Abstract
Hormone receptor positive (HR+) breast cancers are a heterogeneous class with differential prognosis. Although more than half of Indian women present with advanced disease, many such patients do well. We have attempted identification of biologically indolent tumors within HR+HER2- tumors based on gene expression using histological grade as a guide to tumor aggression. 144 HR+HER2- tumors were divided into subclasses based on scores derived by using transcript levels of multiple genes representing survival, proliferation, and apoptotic pathways and compared to classification by Ki-67 labeling index (LI). Clinical characters and disease free survival were compared between the subclasses. The findings were independently validated in the METABRIC data set. Using the previously established estrogen receptor (ER) down stream activity equation, 20% of the tumors with greater than 10% HR positivity by immunohistochemistry (IHC) were still found to have inadequate ER function. A tumor aggression probability score was used to segregate the remainder of tumors into indolent (22%) and aggressive (58%) classes. Significant difference in disease specific survival was seen between the groups (P = .02). Aggression probability based subclassification had a higher hazard ratio and also independent prognostic value (P < .05). Independent validation of the gene panel in the METABRIC data set showed all 3 classes; indolent (24%), aggressive (68%), and insufficient ER signaling (7%) with differential survival (P = .01). In agreement with other recent reports, biologically indolent tumors can be identified with small sets of gene panels and these tumors exist in a population with predominantly late stage disease.
Introduction
Breast cancer patients with hormone receptor positive (HR+) HER2- tumors tend to have the best outcomes [1]. However, there are a significant proportion of women with HR+HER2− breast cancer who have bad outcomes despite having received “standard of care” treatments [2], [3], and a small proportion of late stage tumors do well despite detection in late stages [4], [5].
Gene expression based analysis of breast cancer tumors has established the presence of two sub classes amongst HR+HER2- tumors termed luminal A and luminal B, largely based on differential levels of expression of proliferation genes [6], [7]. In an attempt to spare patients with luminal A type of tumors unnecessary chemotherapy, gene expression based tests that can tell these tumors apart from luminal B tumors have been developed. Oncotype DX, a 21-gene based test; MammaPrint, a 70-gene microarray signature; and PAM-50, a 50-gene based assay, all help identify early HER2 negative ER positive breast cancer with 3 or fewer positive nodes with a higher risk of recurrence [8], [9], [10], [11]. These tests have provided support for the use of genomic assays in making therapeutic decisions but are limited largely to early stage lymph node negative tumors.
Indian women seek medical attention at later stages with locally advanced disease compared to the women in advanced Western economies [12], [13], [14]. Hence, a majority of the tumors have already spread to the regional lymph nodes (LNs). LN status has been the most important determinant amongst all other clinical variables for overall survival in breast cancer. The current clinical approach is to offer chemotherapy to all patients with 4 or more positive nodes at first presentation. However, there are both anecdotal as well as outcome data that indicate that even amongst women with nodal spread, a small but significant proportion do not have adverse outcomes despite being spared chemotherapy [5]. The biological behavior of these tumors seems to be the inverse of HR negative tumors that spread metastatically despite being detected and treated when they are still under 2 cm and confined to the breast tissue [15].
Although the 4 immunohistochemistry (IHC) based panel that includes ER, PR, HER2 and Ki-67 is now part of the routine testing for women with breast cancer in the established tertiary care centers in the country, establishing the predictive or even prognostic utility of Ki-67 for subclassification of luminal tumors and deriving a specific cut off have proven to be an intractable problem [16], [17], [18].
Features of tumor aggression which have been conventionally measured in the form of histological grade (HG) are used as a guide to select for markers that best identify aggressive tumors. Using the lead of others [19], [20], we have attempted to dissect the biological heterogeneity mediated by pathways of differentiation, proliferation and survival in distinguishing subclasses with differential prognosis within HR + HER2- tumors using histological grade as the guide. HG in breast cancer is an integrative measure of cellular differentiation and replicative potential which stratifies tumors based on the degree of aggressiveness. Multivariate analysis of large cohorts of breast cancer patients has shown the prognostic ability of HG as a single marker [21]. Although HG suffers from significant interobserver variability, the outcomes in the extreme classes; grade 1 and grade 3, are significantly different when compared to the heterogenous grade 2 category which forms 50% of all the tumors [22]. In common with other studies [20], we have tried to overcome this drawback of HG by evaluating the gene expression (GE) levels of multiple markers and integrating their values to derive a probability equation for evaluation of the level of “aggression” in HR + HER2- tumors. When used in conjunction with our previously published method [23] to quantitate the ER downstream function, we are able to definitively identify the biologically indolent HR + HER2- tumors in a case series where two thirds of patients are LN positive.
Materials and Methods
Patients and samples for this study were obtained from our ongoing observational prospective cohort study of breast cancer patients from two tertiary care hospitals in Bangalore, India. The study was approved by the ethical review boards of the hospitals involved. Informed consent for use of the specimens for research was obtained from all patients. Our observational cohort of 446 patients was recruited at diagnosis over a 5 year period, between June 2008 and February 2013, and are being actively followed up through a dedicated breast cancer support group, “Aadhara”. This cohort has completed median of 65 months with 97% follow up as of 30 June 2016. Tumors from 140 patients (including 4 bilateral) were used for the final analysis and the process of progressive exclusion of samples from the analysis is shown in Supplementary Figure S1. Only blocks with greater than 50% cancer epithelial cells which passed our molecular QC [24] were used for the analysis. Information on the hormone receptor status like ER, PR and HER2 was obtained from hospital records.
The METABRIC data (normalized microarray information) from 1208 patients were downloaded from European Genomic archive [25] for cross validation of our observations.
Immunohistochemistry
We performed IHC for Ki-67 on each of the tumor blocks selected according to standard procedures. Briefly, sections (5 μm in thickness) were cut from FFPE blocks on poly L-lysine coated slides, subjected to deparaffinization in xylene, and rehydrated in graded alcohol. After blocking endogenous peroxidase with a 3% hydrogen peroxide solution, antigen retrieval was done in 0.01 M EDTA buffer at pH 8 in a microwave at 800 W for 2 minutes, 480 W for 7 minutes, followed by 160 W for 11 minutes. Primary blocking was done with 1% bovine serum albumin (Sigma, Cat # A 3803) for 30 minutes at room temperature. Primary antibody for Ki-67(Clone MIB1, DAKO, 1:100, Cat # M7240) was applied for 1 hour at room temperature. Sections were further incubated with secondary antibody (DAKO REAL EnVision) for 20 minutes as per the kit instructions, followed by development of the color using DAB (DAKO REAL EnVision) for 10 minutes. Sections were counterstained with hematoxylin and mounted after dehydration in graded alcohol and xylene. Appropriate positive and negative controls were run for antibody in all the experiments. Labeling index (LI) for Ki-67 was calculated by counting at least 500 tumor cell nuclei according to the recommendations of Dowsett et al. [16]. LI value of 20% and above was considered as Ki-67 high as recommended by the St. Gallen Breast Cancer Consortium (SG‐BCC) guidelines (2013) and other recent studies [16], [18].
RNA Extraction, Reverse Transcription and qRT PCR for Selected Set of Genes
Total RNA was extracted from two 20 μm sections taken from each patient's tumor block as described before [24]. Briefly, sections were deparaffinized by heat, and then subjected to overnight digestion using proteinase K (Qiagen #19133). Total RNA was then extracted using TRI Reagent protocol according to manufacturer instructions (Sigma Aldrich # T9424). Quantitation of the RNA was done using the Ribogreen dye (Invitrogen # R11490 Quant-iT Ribogreen RNA assay kit) on a fluorescent plate reader (Tecan M200-Pro Infinite Series). 500 ng of total RNA was then reverse transcribed using the ABI high capacity cDNA archive kit (ABI # 4322171) as per the manufacturer's protocol. The methods used for nucleic acid extraction, qRT-PCR, selection of housekeeping genes (HKG), and quality control criteria for inclusion of samples in this analysis have been described in detail in a previous publication from our laboratory [24].
The expression level of selected set of ER regulated genes (ESR1, PR, GATA3, TFF1, FOXA1, XBP1) and a set of selected genes (ANLN, CENPF, UBE2C, CCNB1, FOXM1, BIRC5, and BCL2) which are involved in survival proliferation and differentiation in luminal tumors, was determined along with a panel of 3 reference genes (PUM1, RPLPO, ACTB). Primers for all genes were designed using Primer3Plus and manufactured by Eurofins, Bangalore, India. The reference genes normalize for any variations that may be introduced through variations in sample processing and handling methods which in turn lead to varied levels of RNA preservation in the FFPE blocks. The primer sequences for the ER regulated genes are as given in previous publication [23], and those for other set of genes are given in Supplementary Table S1. Using 5 ng cDNA template per reaction, qRT-PCR was done in duplicate using SYBR Green on the LightCycler 480 II (Roche Diagnostics). Total RNA from normal human mammary gland (Clontech, USA, # 636576) and Universal Human Reference RNA (Agilent, # 740000) was also reverse transcribed and 0.1 ng of this template was run in the assay as a control. Total reaction volume was 10 μl. Pre incubation and initial denaturation of the template cDNA were performed at 95 °C for 10 minutes, followed by amplification for 45 cycles at 95 °C for 15 seconds and 60 °C for 1 minute. Quality control criteria of cycle of threshold (Ct) values less than 32.25 were used for the selection of samples as detailed earlier [24]. Ct values for the test genes were in turn normalized relative to the mean Ct value of the three reference genes for each sample as ∆Ct. The relative normalized units (RNU) of expression of the test genes were calculated as 15- ∆Ct, representing the dynamic range of the assay being 15 Cts.
Statistical Analysis
Descriptive statistics of demographic and clinical variables were obtained for the chosen set of tumors and also within subgroups defined by the IHC and gene expression subclasses.
Derivation of the Tumor Aggression Probability Score: The association between each of the chosen genes (ANLN, CENPF, UBE2C, CCNB1, FOXM1, BIRC5, and BCL2) and the dichotomized categories of high and low, using HG (with grade 3 as high and grade 1 and 2 grouped as low) was determined by binomial logistic regression models within the 144 HR+HER2− tumors. Only 125 specimens out of 144 had accurate grade information and were used for this derivation. Only 2 genes (ANLN and BCL2) with a significant positive correlation with high grade were further used to calculate a probability distribution of aggression which was derived by fitting a binomial logistic regression model using the 2 genes as the predictor and grade 3 as the determinant. This score is referred hereafter as tumor aggression score. Tumors were grouped into low, and high groups using the lowest quartile value in the tumor aggression probability score as the cutoff (0.238).
Derivation of the ER Probability Score: ER probability score was calculated as detailed in a previous publication [23]. We used the complete set of 274 tumor specimens that qualified for the molecular analysis for this derivation. Briefly, using multiple ER regulated genes (ESR1, PR, GATA3, TFF1, FOXA1, XBP1), we fit several binomial logistic regression models with ER status by IHC (>10%) as the determinant and different combinations of test genes as the potential predictors. The different models were compared using the Bayesian information criterion (BIC). The predicted probability from the best fitting model (with lowest BIC) was chosen. Receiver operating characteristics (ROC) curve corresponding to the best fitting model was plotted, and the cutoff on the probability score corresponding to the point where both sensitivity and specificity were maximized was selected to classify patients into fully functional adequate ER and inadequate ER downstream function groups.
Likewise, we derived the probability score for tumor aggression and ER function in METABRIC data series using the same set of genes. 638 tumors which were ER positive and HER2 negative and had information on grade and GE by microarray along with survival were used for the derivation.
Sequential application of ER probability and tumor aggression probability scores was used to categorize tumors into GE based classes. The patient characteristics were compared between the GE based classes using chi‐square test or Kruskal-Wallis test and Mann-Whitney U test.
Kaplan-Meier survival curves and log rank tests were used to compare the survival between the classes defined by GE as per the probability score obtained from the logistic regression models. Cox proportional hazards model was used to examine the effect of probability score based classification on survival.
All statistical analyses were done on statistical software XLSTAT version 2012.3.03 and SPSS software version 18 (Chicago, IL).
Results
Classification of Tumors Based on Tumor Aggression and ER Probability Score
We calculated the ER probability score as detailed previously [23] using ER IHC positivity of greater than 10% cells stained as the determinant and arrived at cutoff of 0.68 by ROC analysis. Tumors with probability values above this number were considered to have fully functional ER downstream signaling activity. Tumors with ER probability score less than 0.68 were classified as the inadequate ER downstream function group.
Using similar methods, we derived a probabilistic measure of tumor aggression that was derived by using HG as the determinant. The final predictors chosen included genes involved in cell growth and migration (ANLN), and inhibition of apoptosis (BCL2). Model parameters are given in Supplementary Table S2. The probability score values ranged from 0.07 to 0.77 with a median of 0.32. Distribution of the tumor aggression probability score amongst HG is shown in Figure 1. The mean values of the aggression probability score were significantly different between G1 and G2 compared to G3 (0.27, 0.31 and 0.40 in G1, G2 and G3, respectively, P = .001).
Figure 1.
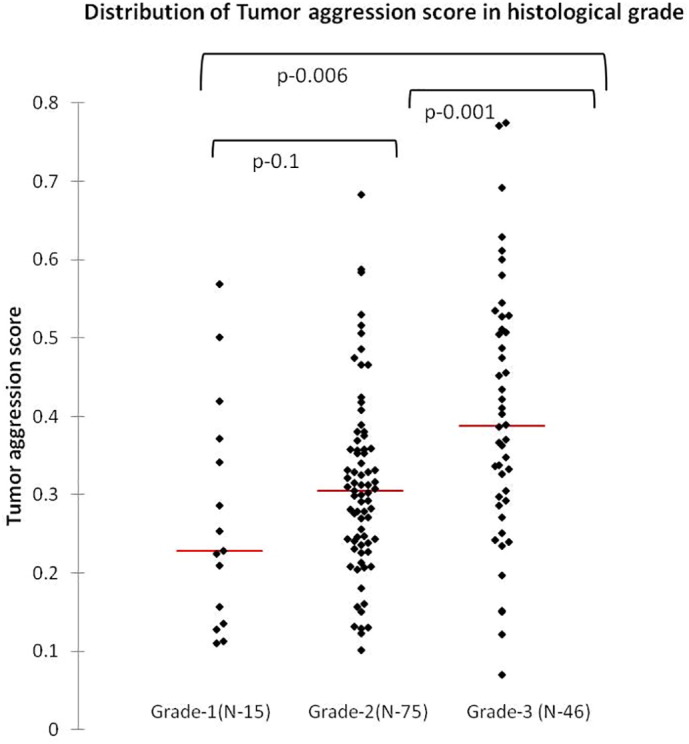
Increasing mean distribution of tumor aggression score in 3 HG.
The probability distribution for aggression was dichotomized at the lowest quartile value (0.24) into low and high groups. The reasoning behind the selection of the lowest quartile for dichotomization was that this cutoff selected for the lowest proportion of HG 3 [7/46 (15%)] tumors in the low aggression category.
Next, using both the probability scores sequentially, we classified the tumors into three groups as below. Tumors with ER probability more than 0.68 and tumor aggression probability less than 0.24 were classified as indolent tumors; those with ER probability more than 0.68 and tumor aggression probability more than 0.24 were classified as aggressive. As stated above, tumors with ER probability score less than 0.68 were classified into the third group as inadequate ER downstream function group.
Only 115/144 (80%) were classified as having adequate ER signaling by the GE based ER probability score and 20% (29/144) were of the inadequate ER downstream function class, suggesting that a small but significant fraction of tumors that were unambiguously HR+ by IHC criteria had only marginal ER regulated down stream activity. This is quite a striking result since the ER probability score was being applied to tumors where nearly 90% had been scored as having >10% of tumor nuclei demonstrating immunoreactivity to ER.
As seen in Table 1 above, the three subgroups of indolent, aggressive and the inadequate ER downstream function class showed striking differences in the classical clinicopathological variables. There was a clear gradation, with the indolent tumors having the most favorable values in Clinicopathological variables including highest median age at diagnosis (P < .001), lowest proportion of HG 3 tumors (Mann-Whitney U indolent Vs inadequate ER, P = .03), and higher proportion of PR positive and tumors in post menopausal women. On the other hand, the inadequate ER downstream function group had the least favorable values for all of these variables and the aggressive had intermediate values. The most commonly used clinical variable LN status however was unable to distinguish the indolent tumors from the aggressive luminal categories (P = .4).
Table 1.
Clinicopathological Features of the Subclasses Identified by Probability Score Are Given Below
| Indolent n (%) | Aggressive n (%) | Inadequate ER n (%) | ||
|---|---|---|---|---|
| n | 32 | 83 | 29 | |
| Age | Median | 64 | 59 | 52 |
| Mean | 66 | 58 | 53 | |
| T size | Mean | 3.2 | 3 | 3.7 |
| Median | 3 | 3 | 3.7 | |
| T1 | 13 (41) | 25 (30) | 5 (17) | |
| T2 | 15 (47) | 50 (60) | 19 (66) | |
| T3 | 4 (12) | 5 (6) | 4 (14) | |
| TX | 0 (0) | 3 (4) | 1 (3) | |
| Grade | I | 7 (22) | 5 (6) | 3 (10) |
| II | 19 (59) | 43 (52) | 13 (45) | |
| III | 5 (16) | 30 (36) | 11 (38) | |
| NA | 1 (3) | 5 (6) | 2 (7) | |
| Lymph node | Negative | 13 (41) | 24 (29) | 11 (38) |
| Positive | 18 (56) | 55 (66) | 17 (59) | |
| Nx | 1 (3) | 4 (5) | 1 (3) | |
| Stage | I | 5 (16) | 11 (13) | 5 (17) |
| II | 18 (56) | 41 (50) | 16 (55) | |
| III | 9 (28) | 26 (31) | 8 (28) | |
| IV | 0 (0) | 5 (6) | 0 (0) | |
| PR | Negative | 3 (9) | 17 (20) | 9 (31) |
| Positive | 29 (91) | 66 (80) | 20 (69) | |
| Menopausal status (n = 140) | Pre | 4 (13) | 18 (22) | 9 (31) |
| Post | 26 (87) | 63 (78) | 20 (69) |
Breast Cancer Subtypes Defined by Ki-67 Labeling Index
Of all 144 tumors, Ki-67 LI could be calculated only in 127 tumors (125 patients with 2 bilateral). The median value of Ki-67 in these 127 tumors was 13, indicating the predominance of low proliferating tumors. We used a cutoff of Ki-67 LI of 20 (SG‐BCC 2013) [18] to subdivide the tumors into luminal A and B and found that 88 (69%) tumors were classified as luminal A and 39 (31%) were luminal B. Their clinical features are presented in Table 2.
Table 2.
Clinical Features in Subclasses Divided by Ki-67-LI
| Luminal A n (%) | Luminal B n (%) | ||
|---|---|---|---|
| n | 88 | 39 | |
| Age | Mean | 58.6 | 56.5 |
| Median | 59 | 56 | |
| T size | T1 | 30 (34) | 9 (23) |
| T2 | 47 (53) | 26 (67) | |
| T3 | 9 (10) | 2 (5) | |
| NA | 3 (3) | 2(5) | |
| LN | Negative | 32 (37) | 11 (28) |
| Positive | 53 (60) | 27 (69) | |
| NA | 3 (3) | 1 (3) | |
| Grade | I | 9 (10) | 2 (5) |
| II | 52 (59) | 17 (43) | |
| III | 22 (25) | 19 (49) | |
| NA | 5 (6) | 1 (3) | |
| MP status | Post | 70 (80) | 29 (74) |
| Pre | 18 (20) | 10 (26) |
As expected, the median age was higher in the luminal A class compared to luminal B group by 3 years. The only other notable difference was in the proportion of tumors that were grade 3 histology. Only a quarter of luminal A tumors were grade 3 compared to almost half of luminal B tumors. The stage distribution between the two classes was almost identical.
GE Based Classification Identifies Biological Classes with Differential Prognosis
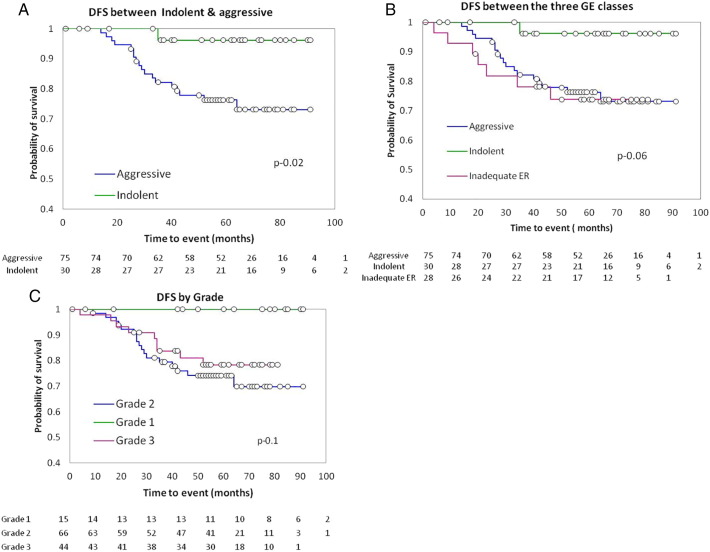
To test if the GE based classes have differential outcomes, we performed the survival analysis between the three classes identified by the probability scores by Kaplan-Meier analysis. Patients with distant metastasis (stage IV) at presentation and lost to follow up were excluded from the survival analysis. As seen in the figure below, a clear separation emerged between the indolent and aggressive tumors (Figure 2A). Indolent tumors had the best survival, and the inadequate ER had the lowest survival (Figure 2B). There was no obvious difference in the treatment history of the patients in the different subgroups that might have contributed to this separation (treatment history in supplementary information).
Figure 2.
The disease free survival (DFS) between the subclasses identified by GE classes. (A) DFS between indolent and aggressive tumors. (B) DFS between 3 classes. (C) DFS by HG.
Similar analysis was done using HG as 1, 2 and 3, and survival was compared to see if grade could differentiate prognosis. Although not statistically significant, a separation emerged between grade 1 tumors when compared to grades 2 and 3 which were further not differentiated. This confirmed the ability of HG in that well differentiated tumors had the best prognosis, but this was limited to a small subset of tumors.
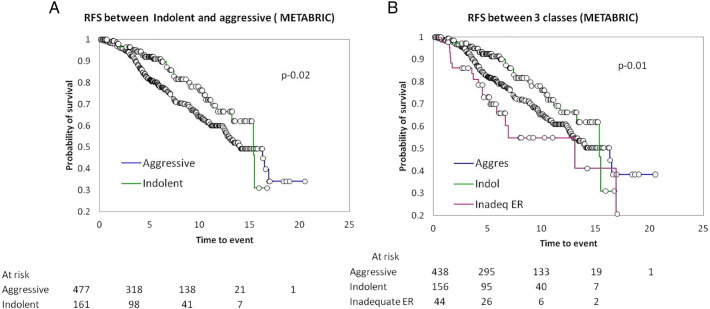
We then performed the analysis on ER + HER2- tumors from the METABRIC data set to derive the probabilities and classify the tumors into similar biological classes. The analysis was limited to 638 tumors which had the information on GE, grade, and survival. Amongst these tumors, 488 (69%) were classified as aggressive, 156 (24%) as indolent and 44 (7%) as inadequate functional ER. Clinical characteristics of these tumors mirror those of ours and are presented in Supplementary Table S3. Our analysis not only identified the three classes but also suggested that the biological behaviors are distinct based on the differential survival pattern within them as shown by Kaplan-Meier survival analysis in Figure 3, A and B.
Figure 3.
The relapse free survival (RFS) in 3 classes identified in METABRIC data series. (A) RFS between the two classes: indolent and aggressive. (B) RFS between the 3 GE classes.
To test if Ki-67 LI based classification of tumors into luminal A and B at cutoff of LI-20 would separate tumors into groups whose survival differed, we performed survival analysis between the two subclasses. (Both the patients with bilateral tumors had at least one luminal B tumor and hence were assigned to the luminal B class for the computation of the Kaplan-Meier survival analysis.)
No clear or significant separation (log-rank test, P = .65) emerged between the two subclasses (Supplementary Figure S2A). This suggests that IHC classification with a Ki-67 LI cutoff of 20 does not have prognostic value in a set of patients where approximately 80% are at stage II/III at first presentation. This is not very surprising because these cutoffs were derived from case series that were comprised almost entirely of early breast cancers (T1 N0 tumors). Therefore, we increased the cutoff of Ki-67 LI to 25 and performed survival analysis again between the subtypes (Supplementary Figure S2B). Even with the increased Ki-67 LI cutoff, there was no significant difference in the survival between the two classes (P = .75).
Probability Scores Have Independent Prognostic Information Irrespective of LN Status
To examine the clinical relevance of our classification based on probability scores, we performed Cox proportional hazards analysis with other known prognostic markers. Univariate analysis done with known prognostic clinical parameters on all cases (Table 3) showed stage III, T size and LN status to be significant predictors of survival along with our classification. Multivariate analysis showed LN, aggression probability and inadequate ER functional class to be significant predictors of outcome with hazard ratio of 4.1 (95% CI, 1.2-13.8), 8.4 (95% CI, 1.1-65.1), and 8.9 (95% CI, 1-69.7), respectively.
Table 3.
Cox Proportional Hazard Models of GE Subclasses with Other Clinical Variables
| Univariate CI |
Multivariate CI |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variable | Reference | HR | Low | High | P | HR | Low | High | P | |
| Age | <50 years | >50 years | 1.64 | 0.73 | 3.68 | .23 | ||||
| T size | T2 | T1 | 1.87 | 0.69 | 5.08 | .22 | ||||
| T3 | 3.16 | 0.85 | 11.80 | .09 | 1.26 | 0.99 | 1.60 | .07 | ||
| LN status | Positive | Negative | 4.62 | 1.39 | 15.40 | .01 | 4.13 | 1.24 | 13.81 | .02 |
| Stage | II | Stage I | 3.13 | 0.40 | 24.49 | .28 | ||||
| III | 8.87 | 1.17 | 67.18 | .03 | ||||||
| Grade | Gr III | Gr I & II | 0.93 | 0.41 | 2.08 | .85 | ||||
| MP status | Pre | Post | 0.98 | 0.39 | 2.44 | .96 | ||||
| GE classification | Aggressive | Indolent | 7.34 | 0.98 | 54.98 | .05 | 8.49 | 1.11 | 65.12 | .04 |
| Inadequate ER | 8.18 | 1.01 | 66.47 | .05 | 8.54 | 1.05 | 69.73 | .05 | ||
Discussion
GE profiling studies have fundamentally changed our conceptual approach to breast cancer. Although IHC derived information is the basis of clinically useful subtype identification, GE signatures give molecular read outs which are quantitative over a broad dynamic range. Clinical validation of multiple such assays in the past decade has shown their utility in selecting the subtypes within HR+HER2− tumors [8], [9], [10], [11]. Using a similar approach, we have tried to dissect the biological heterogeneity within these tumors using HG as a guide.
Amongst the few conventional prognostic indicators in breast cancer, HG estimated based on level of differentiation, nuclear pleomorphism and mitosis is considered second only to LN status [22], [26]. Utility of grade along with stage information in prognosis was established in the pregenomic era [27]. HG estimated by a trained pathologist is the best clinical variable of prognosis in LN negative disease. However, it suffers from high inter and intra observer variability due to subjective evaluation and existence of intermediate category which is a mixture of both high and low risk groups [28]. In an attempt to overcome this drawback, Sortirou et al. developed the genomic grade index (GGI) comprising a 97 gene signature [20] using a discovery cohort of 189 and further validation on 597 tumors with different subtypes [29], [30]. GGI classifies tumors into two types, effectively classifying the G2 into high or low grade, and is further shown to be predictive of recurrence in endocrine treated patients [31], [32]. GGI is now offered as 8 gene panel (including four test and four housekeeping genes) measured by qRT-PCR on HR+HER2− tumors [33], and this test was compared against Ki-67 and mitotic activity index (MAI) with HG and found to be the best predictor [34]. In our methodology for development of tumor aggression probability score, we have used grade as the determinant like in GGI, but without any overlap in the 4 genes used by them. This grade based aggression probability score is able to classify tumors into different prognostic groups as shown in GGI using just 2 genes in HR+HER2− breast cancers.
In our previous work, we had demonstrated that a majority of low ER (1%-10%) tumors in fact behaved like ER negative tumors [23]. In this study, we found that approximately 20% of tumors that met the more stringent cutoff of 10% also did not have adequate ER down stream activity. This is probably due to the fact that ER probability score is a continuous variable and therefore identifies tumors with low downstream activity, which is supported by the poorest prognosis in this subgroup by survival analysis. Only 4/29 of these tumors had less than 10% of the cells immunoreactive to ER protein, but all these were PR positive and hence included as HR positive. None of the patients had stage 4 disease, and less than a third (28%) had stage 3 disease, thus ruling out late stage presentation as the reason for poor prognosis in this group. All but two patients have received antiendocrine therapy as part of routine standard of care treatment. These two patients could not receive antiendocrine therapy as they succumbed early due to the disease, and one was eliminated from survival analysis. Multivariate analysis against all known variables further proved the independent prognostic ability of this classification with a significant hazard ratio.
Although LN involvement is the most validated independent prognostic marker for poor prognosis, approximately 20% of patients with early breast cancer (LN−) have poor prognosis as opposed to a small proportion of late stage tumors with good prognosis [35]. This indicates that although anatomic characteristics such as tumor size and LN involvement have excellent prognostic utility in the majority of patients, these variables might be misleading in a small subset at either end of the spectrum of aggression. Molecular profiling using multiple markers have been shown to be more accurate in predicting outcomes within LN positive tumors [4], [10]. In agreement with other recent reports, our study confirms the presence of biologically indolent tumors that can be identified using small gene panels that capture the critical attributes of proliferation, apoptosis and cell survival. In addition, it provides support for the widespread idea that such tumors exist even in a population with predominantly late stage disease, suggesting that the intrinsic biology of these tumors is one of indolence.
To overcome the limitations of the small numbers tested in this report, we tested the equations derived from the examination of tumors of this series on the METABRIC data set, which represents a multinational archive of 2000 tumors with detailed clinical and molecular profiling with GE and copy number variation data [25]. Not only were we able to identify tumors that had similar gene expression patterns as those seen in our cohort, but the proportions of patients with tumors belonging to the three subclasses was not vastly different, except for the inadequate ER function class which comprised a mere 7% in the METABRIC data set. However, these tumors had the worst overall survival similar to our series, confirming the biological similarity of these tumors.
Ki-67 LI was not of any use in segregating the patients in our cohort into classes based on survival despite using the higher cutoff of 20. We are not sure of how much can be made of this given the known limitations and challenges of deriving useful stratification cutoffs from Ki-67 LI, although it continues to provide useful information on the proliferative state of the tumor. Prognostic significance and appropriate cutoff may have to be derived in large series comprising of late stage tumors with adequate follow up.
Conclusion
The findings of this study show the utility of ER downstream functional assessment in addition to tumor aggression measured against the conventional grade with minimum set of markers for better stratification of the HR+HER2− breast cancers. Further investigations into the pathways mediating such indolence, be it a reduced propensity for tumor stromal interactions or an increased immune activation, might help move such analyses from mere identification of subgroups to identification of actual clinically actionable alterations.
Funding Sources
We gratefully acknowledge Nadathur Estates Private Ltd., Bangalore, India, for their generous funding which enabled this research. The patient follow up was supported by The Bagaria Education Trust, Bangalore, India.
Disclosure of Potential Conflicts of Interest
Authors declare no potential conflicts of interest.
Acknowledgements
We are thankful to Dr. Nandini Dendukuri, Department of Epidemiology, Biostatistics and Occupational Health, McGill University, Montreal, Canada and Dr. Tinku Thomas, Department of Epidemiology and Biostatistics, St. John's Research Institute, for validation and guidance for all statistical analysis.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.tranon.2017.04.011.
Appendix A. Supplementary data
Supplementary materials
References
- 1.Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 2.Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, Cheang MC, Gelmon K, Nielsen TO, Blomqvist C. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 4.Albain KS, Barlow WE, Shak S, Hortobagyi GN, Livingston RB, Yeh IT, Ravdin P, Bugarini R, Baehner FL, Davidson NE. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11:55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gnant M, Sestak I, Filipits M, Dowsett M, Balic M, Lopez-Knowles E, Greil R, Dubsky P, Stoeger H, Rudas M. Identifying clinically relevant prognostic subgroups of postmenopausal women with node-positive hormone receptor-positive early-stage breast cancer treated with endocrine therapy: a combined analysis of ABCSG-8 and ATAC using the PAM50 risk of recurrence score and intrinsic subtype. Ann Oncol. 2015;26:1685–1691. doi: 10.1093/annonc/mdv215. [DOI] [PubMed] [Google Scholar]
- 6.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 7.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bastien RR, Rodriguez-Lescure A, Ebbert MT, Prat A, Munarriz B, Rowe L, Miller P, Ruiz-Borrego M, Anderson D, Lyons B. PAM50 breast cancer subtyping by RT-qPCR and concordance with standard clinical molecular markers. BMC Med Genomics. 2012;5:1755–8794. doi: 10.1186/1755-8794-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardoso F, van't Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, Pierga JY, Brain E, Causeret S, DeLorenzi M. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N Engl J Med. 2016;375:717–729. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 10.Martin M, Brase JC, Calvo L, Krappmann K, Ruiz-Borrego M, Fisch K, Ruiz A, Weber KE, Munarriz B, Petry C. Clinical validation of the EndoPredict test in node-positive, chemotherapy-treated ER+/HER2- breast cancer patients: results from the GEICAM 9906 trial. Breast Cancer Res. 2014;16 doi: 10.1186/bcr3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 12.Agarwal G, Pradeep PV, Aggarwal V, Yip CH, Cheung PS. Spectrum of breast cancer in Asian women. World J Surg. 2007;31:1031–1040. doi: 10.1007/s00268-005-0585-9. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh J, Gupta S, Desai S, Shet T, Radhakrishnan S, Suryavanshi P, Parmar V, Jalali R, Goyal G, Hawaldar R. Estrogen, progesterone and HER2 receptor expression in breast tumors of patients, and their usage of HER2-targeted therapy, in a tertiary care centre in India. Indian J Cancer. 2011;48:391–396. doi: 10.4103/0019-509X.92245. [DOI] [PubMed] [Google Scholar]
- 14.Leong SP, Shen ZZ, Liu TJ, Agarwal G, Tajima T, Paik NS, Sandelin K, Derossis A, Cody H, Foulkes WD. Is breast cancer the same disease in Asian and Western countries? World J Surg. 2010;34:2308–2324. doi: 10.1007/s00268-010-0683-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foulkes WD, Reis-Filho JS, Narod SA. Tumor size and survival in breast cancer--a reappraisal. Nat Rev Clin Oncol. 2010;7:348–353. doi: 10.1038/nrclinonc.2010.39. [DOI] [PubMed] [Google Scholar]
- 16.Dowsett M, Nielsen TO, A'Hern R, Bartlett J, Coombes RC, Cuzick J, Ellis M, Henry NL, Hugh JC, Lively T. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103:1656–1664. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park YH, Im SA, Cho EY, Ahn JH, Woo SY, Kim S, Keam B, Lee JE, Han W, Nam SJ. Validation and comparison of CS-IHC4 scores with a nomogram to predict recurrence in hormone receptor-positive breast cancers. Oncology. 2014;86:279–288. doi: 10.1159/000362281. [DOI] [PubMed] [Google Scholar]
- 18.Untch M, Gerber B, Harbeck N, Jackisch C, Marschner N, Mobus V, von Minckwitz G, Loibl S, Beckmann MW, Blohmer JU. 13th st. Gallen international breast cancer conference 2013: primary therapy of early breast cancer evidence, controversies, consensus - opinion of a german team of experts (zurich 2013) Breast Care (Basel) 2013;8:221–229. doi: 10.1159/000351692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, Lindahl T, Pawitan Y, Hall P, Nordgren H. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 20.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 21.Engstrom MJ, Opdahl S, Hagen AI, Romundstad PR, Akslen LA, Haugen OA, Vatten LJ, Bofin AM. Molecular subtypes, histopathological grade and survival in a historic cohort of breast cancer patients. Breast Cancer Res Treat. 2013;140:463–473. doi: 10.1007/s10549-013-2647-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bloom HJ, Richardson WW. Histological grading and prognosis in breast cancer; a study of 1409 cases of which 359 have been followed for 15 years. Br J Cancer. 1957;11:359–377. doi: 10.1038/bjc.1957.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prabhu JS, Korlimarla A, Desai K, Alexander A, Raghavan R, Anupama C, Dendukuri N, Manjunath S, Correa M, Raman N. A Majority of Low (1-10%) ER Positive Breast Cancers Behave Like Hormone Receptor Negative Tumors. J Cancer. 2014;5:156–165. doi: 10.7150/jca.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korlimarla A, Prabhu JS, Anupama CE, Remacle J, Wahi K, Sridhar TS. Separate quality-control measures are necessary for estimation of RNA and methylated DNA from formalin-fixed, paraffin-embedded specimens by quantitative PCR. J Mol Diagn. 2014;16:253–260. doi: 10.1016/j.jmoldx.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. C. W. Elston & I. O. Ellis. Histopathology 1991; 19; 403-410. Histopathology. 2002;41:151–152. [PubMed] [Google Scholar]
- 27.Henson DE, Ries L, Freedman LS, Carriaga M. Relationship among outcome, stage of disease, and histologic grade for 22,616 cases of breast cancer. The basis for a prognostic index. Cancer. 1991;68:2142–2149. doi: 10.1002/1097-0142(19911115)68:10<2142::aid-cncr2820681010>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 28.Paradiso A, Ellis IO, Zito FA, Marubini E, Pizzamiglio S, Verderio P. Short- and long-term effects of a training session on pathologists' performance: the INQAT experience for histological grading in breast cancer. J Clin Pathol. 2009;62:279–281. doi: 10.1136/jcp.2008.061036. [DOI] [PubMed] [Google Scholar]
- 29.Desmedt C, Giobbie-Hurder A, Neven P, Paridaens R, Christiaens MR, Smeets A, Lallemand F, Haibe-Kains B, Viale G, Gelber RD. The Gene expression Grade Index: a potential predictor of relapse for endocrine-treated breast cancer patients in the BIG 1-98 trial. BMC Med Genomics. 2009;2:40. doi: 10.1186/1755-8794-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, Ellis P, Harris A, Bergh J, Foekens JA. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 31.Liedtke C, Hatzis C, Symmans WF, Desmedt C, Haibe-Kains B, Valero V, Kuerer H, Hortobagyi GN, Piccart-Gebhart M, Sotiriou C. Genomic grade index is associated with response to chemotherapy in patients with breast cancer. J Clin Oncol. 2009;27:3185–3191. doi: 10.1200/JCO.2008.18.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naoi Y, Kishi K, Tanei T, Tsunashima R, Tominaga N, Baba Y, Kim SJ, Taguchi T, Tamaki Y, Noguchi S. High genomic grade index associated with poor prognosis for lymph node-negative and estrogen receptor-positive breast cancers and with good response to chemotherapy. Cancer. 2011;117:472–479. doi: 10.1002/cncr.25626. [DOI] [PubMed] [Google Scholar]
- 33.Toussaint J, Sieuwerts AM, Haibe-Kains B, Desmedt C, Rouas G, Harris AL, Larsimont D, Piccart M, Foekens JA, Durbecq V. Improvement of the clinical applicability of the Genomic Grade Index through a qRT-PCR test performed on frozen and formalin-fixed paraffin-embedded tissues. BMC Genomics. 2009;10:424. doi: 10.1186/1471-2164-10-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertucci F, Finetti P, Roche H, Le Doussal JM, Marisa L, Martin AL, Lacroix-Triki M, Blanc-Fournier C, Jacquemier J, Peyro-Saint-Paul H. Comparison of the prognostic value of genomic grade index, Ki67 expression and mitotic activity index in early node-positive breast cancer patients. Ann Oncol. 2013;24:625–632. doi: 10.1093/annonc/mds510. [DOI] [PubMed] [Google Scholar]
- 35.Carter CL, Allen C, Henson DE. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer. 1989;63:181–187. doi: 10.1002/1097-0142(19890101)63:1<181::aid-cncr2820630129>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials