Abstract
Aim:
There is controversy regarding the potential fitness costs of rifampicin (RIF) resistance-conferring mutations in the Mycobacterium tuberculosis (Mtb) rpoB gene. We characterized the pathogenicity of an Mtb RpoB H526D mutant.
Materials & methods:
A mutant containing the RpoB H526D mutation was isolated from wild-type Mtb grown on RIF-containing plates and complemented for determination of in vitro and in vivo fitness costs.
Results:
The RpoB H526D mutant showed reduced survival relative to control strains during progressive hypoxia and delayed growth following resuscitation from nutrient starvation (p < 0.05), which was associated with reduced expression of the resuscitation-promoting factor genes rpfB, rpfC and rpfE. Relative to the isogenic wild-type strain, the mutant showed significantly attenuated growth and long-term survival as well as reduced inflammation in mouse lungs.
Conclusion & future perspective:
Our data suggest that RpoB H526D mutation confers a fitness cost during growth-limiting conditions in vitro and in mouse lungs.
KEYWORDS : inflammation, murine model, Mycobacterium tuberculosis, nutrient starvation, persistence, progressive hypoxia, resuscitation-promoting factor, Rpf, rpoB, stringent response, virulence
The emergence of multidrug-resistant (MDR) TB, which is resistant to rifampicin (RIF) and isoniazid (INH), poses significant challenges for global TB control efforts [1]. MDR-TB treatment success rates are only 50% globally, leading the WHO to conclude that MDR-TB represents a ‘public health crisis’ [1]. An improved understanding of the pathogenesis of MDR-TB is required to develop new treatment strategies to control its further spread.
RIF is a key sterilizing drug, permitting TB treatment shortening to the current 6 months [2]. RIF binds to the Mycobacterium tuberculosis (Mtb) RNA polymerase β-subunit, inhibiting RNA transcription [3]. More than 95% of RIF-resistant strains contain mutations in the Mtb rpoB gene [4]. An 81-bp fragment of rpoB encoding codons 507–533 (Escherichia coli nomenclature), known as the RIF resistance determining region, harbors most mutations conferring high-level resistance [5]. RIF-resistant Mtb clinical isolates most commonly contain mutations in codons 531, 526 or 516 [6–8]. The mutation RpoB H526D is the second most frequent RIF resistance conferring mutation worldwide [6].
The impact of rpoB mutations on Mtb virulence is controversial. RIF-resistant Mtb strains carrying RpoB mutations at codons 522, 526 or 531 showed reduced growth in macrophages and in nutrient-poor broth [4,9,10], Conversely, RpoB S531L may confer little or no loss of Mtb fitness, and is often associated with compensatory mutations in the rpoA and rpoC genes [9–11], which may be condition dependent [10].
Mtb encounters various stress conditions during host infection, including hypoxia, carbon starvation, and exposure to reactive oxygen and nitrogen species [12]. Mtb clinical strains resistant to multiple antibiotics exhibit reduced growth and impaired inflammation relative to drug-susceptible Mtb in mouse models [13]. However, less is known about the specific contribution of rpoB mutations to these attenuated phenotypes.
The vast majority of immune competent individuals infected with Mtb are able to control bacillary growth in necrotic lung granulomas, leading to lifelong asymptomatic infection (latent TB infection) [14,15]. Although several host factors, such as, HIV infection, TNF blockade and diabetes mellitus, are known risk factors for reactivation disease [15], and specific Mtb virulence factors have been implicated in disease progression [16], the molecular basis underlying Mtb resuscitation from a growth-restricted state remains poorly characterized. Mtb has five rpf genes encoding RpfA–E, a group of secreted lytic transglycosylases sharing a highly conserved 70-residue domain with dramatic potency at picomolar concentrations to stimulate mycobacterial regrowth from dormancy [17–20]. Addition of Rpfs to sputum samples of patients treated with antitubercular drugs enhances the growth of Mtb [21,22]. However, the effect of RIF resistance conferring mutations on Mtb resuscitation from stress conditions and on expression of rpf genes remains to be elucidated.
In the current study, we isolated a spontaneous RIF-resistant Mtb CDC1551 strain containing the RpoB H526D mutation to mimic commonly encountered RIF-resistant clinical isolates, and then reintroduced the native rpoB gene to generate a merodiploid-complemented strain (rpoB comp). The growth and survival of the rpoB mutant was evaluated relative to the parental wild-type and complemented strains during nutrient starvation and progressive hypoxia, and in the lungs of BALB/c mice. In addition, we studied the growth kinetics of each Mtb strain upon resuscitation from nutrient starvation conditions and characterized the transcriptional profile of the five rpf genes and rpoB by quantitative reverse transcription polymerase chain reaction (RT-qPCR).
Materials & methods
• Bacterial strains
Wild-type Mtb CDC1551 [23] was plated on Middlebrook 7H10 agar (BD Difco, MD, USA) containing RIF 1 μg/ml (Sigma-Aldrich, MO, USA) at 37°C for 21 days. Genomic DNA isolated from individual RIF-resistant colonies was used to analyze mutations by DNA sequencing using primers, rpoB-F and rpoB-R (Supplementary Table 1). The MIC of RIF and INH (Sigma-Aldrich) was assessed [24].
• Complementation of the rpoB mutant strain
A 4202-bp DNA fragment containing the entire rpoB gene, including 417 bp of 5′-flanking sequence and 248 bp of 3′-flanking sequence, was PCR-amplified from Mtb CDC1551 genomic DNA using primers, rpoB-1F and rpoB-1R (Supplementary Table 1), which introduced an XbaI site at the 5′ end. After XbaI-digestion, the rpoB PCR product was ligated to XbaI-digested Escherichia coli–Mycobacterium shuttle vector, pMH94 [25], followed by transformation into E. coli DH5α competent cells. The rpoB-containing construct was confirmed by DNA sequencing and electroporated into competent rpoB mutant cells. Genomic DNA was purified from individual colonies on 7H10 agar plates containing hygromycin (50 μg/ml) and RIF (2 μg/ml) and digested with SacI (NE Biolabs, MA USA). Southern blotting was performed [26] using a 561-bp rpoB probe generated by adding digoxigenin (DIG)-dUTP in PCR reactions containing primer pairs rpoB-2F and rpoB-2R (Supplementary Table 1).
• Growth kinetics/survival & resuscitation in vitro
For growth kinetics assays in nutrient-rich broth, each strain was inoculated (final optical density [OD]600 nm ∼0.05) in 10 ml of supplemented Middlebrook 7H9 broth (BD Difco) containing 10% oleic acid-albumin-dextrose-catalase (BD), 0.1% glycerol and 0.05% Tween-80 (supplemented 7H9 broth), in 50 ml conical tubes in a roller incubator at 37°C for 13 days. For nutrient starvation (NS) experiments, bacterial pellets were washed three-times with 1 × phosphate-buffered saline (Quality Biological, Inc., MD USA) containing 0.05% Tween-80 and resuspended in 10 ml of the same medium (at OD600 nm ∼0.1) in 50 ml conical tubes prior to standing incubation at 37°C up to 28 days [27]. For progressive hypoxia studies, each strain was inoculated into Dubos Tween Albumin Broth (BD Difco) supplemented with the hypoxia indicator dye, methylene blue (500 mg/l) [28] for 21 days after dye color change.
For resuscitation studies, pellets of 9-, 14- and 28-day nutrient-starved cultures were centrifuged and resuspended in 7H9 broth (at OD600 nm ∼0.1), followed by incubation in a shaker at 37°C. The growth of these resuscitated cultures was monitored up to 13 days by OD600 nm and CFU.
Samples from various time points were collected from the above cultures, serially diluted in phosphate-buffered saline and plated on 7H10 agar. The plates were incubated at 37°C for 21 days prior to colony counting. To observe colony size, a longer incubation time was used. All samples were prepared in duplicate or triplicate and each experiment was repeated at least twice.
• In vitro competition assays
The wild-type and the RpoB H526D mutant strains were grown in 7H9 broth to OD600 nm = 0.5 (∼5 × 107 CFU/ml) and diluted in 7H9 broth to OD600 nm = 0.01 (∼106 bacteria/ml) for studies in nutrient-rich broth and to OD600 nm = 0.001 (∼105 bacteria/ml) for hypoxia studies. For nutrient-rich conditions, equal volumes of the mutant and wild-type strains (5 ml of ∼106 bacteria/ml of each strain) were mixed prior to inoculation in supplemented 7H9 broth. For the progressive hypoxia model, equal volumes of the mutant and wild-type strains (10 ml of ∼105 bacteria/ml of each strain) were mixed prior to inoculation [28]. Samples from each time point were plated on 7H10 agar plates with and without RIF 1 μg/ml after serial dilutions. The number of wild-type bacteria was calculated by subtracting the number of bacteria growing on RIF-containing plates from the total number of bacteria growing on antibiotic-free plates. The competitive fitness, W of the rpoB mutant relative to the wild-type strain was calculated using the following formula: W = ln (RF/RI)/ln (SF/SI), where RI and SI refer to the number of RIF-resistant and -susceptible bacteria at baseline, respectively, and RF and SF refer to the same bacterial populations at the study end points [29]. Since the starting bacterial density of both wild-type and RpoB H526D mutant in the competition assays was 106 CFU/ml, which is two orders of magnitude lower than that required for spontaneous generation of RIF-resistant mutants in vitro (108 CFU/ml), any potential contribution of RIF-resistant colonies derived from the wild-type strain to the total rifampicin-resistant colony count is expected to be negligible.
• Gene expression by RT-qPCR
After 11 days of nutrient starvation, bacterial pellets were resuspended in supplemented 7H9 broth and allowed to grow until log phase. Total RNA was purified from nutrient-starved cultures and resuscitated cultures (at OD600 nm = 0.2 and OD600 nm = 0.4), and treated with DNAse (Thermo Fisher Scientific, MA USA) [26]. Following cDNA synthesis with random primers and superscript III reverse transcriptase, RT-qPCR was performed using IQ SYBR Green Supermix (Bio-Rad, CA, USA) and iCycler 5.0 (Bio-Rad) with the primer pairs listed in Supplementary Table 1. The cycle threshold value (CT) measured for each gene was normalized to that of the housekeeping gene, sigA (ΔCT) [26]. For rpoB gene expression, the difference in CT value (ΔCT) under each growth condition was used to calculate the fold change in gene expression between the rpoB mutant and the control strains (2∧-ΔCT) [30]. For rpf gene expression, ΔΔCT was calculated by subtracting the CT value of each gene during resuscitation in 7H9 broth from the corresponding CT value of the gene during nutrient starvation. Relative fold change of each gene in each strain was calculated using the formula: 2∧-ΔΔCT [30]. All samples were prepared as duplicates.
• Animal infections
All animals were maintained under pathogen-free conditions, and fed water and chow ad libitum. All protocols and procedures were approved by the JHU Animal Care and Use Committee. Female BALB/c mice (6–8 weeks; Charles River Labs, MA, USA) were aerosol infected using the Inhalation Exposure System (Glas-Col, IN, USA) and calibrated to deliver approximately 102 bacilli of each strain. On days 1, 28 and 135 postinfection, groups of 4–5 mice were sacrificed to remove the lungs and spleens aseptically. Body, lung and spleen weights were recorded. Lung homogenates were plated on Middlebrook 7H11 selective agar plates (Thermo Fisher Scientific) after serial dilution for CFU counts. The upper lobe of the left lung was processed for histological examination by hematoxylin eosin staining [26,31]. The mouse lung slides were scanned using a Roche Ventana iScan HT (Basel, Switzerland) and viewed with the manufacturer's software (Ventana Image Viewer). For each slide, the number of granulomas was counted, followed by measurement of the total area of alveolated lung affected by granulomas using the image analysis software (Image J). The average percentage of involvement for each group was obtained by using R (Statistical Computing). The mouse aerosol infection study was repeated once with similar findings.
• Statistical analysis
Means and standard deviations were calculated for each data set. Differences between calculated means were compared by the Student's t-test. A p-value ≤0.05 was considered statistically significant.
Results
• Isolation of the Mtb RpoB H526D mutant & generation of the complemented strain
In order to investigate the role of the RpoB H526D mutation alone in Mtb virulence, a well-characterized laboratory Mtb strain CDC1551 [27,28] was used to select individual RIF-resistant colonies on 7H10 agar containing RIF (1 μg/ml). DNA sequencing of the rpoB RIF resistance determining region was performed for each colony isolated (data not shown), and a strain containing a C→G mutation at nucleotide position 1333 of the rpoB gene, causing an amino acid substitution of aspartate (D) for histidine (H) at codon 445 (H526D in the E. coli annotation), was selected for further study. The MIC of RIF against the rpoB mutant was determined to be 8 μg/ml, while that of INH was 0.06 μg/ml, indicating resistance and susceptibility to RIF and INH, respectively. Mutations in rpoA and rpoC, and additional mutations in rpoB were not detected by DNA sequencing (data not shown).
In order to demonstrate that phenotypes observed in the rpoB mutant were attributable to the point mutation in the rpoB gene, we generated a merodiploid-complemented strain by introducing a copy of the wild-type rpoB gene at the mycobacterial attB site using site-specific recombination [26]. Successful complementation was confirmed by Southern blot using DIG-labeled rpoB probe after DNA digestion (Supplementary Figure 1A & B). The MIC of RIF and INH against the wild-type CDC1551 and the complemented strain were 0.25 and 0.06 μg/ml, respectively.
• The Mtb RpoB H526D mutant shows wild-type growth & survival in nutrient-rich & nutrient-starved conditions, but reduced survival during progressive hypoxia
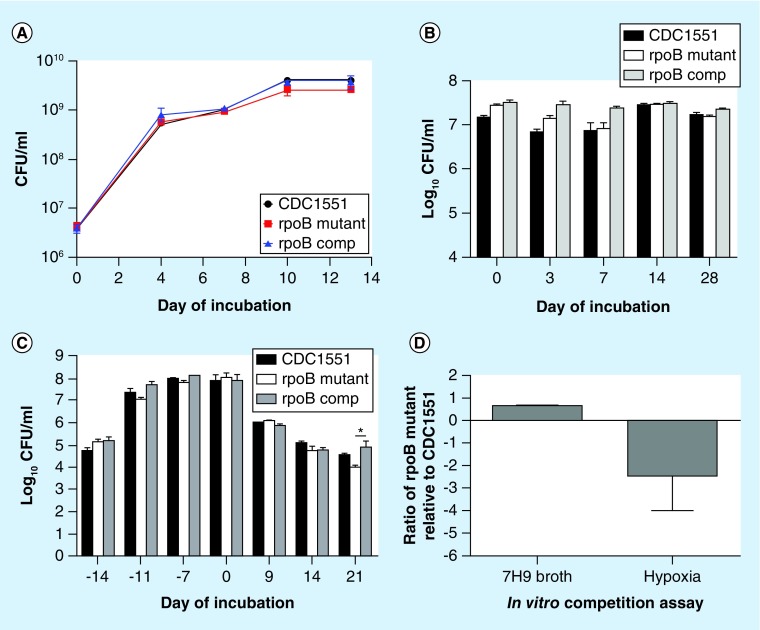
When inoculated into axenic cultures, the RpoB H526D mutant showed no statistically significant difference in growth relative to control strains in nutrient-rich broth and equivalent survival during nutrient starvation (Figure 1A & B). However, after 21 days of exposure to progressive hypoxia [28], the rpoB mutant showed a significant decrease in CFU relative to the wild-type (p > 0.05) and complemented (p < 0.05) strains (Figure 1C). In order to ensure equivalent nutrient availability and oxygen depletion kinetics for each strain, a competitive assay was used to study bacillary survival capacity of the mutant and wild-type in supplemented 7H9 broth and during progressive hypoxia. The methylene blue indicator dye changed color 14 days after inoculation of cultures, indicating reduced oxygen tension and entry of Mtb into nonreplicating persistence stage 2 [28]. A minor fitness cost of the mutant relative to the wild-type was observed in supplemented 7H9 broth (W = 0.7; Figure 1D & Table 1). Despite starting with a higher bacterial density in the hypoxia model, the RpoB H526D mutant showed markedly reduced survival relative to the wild-type after 7 days of nonreplicating persistence stage 2 (relative fitness cost = -2.4; Figure 1D & Table 1).
Figure 1. . In vitro phenotypes associated with Mycobacterium tuberculosis RpoB mutation H526D.
(A) Growth kinetics of each strain in supplemented Middlebrook 7H9 broth. (B) Mycobacterium tuberculosis survival during nutrient starvation. (C) Mycobacterium tuberculosis survival during progressive hypoxia. (D) In vitro competition assay. The rpoB mutant and the wild-type CDC1551 were equally mixed and incubated in supplemented Middlebrook 7H9 broth as well as in the progressive hypoxia model. CFU were counted on 7H10 agar plates with and without 1 μg/ml of Rifampicin on days 0 and 7. The competitive fitness, W of the rpoB mutant relative to the wild-type strain was calculated using the following formula: W = ln (RF/RI)/ln (SF/SI), where RI and SI refer to the number of Rifampicin-resistant and -susceptible bacteria at baseline, respectively, and RF and SF refer to the same bacterial populations at the study end points.
comp: Complemented strain of the RpoB H526D mutant.
Table 1. . In vitro competitive assays reveal fitness cost of the RpoB H526D mutant.
| Strain mixture |
7H9 broth (CFU/ml) |
Progressive hypoxia (CFU/ml) |
||
|---|---|---|---|---|
| Day 0 | Stationary phase (day 10) | Day 0 | 7 days after color change (day 21) | |
| CDC1551 |
1.2 × 106 |
2.5 × 108 |
7 × 104 |
1.3 × 105 |
|
rpoB mutant |
2.2 × 106 |
9.4 × 107 |
1.5 × 105 |
6.5 × 104 |
| Ratio | 0.70 | -2.4 | ||
• Delayed resuscitation of the RpoB H526D mutant from nutrient starvation
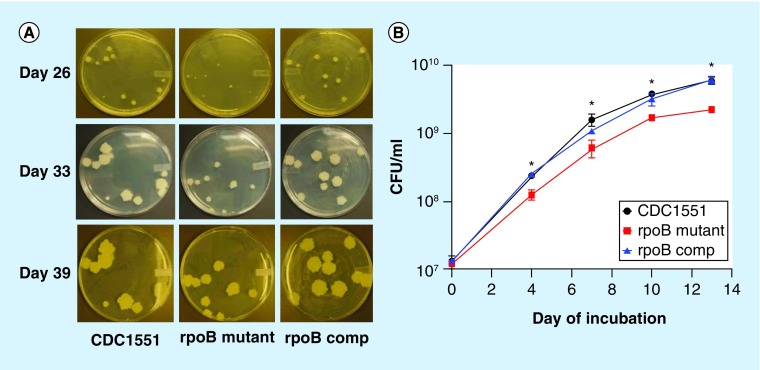
Although the colony size of the RpoB H526D mutant was similar to that of control strains when logarithmically growing cultures in supplemented 7H9 broth were plated, the mutant displayed relatively smaller colonies following plating of cultures from stationary phase or progressive hypoxia (data not shown). Similarly, the RpoB H526D mutant exhibited a small colony phenotype on plates following 28 days of nutrient starvation (Figure 2A). The mutant colonies continued to increase in diameter, such that by 39 days of incubation on solid, nutrient-rich agar, mutant colony size was equivalent to that of control strains. In order to investigate the possibility of a resuscitation defect for the RpoB H526D mutant, nutrient-starved cultures of each strain were pelleted and resuspended in fresh supplemented 7H9 broth following 9, 14 and 28 days of nutrient starvation. Relative to the control strains, the 9-day nutrient-starved mutant showed an approximately 3-day lag in attaining an OD600 nm value of 3.0, and failed to achieve the same growth peak (Supplementary Figure 2). CFU analysis confirmed these results, revealing a significantly reduced density of the mutant relative to the control strains at each time point (p < 0.05; Figure 2B). Similar results were observed during resuscitation of 14- and 28-day nutrient-starved cultures (data not shown).
Figure 2. . Delayed resuscitation phenotype in the Mycobacterium tuberculosis RpoB H526D mutant.
(A) Colony size of 28-day nutrient-starved cultures on Middlebrook 7H10 agar plates after plating for 26, 33 and 39 days. (B) Growth rates of 9-day nutrient-starved cultures in supplemented Middlebrook 7H9 broth, as measured by CFU on Middlebrook 7H10 agar plates.
*p < 0.05.
• RpoB mutation H526D is associated with reduced expression of rpfB, rpfC & rpfE genes during resuscitation from growth-limiting conditions
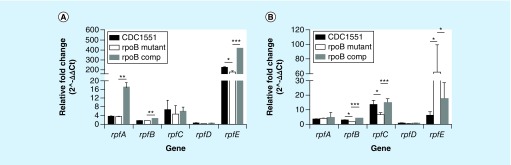
Five mycobacterial resuscitation-promoting factors (Rpfs) have been implicated in the resuscitation of dormant Mtb [32]. We tested the expression level of each of the five rpf genes (rpfA, rpfB, rpfC, rpfD and rpfE) in the rpoB mutant and control strains upon resuscitation from 11 days of nutrient starvation. All five genes were upregulated in resuscitated cultures of each strain relative to nutrient-starved cultures of the same strains (Figure 3A & B). rpfE was dramatically upregulated in the mutant during early resuscitation (OD = 0.2) relative to nutrient starvation, but the level of induction was 50-fold lower than that in the resuscitated wild-type versus the nutrient-starved wild-type (p < 0.05; Figure 3A). On the other hand, rpfE expression was 56-fold more upregulated in the mutant than in the wild-type when comparing gene expression levels of each strain during late resuscitation (OD = 0.4) relative to nutrient starvation (p = 0.01; Figure 3B). Expression of rpfA and rpfD remained relatively stable, with no obvious differences between the mutant and the wild-type during resuscitation (Figure 3A and B). Although rpfB and rpfC were upregulated similarly during initial resuscitation, the level of induction of rpfB and rpfC was 1.5-fold and 6.7-fold, respectively, less than the induction of the same genes in the wild-type during resuscitation relative to nutrient starvation (p < 0.05 and p = 0.002, respectively; Figure 3B). Relative expressions of the rpf genes during resuscitation were restored in the complemented strain (Figure 3).
Figure 3. . Expression of the Mycobacterium tuberculosis rpf genes rpfA–E upon resuscitation from nutrient starvation, as measured by RT-qPCR.
(A) Expression of rpf genes of Mtb strains grown to OD600 nm= 0.2 after resuscitation in 7H9 broth relative to that of corresponding NS cultures; (B) Expression of rpf genes of Mtb strains grown to OD600nm = 0.4 after resuscitation in 7H9 broth relative to that of corresponding NS culture.
RNA was purified from Mtb strains following 11 days of nutrient starvation and after resuscitation in supplemented 7H9 broth at two time points (OD600 nm = 0.2 and OD600 nm = 0.4), and RT-qPCR was performed after cDNA synthesis. After normalization to sigA, ΔΔCT was calculated using the difference in CT value (ΔCT) of each rpf gene upon resuscitation relative to that during nutrient starvation. The fold change in rpf gene expression in the rpoB mutant and the control strains was calculated by 2∧-ΔΔCT.
*p < 0.05; **p < 0.05; ***p < 0.001. CT: Cycle threshold; OD: Optical density.
• The RpoB H526D mutation does not alter rpoB gene expression during nutrient starvation or upon resuscitation
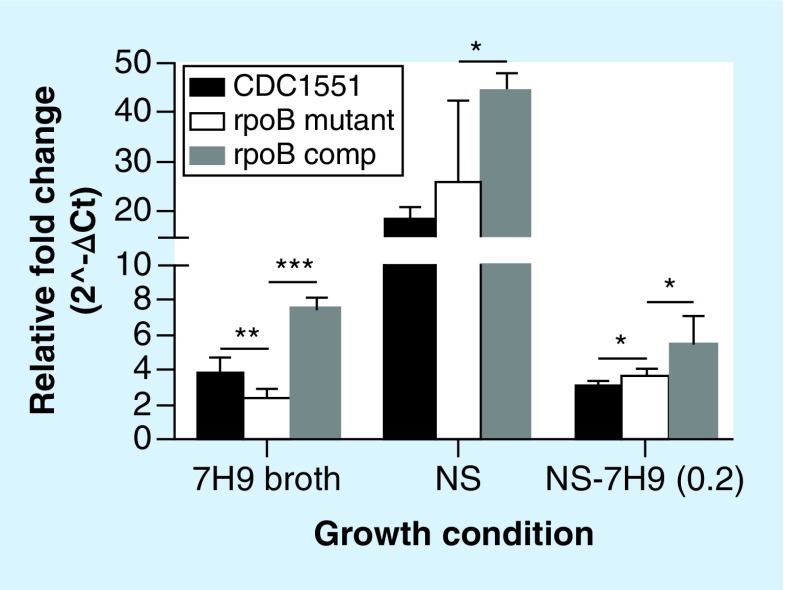
In order to determine the effect of the RpoB H526D mutation on rpoB gene expression, we used RT-qPCR to evaluate rpoB expression in the mutant under different growth conditions. During nutrient starvation and upon resuscitation, the expression of rpoB in the rpoB mutant was equal to or more than that in the wild-type (p < 0.05; Figure 4). Interestingly, rpoB was at least 4.5-fold more highly expressed in the mutant and wild-type strains during nutrient starvation relative to nutrient-rich conditions (p < 0.0001; Figure 4). However, rpoB expression was lower in the rpoB mutant than in the wild-type during exponential growth in nutrient-rich broth (p < 0.01; Figure 4). Relative overexpression of the rpoB gene was seen in the complemented strain relative to the wild-type under all the conditions tested (p < 0.01 or p = 0.0001; Figure 4), perhaps due to the presence of two copies of the gene in this strain.
Figure 4. . Expression of the Mycobacterium tuberculosis rpoB gene during exponential growth in nutrient-rich broth, during nutrient starvation and upon resuscitation, as measured by RT-qPCR.
The CT of rpoB gene was normalized to that of sigA under each growth condition to calculate ΔCT followed by conversion of relative fold change (2∧-ΔCT) in the rpoB mutant and control strains.
*p < 0.05; **p < 0.01; ***p < 0.001.
comp: Complemented strain of the RpoB H526D mutant; CT: Cycle threshold; mutant: RpoB H526D mutant; NS: Nutrient-starved; NS-7H9 (0.2): Resuscitated NS cultures in 7H9 broth at OD600nm = 0.2.
• The RpoB H526D mutation is associated with reduced Mtb survival in mice
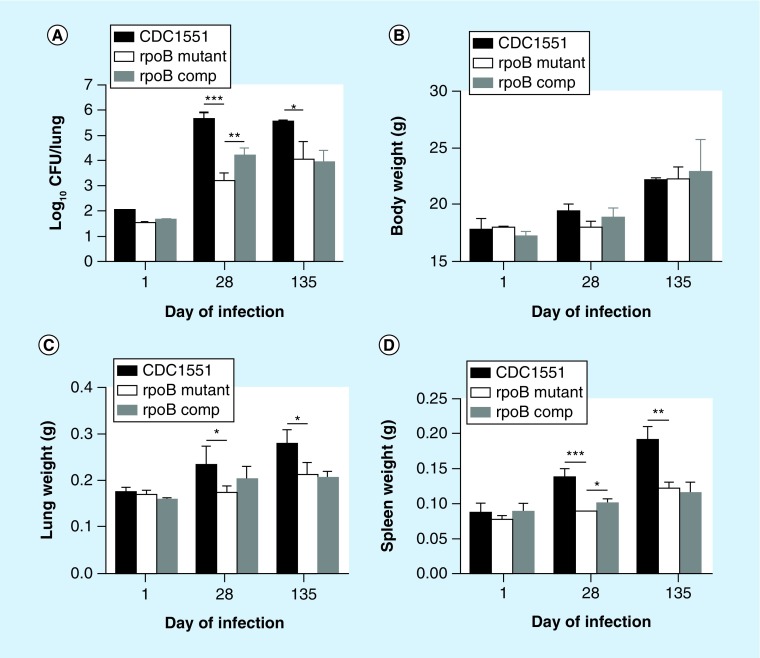
In order to investigate the effect of the RpoB H526D mutation on Mtb virulence in vivo, we aerosol-infected separate groups of BALB/c mice with equal inocula of each strain. By 28 days postinfection, the bacterial count in the lungs of mice infected with the RpoB H526D mutant was 2.4 log10 lower than that in mice infected with the wild-type (p < 0.0001; Figure 5A) and 0.9 log10 lower than that in mice infected with the complemented strain (p < 0.006). Although the mean lung bacillary burden of mice infected with the control strains remained relatively stable until day 135 following aerosol infection, that of mutant-infected mice increased nonsignificantly relative to the corresponding value on day 28 (p = 0.14). Bacillary counts in the lungs of mutant-infected mice remained significantly lower than those of mice infected with the wild-type strain after 135 days of infection (p = 0.02), but were not significantly different from those of the complemented strain (p = 0.7). When assessed 28 days after incubation on solid agar, the colony size of each strain harvested from mouse lungs was equivalent on the day after aerosol infection, but the RpoB H526D mutant colonies were much smaller in diameter than those of the wild-type from lungs harvested at day 135 postinfection (data not shown). The wild-type colony size was not fully restored in the complemented strain. No mutations were detected in the rpoA and rpoC genes or in the rest of the rpoB gene by DNA sequencing of the five mutant colonies randomly selected from agar plated with day 135 lung homogenates (data not shown).
Figure 5. . Mutation RpoB H526D impairs Mycobacterium tuberculosis growth in mouse lungs.
BALB/c mice were aerosol infected with equal inocula of the RpoB H526D mutant, wild-type or complemented strains. (A) Log 10 CFU/lung (*p = 0.02; **p = 0.006; ***p < 0.0001); (B) Bodyweights of Mycobacterium tuberculosis (Mtb)-infected mice. (C) Lung weights of Mtb-infected mice (*p = 0.05). (D) Spleen weights of Mtb-infected mice (*p = 0.03; **p = 0.02; ***p = 0.0002).
comp: Complemented strain of the RpoB H526D mutant.
• The RpoB H526D mutation is associated with reduced Mtb-induced inflammation in mice
Mice in all groups gained weight equivalently throughout the study (Figure 5B). On days 28 and 135 post-infection, the mean lung weights of mice infected with the wild-type were greater than those of the mutant (p = 0.05; Figure 5C). Similarly, on days 28 and 135, the mean spleen weights of mice infected with the wild-type were greater than those of the mutant (p = 0.002 and p = 0.02, respectively; Figure 5D). Wild-type mean organ weights were partially restored in the complemented strain on day 28 (p = 0.03 for spleen weight, Figure 5D), but not on day 135 postinfection (Figure 5C & D).
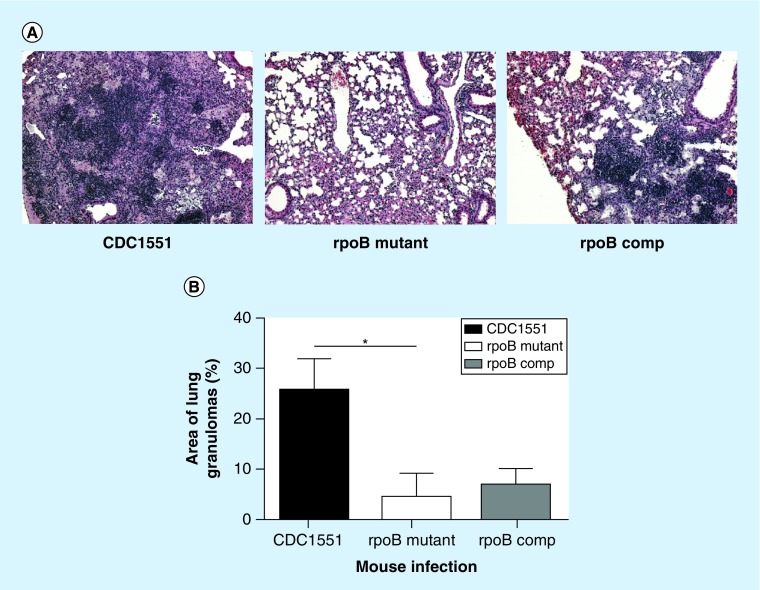
Histopathological examination of mouse lung samples was performed on days 28 (data not shown) and 135 postinfection. On day 135, the lungs of wild-type-infected mice displayed non-necrotizing granulomas consisting of histiocytes, large foamy macrophages and lymphocytic inflammation. These granulomas were primarily associated with small airways and were confluent with areas of lymphocytic bronchiolitis (Figure 6A). Two of the four mutant-infected lungs showed small, scattered granulomas (Figure 6A). Conversely, all of the lungs infected with the complemented strain contained multiple granulomas and confluent bronchiolitis (Figure 6A). By morphometric analysis, the percent surface area of lungs occupied by granulomas was 0.25, 0.04 (p = 0.01 relative to wild-type) and 0.07 (p = 0.3, relative to the mutant) for the wild-type, mutant and complemented strains, respectively (Figure 6B). The murine model study was repeated, yielding similar findings in CFU, total body and organ weights, and histopathology.
Figure 6. . Reduced mouse lung inflammation associated with mutation RpoB H526D.
(A) Histology of mouse lungs on day 135 postinfection; hematoxylin/eosin staining, 200 × magnification. (B) Histological analysis of lung surface area (%) involved by granulomas (*p = 0.01). The mouse lung slides were scanned using a Roche Ventana iScan HT and viewed with the manufacturer's software (Ventana Image Viewer). For each slide, the number of granulomas was counted, followed by measurement of the total area of alveolated lung affected by granulomas using the image analysis software (Image J). The average percentage of involvement for each group was obtained by using R (Statistical Computing). *p < 0.05.
comp: Complemented strain of the RpoB H526D mutant.
Discussion
Mtb fitness during the natural course of infection is a function of various virulence properties, including the abilities to infect a susceptible host following aerosol exposure, proliferate and persist in the lungs and other tissues, develop pulmonary cavities and transmit to a secondary host. These characteristics are difficult to study in humans directly and are often overshadowed by host susceptibility and/or environmental factors. Previous studies characterized the Mtb fitness costs associated with multiple drug resistance mutations under various in vitro stress conditions [10] and in the murine model [13]. However, because a drug-susceptible control strain with identical genetic background was not available in these studies, and because of the presence of multiple resistance mutations, it is not possible to ascribe specific fitness costs to individual resistance mutations. In the current study, we focused on the Mtb fitness costs associated with RIF monoresistance. To our knowledge, our study is the first to show that the commonly observed RpoB mutation H526D is associated with reduced Mtb virulence in mice and delayed resuscitation from nutrient starvation.
Mtb encounters various stress conditions during host infection, including hypoxia, carbon starvation, and exposure to reactive oxygen and nitrogen species [12]. The RpoB H526D mutant in the current study had a statistically insignificant fitness cost in nutrient-rich broth, which was less marked than that observed for Mtb strains containing point mutations in the same codon in earlier studies [29,33]. On the other hand, our in vitro competition assays revealed significantly reduced survival of the RpoB H526D mutant during progressive hypoxia. The RpoB S531L mutation has been reported to confer an Mtb fitness cost during carbon limitation and starvation for nutrients, including biotin and iron, and the compensatory change V483G in RpoC restores wild-type growth under these stringent conditions [10]. Conversely, we found that the Mtb RpoB H526D mutant showed similar survival to control strains during nutrient starvation. This discrepancy may be explained by the differences in amino acid substitutions in RpoB, which may differentially alter the transcriptional function of RpoB and resulting Mtb phenotypes [10,29] as well as the differences in experimental conditions between the two studies. We observed equivalent rpoB induction in the RpoB H526D mutant and control strains during nutrient starvation relative to nutrient-rich conditions, suggesting a functional transcription apparatus in the nutrient-starved mutant.
Despite recent advances in our understanding of Mtb resuscitation in the context of TB clinical outcomes [14,21,34], the role of RIF-resistant mutations on Mtb resuscitation has not been studied previously. For the first time, we revealed that RpoB mutation H526D was associated with defective Mtb resuscitation from growth-limiting conditions, leading to a small colony phenotype, which was also observed for an RpoB S531L mutant and was partially and completely reversed by RpoC mutations F452L and V483G, respectively [10]. The Rpfs, RpfA–E have been implicated in Mtb resuscitation [17–20] through hydrolysis of the mycobacterial peptidoglycan with partnering proteins, which is crucial for cell elongation and cell division [35,36]. Consistent with previous work [32], we observed upregulation of all five rpf genes to varying degrees upon resuscitation of both the wild-type and the mutant. Our gene expression data revealed that the defective resuscitation of the nutrient-starved mutant may be due to lower induction of rpfB, rpfC and rpfE relative to the wild-type upon repletion of nutrients. Although individual rpf genes are dispensable for Mtb resuscitation, complementation of an Mtb mutant lacking all five genes with either rpfB or rpfE is sufficient to reverse delayed colony formation upon removal from stress conditions [37]. An Mtb mutant lacking the rpfB gene was found to have defective resuscitation in mice [38] and rpfE was shown to be involved in the switch from slow to rapid mycobacterial growth in a chemostat [39]. The hierarchy of RpfB and RpfE in Mtb resuscitation may potentially be explained by the observation that the partnering peptidoglycan hydrolase, Rpf interacting protein A, appears to interact with these two Rpfs but not with RpfA, RpfC or RpfD for cell division [35,40]. It is possible that the mutation RpoB H526D alters the expression of rpfB and rpfE as well as that of other genes involved in Mtb resuscitation, leading to delayed Mtb growth upon repletion of nutrients. Interestingly, during late-stage resuscitation, rpfE expression was induced to a greater extent in the mutant than in the control strains, potentially compensating for deficiency of rpfB in the mutant. Our finding paves the way for future mechanistic studies to test the role of Rpfs in the pathogenesis of RIF-resistant TB. For example, rpf genes of interest could be conditionally overexpressed in the RpoB H526D mutant or recombinant Rpf proteins could be added to the media to determine whether resuscitation of the rpoB mutant from stress conditions can be accelerated.
The markedly reduced growth phenotype observed in vivo may be associated with reduced level of rpoB expression as well as deficiency in RpoB function due to the mutation H526D. Interestingly, we found decreased Mtb-induced lung inflammation in mice infected with the Mtb RpoB H526D mutant, which may be explained at least partially by the reduced bacterial burden in these animals and/or by altered expression of key antigens in the setting of rpoB mutation [41]. In support of the latter hypothesis, a recent report also found broad remodeling of cell wall lipids, including altered levels of mycobactin and carboxymycobactin siderophores and acylated sulfoglycolipids, in an RpoB H526Y mutant and other RIF-resistant Mtb strains [33]. Mycobactin-mediated iron acquisition is required for Mtb growth in macrophages [42] and sulfoglycolipids are associated with Mtb virulence in the guinea pig model of infection [43]. Consistent with our findings, reduced bacillary growth and lung inflammation were observed in mice infected with MDR and extensively drug-resistant (XDR) Mtb strains relative to drug-susceptible Mtb [13].
Despite the fitness costs associated with mutation RpoB H526D, our data and clinical studies show that strains containing this mutation can establish chronic infection in mice and frequently cause disease in humans. Although we did not detect the well-described compensatory mutations in rpoA and rpoC, it is possible that our mutant strain harbors compensatory mutations in other genes, which could be identified by whole-genome sequencing, potentially contributing to persistence in vivo. It remains to be determined whether this RpoB mutation H526D confers reduced virulence in humans, and to what extent its presence contributes to bacterial clearance during the prolonged treatment with the more costly and toxic, but less effective, second-line drugs required to cure MDR TB [13,44]. Interestingly, the presence of certain gyrA mutations conferring fluoroquinolone resistance has been postulated to compensate for rpoB mutations [45], perhaps due to the link between transcription rate and DNA supercoiling [46], suggesting a plausible mechanism favoring the transformation of MDR TB into XDR TB, defined as MDR TB with additional resistance to the fluoroquinolones and at least one injectable agent, or pre-XDR TB. Another measure of Mtb virulence is transmissibility. In our study, the RpoB H526D mutant was implanted into the mouse lungs at the expected inoculum dose, although very little is known about any effects of this mutation on strain transmissibility in humans.
Recent studies suggest that HIV coinfection is not associated with the evolution of Mtb drug resistance and transmission of MDR-TB, but HIV patients are more susceptible to acquiring Mtb infection and progression to active TB [47]. However, it is not known whether HIV-related cellular and other immune defects negate or mitigate against any fitness costs associated with Mtb containing RpoB mutation H526D or other RpoB mutations. Although reversibility of resistance mutations to wild-type alleles is well described in bacteria [48], it is not known to what extent this occurs in RIF-resistant Mtb strains during natural infection.
Wild-type virulence vis-à-vis lung bacterial burden and pathology was only partially restored by the complemented strain. Incomplete complementation may be due to the fully functional copy of the rpoB gene acting in trans with the mutant gene encoding the (presumably) partially functional RpoB H526D mutant protein. Our RT-qPCR data revealed higher expression of rpoB in the complemented strain relative to the mutant and wild-type strains during exponential growth in nutrient-rich broth as well as during nutrient starvation, which may have contributed to the distinct phenotypes between the merodiploid-complemented strain and the wild-type strain in vivo. In addition, we cannot exclude the possible contribution of a secondary mutation, which could be identified by whole-genome sequencing, on the attenuated phenotype of the mutant in vivo. An alternative approach, which we will adopt in the future, is to use allelic replacement to generate the point mutation in the rpoB gene associated with the amino acid change at codon H526, in the absence of RIF selection to minimize the likelihood of secondary mutations.
Conclusion & future perspective
We found that the RpoB H526D mutation is associated with reduced Mtb fitness in vitro and in the standard murine model of chronic TB. Delayed resuscitation of the RpoB H526D mutant from growth-limiting conditions was associated with reduced early expression of rpfB, rpfC and rpfE. Our findings may motivate further studies on the contribution of individual drug resistance conferring mutations in the pathogenesis of MDR and XDR TB, leading to improved therapeutic strategies. The precise role of Rpfs and other related Mtb factors in the reactivation of latent infection with drug-susceptible or drug-resistant Mtb strains deserves further investigation. Future studies are also needed to characterize the virulence of drug-resistant Mtb strains containing mutations in rpoB and other genes in the immune-deficient host.
SUMMARY POINTS.
The fitness cost of the RpoB H526D mutation on Mycobacterium tuberculosis (Mtb) growth and survival is condition-dependent. The mutant showed reduced survival relative to control strains during progressive hypoxia, but not during nutrient starvation.
Relative to wild-type, the Mtb RpoB H526D mutant showed a delay in resuscitation from nutrient starvation, associated with reduced expression of the resuscitation-promoting factor genes rpfB, rpfC and rpfE.
Relative to the isogenic wild-type strain, the Mtb RpoB H526D mutant showed significantly attenuated growth and long-term survival as well as reduced inflammation, in mouse lungs.
Supplementary Material
Footnotes
Author contributions
PC Karakousis, D Rifat and Y Yu are responsible for the research concept and design. D Rifat and PC Karakousis wrote and revised the article. D Rifat, VL Campodónico, J Tao and A Alp conducted the experiments, data analysis and interpretation. JA Miller performed examination of histopathology of infected mouse lungs.
Financial & competing interests disclosure
This work was supported by the State Key Development Programs for Basic Research of China (973 program no. 2015CB554203) and the National Natural Science Foundation of China (no. 81361120383) to Y Yao and NIH grant R01AI106613 to PC Karakousis. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
- 1.Global tuberculosis report 2015. WHO, Geneva, Switzerland. 2015. www.who.int/tb/publications/global_report/en/
- 2.A controlled trial of six months chemotherapy in pulmonary tuberculosis. Second report: results during the 24 months after the end of chemotherapy. British Thoracic Association. Am. Rev. Respir. Dis. 1982;126(3):460–462. doi: 10.1164/arrd.1982.126.3.460. [DOI] [PubMed] [Google Scholar]
- 3.Campbell EA, Korzheva N, Mustaev A, et al. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104(6):901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 4.Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 1998;79(1):3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 5.Heep M, Brandstatter B, Rieger U, et al. Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 2001;39(1):107–110. doi: 10.1128/JCM.39.1.107-110.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Musser JM. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 1995;8(4):496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandis G, Hughes D. Genetic characterization of compensatory evolution in strains carrying rpoB Ser531Leu, the rifampicin resistance mutation most frequently found in clinical isolates. J. Antimicrob. Chemother. 2013;68(11):2493–2497. doi: 10.1093/jac/dkt224. [DOI] [PubMed] [Google Scholar]
- 8.Qian L, Abe C, Lin TP, et al. rpoB genotypes of Mycobacterium tuberculosis Beijing family isolates from East Asian countries. J. Clin. Microbiol. 2002;40(3):1091–1094. doi: 10.1128/JCM.40.3.1091-1094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mariam DH, Mengistu Y, Hoffner SE, Andersson DI. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis . Antimicrob. Agents Chemother. 2004;48(4):1289–1294. doi: 10.1128/AAC.48.4.1289-1294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song T, Park Y, Shamputa IC, et al. Fitness costs of rifampicin resistance in Mycobacterium tuberculosis are amplified under conditions of nutrient starvation and compensated by mutation in the beta’ subunit of RNA polymerase. Mol. Microbiol. 2014;91(6):1106–1119. doi: 10.1111/mmi.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandis G, Wrande M, Liljas L, Hughes D. Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Mol. Microbiol. 2012;85(1):142–151. doi: 10.1111/j.1365-2958.2012.08099.x. [DOI] [PubMed] [Google Scholar]
- 12.Fenton MJ, Vermeulen MW. Immunopathology of tuberculosis: roles of macrophages and monocytes. Infect. Immun. 1996;64(3):683–690. doi: 10.1128/iai.64.3.683-690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith KL, Saini D, Bardarov S, et al. Reduced virulence of an extensively drug-resistant outbreak strain of Mycobacterium tuberculosis in a murine model. PLoS ONE. 2014;9(4):e94953. doi: 10.1371/journal.pone.0094953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dooley KE, Lahlou O, Ghali I, et al. Risk factors for tuberculosis treatment failure, default, or relapse and outcomes of retreatment in Morocco. BMC Public Health. 2011;11:140. doi: 10.1186/1471-2458-11-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutta NK, Karakousis PC. Latent tuberculosis infection: myths, models, and molecular mechanisms. Microbiol. Mol. Biol. Rev. 2014;78(3):343–371. doi: 10.1128/MMBR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coscolla M, Gagneux S, Does M. Does M. tuberculosis genomic diversity explain disease diversity? Drug. Discov. Today Dis. Mech. 2010;7(1):e43–e59. doi: 10.1016/j.ddmec.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaprelyants AS, Mukamolova GV, Ruggiero A, et al. Resuscitation-promoting factors (Rpf): in search of inhibitors. Protein Pept. Lett. 2012;19(10):1026–1034. doi: 10.2174/092986612802762723. [DOI] [PubMed] [Google Scholar]
- 18.Mukamolova GV, Kaprelyants AS, Young DI, Young M, Kell DB. A bacterial cytokine. Proc. Natl Acad. Sci. USA. 1998;95(15):8916–8921. doi: 10.1073/pnas.95.15.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukamolova GV, Turapov OA, Kazarian K, et al. The rpf gene of Micrococcus luteus encodes an essential secreted growth factor. Mol. Microbiol. 2002;46(3):611–621. doi: 10.1046/j.1365-2958.2002.03183.x. [DOI] [PubMed] [Google Scholar]
- 20.Kana BD, Mizrahi V. Resuscitation-promoting factors as lytic enzymes for bacterial growth and signaling. FEMS Immunol. Med. Microbiol. 2010;58(1):39–50. doi: 10.1111/j.1574-695X.2009.00606.x. [DOI] [PubMed] [Google Scholar]
- 21.Loraine J, Pu F, Turapov O, Mukamolova GV. Development of an in vitro assay for detection of drug-induced resuscitation-promoting-factor-dependent mycobacteria. Antimicrob. Agents Chemother. 2016;60(10):6227–6233. doi: 10.1128/AAC.00518-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Therese KL, Gayathri R, Dhanurekha L, et al. Detection of Mycobacterium tuberculosis directly from sputum specimens & phenotypic drug resistance pattern of M. tuberculosis isolates from suspected tuberculosis patients in Chennai. Indian J. Med. Res. 2012;135(5):778–782. [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmad Z, Nuermberger EL, Tasneen R, et al. Comparison of the ‘Denver regimen’ against acute tuberculosis in the mouse and guinea pig. J. Antimicrob. Chemother. 2010;65(4):729–734. doi: 10.1093/jac/dkq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chuang YM, Belchis DA, Karakousis PC. The polyphosphate kinase gene ppk2 is required for Mycobacterium tuberculosis inorganic polyphosphate regulation and virulence. MBio. 2013;4(3):e00039–e00013. doi: 10.1128/mBio.00039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klinkenberg LG, Lee JH, Bishai WR, Karakousis PC. The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. J. Infect. Dis. 2010;202(9):1397–1404. doi: 10.1086/656524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rifat D, Belchis DA, Karakousis PC. senX3-independent contribution of regX3 to Mycobacterium tuberculosis virulence. BMC Microbiol. 2014;14:265. doi: 10.1186/s12866-014-0265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karakousis PC, Williams EP, Bishai WR. Altered expression of isoniazid-regulated genes in drug-treated dormant Mycobacterium tuberculosis . J. Antimicrob. Chemother. 2008;61(2):323–331. doi: 10.1093/jac/dkm485. [DOI] [PubMed] [Google Scholar]
- 28.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 1996;64(6):2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJ. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis . Science. 2006;312(5782):1944–1946. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]
- 30.Ren J, Sang Y, Tan Y, et al. Acetylation of lysine 201 inhibits the DNA-binding ability of PhoP to regulate Salmonella virulence. PLoS Pathog. 2016;12(3):e1005458. doi: 10.1371/journal.ppat.1005458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klinkenberg LG, Sutherland LA, Bishai WR, Karakousis PC. Metronidazole lacks activity against Mycobacterium tuberculosis in an in vivo hypoxic granuloma model of latency. J. Infect. Dis. 2008;198(2):275–283. doi: 10.1086/589515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta RK, Srivastava BS, Srivastava R. Comparative expression analysis of rpf-like genes of Mycobacterium tuberculosis H37Rv under different physiological stress and growth conditions. Microbiology. 2010;156(Pt 9):2714–2722. doi: 10.1099/mic.0.037622-0. [DOI] [PubMed] [Google Scholar]
- 33.Lahiri N, Shah RR, Layre E, et al. Rifampin resistance mutations are associated with broad chemical remodeling of Mycobacterium tuberculosis . J. Biol. Chem. 2016;291(27):14248–14256. doi: 10.1074/jbc.M116.716704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panjabi R, Comstock GW, Golub JE. Recurrent tuberculosis and its risk factors: adequately treated patients are still at high risk. Int. J. Tuberc. Lung Dis. 2007;11(8):828–837. [PubMed] [Google Scholar]
- 35.Hett EC, Chao MC, Steyn AJ, Fortune SM, Deng LL, Rubin EJ. A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis . Mol. Microbiol. 2007;66(3):658–668. doi: 10.1111/j.1365-2958.2007.05945.x. [DOI] [PubMed] [Google Scholar]
- 36.Hett EC, Chao MC, Deng LL, Rubin EJ. A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLoS Pathog. 2008;4(2):e1000001. doi: 10.1371/journal.ppat.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kana BD, Gordhan BG, Downing KJ, et al. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro . Mol. Microbiol. 2008;67(3):672–684. doi: 10.1111/j.1365-2958.2007.06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tufariello JM, Mi K, Xu J, et al. Deletion of the Mycobacterium tuberculosis resuscitation-promoting factor Rv1009 gene results in delayed reactivation from chronic tuberculosis. Infect. Immun. 2006;74(5):2985–2995. doi: 10.1128/IAI.74.5.2985-2995.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beste DJ, Espasa M, Bonde B, Kierzek AM, Stewart GR, Mcfadden J. The genetic requirements for fast and slow growth in mycobacteria. PLoS ONE. 2009;4(4):e5349. doi: 10.1371/journal.pone.0005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruggiero A, Tizzano B, Pedone E, Pedone C, Wilmanns M, Berisio R. Crystal structure of the resuscitation-promoting factor (DeltaDUF)RpfB from M. tuberculosis . J. Mol. Biol. 2009;385(1):153–162. doi: 10.1016/j.jmb.2008.10.042. [DOI] [PubMed] [Google Scholar]
- 41.Bisson GP, Mehaffy C, Broeckling C, et al. Upregulation of the phthiocerol dimycocerosate biosynthetic pathway by rifampin-resistant, rpoB mutant Mycobacterium tuberculosis . J. Bacteriol. 2012;194(23):6441–6452. doi: 10.1128/JB.01013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siegrist MS, Unnikrishnan M, McConnell MJ, et al. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc. Natl Acad. Sci. USA. 2009;106(44):18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goren MB, Brokl O, Schaefer WB. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: correlation of virulence with elaboration of sulfatides and strongly acidic lipids. Infect. Immun. 1974;9(1):142–149. doi: 10.1128/iai.9.1.142-149.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinto L, Menzies D. Treatment of drug-resistant tuberculosis. Infect. Drug Resist. 2011;4:129–135. doi: 10.2147/IDR.S10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borrell S, Teo Y, Giardina F, et al. Epistasis between antibiotic resistance mutations drives the evolution of extensively drug-resistant tuberculosis. Evol. Med. Public Health. 2013;1:65–74. doi: 10.1093/emph/eot003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rovinskiy N, Agbleke AA, Chesnokova O, Pang Z, Higgins NP. Rates of gyrase supercoiling and transcription elongation control supercoil density in a bacterial chromosome. PLoS Genet. 2012;8(8):e1002845. doi: 10.1371/journal.pgen.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eldholm V, Rieux A, Monteserin J, et al. Impact of HIV co-infection on the evolution and transmission of multidrug-resistant tuberculosis. eLife. 2016;5:e16644. doi: 10.7554/eLife.16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 2010;8(4):260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.