Abstract
Background
Follicular lymphoma (FL) is an indolent non-Hodgkin lymphoma that has a risk of transformation to more aggressive lymphoma. Relatively little is known about the non-malignant B-cell and T-cell subset composition within the tumor microenvironment and whether altered phenotypes are associated with patterns of lymphoma B-cell heterogeneity.
Methods
Two mass cytometry (CyTOF) panels were designed to immunophenotype B and T cells in FL tumors. Populations of malignant B cells, non-malignant B cells, and T cells from each FL tumor were identified and their phenotypes compared to B and T cells from healthy human tonsillar tissue.
Results
Diversity in cellular phenotype between tumors was greater for the malignant B cells than for non-malignant B or T cells. The malignant B-cell population bore little phenotypic similarity to any healthy B-cell subset, and unexpectedly clustered closer to naïve B-cell populations than GC B-cell populations. Among the non-malignant B cells within FL tumors, a significant lack of GC and plasmablast B cells was observed relative to tonsil controls. In contrast, non-malignant T cells in FL tumors were present at levels similar to their cognate tonsillar T-cell subsets.
Conclusion
Mass cytometry revealed that diverse HLA-DR expression on FL cells within individual tumors contributed greatly to tumor heterogeneity. Both malignant and non-malignant B cells in the tumor bore little phenotypic resemblance to healthy GC B cells despite the presence of T follicular helper cells in the tumor. These findings suggest that ongoing signaling interactions between malignant B cells and intra-tumor T cells shape the tumor microenvironment.
Keywords: Follicular lymphoma, mass cytometry (CyTOF), human tissue, tumor-infiltrating lymphocyte
Introduction
Follicular Lymphoma (FL) is a B-cell malignancy and the second most common non-Hodgkin lymphoma. Although patient overall survival is now measured in decades, FL is considered incurable and multiple relapses are common (1). Even with the use of the life-extending anti-CD20 drug Rituximab, FL has remained largely incurable and maintains a 3% per-year transformation rate to the more aggressive diffuse large B cell lymphoma (DLBCL). FL is named for its follicle-like appearance and is thought to arise from mature germinal center (GC) B cells (2). Malignant FL B cells are characterized by light-chain restriction and the t(14;18) translocation that leads to overexpression of B cell CLL/lymphoma 2 (BCL-2) (3). As the disease evolves, more mutations are acquired, leading to genetic heterogeneity in the tumor (4,5). The high rate of relapse and the identification of negative prognostic cells present at high levels within some FL tumors (6) has highlighted the need to understand the biology of FL tumor cell heterogeneity and interaction with the microenvironment.
Much of the past FL research has focused on genetics (1,2,5) and flow cytometric analysis with a limited number of markers measured per single cell (6,7). The introduction of mass cytometry and the associated field of computational high-dimensional analysis have paved the way for in-depth analysis of single cell phenotype (8,9). Here, malignant and non-malignant tumor cells were characterized at the single cell protein level and compared to the lymphocytes from healthy tonsils. Non-malignant healthy donor tonsil is an accessible lymphoid tissue that contains mature B cells that are pre-, during-, and post-germinal center reaction (10) and that has been studied previously by mass cytometry (11-13). While differences exist in the phenotype and organization of the cells in human secondary lymphoid organs including spleen, lymph nodes, and tonsils (10,14,15), both tonsils and reactive lymph nodes are valuable comparison points for studies of tumor-involved lymph nodes and follicular lymphoma tumors. In this exploratory study, mass cytometry was used to study malignant B cells and non-malignant B and T cells in the tumor microenvironment with a focus on intra- and inter-tumor heterogeneity, and on changes in the composition of immune cells within tumors. Mass cytometry was selected due to the ability to characterize 35 or more features of individual cells and the ability to reveal unexpected malignant and non-malignant cell subsets with unusual phenotypes (16-20).
Materials and Methods
Tissue Sample Collection
FL samples were selected from the biobank at The Norwegian Radium Hospital, Oslo University Hospital. Tonsils were obtained from patients undergoing tonsillectomy at Agroklinikken, Asker, Norway. All samples were obtained with patient consent in accordance with the Declaration of Helsinki. The study was approved by the regional committee for research ethics, Oslo Norway, and the Vanderbilt institutional review board (IRB).
Mass cytometry
Cryopreserved single cells were thawed, pelleted, and rested in RPMI-1640 with 10% FBS for 45 minutes before staining. Viability was determined by trypan blue staining and manual cell counting. Samples with less than 50% viability were excluded from the study. At least one million live cells per sample were stained with surface antibodies (listed in Table S1) for 30 minutes, then washed and fixed in 1.6% PFA for 5 minutes followed by cell membrane permeabilization with >90% cold methanol. Cells were stored in methanol at -80°C for up to two weeks. Samples were washed twice and stained with intracellular antibodies (listed in Table S1) for 30 minutes. Cells were then incubated in iridium cell tracker (Fluidigm) at the recommended concentration for 20 minutes. Stained and intercalated samples were collected on a CyTOF 1 (Fluidigm) at the Vanderbilt flow cytometry core.
Data analysis
The cloud based flow cytometry platform, Cytobank (21), was used for file storage and data analysis including biaxial data display and gating, viSNE (22), SPADE (23), histograms, heat maps, and statistics consistent with standard computational analysis workflow (24). The statistical language R with RStudio was used for hierarchical clustering (25,26).
A computational light-chain channel was added to the B-cell panel files post data collection. The channel was named “light_chain” and was created by selecting the value of either IgL or IgK, whichever was higher, for each cell event. This new light chain channel reported the light chain level for each B cell regardless of isotype and was used in computational analysis of phenotype.
The dimensionality reduction similarity mapping tool viSNE (22) was used to create two-dimensional t-SNE visualizations of multidimensional cellular phenotypes (9,24,27). Cells that had similar phenotypes were placed close together on viSNE maps created based on analysis of 20 cellular measurements. T cells from all tonsil and tumor samples were analyzed together by viSNE using 20 markers from the T-cell panel (Fig. 1 and in depth in Fig. 2). A total of three B cell viSNE analyses were performed each using the same 20 B-cell panel markers. All B cells were initially analyzed together (Fig. 1), and that map was used to gate for malignant and non-malignant B cells. In the two subsequent B cell viSNE analyses, the non-malignant B cells from FL samples and all tonsillar B cells were analyzed together in a “non-malignant B cell” viSNE (Fig. 3), and the malignant B cells were analyzed together in a “malignant B cell” viSNE (Fig. 4). SPADE was applied to the t-SNE axes created by viSNE (Fig. 5) to cluster cells into cell subsets (16,24,28). Variance in marker expression between clusters was calculated for nodes containing at least 100 cells using transformed median values (arcsinh scale, cofactor of 15, as in (6)).
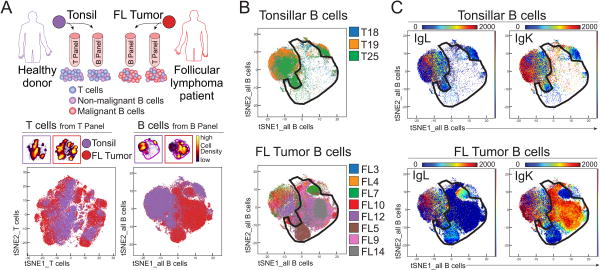
Figure 1. High dimensional phenotyping of lymphoma B cells, non-malignant B cells, and tumor-infiltrating T cells.
A) FL tumor samples and tonsils from healthy donors were studied by mass cytometry. Each sample was split in two and analyzed by two antibody panels, one focused on B cells and the other on T cells, to identify three main populations of cells in follicular lymphoma (FL) tumor samples (malignant B cells, non-malignant B cells, and T cells) as shown in the cartoon. CD3+ T cells (bottom left) and CD19+ B cells (bottom right) from tonsils and FL samples were analyzed together in a viSNE based on 20 markers from the T-cell panel and 20 markers from the B-cell panel, respectively. B-C) Malignant B cells were gated in an area of the viSNE map where tonsillar B cells were mostly absent (B) and the cells were light-chain restricted (C). Top row shows all tonsil samples combined, bottom row shows all FL samples combined.
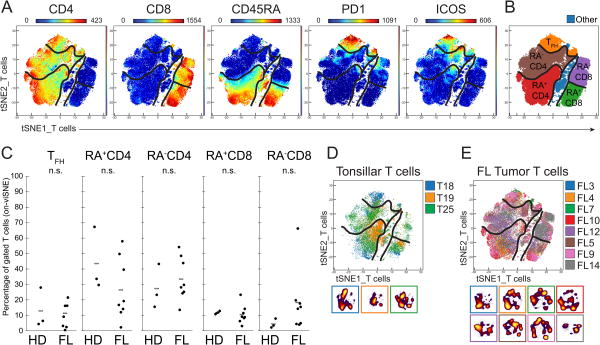
Figure 2. The abundance and phenotypes of tumor infiltrating T-cell subsets are comparable to those of healthy tonsillar T cells.
A) Expression of measured proteins is shown as a heat plot on t-SNE axes (tSNEs_T cells). Markers shown were used to make the expert-gated populations shown in (B). Populations are denoted by black lines. B) T-cell populations were gated in viSNE based on markers shown in A. C) Distribution of T-cell subsets across samples. Figure shows all healthy donor (HD) tonsil samples (D) and all FL samples (E) overlaid with individual cell-density plots beneath.
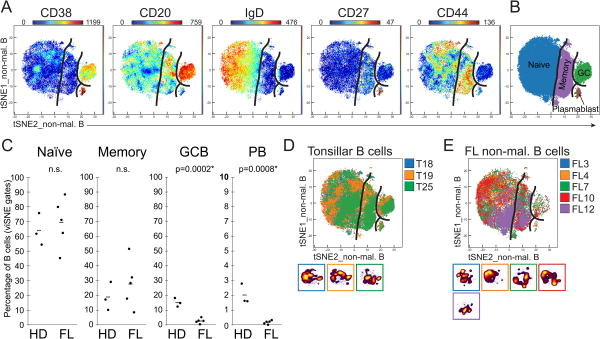
Figure 3. Disruption of germinal center and plasmablast populations is observed in tumor-infiltrating non-malignant B cells.
Non-malignant B cells, identified in Figure 1, were analyzed in a new viSNE (tSNEs_non-mal. B). FL samples with 90% or more malignant cells were excluded from the analysis. A) Expression of measured proteins is shown as a heat plot on t-SNE axes. B) Markers shown in (A) were used to gate established B-cell populations. C) Distribution of B-cell subsets across samples. Figure shows all healthy donor (HD) tonsil samples (D) and all FL samples (E) overlaid and with color coded individual cell density plots beneath.
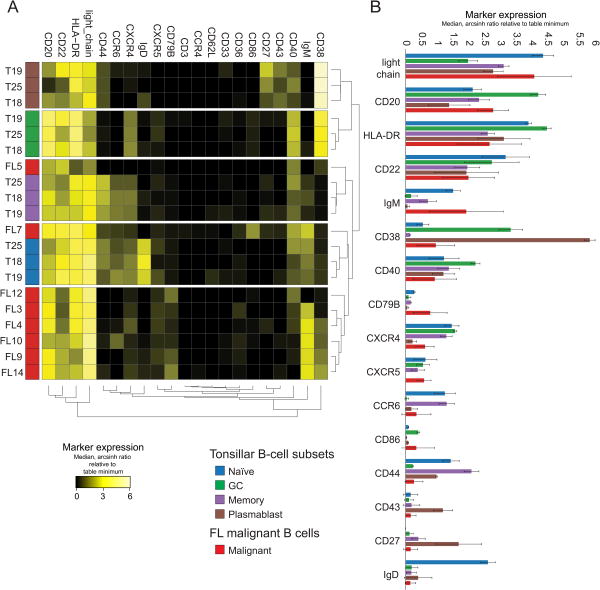
Figure 4. Hierarchical clustering grouped most malignant B-cell populations and revealed malignant B cells are more phenotypically similar to naive and memory B cells than to GC B cells.
A) Dendrogram shows hierarchical clustering of healthy tonsillar B cell subsets and FL malignant B cells (as gated in Figure 3 and Figure 1 respectively) based on median marker expression. B) Average median marker expression of healthy tonsillar B cell subsets and FL malignant B cells. Markers are ordered by FL marker median mass intensity (MMI) from top to bottom.
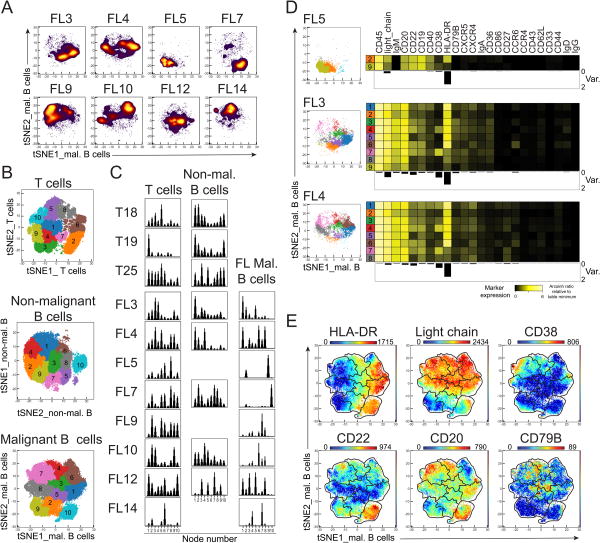
Figure 5. Intratumor heterogeneity differed between the FL samples and HLA-DR was the most variable marker across samples.
Malignant B cells, gated in Figure 1, were analyzed in a new viSNE. Individual density plots are shown. B) SPADE analysis with 10 nodes were analyzed on the viSNE of T cells (top), non-malignant B cells (middle), and malignant B cells (bottom). Figure shows each numbered node as a separate color on a viSNE map of aggregated files. C) Histograms display number of events in each SPADE node. D) Heat map of median marker expression for each SPADE node with more than 100 cells for the FL samples. Below each heat map, the variance in each marker between the SPADE nodes are displayed as bar graphs. E) Expression of measured proteins is shown as a heat plot on t-SNE axes (malignant B cells viSNE) for some of the markers with high variance across several samples. Gates represent SPADE nodes.
Results
FL malignant B cells were phenotypically distinct from tonsillar B cells
Eight FL lymph node tumor samples were phenotyped by mass cytometry. Each sample was stained with two different panels, one focused on T-cell makers and one on B-cell markers. Three non-malignant healthy donor tonsils were stained with the same panels to serve as healthy controls (Fig. 1). After initial gating to identify singlet T cells or B cells (Supplementary Fig. 1A), viSNE analyses were performed. CD45+ CD3+ CD19- cells from the 11 T-cell panel files were analyzed together in the “T cells” viSNE, (Fig 1A; tSNEs_T cells). The panel was designed to identify and characterize T-cell subsets, and included for viSNE analysis the following 20 markers: CCR6, CCR5, CD4, CD8, CCR4, CD43, ICOS, TCRγδ, CD45RA, CXCR3, CCR7, CD69, CD44, CD27, CTLA4, CD25, CXCR5, CD57, PD1, CXCR4. These markers were not used to gate for T cells.
CD45+ CD19+ and/or CD20+ CD3- B cells were analyzed together in a similar fashion for the “all B cells” viSNE analysis (Fig. 1A; tSNEs_all B cells). The panel was designed to identify and characterize B cell subsets, and included for viSNE analysis the following 20 markers: CCR6, IgD, CD20, CCR4, CD43, CD36, CD62L, CD86, CD33, CD22, CD79B, CD40, CD44, CD27, CD38, CD3, CXCR5, HLA-DR, CXCR4, light_chain. The tonsillar B cells mapped almost entirely to the periphery of the all B cells viSNE map, leaving a large space in the center of the map where the majority of B cells from the eight FL tumors fell (Fig. 1, Supplementary Fig. 2). The central area on the viSNE map where the tonsillar B cells were primarily absent was gated as malignant B cells (Fig. 1B,C). The viSNE map was made without the use of IgLambda (IgL) and IgKappa (IgK) to guide the separation of malignant cells; instead, a computational light-chain channel was used to indicate positivity of total light chain. IgL and IgK expression were used afterward to confirm the isotype exclusion of the malignant B cells for each tumor sample (Supplementary Fig. 3). The area outside the malignant gate, the non-malignant B-cell area, for tonsil and FL tumors showed a mixture of light-chain isotypes, as expected (Fig. 1C). Two IgL lymphomas (FL5 and FL7) were clearly identifiable on the viSNE map as IgK- and IgL+ (bottom, Fig. 1C). The intensities of IgL and IgK showed that the malignant area contained light-chain-restricted cells whereas the non-malignant area contained a mix of IgK and IgL cells, similar to the tonsillar B cells (Fig. 1C). A key finding here is the fact that light-chain-restricted cells fell into the malignant area regardless of light-chain isotype. The FL samples contained cells that mapped in the non-malignant area, but to varying degrees. These cells were interpreted as non-malignant B cells present within the tumors.
T-cell distribution trended toward an activated state
Key T-cell markers and the T cells viSNE map (Fig. 2A) were used to subset the T-cell panel samples into five populations (Fig. 2B). For comparison, traditional biaxial gating were performed in parallel (Supplementary Fig. 4A,B). The eight FL tumors and the three tonsils had a high degree of overlap in the T cell viSNE map (Fig. 1D-E), suggesting a similar phenotype of tumor-associated T cells and healthy tonsillar T cells. There were no significant differences between T-cell subset distributions in FL tumors and tonsils (Fig. 2C, Supplementary Fig. 5A). T cells from most samples were distributed throughout the viSNE map with the exception of FL14 (Fig. 2D-E). FL14 had a restricted distribution and contained predominantly CD8+ T cells.
Although there were no significant differences between tonsillar T cell and FL tumor T cell-subset proportions, FL tumors tended to have fewer CD45RA+ or more CD45RA- T cells, suggesting fewer naïve cells were present (Fig. 2C). FL tumors also displayed a skewing towards CD8+ T cells (Supplementary Fig. 6).
Diminished GC and plasmablast populations among non-malignant B cells in FL tumors
Non-malignant tumor-infiltrating B cells were next investigated by analyzing them in a new viSNE together with the tonsillar B cells (tSNEs_non-mal. B) using the same markers as the initial B cell viSNE (Fig. 3). In Figure 1, B cells positioned outside the malignant gate were considered non-malignant due to their overlapping with tonsillar B cells in the viSNE map and the mixture of IgK and IgL expressing cells. Three of the FL tumors contained less than 10% non-malignant B cells and were excluded from this analysis (Supplementary Fig. 2). Key B-cell markers were used to draw expert gates on the non-mal. B viSNE map to divide the B cells into four mature B-cell subsets; naïve, memory, germinal center (GC), and plasmablasts (Fig. 3A-B, Supplementary Fig. 4C,D). The FL tumor non-malignant B cells and the tonsillar B cells showed a high degree of overlap in the B cell viSNE map (Fig. 3D-E), suggesting a similar phenotype of non-malignant B cells and healthy tonsillar B cells. There were significant differences in B cell-subset distributions between tonsil and FL tumor for GC B cells and plasmablasts (Fig. 3C, Supplementary Fig. 5B) with average percentages of 15% and 2.8% for GC B cells and 2% and 0.2% for plasmablasts, respectively.
FL malignant B cells were not germinal-center like in phenotype
The (non-malignant) tonsillar B-cell populations for each tonsil (4 populations from 3 tonsils) from Figure 3 were compared to the malignant B-cell population from each FL tumor sample using median intensity of the same 20 markers used in the B cells viSNE analyses. The populations were hierarchically clustered by median marker intensities, thereby creating a dendrogram that showed how phenotypically similar the populations were. The malignant B cells primarily clustered together, but had a phenotype more similar to naïve and memory B cells than GC and plasmablasts (Fig. 4A). There was no single marker that separated out the malignant B cells (Fig. 4B). Malignant B cells had higher levels of CD79B and IgM than tonsillar B-cell subsets suggesting strong dependence of BCR signaling. However, in contrast to naive B cells, all malignant B cells were IgD-. The malignant B cells also showed downregulation of the co-receptor CD40.
Mass cytometry characterized high variability of intra-tumor phenotypic heterogeneity in malignant B cells
The malignant B cells from FL samples were analyzed in a separate viSNE analysis (tSNEs_mal. B cells). In contrast to the non-malignant B cells and T cells, malignant B cell viSNE maps varied between the samples with cells occupying different areas of the map (Fig. 5A). To quantify this variation, unsupervised clustering using SPADE was utilized to make 10 unbiased clusters of cells for each of the viSNE maps (T cells, malignant B, and non-malignant B; Fig. 5B). Cell distribution between the nodes could then be studied within a sample and across samples (Fig. 5C). There were some variation in the distribution throughout SPADE nodes for all the three cell types, but with greater variability for the malignant B cells than the non-malignant B and T cells (Fig. 5C). For the malignant B cells, some FL samples were dominated by a few SPADE nodes (FL5, FL7, and FL14) whereas others spread out in the viSNE map (FL3, FL4, FL12). The markers that varied the most within tumors were HLA-DR, light_chain, and CD38 (Fig. 5D-E). These results demonstrated variable phenotypic heterogeneity among the malignant B cells within a single tumor.
Discussion
Minimally biased, automated computational analysis using viSNE accurately separated malignant B cells from non-malignant B cells without using BCL-2 expression or light-chain restriction when distinguishing these cell subsets. Light-chain isotype was found to be restricted within the computationally defined malignant cell area, as expected for FL (29-32). Thus, automated computational analysis was effective at identifying malignant and non-malignant cells despite the significant heterogeneity of lymphoma tumors. viSNE also successfully separated populations of non-malignant B cells and T cells.
Multiple viSNE analyses were performed with and without the combined light-chain channel and the malignant cells plotted separately from the tonsillar B cells with both strategies. This result suggested that markers other than light-chain restriction distinguish FL malignant B cells from healthy B cells. CD79B, the signaling subunit of the B-cell receptor, was expressed at higher levels in malignant B cells than in all non-malignant B-cell populations. Similarly, malignant B cells typically displayed high per-cell expression of Ig light chain and IgM. This high level of BCR protein expression on the lymphoma B cells is consistent with a continued dependence on BCR signaling in FL (6). Activation of BCR signaling by auto-antigen might be one of the initial driving forces in FL, and several auto-antigens have been identified (33-35). Furthermore, high BCR expression, potentially followed by BCR-induced activation, aligns with the observed skewing of the non-malignant B-cell populations and suggests that the malignant B cells outcompete the non-malignant B cells that are most dependent on support from the microenvironment for survival and selection, such as GC B cells and plasmablasts (36). Together with the survival advantage provided by overexpression of anti-apoptotic BCL-2, this may explain the deficit in non-malignant GC and plasma B cell populations observed here in FL tumors.
FL is traditionally considered to be a germinal center malignancy. FL malignant B cells contain evidence of SHM in their immunoglobulin genes (37), and exhibit a GC-like gene expression pattern (38,39). However, key signaling receptors differed between GC and malignant B cells and the protein expression pattern of malignant B cells was not GC-like. Critically, higher per-cell expression of BCR subunits CD79B, light-chain, and heavy-chain provided a clear distinction between malignant B cells and GC B cells, which expressed less surface BCR subunit proteins than naïve healthy B cells and malignant B cells. The malignant B cells were lower than GC B cells for other markers, including CD20 and CD38, as shown previously (7). However, malignant B cells did not phenotypically match naïve B cells or any other subset of non-malignant B cells. For example, both malignant B cells and healthy tonsillar GC B cells expressed lower levels of CD44 than naïve B cells. Thus, the expression profile of signaling receptors on malignant B cells distinguished them from all subsets of non-malignant B cells, including GC B cells.
This mass cytometry study extends knowledge of intra- and intertumor heterogeneity that has been observed in prior studies of follicular lymphoma cell genetics, phenotypes, and functional capabilities. Prior studies of clonal evolution in FL have revealed genetic heterogeneity (4,5,40). Single cell analysis of BCR signaling and patterns of protein expression with fluorescence flow cytometry revealed a lymphoma negative prognostic (LNP) cells that exists at diagnosis in patients with poor overall survival (6). Here, we observed similar patterns of CD20 expression as in these prior studies and identified additional proteins that are highly variable among lymphoma cells from the same tumor. In particular, mass cytometry revealed HLA-DR expression as one of the most variable features of FL. Green et al. have previously found CREBBP to be commonly mutated in FL, a mutation that is associated with decreased antigen presentation and expression of HLA-DR on FL B cells (40). Furthermore, this intratumor as well as intertumor variation in HLA-DR is significant as HLA-DR expression has been previously reported in other cancers to be associated with a positive response to anti-PD1 checkpoint inhibitor therapy (41).
In conclusion, the use of mass cytometry to obtain deep profiling of cell subsets enabled identification of biologically important features, such as tumor heterogeneity and loss of non-malignant B-cell subsets.
Supplementary Material
Acknowledgments
Funding sources: Study and researchers were supported by NIH/NCI R00 CA143231 (J.M.I.), F31 CA199993 (A.R.G.), the Vanderbilt-Ingram Cancer Center (VICC, P30 CA68485), a VICC Ambassadors award (J.M.I.), and The Norwegian Cancer Society (K.H.).
Footnotes
Conflict-of-interest disclosure: J.M.I. is co-founder and board member at Cytobank Inc. and received research support from Incyte Corp.
References
- 1.Dave SS. Gene expression signatures and outcome prediction in mature B-cell malignancies. Current Treatment Options in Oncology. 2006;7:261–269. doi: 10.1007/s11864-006-0035-4. [DOI] [PubMed] [Google Scholar]
- 2.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, Fisher RI, Braziel RM, Rimsza LM, Grogan TM, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–69. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 3.Ngan BY, Chen-Levy Z, Weiss LM, Warnke RA, Cleary ML. Expression in Non-Hodgkin's Lymphoma of the bcl-2 Protein Associated with the t(14;18) Chromosomal Translocation. New England Journal of Medicine. 1988;318:1638–1644. doi: 10.1056/NEJM198806233182502. [DOI] [PubMed] [Google Scholar]
- 4.Eide MB, Liestøl K, Lingjærde OC, Hystad ME, Kresse SH, Meza-Zepeda L, Myklebost O, Trøen G, Aamot HV, Holte H, et al. Genomic alterations reveal potential for higher grade transformation in follicular lymphoma and confirm parallel evolution of tumor cell clones. Blood. 2010;116:1489–1497. doi: 10.1182/blood-2010-03-272278. [DOI] [PubMed] [Google Scholar]
- 5.Green MR, Gentles AJ, Nair RV, Irish JM, Kihira S, Liu CL, Kela I, Hopmans ES, Myklebust JH, Ji H, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121:1604–11. doi: 10.1182/blood-2012-09-457283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irish JM, Myklebust JH, Alizadeh AA, Houot R, Sharman JP, Czerwinski DK, Nolan GP, Levy R. B-cell signaling networks reveal a negative prognostic human lymphoma cell subset that emerges during tumor progression. Proceedings of the National Academy of Sciences. 2010 doi: 10.1073/pnas.1002057107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mantei KW, Brent L. Flow cytometric evaluation of CD38 expression assists in distinguishing follicular hyperplasia from follicular lymphoma. Cytometry Part B: Clinical Cytometry. 2009;76B doi: 10.1002/cyto.b.20477. [DOI] [PubMed] [Google Scholar]
- 8.Greenplate AR, Johnson DB, Ferrell PB, Irish JM. Systems immune monitoring in cancer therapy. Eur J Cancer. 2016;61:77–84. doi: 10.1016/j.ejca.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saeys Y, Gassen SV, Lambrecht BN. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat Rev Immunol. 2016;16:449–62. doi: 10.1038/nri.2016.56. [DOI] [PubMed] [Google Scholar]
- 10.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polikowsky HG, Wogsland CE, Diggins KE, Huse K, Irish JM. Cutting Edge: Redox Signaling Hypersensitivity Distinguishes Human Germinal Center B Cells. J Immunol. 2015;195:1364–7. doi: 10.4049/jimmunol.1500904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sen N, Mukherjee G, Sen A, Bendall SC, Sung P, Nolan GP, Arvin AM. Single-cell mass cytometry analysis of human tonsil T cell remodeling by varicella zoster virus. Cell Rep. 2014;8:633–45. doi: 10.1016/j.celrep.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong MT, Chen J, Narayanan S, Lin W, Anicete R, Kiaang HT, De Lafaille MA, Poidinger M, Newell EW. Mapping the Diversity of Follicular Helper T Cells in Human Blood and Tonsils Using High-Dimensional Mass Cytometry Analysis. Cell Rep. 2015;11:1822–33. doi: 10.1016/j.celrep.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Brachtel EF, Washiyama M, Johnson GD, Tenner-Racz K, Racz P, MacLennan IC. Differences in the germinal centres of palatine tonsils and lymph nodes. Scand J Immunol. 1996;43:239–47. [PubMed] [Google Scholar]
- 15.Vidal-Rubio B, Sanchez-Carril M, Oliver-Morales J, Gonzalez-Femandez A, Gambon-Deza F. Changes in human lymphocyte subpopulations in tonsils and regional lymph nodes of human head and neck squamous carcinoma compared to control lymph nodes. BMC Immunol. 2001;2:2. doi: 10.1186/1471-2172-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KW, Low D, Ruedl C, Riccardi-Castagnoli P, Poidinger M, et al. High-dimensional analysis of the murine myeloid cell system. Nat Immunol. 2014;15:1181–9. doi: 10.1038/ni.3006. [DOI] [PubMed] [Google Scholar]
- 17.Bendall SC, Davis KL, Amir el AD, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP, Pe'er D. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–25. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irish JM, Doxie DB. High-dimensional single-cell cancer biology. Curr Top Microbiol Immunol. 2014;377:1–21. doi: 10.1007/82_2014_367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leelatian N, Doxie DB, Greenplate AR, Mobley BC, Lehman JM, Sinnaeve J, Kauffmann RM, Werkhaven JA, Mistry AM, Weaver KD, et al. Single cell analysis of human tissues and solid tumors with mass cytometry. Cytometry B Clin Cytom. 2016 doi: 10.1002/cyto.b.21481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenplate AR, Johnson DB, Roussel M, Savona MR, Sosman JA, Puzanov I, Ferrell PB, Irish JM. Myelodysplastic Syndrome Revealed by Systems Immunology in a Melanoma Patient Undergoing Anti-PD-1 Therapy. Cancer Immunol Res. 2016 doi: 10.1158/2326-6066.CIR-15-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotecha N, Krutzik PO, Irish JM. Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom. 2010;Chapter 10(Unit10):17. doi: 10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amir EaD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe'er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotech. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu P, Simonds EF, Bendall SC, Gibbs KD, Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–91. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diggins KE, Ferrell PB, Jr, Irish JM. Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods. 2015;82:55–63. doi: 10.1016/j.ymeth.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. URL https://www.R-project.org. [Google Scholar]
- 26.RStudio Team. RStudio: Integrated Development for R. Boston, MA: RStudio, Inc.; 2015. URL http://www.rstudio.com/ [Google Scholar]
- 27.Newell EW, Cheng Y. Mass cytometry: blessed with the curse of dimensionality. Nat Immunol. 2016;17:890–5. doi: 10.1038/ni.3485. [DOI] [PubMed] [Google Scholar]
- 28.Shekhar K, Brodin P, Davis MM, Chakraborty AK. Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding (ACCENSE) Proceedings of the National Academy of Sciences. 2014;111:202–207. doi: 10.1073/pnas.1321405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horna P, Olteanu H, Kroft SH, Harrington AM. Flow cytometric analysis of surface light chain expression patterns in B-cell lymphomas using monoclonal and polyclonal antibodies. Am J Clin Pathol. 2011;136:954–9. doi: 10.1309/AJCP3C2QZZBPTMLB. [DOI] [PubMed] [Google Scholar]
- 30.Davidson B, Risberg B, Berner A, Smeland EB, Torlakovic E. Evaluation of lymphoid cell populations in cytology specimens using flow cytometry and polymerase chain reaction. Diagn Mol Pathol. 1999;8:183–8. doi: 10.1097/00019606-199912000-00003. [DOI] [PubMed] [Google Scholar]
- 31.Irish JM, Czerwinski DK, Nolan GP, Levy R. Altered B-cell receptor signaling kinetics distinguish human follicular lymphoma B cells from tumor-infiltrating nonmalignant B cells. Blood. 2006;108:3135–42. doi: 10.1182/blood-2006-02-003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Craig FE, Foon KA. Flow cytometric immunophenotyping for hematologic neoplasms. Blood. 2008;111:3941–67. doi: 10.1182/blood-2007-11-120535. [DOI] [PubMed] [Google Scholar]
- 33.Cha SC, Qin H, Kannan S, Rawal S, Watkins LS, Baio FE, Wu W, Ong J, Wei J, Kwak B, et al. Nonstereotyped Lymphoma B Cell Receptors Recognize Vimentin as a Shared Autoantigen. The Journal of Immunology. 2013;190:4887–4898. doi: 10.4049/jimmunol.1300179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coelho V, Krysov S, Ghaemmaghami AM, Emara M, Potter KN, Johnson P, Packham G, Martinez-Pomares L, Stevenson FK. Glycosylation of surface Ig creates a functional bridge between human follicular lymphoma and microenvironmental lectins. Proceedings of the National Academy of Sciences. 2010;107:18587–18592. doi: 10.1073/pnas.1009388107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sachen KL, Strohman MJ, Singletary J, Alizadeh AA, Kattah NH, Lossos C, Mellins ED, Levy S, Levy R. Self-antigen recognition by follicular lymphoma B-cell receptors. Blood. 2012;120:4182–4190. doi: 10.1182/blood-2012-05-427534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Victora GD, Nussenzweig MC. Germinal Centers. Annual Review of Immunology. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 37.Zhu D, McCarthy H, Ottensmeier CH, Johnson P, Hamblin TJ, Stevenson FK. Acquisition of potential N-glycosylation sites in the immunoglobulin variable region by somatic mutation is a distinctive feature of follicular lymphoma. Blood. 2002;99:2562–2568. doi: 10.1182/blood.v99.7.2562. [DOI] [PubMed] [Google Scholar]
- 38.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 39.Elenitoba-Johnson KSJ, Jenson SD, Abbott RT, Palais RA, Bohling SD, Lin Z, Tripp S, Shami PJ, Wang LY, Coupland RW, et al. Involvement of multiple signaling pathways in follicular lymphoma transformation: p38-mitogen-activated protein kinase as a target for therapy. Proceedings of the National Academy of Sciences. 2003;100:7259–7264. doi: 10.1073/pnas.1137463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Green MR, Kihira S, Liu CL, Nair RV, Salari R, Gentles AJ, Irish J, Stehr H, Vicente-Dueñas C, Romero-Camarero I, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci U S A. 2015;112:E1116–25. doi: 10.1073/pnas.1501199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson DB, Estrada MV, Salgado R, Sanchez V, Doxie DB, Opalenik SR, Vilgelm AE, Feld E, Johnson AS, Greenplate AR, et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat Commun. 2016;7:10582. doi: 10.1038/ncomms10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.