3 times with PBS and fixed with 4% Formaldehyde (Electron Microscopy Sciences, Hatfield PA) for 20 min. The formaldehyde was removed, 200 µl PBS added to each well, and the plate stored at 4°C until imaging on a Perkin Elmer Operetta with guidance from the High Throughput and High Content Screening Core Facility (UCD). Harmony Software version 3.5 was then used for analyzing and imaging the nucleus with Hoechst staining and phase-contrast compared to the cells that had internal Zymosan particles. Phagocytic index = total Zymosan ingestion/total macrophages × 100.
For the efferocytosis assay, Jurkat cells (ATCC, Manassas VA) were radiated with ultraviolet radiation at 302 nm for 12.5 min, then incubated at 37°C for 2.5 hr. The irradiated Jurkat cells were added to 50,000 BALF cells (plated the same as those described for phagocytosis to remove all other cell types, thus macrophages remain) with macrophage incomplete media and incubated at 37°C for 2 hr. The cells were washed 3 times with PBS then stained with Hema 3 Solution II stain (Fisher Scientific, Kalamazoo MI), similar to Wright’s Giemsa (45). Cells were then counted on an Olympus CKX41 Microscope at 40x magnification. Efferocytic index = total apoptotic ingestions/total macrophages × 100.
RNA isolation and cDNA synthesis for mRNA analyses from BALF immune cells
Total RNA was isolated from BALF immune cells (primarily macrophages, ∼90%, 10% lymphocytes) utilizing the Macherey-Nagel NucleoSpin RNA II kit (Clontech Laboratories, Mountain View CA) following kit specifications. cDNA for the qRTPCR was prepared from the total RNA via whole transcriptome amplification using the QuantiTect Whole Transcriptome Kit (Qiagen, Valencia CA) following kit specifications.
Affymetrix Micro Array Mouse Gene 2.0 array analysis
Total RNA was quality tested using an Agilent Tapestation Bioanalyzer (Agilent) and only those samples passing with a RIN of 7 or higher were used for the microarray studies and validation using qRTPCR analysis. Gene expression analysis was conducted using Affymetrix Mouse Gene 2.0 ST arrays, similar to previous studies (29). All array studies, including quality testing, amplification of total RNA, hybridization, and scanning of the arrays were performed at the Genomics and Microarray Core at the UCD Cancer Center, similar to that described in Bauer et al., 2009 (29). Microarray analysis was performed using Partek Genomics Suite 6.6 software to determine gene expression differences between strains and treatment. The CEL files were all RMA normalized. The samples were then normalized to the BALB oil treatment group and ≥ 1.3-fold changes in genes with increased or decreased expression identified in BALBLpsd macrophages compared to BALB BHT samples using a 2-way analysis of variance (ANOVA; p<0.05) followed by the Benjamini and Hochberg False Discovery Rate test for multiple comparisons p<0.05. Number of animals per treatment group was: n=4 for BALB mice; n=2 for oil treated BALBLpsd mice and n=3 for BALBLpsd BHT treated mice. The raw data discussed in this publication have been deposited into NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/, Series GSE68067).
Quantitative real time PCR (qRTPCR)
qRTPCR was done using Sybr green assays on an Eppendorf Mastercycler ep realplex4 (Eppendorf, Westbury NY) as follows: 10 µl 2X SYBR Green PCRMaster Mix, 250 nM forward and reverse primers, 3.0 µl nuclease-free H2O and 2 µl of cDNA per 20 µl reaction. The reverse and forward primers and the PCR reaction conditions can be found in the Table S1. L32 was used to normalize all the genes (46).
Enzyme linked immunosorbent assay (ELISA)
A KC (Cxcl1) DuoSet ELISA (RnD Systems, Minneapolis MN) was used to determine KC content in whole lung cell homogenates (50 µg). Raybiotech IGF-1 ELISA (100 µg) and Mouse Cytokine Arrays (50 µg; QAM-CAA-1000; RayBiotech, Inc., Norcross, VA) were also used to detect cytokine, chemokine, and growth factor content within whole lung tissue homogenates. The results were analyzed using Prism 6.0 (GraphPad, La Jolla CA), with the exception of the Raybiotech Mouse Cytokine Arrays which were analyzed at RayBiotech, Inc.
Bronchiolar Club (Clara) cell enrichment
Club cells enrichment used an established method (47). Briefly, lungs were digested with intratracheally instilled fresh elastase (Worthington Biochemical, Freehold, NJ) to detach Club cells, minced, and then filtered. Club cells were separated from macrophages by differential adherence to an IgG (Sigma, St. Louis, MO) -coated plate. Club cell purity was >80% and the cells were >95% viable. Cells were then counted using a hemocytometer to determine the total number of cells per treatment and strain.
Wound healing assay using a co-culture system
C10 cells, a non-tumorigenic alveolar type II cell pneumocyte cell line derived from BALB lung (48), were grown in 12-well dishes to confluence using C10 complete media (CMRL Media (Invitrogen), 10% Fetal Bovine Serum,1% L-Glutamine; 1% Penicillin streptomycin). Bone marrow cells were flushed with bone marrow derived macrophage media ((BMDM) media: RPMI medium 1640 (Invitrogen); 1% Sodium Pyruvate (Invitrogen); 175 µl β-mercaptoethanol; 10% Fetal Bovine Serum; 1% L-Glutamine; 1% Penicillin streptomycin) from femurs of previously treated experimental mice, washed, and counted. Flasks were seeded and incubated overnight at 37°C for stromal elimination. After 7 – 10 days of washing, media changes, and addition of macrophage-colony stimulating factor (M-CSF; R&D Systems, Minneapolis, MN), cells were differentiated, as previously described (49), and verified using the F4/80 antibody used above. At a 1:5 ratio of macrophage:epithelial cell, BMDMs were then placed into 0.4 µm inserts (USA Scientific, Ocala, FL) and following C10 cells confluency, placed in the same well as the C10 cells for co-culture overnight for incubation at 37°C. A wound was made the following day in the center of the C10 cell using a 200 µl pipette tip to press firmly against the plate and media replaced (50).The plate was washed with C10 incomplete media (complete media minus the FBS) and C10 complete media (with 2.5% FBS instead of 10%) was added to the wells and inserts. The plate was imaged at 6, 12, and 24 hr using the Nikon Eclipse Ti inverted research microscope (Nikon Corporation, Tokyo, Japan) to determine any changes. Areas were determined using NIH Image J Software.
Statistical analysis
Statistics was performed using a 2-way ANOVA (factors: strain and treatment) and comparisons of means used Student-Newman-Keuls (SNK) a posteriori test). Data presented are all mean ± SEM. Sigma Plot (12.3) software was used for all graphs and statistics. For all analyses, p<0.05 was considered statistically significant.
Results
Immunophenotyping of pulmonary immune cells in response to BHT
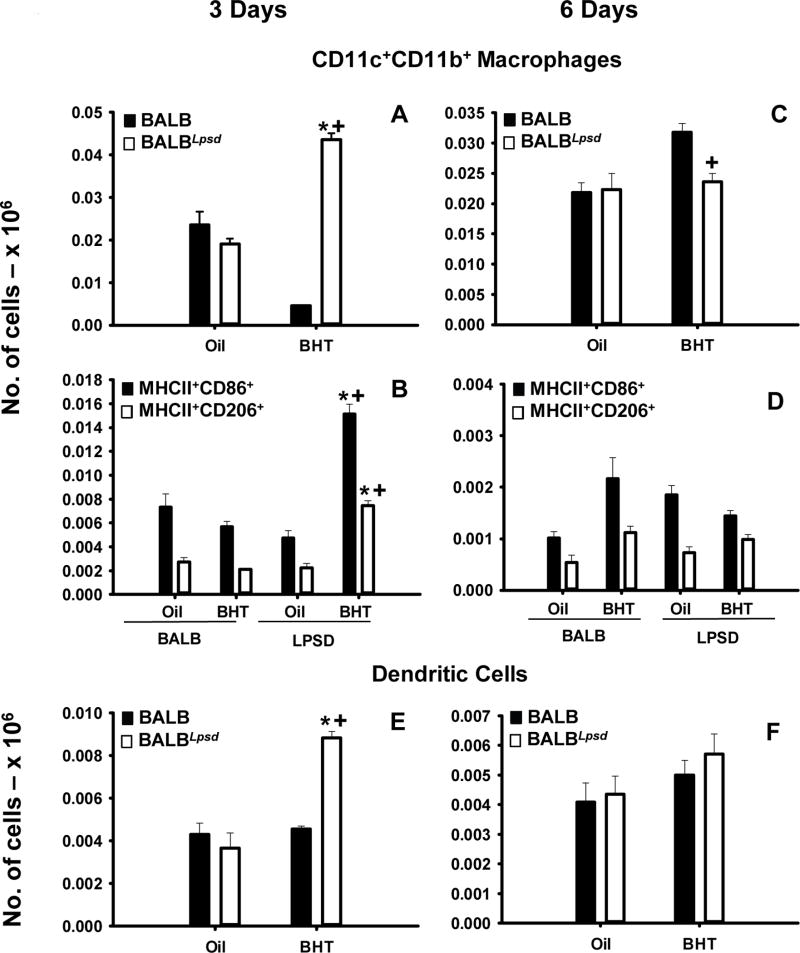
These studies utilized flow cytometric staining to immunophenotype specific pulmonary immune cell populations between strains at two previously identified key time points for peaks in macrophage infiltration into BALF (day 3 and 6 following BHT) (28). BALF cell differentials were significantly different (macrophages and lymphocytes) between the BALB and BALBLpsd mice in response to BHT and validated previous findings (Table S2) (6, 28, 30). Several cell surface markers were used and macrophages segregated into three subtypes: alveolar macrophages (AM) – CD11c+CD11b− F4/80+Ly6G−, interstitial macrophages (IM) – CD11c−CD11b+F4/80+Ly6G− and double positive (DP, activated) macrophages CD11c+CD11b+F4/80+Ly6G− macrophages (Fig 1 A,C,E,G and Fig 2 A, C). In all three macrophage subtypes, there were significant increases in the numbers of macrophages in the BalbLPSD BHT-treated mice in comparison to the Tlr4-sufficient mice (BALB) and control (oil treated) BalbLpsdmice 3 days following BHT (P <0.05). There were expected differences in the numbers of alveolar macrophages in comparison to the other two subtypes identified, which has been previously demonstrated in the literature (37). Further characterization of the macrophages utilizing MHCII, CD86 – “M1” and CD206 – ‘mannose receptor, “M2” markers (Fig 1B,D,F,H and Fig 2 B, D), demonstrated similar significant trends in responses, whereby there were increased numbers of both types of macrophage programming in the Tlr4-mutant mice in comparison to the other treated mice. Within the AM subtype, there were few differences between these markers compared to the IM and the DP macrophages, which exhibited more CD86 staining, albeit with significant increases in the CD206 staining macrophages as well. Interestingly, another antigen presenting cell type, dendritic cells (DCs; CD11b+CD11c+F4/80−Ly6g−), were also significantly elevated in the BALBLpsd mice with BHT treatment compared to BALB mice (Fig 2E). These observed changes were all at 3 days following BHT; no macrophage or DC differences were observed at the 6 day time point with flow, however, BALF macrophages were significantly elevated in the BALBLpsd mice compared to BALB with BHT treatment.
Figure 2. Pulmonary CD11b+CD11c+ innate immune cell populations differ between BALBLpsd and BALB mice following BHT.
Lung cells were stained with CD11c, CD11b, F4/80, Ly6G, MHCII, CD86 and CD206 for immunophenotyping. A) Activated double positive (DP) macrophages - CD11c+CD11b+F4/80+Ly6G 3 days following BHT or oil ; B) Additional markers for DP activated macrophages at the 3 day time point - MHCII+CD86+ or CD206+; C) Activated double positive (DP) macrophages - CD11c+CD11b+F4/80+Ly6G 6 days following BHT or oil ; D) Additional markers for DP activated macrophages at the 6 day time point - MHCII+CD86+ or CD206+;Dendritic cells- CD11c+CD11b+F4/80Ly6G 3 (E) and 6 (F) days following BHT or oil. Mean ± SEM presented; n = 5; repeated 2 times. *, P<0.05 compared to oil controls; +, p<0.05 compared to BALB mice.
We also evaluated several other immune cell types within the lungs of these mice including lymphocyte populations (Table 1), myeloid-derived suppressor cells (MDSC; Fig 3), and PMNs (Fig 3). At 3 days, we observed increases in CD3+CD4+CD8− T cells and B cells (B220+), but not CD3+CD4−CD8+ T cells, in both strains with BHT (Table 1). Mature natural killer (NK) cells (B220−CD49+) were also significantly increased in BALBLpsd mice 3 days following BHT, over that observed in any other treatment, however immature NK cells (B220+CD49+) were unchanged. At 6 days, CD4+ T cells, CD8+ T cells, B cells, and NK cells were all unchanged. Additionally, we observed significant differences in the granulocytic (gMDSCs; CD11bhiLy6G+F4/80−) and monocytic MDSCs (mMDSCs; CD11bhiLy6G−F4/80+) at the 3 day time point (P< 0.05), and PMNs (CD11c−CD11b+Ly6G+F4/80−) and gMDSCs at 6 days in the Tlr4-mutant mice only (Fig 3).
Table 1.
Pulmonary lymphoid cell populations in BALB and BALBLpsd mice following BHT analyzed by flow cytometry.
| Time point | Cell Type* | BALB | BALBLpsd | ||
|---|---|---|---|---|---|
| Oil | BHT | Oil | BHT | ||
| (× 104 cells) | (× 104 cells) | ||||
| 3 day BHT | B | 1.2 ± 0.002 | 2.0 ± 0.001 † | .3 ± .04 | 1.0 ± 0.2 † |
| CD4+ T | 1.0 ± 0.1 | 2.8 ± 0.1 † | 0.3 ± 0.04 | 2.1 ± 0.3 † | |
| CD8+ T | 0.2 ± 0.1 | 0.3 ± 0.04 | 0.04 ± 0.01 | 0.3 ± 0.04 | |
| Mature NK | 0.1 ± 0.01 | 0.01 ± 0.02 | 0.03 ± 0.01 | 0.2 ± 0.02 †‡ | |
| Immature NK | 0.0003 ± 0.0002 | 0.0004 ± 0.0002 | 0.0004 ± 0.0001 | 0.0002 ± 0.0001 | |
| 6 day BHT | B | 3.0 ± 0.7 | 1.6 ± 0.2 | 1.2 ± 0.4 | 1.4 ± 0.1 |
| CD4+ T | 0.5 ± 0.1 | 2.8 ± 0.3 | 1.4 ± 0.3 | 2.0 ± 0.1 | |
| CD8+ T | 0.1 ± 0.01 | 0.1 ± 0.01 | 0.05 ± 0.01 | 0.2 ± 0.01 | |
| Mature NK | 0.1 ± 0.02 | 0.1 ± 0.01 | 0.1 ± 0.01 | 0.1 ± 0.01 | |
| Immature NK | 0.007 ± 0.002 | 0.003 ± 0.001 | 0.0004 ± 0.0001§ | 0.0002 ± 0.00004§ | |
Cell surface markers used B cells (B220+CD49 cells); CD4+ T cells (CD3+CD4+CD8 cells), CD8+ T cells (CD3+CD4CD8+ cells); mature Natural Killer (NK) cells (CD49+B220); immature NK cells (CD49+B220+).
p<0.05 compared to oil controls;
P<0.05 compared to BALB BHT treatment;
p<0.05 compared to BALB.
Functional differences in BALF macrophages
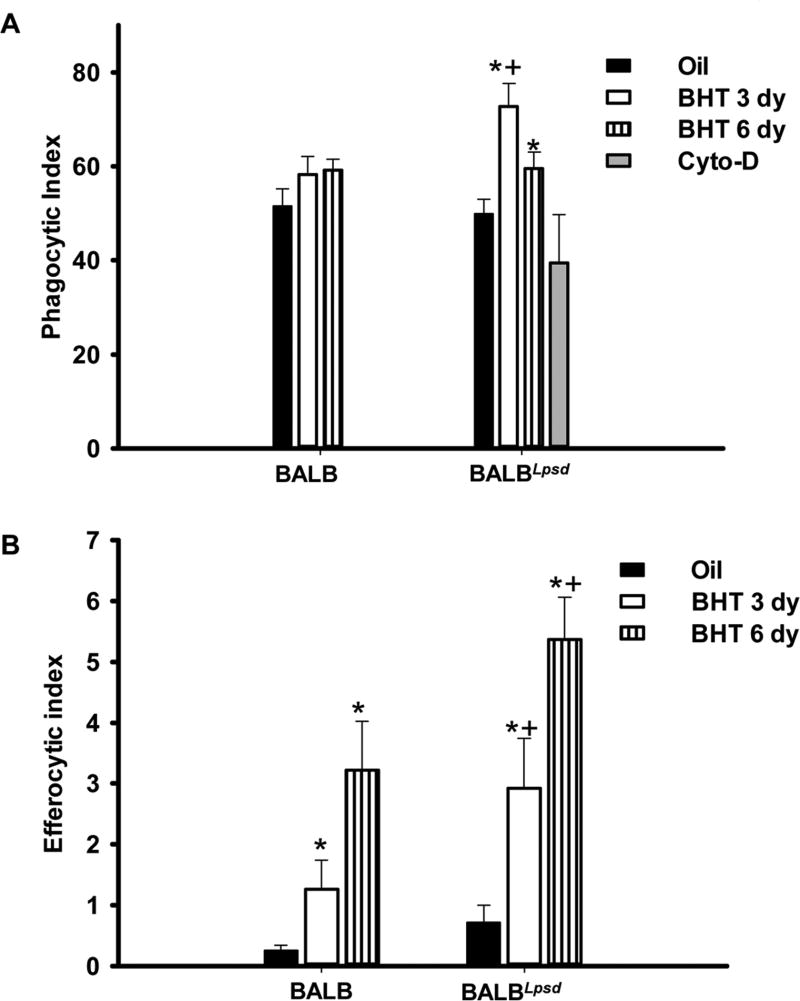
In order to comprehend these BALF macrophage strain differences and determine if these differences relate to macrophage functionality, we measured both general phagocytosis (Zymosan particles) and the phagocytosis of apoptotic cells known as efferocytosis. These functions typically indicate increased inflammation and apoptotic cells. Eighteen hr following plating of BALF cells with a macrophage specific media, only macrophages remained for functional assays. Our results (Fig 4) showed that there was a significant increase of phagocytic and efferocytic function in the BHT-treated BALBLpsd BALF macrophages when compared to the BALB BHT-treated macrophages at both time points. However, it is unclear whether functional differences exist in the IM or DP macrophage populations, although some DP activated macrophages are likely present in the alveolar regions (hence BALF).
Figure 4. Functional differences exist in BALF macrophages following BHT treatment.
A) Phagocytic index in BALB versus BALBLpsd mice 3 and 6 days post-BHT; B) Efferocytic index in BALB versus BALBLpsd mice 3 and 6 days post-BHT. Phagocytic (zymosan) and efferocytic (apoptotic cells) index was calculated by number of positive cells/total number of macrophages × 100. Mean ± SEM presented; n = 3 per treatment group; repeated three times. *P < 0.05 for BALBLpsd vs. oil; +p<0.05 for BALBLpsd vs. all other treatment groups.
Analysis of BALF immune cells and macrophage programming using transcriptomics
BALF immune cells, >90% macrophages, were used to examine gene signature differences between strains. 240 genes were identified as ≥ 1.3-fold significantly altered in Tlr4-mutant mice compared wild-type mice, 142 increased and 98 decreased genes (see Table S3). Hierarchical clustering between strains and treatment depicts the distinct differences with BHT treatment in the BALBLpsd mice compared to BALB mice; BALB mice exposed to BHT are similar in appearance to their controls (Fig. S1A). Following gene ontology (GO) functional analysis (Fig. S1B), the major significant GO categories identified for genes increased in BALBLpsd compared to BALB macrophages were immune system process (e.g. activation of immune response, leukocyte activation, and antigen processing and presentation; see Fig. S1C), biological adhesion, locomotion, and biological regulation. Those GO categories for genes decreased were biogenesis and cellular process. Partek Pathway analysis revealed several interesting pathways that were significantly enriched (p<0.05, see Table S3) in the BALBLpsd mice compared to BALB including chemokine signaling pathway, complement coagulation cascades, phagosome, cell adhesion molecules, Fcgamma receptor-mediated phagocytosis, and leukocyte transendothelial migration from the 142 genes significantly increased in the BALBLpsd mice treated with BHT. These pathways are intriguing based on the increased phagocytosis observed in these macrophages (Fig 4) and support the functional data.
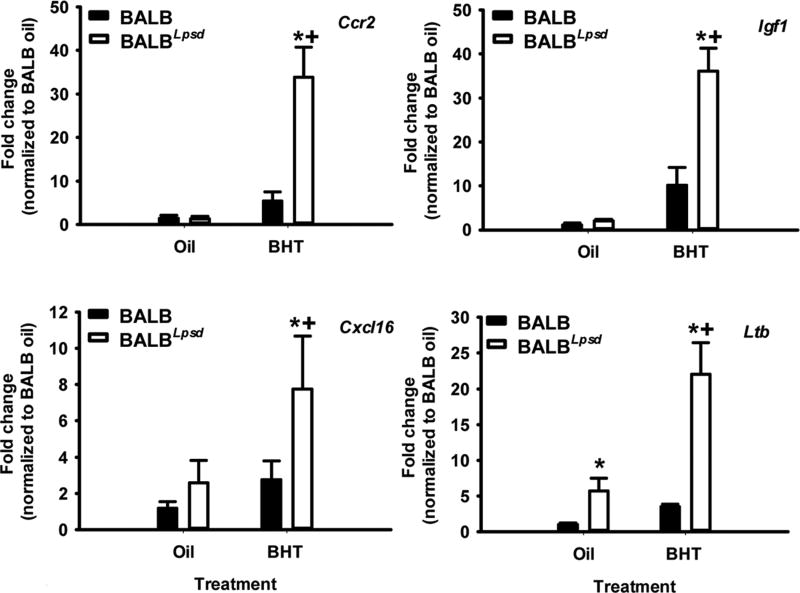
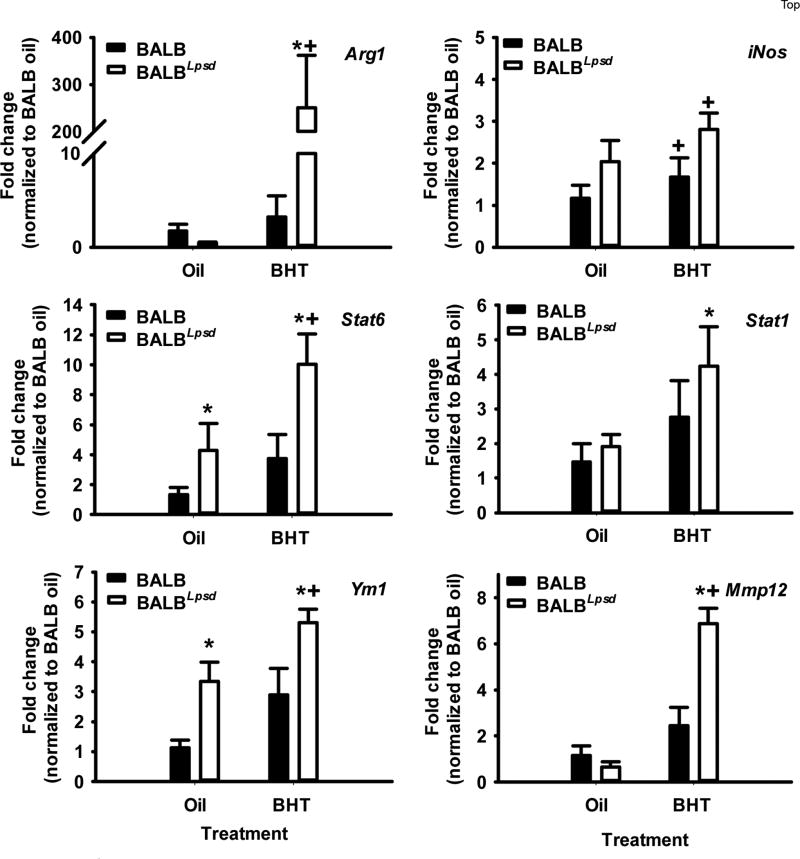
We validated a subset of genes identified from the transcriptomic analysis by qRTPCR (Fig 5). Chemokine receptor 2 (Ccr2), insulin-like growth factor 1 (Igf1), CXC chemokine ligand 16 (Cxcl16), and lymphotoxin β (Ltb) were increased in the BALBLpsd mice in comparison to the BALB BHT-treated mice. Ccr2, for example, is a gene under the phagocytosis pathway. We then used additional macrophage programming markers to further delineate differences between these two strains with treatment. Because recent research suggests plasticity between macrophage programming and thus macrophages are not always clearly delineated “M1” and “M2”, we also looked at genes that have previously been designated as M1 – Stat 1 and inducible nitric oxide synthase (51) or M2 – arginase 1 (Arg1), matrix metalloproteinase (Mmp12), Stat6, and Ym1 (Chil3; chitinase-like 3) (Fig 6). Overall, an “M2” gene signature dominated where the “M2” genes were significantly higher in the BHT-treated BALBLpsd mice compared to similarly treated BALB mice and “M1” genes were slightly elevated, although not to the same extent as the “M2” genes.
Figure 5. Validation of transcriptomics analysis using quantitative RTPCR.
BALF immune cell RNA was used to confirm the results identified using the Affymetrix array. All treatment groups were normalized to BALB oil. Mean ± SEM presented; n = 3; repeated 2 times. *P < 0.05 for BALBLpsd vs. oil; +p<0.05 for BALBLpsd vs. all other treatment groups. Ccr2, CC chemokine receptor 2; Igf1, insulin-like growth factor 1; Ltb, lymphotoxin beta; Cxcl16, C-X-C motif ligand 16.
Figure 6. BALF macrophage programming between strains demonstrates differential responsiveness in M1 versus M2 genes.
BALF macrophages from BALBLpsd and BALB mice 3 days following treatment with BHT or oil vehicle control. All treatment groups were normalized to BALB oil. Mean ± SEM presented; n = 3; repeated 2 times. *P < 0.05 for BALBLpsd vs. oil; +p<0.05 for BALBLpsd vs. all other treatment groups. Arg1, arginase 1; Stat6, signal transducer and activator of transcription 6; Ym1 (Chil3; chitinase-like 3); iNOS, inducible nitric oxide synthase; Mmp12, metalloproteinase 12.
Differential protein analysis using protein arrays and ELISAs
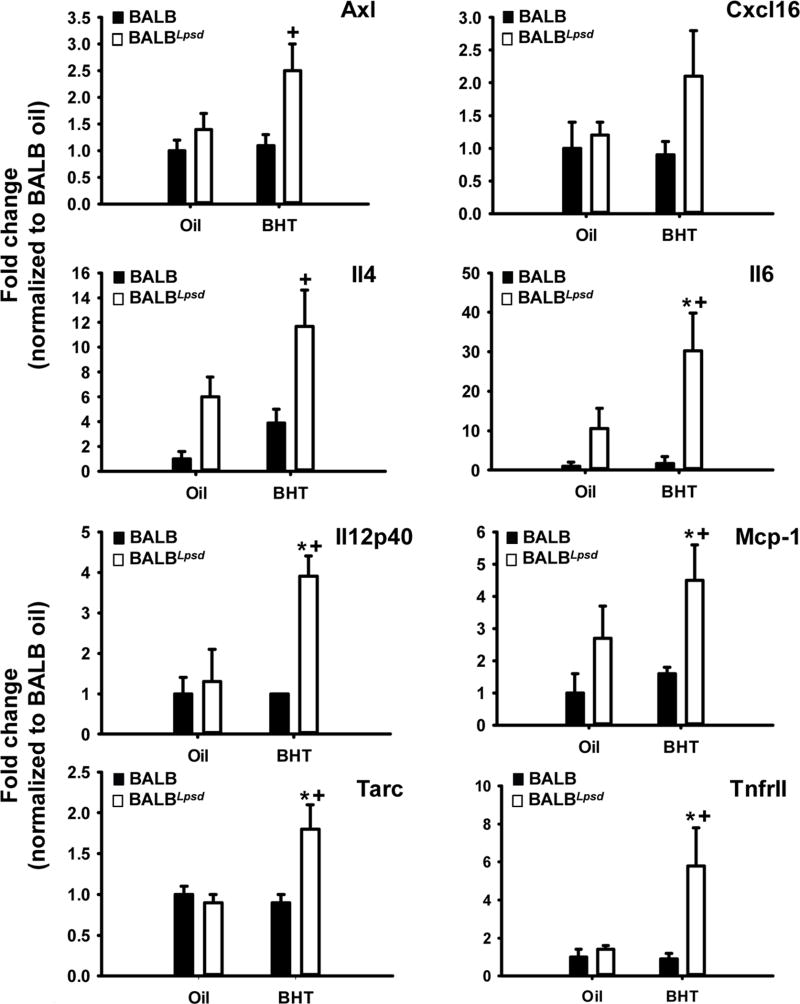
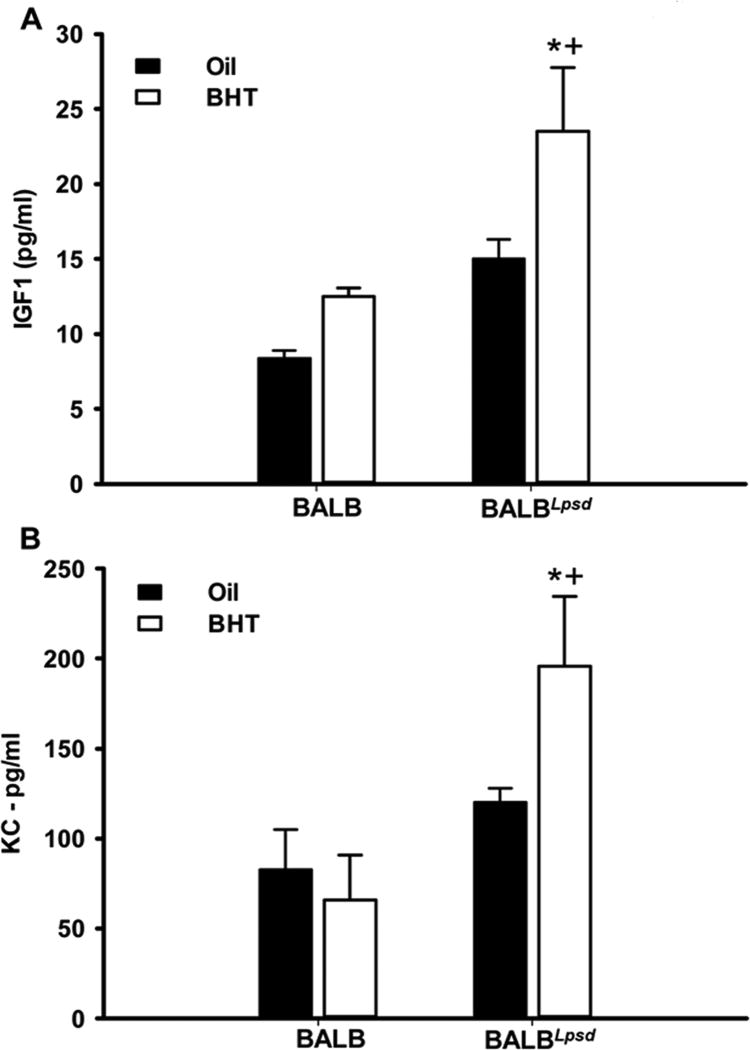
We then used Raybiotech mouse cytokine arrays to evaluate strain differences in whole lung protein homogenates to determine differences in cytokine and chemokine protein levels in the pulmonary microenvironment 3 days following BHT. Similar to the results observed with gene expression and immune cell populations, Tlr4-deficiency led to increased cytokine production in the BHT-treated lungs, over the control mice (BALB and oil controls). The up-regulation of these cytokines, chemokines, their receptors, and their signaling mediators, such as macrophage chemotactic protein-1 (Mcp-1), Tarc (Ccl17), osteopontin (Opn), and tumor necrosis factor receptor (TNFR)II, are indicative of increased migration, recruitment, and inflammation of macrophages in Tlr4-mutant mice (Fig 7 or data not shown). Some of the same cytokines (Cxcl16; Opn) or their receptors (Ccr2) were also up-regulated in the transcriptome studies above or those already published, and individual ELISAs in the BHT-treated BALBLpsd mice compared to all other experimental groups (data not shown) (29). The increased up-regulation of both “M1” - Il12p40, Il2 and “M2” - Il4 skewing cytokines, as well as pro-inflammatory pathways (Il6, Fig 7) once again re-iterates that the macrophage polarization observed during BHT-induced tumor promotion is neither M1 or M2 but instead displays a more plastic signature. Additionally, Axl (Fig 7), a tyrosine kinase receptor involved in efferocytosis, and other pro-inflammatory markers (IL1β, GITR) were also significantly elevated in the BHT-treated BALBLpsd mice (data not shown). Igf-1 ELISAs also further supported the gene expression observed at 3 days, with significant increases in the BALBLpsd mice following BHT exposure compared to all other treatments (Fig 8A).
Figure 7. BHT upregulates protein expression of pro-inflammatory, Th1 and Th2 cytokines in Tlr4-mutant compared to -sufficient mice.
Whole lung tissue protein was analyzed via Raybiotech cytokine array for differences in protein expression. All treatment groups were normalized to BALB oil. Mean fold-change ± SEM presented; n = 3 per treatment group. *P < 0.05 for BALBLpsd vs. oil; +p<0.05 for BALBLpsd vs. all other treatment groups. Axl, Axl receptor tyrosine kinase; Cxcl16, chemokine (CXC motif) ligand 16; IL4, interleukin 4; Mcp1 (Ccl1),macrophage chemoattractant protein 1; Tarc (Ccl17), thymus and activation regulated chemokine; Tnfr11, tumor necrosis factor receptor II.
Figure 8. Differences in Igf1 and Kc between BALBLpsd and BALB mice.
A) Igf1 ELISA on whole lung tissue from both strains 3 days following BHT. B) Kc ELISA on whole lung tissue from both strains 6 days following BHT. All treatment groups were normalized to BALB oil. Mean ± SEM presented; n = 3 per treatment group. *P < 0.05 for BALBLpsd vs. oil; +p<0.05 for BALBLpsd vs. all other treatment groups.
We also performed Kc ELISAs on lung homogenates to provide further evidence for the PMN response at 6 days in the BALBLpsd mice. Significant increases in KC were observed in the BALBLpsd mice over the controls and BALB mice (Fig 8B).
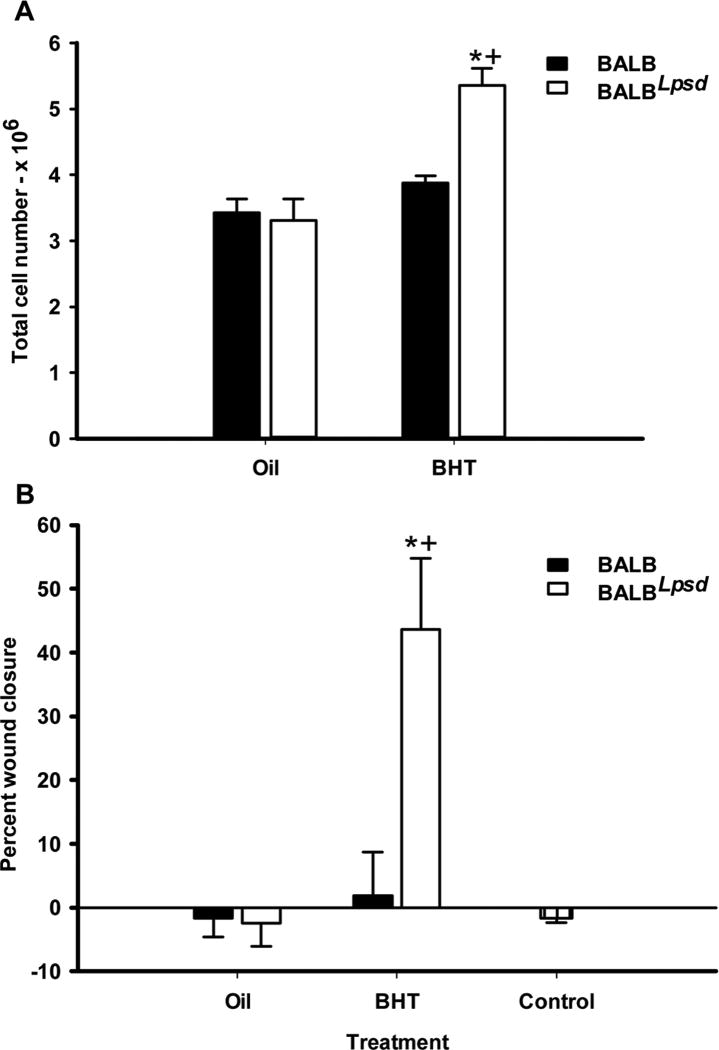
Progenitor cell effects: bronchiolar Club Cell and co-culture studies using bone marrow-derived macrophages (BMDM) with an alveolar type II cell line
Six days following BHT or control treatment, Club cells were isolated from both strains to determine differences in total cell numbers based on previous studies demonstrating proliferation of type II and Club cells at this time point (12). We observed significant increases in cell numbers in the BALBLpsd mice compared to BALB mice (Fig 9A), further supporting phenotype differences between strains described above. BMDM were then used from both strains (BALB and BALBLpsd) isolated 3 days following BHT treatment and differentiated. These BMDM were then used in co-culture studies with C10 cells (an alveolar type II cell line, (48)) for a wound healing assay to determine differences in the ability of the epithelial cells to close the wound, by migration and proliferation of epithelial cells. We did not observe any differences between strain or treatment prior to 24 hr in these studies. Fig 9B demonstrates the percent closure in C10 cells in co-culture with the BMDM from the BALBLpsd vs BALB mice, with significant increases in the BALBLpsd mice treated with BHT compared to all other treatment groups, including C10 cells grown in the absence of BMDMs (Fig. 9B control).
Figure 9. Progenitor cell effects were only observed in BALBLpsd mice in response to BHT.
A) Isolated Club cell numbers were increased 6 days following BHT in BALBLpsd mice only. B) Bone marrow-derived macrophages (BMDM) from previously BHT-treated BALBLpsd mice grown in co-culture with C10 cells (epithelial) induce significant wound healing in C10 cells compared to other treatment groups. Control treatment group were C10 cells in the absence of BMDMs. Mean ± SEM presented; n = 3–8 per treatment group; repeated three times. *P < 0.05 for BALBLpsd vs. oil; +p<0.05 for BALBLpsd vs. all other treatment groups.
Discussion
In these studies, we demonstrate for the first time that there are distinct pulmonary innate immune cell populations, including macrophages, PMNs, DCs, NK cells, CD4+ T cells, and MDSCs, in the lung that are likely influencing the increased amount of promotion observed in the Tlr4-mutant mice (Figs 1–3, Table 1). The macrophage populations varied in numbers with alveolar > activated> interstitial, not unlike previous studies in other chronic lung diseases, such as fibrosis (33, 37). BALBLpsd mice had elevations in every macrophage population compared to BALB mice. Both IM and DP activated macrophages also displayed specific cell surface marker differences, namely, both CD86+ and CD206+ macrophages, two commonly used markers to distinguish between “M1 vs M2” macrophages (35, 39) and support the concept of macrophage phenotype plasticity in our model. Between the two strains treated with BHT, the BALBLpsd had significant increases in both CD86+ and CD206+ populations over that observed in the BALB mice. However, there are caveats for these markers such as CD86, a co-stimulatory molecule, can be expressed on some M2 macrophage populations (M2b) and CD206, mannose receptor 1, is also expressed on other macrophage populations (36, 39). It is important to note that these two markers do not equate to the total cell content per macrophage subtype, therefore there are other types of macrophages within these populations that are unaccounted for with these markers. Given that these are only two markers, we chose to assess gene expression as well in the BALF macrophages to further identify macrophage gene signatures for BHT responsiveness between strains. Multiple genes were identified from the transcriptomic analysis, such as Igf1, as well as significant pathways, such as phagocytosis, that were supported by both protein analysis (e.g. Igf1 ELISA; Fig 8A) and functional data (phagocytosis; Fig 4A). Additional macrophage programming genes (eg. Arg1, iNos, Stat-6; Fig 6) were investigated to provide more support for a specific type of programming. Overall, based on these studies and previous data in whole lung, we observed more “M2 vs M1” gene expression patterns in the BALF macrophages and immune cells that we hypothesize are involved in facilitating increased tumor promotion observed in the BALBLpsd compared to BALB mice. It is not the case that “M1” polarized macrophages are not present in these lungs, but that overall, at least for the BALF macrophages (primarily AM and some DP), there appears to be a more consistent “M2” gene signature.
Functional analysis using phagocytosis and efferocytosis also suggests that a mixed population of macrophages exist in the BALBLpsd lungs following BHT treatment. Phagocytosis is a more “M1” macrophage function and efferocytosis an “M2” macrophage function, although evidence exists for both populations of macrophages to elicit these functions (52). While the numbers of macrophage populations were not different at 6 days in whole lung by flow cytometry, the BALBLpsd BALF macrophages were increased in number and more efferocytic. Additionally, the function of the other subsets of macrophages is unclear, although the DP activated macrophages are likely in the BALF and influencing the functional responses, at least for the 3 day time point. Some portion of these pulmonary macrophage populations may also be similar to the restorative macrophages that accumulate during liver fibrosis and possibly lung fibrosis (37, 53). Restorative macrophages express MMPs, IGF1, among other markers, that make them unique and outside the typical M1/M2 classification. Based on our results, including expression of markers such as MMP12 and IGF1, this sort of restorative macrophage appears closer to those accumulating in the BALBLpsd lungs than simply a mixed M1/M2 phenotype. However, timing of these responses is another important factor and at later time points, those macrophages present early may adapt further to support the growth of the tumors in the BALBLpsd mice, similar to Fritz et al., 2014 (17).
Significant increases in DCs, specific lymphocyte populations, PMNs and MDSCs following BHT treatment in the BALBLpsd mice are also likely influencing the lung microenvironment. Pulmonary DCs were also highly elevated in Tlr4-mutant mice in an allergic inflammation model, which supports our study, as well as several inflammatory markers typically increased by activated DCs in lungs of BALBLspd mice treated with BHT, such as costimulatory molecule (CD40, data not shown) and Ccl17 (Tarc) (Fig 7) (42, 54, 55). The DCs can also lead to NK cell activation, which we observed increased cell numbers, with potential to secrete cytokines/chemokines that activate specific types of macrophages (e.g. Ifnγ and M1 macrophages).
Several recent studies demonstrated PMN involvement during lung tumor promotion (15, 56, 57). PMNs can also be categorized as “N1 vs. N2”, with N2 types of neutrophils considered pro-tumorigenic (58). Ccl2 (Mcp-1) is a major marker for N2 types of PMNs (58). We also observed increased levels of Mcp-1 and Ccr2, the receptor for Mcp-1, in the BALBLpsd mice exposed to BHT, which supports protumorigenic PMNs in our model. Additionally, blockade of Cxcr2, the major receptor for Kc, in a COPD-like promotion model, demonstrated decreased lung tumorigenesis (57). Kc was elevated in our model as well, which is likely the primary PMN chemoattractant based on our other cancer or lung inflammation models (e.g. urethane-induced carcinogenesis) (20, 56, 59), and has been shown to recruit MDSCs (60). These MDSCs are another heterogeneous immunosuppressive cell type and are known to create a pro-tumorigenic environment, by increasing growth factors (e.g. TGFβ), MMPs, and cytokine secretion (IL1, TNF, IL6) followed by suppressing normal antitumor immunity (61). Targeting these MDSCs therapeutically in a lung cancer model using a gemcitabine/superoxide dismutase combination was successful in increasing effector and memory CD8+ T cells to inhibit MDSC suppressive effects (62). Thus, inhibition or blockade of PMNs, MDSCs, or macrophages have all been shown to be efficacious against lung tumor development, although only PMNs and macrophage inhibition during the promotion stage (13, 15, 17, 57).
During this very early stage of tumorigenesis, we also demonstrated significant increases in progenitor cell numbers (Club cells; Fig 9A) in BALBLpsd mice only, which supports potential proliferation during these early events in promotion. Interestingly, the BMDM from BALBLpsd mice previously treated with BHT in vivo were able to significantly heal a wound in the C10 epithelial cells, suggesting that these macrophages were adapted in some manner in response to BHT and produce factors that influence growth and migration of these epithelial cells (Fig 9B). Therefore these BMDM results imply that the macrophage progenitor cells in the bone marrow themselves are programmed before entering the lung microenvironment and supports that Tlr4-mutant BM monocyte infiltration into the lung promotes the transition of the epithelial cells from initiated cell to adenoma development, although do not rule out the importance of the other cell types investigated. Previous studies also observed similar phenomena in that bone marrow monocyte polarization matched that of the alveolar macrophage polarization in urethane treated mice (38). Recent evidence also suggests that these pulmonary macrophage populations may be derived from different locales; resident macrophages are thought to originate from yolk sac (embryonic, F4/80hi) and can themselves proliferate versus those derived from bone marrow that can infiltrate into the lung during injury and insult (75). These BMDM observations herein support bone marrow origins in the macrophages influencing these responses, similar to previous studies (76, 77). Differences in origin of these cells may help to account for the other population of macrophages identified by flow cytometry that did not fall into the markers studied (eg.F4/80+, CD86, CD204), why there are mixed populations, and why we observed differences with the BMDMs. Identifying the source of origin for these macrophages could provide additional targets for therapy. We also previously demonstrated decreased gap junction intercellular communication in BALF fromTlr4-mutant mice treated with BHT using a novel ex vivo approach with C10 cells (see Hill et al., 2013, http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4098145/figure/F3/) (28–30). Gap junction inhibition is a key mechanism used by epithelial cells to decrease cell-cell communication and is typically inhibited during the tumor promotion stage of carcinogenesis (47). Additional exploration into the role of the macrophages and other innate immune cells identified in eliciting factors that inhibit these gap junctions are important future directions and potential therapeutic targets.
Lastly, the growth factors and cytokines/chemokines regulating the BHT responses are not clear, however, several potential candidates were identified in our study, such as Igf1 and Kc, described above in the lung tissue. We also observed increases in Igf1 and Kc in the medium from the BALBLpsd BHT-treated BMDM in coculture with C10 cells (data not shown). IGF1 blockade inhibited macrophage conditioned medium-induced proliferation of several lung cell types, including E10 cells (alveolar type II cell) and LM2 cells, a line derived from a papillary ADC in mice (63). Further, a recent study showed the IGF1 receptor inhibitor (picropodophyllin) was an effective lung ADC chemopreventive agent (64). Thus future studies will further investigate the role of these mediators and potential biomarkers in eliciting promotion in mice lacking Tlr4.
The TLR4 pathway is a double-edged sword in the lung due its involvement in pro-inflammatory and protective responses. In some models, TLR4 induction leads to increased amounts of inflammation, although in lung, these are often acute models of lung injury or disease (19, 21). However, this is not the case in other models, including more chronic models in lung, such as infection models, asthma, and emphysema (23, 24, 54). In Hollingsworth et al., the authors demonstrated that Tlr4-mutant mice were more susceptible than wildtype mice to eosinophilia and had increases in cells such as dendritic cells in a model of ongoing allergic asthma. However, similar to our results, Th1 responses were not increased with reductions in Th2 responses. In another study, Zhang et al.(65), a lack of Tlr4 led to symptoms of emphysema as the mice aged in a mechanism hypothesized to be the result of the requirement of TLR4 in lung structural cells important for maintenance of normal lung architecture through modulation of oxidant regulation (65). Thus, in other lung models, TLR4 signaling appears to stimulate a protective effect. We would hypothesize that in our model similar to the asthma model described above, that dendritic cells are an important cell type as well as macrophages, PMNs, and MDSCs. We are not clear on the specific DAMP(s) stimulating the response, similar to other models (54) but have been focusing on the downstream effectors.
Both results in the asthma model and in our cancer model are consistent with epidemiologic data. Low doses of endotoxin exposure, particularly in children, reduce the risk of developing asthma (64). Epidemiologic studies on endotoxin exposure in agricultural occupations (e.g. milking parlors) and some textile workers reduced lung cancer risk (26, 27, 66), although the amount of exposure and type is endotoxin is likely critical to the response observed. TLR4 is also protective in other cancer models (eg. skin, breast, liver) including both mouse and human (67–72). For example, in skin carcinogenesis, the Th1 cytokines were reduced and IL17 increased in TLR4-deficient mice. In more advanced breast cancer models, those patients with a loss-of-function TLR4 allele relapse more quickly than those with a normal TLR4 allele (73, 74). In a chemically-induced hepatocellular carcinogenesis model, TLR4 mutant mice developed significantly more tumors in part through decreased DNA repair mechanisms such as altered Ku70/80 (71). Lastly in a small pilot study with lung ADC patients (McLaren Regional Medical Center, Flint, MI.), we demonstrated that TLR4 expression was significantly decreased in tumor tissue compared to adjacent, non-neoplastic lung tissue (unpublished data, A.K.Bauer). Collectively, these cancer studies support our findings and suggest the observed proliferative responses in our model may result from downstream effects that will be investigated in future studies. Therefore, TLR4 appears to have both pro- and anti-tumorigenic properties in lung cancer that may depend on stage of tumor development.
Conclusions
In summary, based on our novel cell-specific data presented in these studies, supported by our previous data in whole lung (see Bauer et al., 2005, http://jnci.oxfordjournals.org/content/97/23/1778.long; Bauer et al., 2009, http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2785769/figure/F3/), we suggest that the pulmonary microenvironment in Tlr4-mutant mice provides a preferential mélange of innate immune cell types (AM, IM, DP macrophages, DCs, PMNs, MDSCs, NK cells, CD4 T cells), inflammatory mediators (KC, MCP-1, MMP12), and growth factors (IGF1) that enhances the transition from normal to tumorigenic cells (Fig 10). The cell-specific gene signatures identified in these studies, which differ greatly from whole lung, also indicate a more plastic nature of these macrophages and innate immune cells and provide several potential predictive candidates/biomarkers that may be driving the proliferative epithelial responses observed in this early stage of tumor development (e.g. IGF1, KC, MMP12, STAT6, CCR2, CCL17, CXCL16, CD206, CD86). Lastly, the BMDM were adapted in mice treated with BHT that led to increased wound healing, which further supports the increased proliferative responses in Tlr4-mutant mice, providing an overall pro-tumorigenic pulmonary microenvironment.
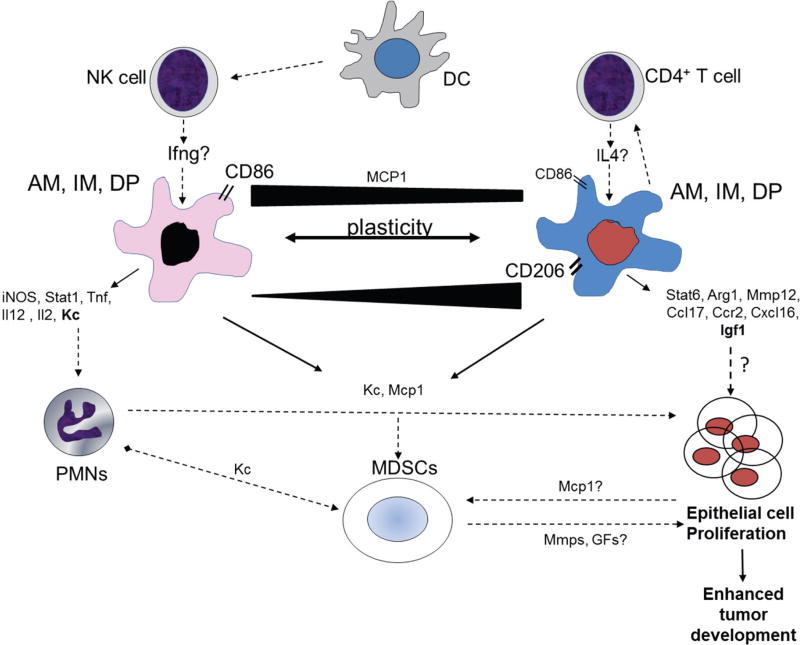
Figure 10. Proposed working hypothesis for how innate immune cells influence tumor promotion in Tlr4-mutant mice.
Alveolar (AM), interstitial (IM), and double positive activated (DP) macrophages were elevated in the lungs of BALBLpsd compared to BALB mice, as well as neutrophils (PMNs), dendritic cells (DC), mature natural killer cells (NK), myeloid-derived suppressor cells (MDSC)s, and CD4+ T cells (for both strains with BHT). We propose that DCs activate the NK cells leading to more “M1-like” macrophages and CD4+ T cells activate the more “M2-like” macrophages. Both CD86 and CD206 markers were increased on these macrophages as well, however, other studies demonstrate that these markers are more plastic between different macrophage subtypes (black bar indicates amounts of expression on M1 vs. M2 cells)(39). Multiple genes are expressed in BALF macrophages (both AM and DP) in response to BHT that likely influence increased promotion of initiated progenitor cells (Club cells and type II cells). In particular, based on our findings and others (57, 59), both KC and IGF1 are potential candidates that drive proliferation of epithelial cells in these lungs, likely produced by PMNs and macrophages and involved in both PMN and MDSC recruitment. Lastly, the MDSCs may either directly or indirectly influence proliferation through chemokines and MMPs, both of which were elevated in these mice (61). Black lines with arrows indicate that we have evidence for these events in this model; dotted lines indicate potential mechanistic links for cells that are increased in this model; red line indicates inhibitory event previously described (30). Abbreviations: KC, keratinocyte factor; MCP-1, macrophage chemotactic protein 1; iNOS, inducible nitric oxide synthase; arg1, arginase 1; Tnf, tumor necrosis factor; IL2, interleukin 2; MMP12, metalloproteinase 12; MDSCs, myeloid derived suppressor cells.
The vast complexity of the lung microenvironment often discourages identification of cell types driving disease mechanisms, however, to discover new treatment approaches in both cancer prevention as well as therapeutics, a focus on cell specificity is critical. Collectively, these findings all emphasize the need for additional research using this model of tumor promotion to further delineate 1) the role of TLR4 using current TLR4 agonists, such as monophosphoryl lipid A (MPL), the biologically active component of endotoxin that is less toxic, immunostimulatory, and in current use in Europe for asthma (78); 2) adoptive bone marrow transfer studies in BALB and BALBLpsd mice; and 3) future studies to target potential therapeutic pathways or test potential biomarkers, such as IGF1 and KC, as well as future studies to target specific cell types depending on locale (yolk sac versus bone marrow derived). Lastly, studies to assess single nucleotide polymorphisms and protein expression of these specific pathways (eg. TLR4, IGF1, KC) and their association with different stages of lung neoplasia in human pulmonary ADC patients are warranted.
Supplementary Material
A) Depiction of the clustering of 240 genes that significantly differed between BALBLpsd and BALB mice 3 days following BHT treatment. The color bar indicates the expression (red increased; blue decreased). B) The gene ontology (GO) functional analysis for the genes increased 142 genes out of 240 genes. Enrichment score of 3 and higher are considered significant. C) Example of immune system processes expanded from the GO analysis presented in B. Again, any functions that have an enrichment score above 3 are considered significant.
A) In this table we list all the 240 genes on the microarray that were significantly different between the BALBLpsd and BALB mice 3 days following treatment with chronic BHT (P<0.05 and ≥1.3-fold changes, both increased and decreased). B) These are the pathways identified in genes that were increased in the BALBLpsd mice compared to BALB mice, analyzed in Partek Pathway Analysis. C) These are the pathways identified in genes that were decreased in the BALBLpsd mice compared to BALB mice, analyzed in Partek Pathway Analysis. Pathways with significance at p<0.05 or below are presented.
Acknowledgments
We would like to thank Dr. William Janssen, his laboratory, and Dr. Elizabeth Redente for guidance with the flow cytometry studies as well as Dr. Jonathan Shanahan for critically reviewing the manuscript.
Funding
This research was funded by the American Cancer Society (RSG-10-162-01-LIB, AKB). The American Cancer Society had no involvement in the design or in conducting this research.
Footnotes
Declaration of Interests
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer Journal international du cancer. 2015;136(5):E359–86. doi: 10.1002/ijc.29210. Epub 2014/09/16. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. [cited 2015 April 2]; Available from: http://www.cancer.gov.
- 3.Schottenfeld D. Etiology and Epidemiology of Lung Cancer. In: Pass HI, Mitchell JB, Johnson DH, Turrisi AT, Minna JD, editors. Lung Cancer-Principles and Practice. 2. Philadelphia: Lippincott Williams and Wilkins; 2000. pp. 367–88. [Google Scholar]
- 4.Bach PB, Mirkin JN, Oliver TK, Azzoli CG, Berry DA, Brawley OW, et al. Benefits and harms of CT screening for lung cancer: a systematic review. Jama. 2012;307(22):2418–29. doi: 10.1001/jama.2012.5521. Epub 2012/05/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nature medicine. 2007;13(3):361–6. doi: 10.1038/nm1556. Epub 2007/03/06. [DOI] [PubMed] [Google Scholar]
- 6.Bauer AK, Rondini EA. Review paper: the role of inflammation in mouse pulmonary neoplasia. Veterinary pathology. 2009;46(3):369–90. doi: 10.1354/vp.08-VP-0217-B-REV. [DOI] [PubMed] [Google Scholar]
- 7.Malkinson AM. Role of inflammation in mouse lung tumorigenesis: a review. Exp Lung Res. 2005;31(1):57–82. doi: 10.1080/01902140490495020. Epub 2005/03/16. [DOI] [PubMed] [Google Scholar]
- 8.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homma S, Sagawa Y, Komita H, Koido S, Nagasaki E, Ryoma Y, et al. Mechanism of antitumor effect on mouse hepatocellular carcinoma by intratumoral injection of OK-432, a streptococcal preparation. Cancer immunology, immunotherapy: CII. 2007;56(8):1265–74. doi: 10.1007/s00262-006-0277-9. Epub 2007/01/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer AK, Malkinson AM, Kleeberger SR. Susceptibility to neoplastic and non-neoplastic pulmonary diseases in mice: genetic similarities. American journal of physiology. 2004;287(4):L685–703. doi: 10.1152/ajplung.00223.2003. [DOI] [PubMed] [Google Scholar]
- 11.Malkinson AM. Molecular comparison of human and mouse pulmonary adenocarcinomas. ExpLung Res. 1998;24(4):541–55. doi: 10.3109/01902149809087385. [DOI] [PubMed] [Google Scholar]
- 12.Bauer AK, Dwyer-Nield LD, Hankin JA, Murphy RC, Malkinson AM. The lung tumor promoter, butylated hydroxytoluene (BHT), causes chronic inflammation in promotion-sensitive BALB/cByJ mice but not in promotion-resistant CXB4 mice. Toxicology. 2001;169(1):1–15. doi: 10.1016/s0300-483x(01)00475-9. [DOI] [PubMed] [Google Scholar]
- 13.Bauer AK, Dwyer-Nield LD, Keil K, Koski K, Malkinson AM. Butylated hydroxytoluene (BHT) induction of pulmonary inflammation: a role in tumor promotion. Exp Lung Res. 2001;27(3):197–216. doi: 10.1080/019021401300053948. [DOI] [PubMed] [Google Scholar]
- 14.Malkinson AM, Koski KM, Evans WA, Festing MF. Butylated hydroxytoluene exposure is necessary to induce lung tumors in BALB mice treated with 3-methylcholanthrene. Cancer Res. 1997;57(14):2832–4. [PubMed] [Google Scholar]
- 15.Vikis HG, Gelman AE, Franklin A, Stein L, Rymaszewski A, Zhu J, et al. Neutrophils are required for 3-methylcholanthrene-initiated, butylated hydroxytoluene-promoted lung carcinogenesis. Molecular carcinogenesis. 2012;51(12):993–1002. doi: 10.1002/mc.20870. Epub 2011/10/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witschi H, Malkinson AM, Thompson JA. Metabolism and pulmonary toxicity of butylated hydroxytoluene (BHT) Pharmacol Ther. 1989;42(1):89–113. doi: 10.1016/0163-7258(89)90023-5. [DOI] [PubMed] [Google Scholar]
- 17.Fritz JM, Tennis MA, Orlicky DJ, Lin H, Ju C, Redente EF, et al. Depletion of tumor-associated macrophages slows the growth of chemically induced mouse lung adenocarcinomas. Frontiers in immunology. 2014;5:587. doi: 10.3389/fimmu.2014.00587. Epub 2014/12/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keith RL, Miller YE. Lung cancer chemoprevention: current status and future prospects. Nature reviews Clinical oncology. 2013;10(6):334–43. doi: 10.1038/nrclinonc.2013.64. Epub 2013/05/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poltorak A, He X, Smirnova I, Liu MY, Huffel CV, Du X, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282(5396):2085–8. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 20.Bauer AK, Rondini EA, Hummel KA, Degraff LM, Walker C, Jedlicka AE, et al. Identification of candidate genes downstream of TLR4 signaling after ozone exposure in mice: a role for heat-shock protein 70. Environmental health perspectives. 2011;119(8):1091–7. doi: 10.1289/ehp.1003326. Epub 2011/05/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleeberger SR, Reddy S, Zhang LY, Jedlicka AE. Genetic susceptibility to ozone-induced lung hyperpermeability: role of toll-like receptor 4. American journal of respiratory cell and molecular biology. 2000;22(5):620–7. doi: 10.1165/ajrcmb.22.5.3912. [DOI] [PubMed] [Google Scholar]
- 22.Kleeberger SR, Reddy SP, Zhang LY, Cho HY, Jedlicka AE. Toll-like receptor 4 mediates ozone-induced murine lung hyperpermeability via inducible nitric oxide synthase. American journal of physiology. 2001;280(2):L326–33. doi: 10.1152/ajplung.2001.280.2.L326. [DOI] [PubMed] [Google Scholar]
- 23.Faure K, Sawa T, Ajayi T, Fujimoto J, Moriyama K, Shime N, et al. TLR4 signaling is essential for survival in acute lung injury induced by virulent Pseudomonas aeruginosa secreting type III secretory toxins. Respir Res. 2004;5(1):1. doi: 10.1186/1465-9921-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Shan P, Qureshi S, Homer R, Medzhitov R, Noble PW, et al. Cutting edge: TLR4 deficiency confers susceptibility to lethal oxidant lung injury. J Immunol. 2005;175(8):4834–8. doi: 10.4049/jimmunol.175.8.4834. [DOI] [PubMed] [Google Scholar]
- 25.Braun-Fahrlander C, Riedler J, Herz U, Eder W, Waser M, Grize L, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med. 2002;347(12):869–77. doi: 10.1056/NEJMoa020057. Epub 2002/09/20. [DOI] [PubMed] [Google Scholar]
- 26.Astrakianakis G, Seixas NS, Ray R, Camp JE, Gao DL, Feng Z, et al. Lung cancer risk among female textile workers exposed to endotoxin. Journal of the National Cancer Institute. 2007;99(5):357–64. doi: 10.1093/jnci/djk063. [DOI] [PubMed] [Google Scholar]
- 27.Lundin JI, Checkoway H. Endotoxin and cancer. Environmental health perspectives. 2009;117(9):1344–50. doi: 10.1289/ehp.0800439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauer AK, Dixon D, DeGraff LM, Cho HY, Walker CR, Malkinson AM, et al. Toll-like receptor 4 in butylated hydroxytoluene-induced mouse pulmonary inflammation and tumorigenesis. Journal of the National Cancer Institute. 2005;97(23):1778–81. doi: 10.1093/jnci/dji403. [DOI] [PubMed] [Google Scholar]
- 29.Bauer AK, Fostel J, Degraff LM, Rondini EA, Walker C, Grissom SF, et al. Transcriptomic analysis of pathways regulated by toll-like receptor 4 in a murine model of chronic pulmonary inflammation and carcinogenesis. Molecular cancer. 2009;8:107. doi: 10.1186/1476-4598-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill T, 3rd, Osgood RS, Velmurugan K, Alexander CM, Upham BL, Bauer AK. Bronchoalveolar Lavage Fluid Utilized Ex Vivo to Validate In Vivo Findings: Inhibition of Gap Junction Activity in Lung Tumor Promotion is Toll-Like Receptor 4-Dependent. Journal of molecular biomarkers & diagnosis. 2013;5(1) doi: 10.4172/2155-9929.1000160. Epub 2014/07/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds SD, Malkinson AM. Clara cell: progenitor for the bronchiolar epithelium. The international journal of biochemistry & cell biology. 2010;42(1):1–4. doi: 10.1016/j.biocel.2009.09.002. Epub 2009/09/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature immunology. 2012;13(11):1118–28. doi: 10.1038/ni.2419. Epub 2012/10/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janssen WJ, Barthel L, Muldrow A, Oberley-Deegan RE, Kearns MT, Jakubzick C, et al. Fas determines differential fates of resident and recruited macrophages during resolution of acute lung injury. American journal of respiratory and critical care medicine. 2011;184(5):547–60. doi: 10.1164/rccm.201011-1891OC. Epub 2011/04/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang SH, Mirabolfathinejad SG, Katta H, Cumpian AM, Gong L, Caetano MS, et al. T helper 17 cells play a critical pathogenic role in lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(15):5664–9. doi: 10.1073/pnas.1319051111. Epub 2014/04/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills CD, Ley K. M1 and M2 macrophages: the chicken and the egg of immunity. Journal of innate immunity. 2014;6(6):716–26. doi: 10.1159/000364945. Epub 2014/08/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nature reviews Immunology. 2011;11(11):723–37. doi: 10.1038/nri3073. Epub 2011/10/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redente EF, Keith RC, Janssen W, Henson PM, Ortiz LA, Downey GP, et al. Tumor necrosis factor-alpha accelerates the resolution of established pulmonary fibrosis in mice by targeting profibrotic lung macrophages. American journal of respiratory cell and molecular biology. 2014;50(4):825–37. doi: 10.1165/rcmb.2013-0386OC. Epub 2013/12/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Redente EF, Orlicky DJ, Bouchard RJ, Malkinson AM. Tumor signaling to the bone marrow changes the phenotype of monocytes and pulmonary macrophages during urethane-induced primary lung tumorigenesis in A/J mice. The American journal of pathology. 2007;170(2):693–708. doi: 10.2353/ajpath.2007.060566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000prime reports. 2014;6:13. doi: 10.12703/P6-13. Epub 2014/03/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korns D, Frasch SC, Fernandez-Boyanapalli R, Henson PM, Bratton DL. Modulation of macrophage efferocytosis in inflammation. Frontiers in immunology. 2011;2:57. doi: 10.3389/fimmu.2011.00057. Epub 2011/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel SN, Wax JS, Perera PY, Padlan C, Potter M, Mock BA. Construction of a BALB/c congenic mouse, C.C3H–Lpsd, that expresses the Lpsd allele: analysis of chromosome 4 markers surrounding the Lps gene. Infect Immun. 1994;62(10):4454–9. doi: 10.1128/iai.62.10.4454-4459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ait Yahia S, Azzaoui I, Everaere L, Vorng H, Chenivesse C, Marquillies P, et al. CCL17 production by dendritic cells is required for NOD1-mediated exacerbation of allergic asthma. American journal of respiratory and critical care medicine. 2014;189(8):899–908. doi: 10.1164/rccm.201310-1827OC. Epub 2014/03/26. [DOI] [PubMed] [Google Scholar]
- 43.Steele VE, Holmes CA, Hawk ET, Kopelovich L, Lubet RA, Crowell JA, et al. Potential use of lipoxygenase inhibitors for cancer chemoprevention. Expert Opinion On Investigational Drugs. 2000;9(9):2121–38. doi: 10.1517/13543784.9.9.2121. [DOI] [PubMed] [Google Scholar]
- 44.Keith RC, Powers JL, Redente EF, Sergew A, Martin RJ, Gizinski A, et al. A novel model of rheumatoid arthritis-associated interstitial lung disease in SKG mice. Exp Lung Res. 2012;38(2):55–66. doi: 10.3109/01902148.2011.636139. Epub 2011/12/22. [DOI] [PubMed] [Google Scholar]
- 45.Richens TR, Linderman DJ, Horstmann SA, Lambert C, Xiao YQ, Keith RL, et al. Cigarette smoke impairs clearance of apoptotic cells through oxidant-dependent activation of RhoA. American journal of respiratory and critical care medicine. 2009;179(11):1011–21. doi: 10.1164/rccm.200807-1148OC. Epub 2009/03/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pryhuber GS, Huyck HL, Baggs R, Oberdorster G, Finkelstein JN. Induction of chemokines by low-dose intratracheal silica is reduced in TNFR I (p55) null mice. Toxicological sciences : an official journal of the Society of Toxicology. 2003;72(1):150–7. doi: 10.1093/toxsci/kfg018. Epub 2003/02/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malkinson AM, Miley FB, Chichester CH, Plopper CG. Isolation of Nonciliated bronchiolar (Clara) epithelial cells from mouse lung. Methods in Toxicology. 1993;1A:123–33. [Google Scholar]
- 48.Malkinson AM, Dwyer-Nield LD, Rice PL, Dinsdale D. Mouse lung epithelial cell lines--tools for the study of differentiation and the neoplastic phenotype. Toxicology. 1997;123(1–2):53–100. doi: 10.1016/s0300-483x(97)00108-x. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. In: Coligan John E, et al., editors. Current protocols in immunology. 2008. Chapter 14:Unit 14 1. Epub 2008/11/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Justus CR, Leffler N, Ruiz-Echevarria M, Yang LV. In vitro cell migration and invasion assays. Journal of visualized experiments : JoVE. 2014;(88) doi: 10.3791/51046. Epub 2014/06/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karabela SP, Psallidas I, Sherrill TP, Kairi CA, Zaynagetdinov R, Cheng DS, et al. Opposing effects of bortezomib-induced nuclear factor-kappaB inhibition on chemical lung carcinogenesis. Carcinogenesis. 2012;33(4):859–67. doi: 10.1093/carcin/bgs024. Epub 2012/01/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geiser M, Lay JC, Bennett WD, Zhou H, Wang X, Peden DB, et al. Effects of ex vivo gamma-tocopherol on airway macrophage function in healthy and mild allergic asthmatics. Journal of innate immunity. 2013;5(6):613–24. doi: 10.1159/000350234. Epub 2013/05/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramachandran P, Pellicoro A, Vernon MA, Boulter L, Aucott RL, Ali A, et al. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(46):E3186–95. doi: 10.1073/pnas.1119964109. Epub 2012/10/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hollingsworth JW, Whitehead GS, Lin KL, Nakano H, Gunn MD, Schwartz DA, et al. TLR4 signaling attenuates ongoing allergic inflammation. J Immunol. 2006;176(10):5856–62. doi: 10.4049/jimmunol.176.10.5856. [DOI] [PubMed] [Google Scholar]
- 55.Ferlazzo G, Morandi B. Cross-Talks between Natural Killer Cells and Distinct Subsets of Dendritic Cells. Frontiers in immunology. 2014;5:159. doi: 10.3389/fimmu.2014.00159. Epub 2014/05/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rondini EA, Walters DM, Bauer AK. Vanadium pentoxide induces pulmonary inflammation and tumor promotion in a strain-dependent manner. Particle and fibre toxicology. 2010;7:9. doi: 10.1186/1743-8977-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gong L, Cumpian AM, Caetano MS, Ochoa CE, De la Garza MM, Lapid DJ, et al. Promoting effect of neutrophils on lung tumorigenesis is mediated by CXCR2 and neutrophil elastase. Molecular cancer. 2013;12(1):154. doi: 10.1186/1476-4598-12-154. Epub 2013/12/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: "N1" versus "N2" TAN. Cancer cell. 2009;16(3):183–94. doi: 10.1016/j.ccr.2009.06.017. Epub 2009/09/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bauer AK, Cho HY, Miller-Degraff L, Walker C, Helms K, Fostel J, et al. Targeted deletion of Nrf2 reduces urethane-induced lung tumor development in mice. PloS one. 2011;6(10):e26590. doi: 10.1371/journal.pone.0026590. Epub 2011/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150(1):165–78. doi: 10.1016/j.cell.2012.04.042. Epub 2012/07/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sevko A, Umansky V. Myeloid-derived suppressor cells interact with tumors in terms of myelopoiesis, tumorigenesis and immunosuppression: thick as thieves. Journal of Cancer. 2013;4(1):3–11. doi: 10.7150/jca.5047. Epub 2013/02/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sawant A, Schafer CC, Jin TH, Zmijewski J, Tse HM, Roth J, et al. Enhancement of antitumor immunity in lung cancer by targeting myeloid-derived suppressor cell pathways. Cancer Res. 2013;73(22):6609–20. doi: 10.1158/0008-5472.CAN-13-0987. Epub 2013/10/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fritz JM, Dwyer-Nield LD, Malkinson AM. Stimulation of neoplastic mouse lung cell proliferation by alveolar macrophage-derived, insulin-like growth factor-1 can be blocked by inhibiting MEK and PI3K activation. Molecular cancer. 2011;10:76. doi: 10.1186/1476-4598-10-76. Epub 2011/06/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Q, Pan J, Lubet RA, Wang Y, You M. Targeting the insulin-like growth factor-1 receptor by picropodophyllin for lung cancer chemoprevention. Molecular carcinogenesis. 2015;54(Suppl 1):E129–37. doi: 10.1002/mc.22206. Epub 2014/08/29. [DOI] [PubMed] [Google Scholar]
- 65.Zhang X, Shan P, Jiang G, Cohn L, Lee PJ. Toll-like receptor 4 deficiency causes pulmonary emphysema. The Journal of clinical investigation. 2006;116(11):3050–9. doi: 10.1172/JCI28139. Epub 2006/10/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lenters V, Basinas I, Beane-Freeman L, Boffetta P, Checkoway H, Coggon D, et al. Endotoxin exposure and lung cancer risk: a systematic review and meta-analysis of the published literature on agriculture and cotton textile workers. Cancer causes & control : CCC. 2010;21(4):523–55. doi: 10.1007/s10552-009-9483-z. Epub 2009/12/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yusuf N, Nasti TH, Long JA, Naseemuddin M, Lucas AP, Xu H, et al. Protective role of Toll-like receptor 4 during the initiation stage of cutaneous chemical carcinogenesis. Cancer Res. 2008;68(2):615–22. doi: 10.1158/0008-5472.CAN-07-5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yusuf N, Nasti TH, Meleth S, Elmets CA. Resveratrol enhances cell-mediated immune response to DMBA through TLR4 and prevents DMBA induced cutaneous carcinogenesis. Molecular carcinogenesis. 2009 doi: 10.1002/mc.20517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hold GL, Rabkin CS, Chow WH, Smith MG, Gammon MD, Risch HA, et al. A functional polymorphism of toll-like receptor 4 gene increases risk of gastric carcinoma and its precursors. Gastroenterology. 2007;132(3):905–12. doi: 10.1053/j.gastro.2006.12.026. [DOI] [PubMed] [Google Scholar]
- 70.Muller-Decker K, Manegold G, Butz H, Hinz DE, Huttner D, Richter KH, et al. Inhibition of cell proliferation by bacterial lipopolysaccharides in TLR4-positive epithelial cells: independence of nitric oxide and cytokine release. The Journal of investigative dermatology. 2005;124(3):553–61. doi: 10.1111/j.0022-202X.2004.23598.x. Epub 2005/03/02. [DOI] [PubMed] [Google Scholar]
- 71.Wang Z, Yan J, Lin H, Hua F, Wang X, Liu H, et al. Toll-like receptor 4 activity protects against hepatocellular tumorigenesis and progression by regulating expression of DNA repair protein Ku70 in mice. Hepatology. 2013;57(5):1869–81. doi: 10.1002/hep.26234. Epub 2013/01/10. [DOI] [PubMed] [Google Scholar]
- 72.Cheng I, Plummer SJ, Casey G, Witte JS. Toll-like receptor 4 genetic variation and advanced prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16(2):352–5. doi: 10.1158/1055-9965.EPI-06-0429. [DOI] [PubMed] [Google Scholar]
- 73.Apetoh L, Ghiringhelli F, Tesniere A, Criollo A, Ortiz C, Lidereau R, et al. The interaction between HMGB1 and TLR4 dictates the outcome of anticancer chemotherapy and radiotherapy. Immunological reviews. 2007;220:47–59. doi: 10.1111/j.1600-065X.2007.00573.x. [DOI] [PubMed] [Google Scholar]
- 74.Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nature medicine. 2007;13(9):1050–9. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- 75.Balkwill F. Tumour necrosis factor and cancer. Nature reviews Cancer. 2009;9(5):361–71. doi: 10.1038/nrc2628. Epub 2009/04/04. [DOI] [PubMed] [Google Scholar]
- 76.Cho HJ, Jung JI, Lim do Y, Kwon GT, Her S, Park JH, et al. Bone marrow-derived, alternatively activated macrophages enhance solid tumor growth and lung metastasis of mammary carcinoma cells in a Balb/C mouse orthotopic model. Breast cancer research: BCR. 2012;14(3):R81. doi: 10.1186/bcr3195. Epub 2012/05/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Redente EF, Dwyer-Nield LD, Merrick DT, Raina K, Agarwal R, Pao W, et al. Tumor progression stage and anatomical site regulate tumor-associated macrophage and bone marrow-derived monocyte polarization. The American journal of pathology. 2010;176(6):2972–85. doi: 10.2353/ajpath.2010.090879. Epub 2010/05/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mastrangelo G, Fadda E, Cegolon L. Endotoxin and cancer chemoprevention. Cancer epidemiology. 2013;37(5):528–33. doi: 10.1016/j.canep.2013.04.008. Epub 2013/05/23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Depiction of the clustering of 240 genes that significantly differed between BALBLpsd and BALB mice 3 days following BHT treatment. The color bar indicates the expression (red increased; blue decreased). B) The gene ontology (GO) functional analysis for the genes increased 142 genes out of 240 genes. Enrichment score of 3 and higher are considered significant. C) Example of immune system processes expanded from the GO analysis presented in B. Again, any functions that have an enrichment score above 3 are considered significant.
A) In this table we list all the 240 genes on the microarray that were significantly different between the BALBLpsd and BALB mice 3 days following treatment with chronic BHT (P<0.05 and ≥1.3-fold changes, both increased and decreased). B) These are the pathways identified in genes that were increased in the BALBLpsd mice compared to BALB mice, analyzed in Partek Pathway Analysis. C) These are the pathways identified in genes that were decreased in the BALBLpsd mice compared to BALB mice, analyzed in Partek Pathway Analysis. Pathways with significance at p<0.05 or below are presented.