Abstract
Male infertility is a multifactorial syndrome encompassing a wide variety of disorders. In recent years, several genome-wide single-nucleotide polymorphism (SNP) association studies (GWAS) have been performed on azoospermia and/or oligozoospermia in different populations including two GWAS on nonobstructive azoospermia in China; however, the association of SNPs with idiopathic male infertility, especially asthenozoospermia and oligozoospermia, and their correlation with semen parameters are still not clear. To investigate genetic variants associated with idiopathic male infertility (asthenozoospermia, oligozoospermia, and oligoasthenozoospermia) in Chinese Han people, 20 candidate SNPs were selected from GWAS results and genotyped using the Sequenom MassARRAY assay. A total of 136 subfertile men and 456 healthy fertile men were recruited. rs6476866 in SLC1A1 (P = 1.919E-4, OR = 0.5905, 95% CI: 0.447–0.78) and rs10129954 in DPF3 (P = 0.0023, OR = 2.199, 95% CI: 1.311–3.689) were strongly associated with idiopathic male infertility. In addition, positive associations were observed between asthenozoospermia and rs215702 in LSM5 (P = 0.0016, OR = 1.479, 95% CI: 1.075–2.033) and between oligoasthenozoospermia and rs2477686 in PEX10 (P = 0.0011, OR = 2.935, 95% CI: 1.492–5.775). In addition, six SNPs (rs215702 in LSM5, rs6476866 in SLC1A1, rs10129954 in DPF3, rs1801133 in MTHFR, rs2477686 in PEX10, and rs10841496 in PED3A) were significantly correlated with semen quality alterations. Our results suggest that idiopathic male infertility in different ethnic groups may share the same mechanism or pathway. Cohort expansion and further mechanistic studies on the role of genetic factors that influence spermatogenesis and sperm progressive motility are suggested.
Keywords: candidate SNPs, Han Chinese, idiopathic male infertility, semen quality
INTRODUCTION
Infertility affects approximately 10%–15% of couples worldwide, of which 20%–25% are idiopathic male infertility including oligozoospermia, asthenozoospermia, teratozoospermia, and azoospermia.1 Male infertility is a multifactorial syndrome affected by intricate genetic-environmental interactions which make the study of causes for male infertility very difficult and complicated.2 Although many studies have explored gene mutations or polymorphisms related to male infertility, only a few genetic variants, such as Y-chromosome microdeletions, autosomal chromosome polymorphisms, and sex chromosome copy number variants, have been shown to be associated with abnormal spermatogenesis and semen quality.3 In recent years, several genome-wide single-nucleotide polymorphism (SNP) association studies (GWAS) have been performed on azoospermia and/or oligozoospermia3,4,5,6 in different populations which found candidate polymorphisms associated with male infertility that have provided valuable information on the pathogenesis of azoospermia and oligozoospermia and may be useful for reproductive therapies. GWAS in a Caucasian population found 21 SNPs significantly associated with azoospermia or oligozoospermia.4 Another GWAS in Hutterite population in the United States found 41 SNPs associated with reduced family size and reduced birth rate, and six of these loci (rs7867029 in phosphohydroxythreonine aminotransferase [PSAT1] gene, rs12870438 in epithelial stromal interaction 1 [breast] [EPSTI1] gene, rs7174015 in gene encoding ubiquitin-specific peptidase [USP8], rs10129954 in D4, zinc and double PHD finger, family 3 [DFP3] gene, rs11236909 near the gene for tsukushi, small leucine-rich proteoglycan [TSKU], and rs724078 near the ubiquitin D [UBD] gene) were also correlated with multiple semen parameters in Chicago men.6 In addition, GWAS on nonobstructive azoospermia (NOA) in Han Chinese identified three risk loci for NOA in Chinese men (rs12097821 in protein arginine N-methyltransferase 6 [PRMT6] gene, rs2477686 in peroxisome biogenesis factor 10 [PEX10] gene, and rs10842262 in Sex Determining Region Y [SRY]-Box 5 [SOX5] gene) although follow-up studies in Japanese and Chinese men failed to replicate these results.7,8 However, another GWAS involving NOA in Han Chinese showed that human leukocyte antigen (HLA) regions were associated with NOA (rs3129878 in major histocompatibility complex, class II, DR alpha [HLA-DRA], and rs498422 in chromosome 6 open reading frame 10 [c6orf10] and butyrophilin-like 2 [BTNL2]).9
As described above, genetic associations in Chinese idiopathic male infertility, especially asthenozoospermia and/or oligozoospermia, are still not clear. The same disease symptoms may share the same mechanisms or pathways in different ethnic groups. Therefore, additional candidate polymorphisms will need to be identified to reveal related functional genes and provide more biomarkers for medical genetic practice in different populations.
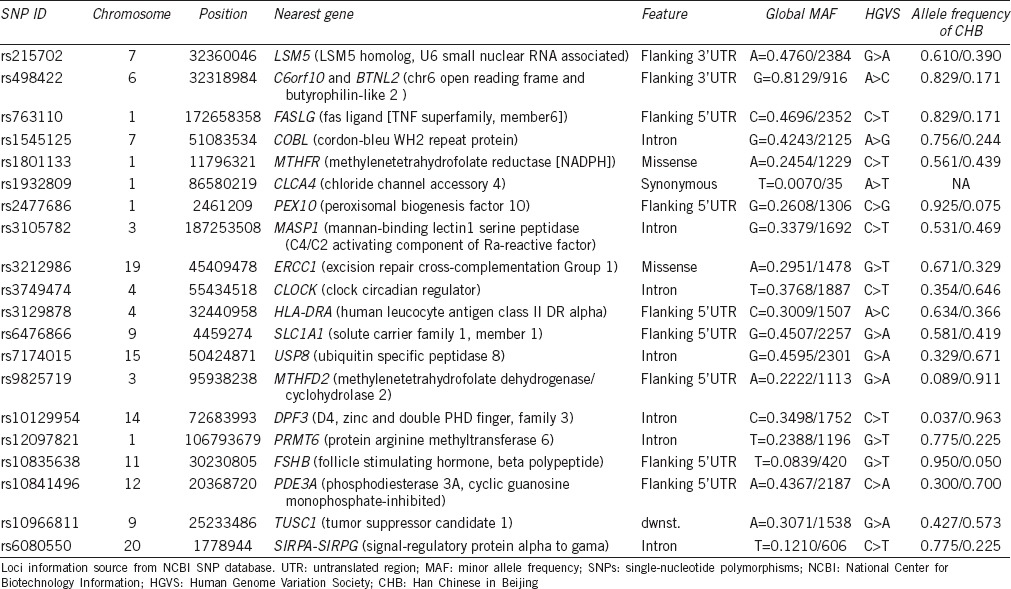
To investigate potential SNPs associated with Chinese idiopathic male infertility, a Sequenom MassARRAY assay was used to determine the association between 20 candidate SNPs and idiopathic male infertility (asthenozoospermia, oligozoospermia, and/or oligoasthenozoospermia) in Chinese Han people. The candidate SNPs were chosen based on their identification – positively associated with male infertility in other populations in some GWAS3,4,5,6,8,9,10 and include the following (Table 1): rs215702 in gene for U6 snRNA-associated Sm-like protein LSm5 (LSM5), rs498422 in C6orf10 and BTNL2, rs763110 in Fas ligand gene (TNF Superfamily, Member 6; FASLG), rs1545125 in cordon-bleu gene (COBL), rs1801133 in gene for methylenetetrahydrofolate reductase (MTHFR), rs1932809 in gene coding chloride channel, calcium activated, family member 4 (CLCA4), rs2477686 in PEX10, rs3105782 in mannan-binding lectin serine peptidase 1 (MASP1), rs3212986 in excision repair cross-complementation Group 1 (ERCC1), rs3749474 in circadian locomotor output cycles kaput (CLOCK), rs3129878 in HLA-DRA, rs6476866 in solute carrier family 1 (SLC1A1), rs7174015 in USP8, rs9825719 in gene for bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial (MTHFD2), rs10129954 in D4, zinc and double PHD finger, family 3 (DPF3), rs12097821 in PRMT6, rs10835638 in gene coding follicle stimulating hormone, beta polypeptide (FSHB), rs10841496 in PDE3A, rs10966811 in gene coding tumor suppressor candidate 1 (TUSC1), and rs6080550 in signal regulatory protein alpha-signal regulatory protein gamma (SIRPA-SIRPG) gene.
Table 1.
Chromosomal location and identity of candidate SNPs
MATERIALS AND METHODS
Subjects and semen analysis
A total of 592 Chinese Han men attending the Reproductive Medical Research Centre of People's Hospital of Shiyan in Hubei Province for assisted reproductive therapies were recruited in this study. Semen samples were collected from each participant under informed consent procedure. This study was approved by the Ethics Review Board of the Institute of Medical Biology, Chinese Academy of Medical Sciences, Kunming, China.
All men recruited in this study underwent a basic andrological examination including medical history, physical examination, semen analysis, hormone analysis, scrotal ultrasound, karyotype testing and Y-chromosome microdeletion screening, abstinence time, duration of marriage, and demographic information. Semen samples were collected by masturbation and examined after liquefaction for 30 min at 37°C. Semen analysis was performed using the computer-assisted semen analysis system (WLJY-9000; Weili New Century Science and Tech Development, Beijing, China) with semen parameters determined according to the guidelines of World Health Organization 2010.11
Among participants, 136 men were identified as idiopathic infertility patients, who had no known clinical and genetic causes of infertility such as testicular dysplasia, hormonal abnormalities, infections, history of diseases affecting fertility (e.g., varicocele), genital tract pathologies, karyotyping anomalies, or Y-chromosome deletions. Men with other factors that could have caused infertility (fever, smoking, exposure to toxins, alcoholism, and drug addiction) were excluded. Eligible cases were divided into the following categories: asthenozoospermia (<32% progressive motile spermatozoa and normal concentration, n = 96), oligoasthenozoospermia (<15 × 106 ml−1 spermatozoa and <32% progressive motile spermatozoa, n = 22), and oligozoospermia (<15 × 106 ml−1 spermatozoa, n = 18). Azoospermic and severe oligozoospermic patients were excluded.
The control group consisted of 456 fertile men who had fathered at least 1 child. All of the fertile controls had normal semen parameters (sperm count >15 × 106 ml−1, total sperm count >39 × 106, total motility >40% motile sperm, progressive motility > 32% (Grade a + b) motile sperm, vitality >58% living sperm, sperm with normal morphology >4%, and leukocyte count <1.0 × 106 ml−1). Men who exhibited normal semen parameters but with unknown fertilization status were not included in this study.
Genotyping
Genomic DNA was extracted from sperm using the standard hydroxybenzene-chloroform protocol. Spermatozoa were collected and lysed using 10% SDS, proteinase K, and dithiothreitol overnight, followed by isometric hydroxybenzene and chloroform extraction and DNA precipitation with isopropanol. More than 50 μg of spermic whole genomic DNA with very good quality was purified per sample. Genotyping of SNPs was performed using the Sequenom MassARRAY platform (Sequenom iPLEX assay, San Diego, CA, USA; Biomiao Biology Corporation, Beijing, China). PCR and single base extension (SBE) primers were designed using the MassARRAY Designer software, version 3.1 (Sequenom, San Diego, CA, USA).
PCR was performed in a 5 μl reaction volume containing 15 ng of genomic DNA, 1X PCR buffer (Mg2+-free), 250 μmol l−1 of each dNTP, 2 mmol l−1 MgCl2, 2.5 U of AmpliTaq Gold Polymerase (Applied Biosystems, Foster City, CA, USA), and 10 pmol of each primer. PCR was performed in an GeneAmp® PCR system 9700 thermal cycler (Applied Biosystems, Foster City, CA, USA) with the following PCR thermal cycling profile: initial denaturation at 94°C for 15 min; 45 cycles of 94°C for 20 s, 56°C for 30 s, and 72°C for 1 min; and final extension for 3 min at 72°C. The PCR products were then treated with 0.5 U shrimp alkaline phosphatase (SAP) and incubated at 37°C for 40 min, followed by enzyme inactivation at 85°C for 5 min.
The SBE reaction was carried out in a 9 μl reaction volume containing 7 μl SAP-treated PCR products and 2 μl iPLEX® pro mix (Applied Biosystems, CA, USA). The iPLEX® pro mix contained 0.2 μl iPLEX buffer, 1 μl iPLEX termination mix, 1 μl iPLEX enzyme, and the SBE primer mix. The iPLEX extension reaction was performed in the GeneAmp® PCR system 9700 thermal cycler with the following PCR thermal cycling profile: initial denaturation at 94°C for 30 s; 40 cycles of 94°C for 5 s, 52°C for 5 s, and 80°C for 5 s; and final extension for 3 min at 72°C.
After desalting of SBE products using SpectroCLEAN resin (Sequenom) according to the manufacturer's protocol, cleaned SBE products were spotted twice on the SpectroCHIP array (Sequenom) using an RS1000 Nanospotter (Sequenom). Finally, the array was detected on a MassARRAY Compact 96 mass spectrometer (Sequenom) and results were visualized using the MassARRAY® analyzer 4 system. At least, two experiments were performed for each sample. SNP calls were analyzed using MassARRAY genotyping software, version 4.0.5 (Sequenom, San Diego, CA, USA).
Statistical analysis
The Hardy–Weinberg equilibrium (HWE) for the control group with a threshold of 0.05 was assessed by PLINK (version 1.07; Harvard & MIT http://pngu.mgh.harvard.edu/~purcell/plink/). Association tests for allelic and genotypic frequencies, as well as dominant and recessive genetic model tests in infertility and control groups (calculated by both Fisher's exact test and χ2 statistics), were also performed by PLINK. Infertility risks were estimated by odds ratios (OR) and 95% confidence intervals (95% CI). Analysis of variance (ANOVA) was used to determine statistical differences in age, abstinence time, and semen parameters. Linear regression analysis was used to determine the correlations of semen concentration and progressive motility with different genotypes using SPSS (version 17.0; SPSS Inc., Chicago, IL, USA) and regressions were plotted by GraphPad Prism 4.0 for Windows (GraphPad software, La Jolla, CA, USA). P <0.05 was considered statistically significant, with Bonferroni corrections for multiple testing calculated as 0.05/n.
RESULTS
Subjects
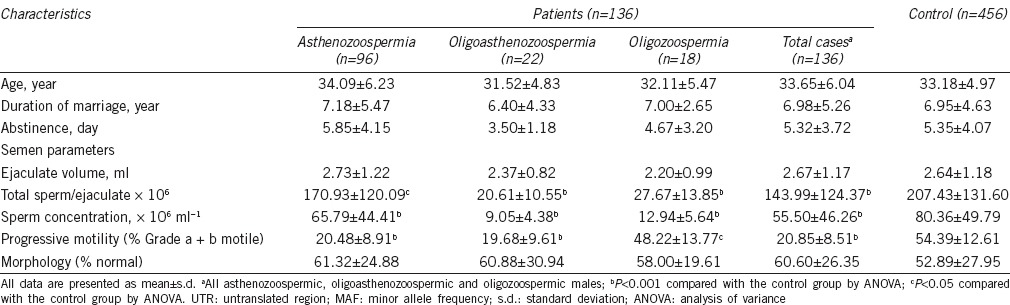
There was no statistical difference between case and control groups in individual characteristics, including age, duration of marriage, and abstinence time (Table 2). However, semen concentration, total semen count per ejaculate, and sperm progressive motility (Grade a + b) were significantly lower in subfertile group (P < 0.001; Table 2).
Table 2.
Comparison of individual characteristics and semen parameters in study groups
The association between candidate SNPs and idiopathic male infertility
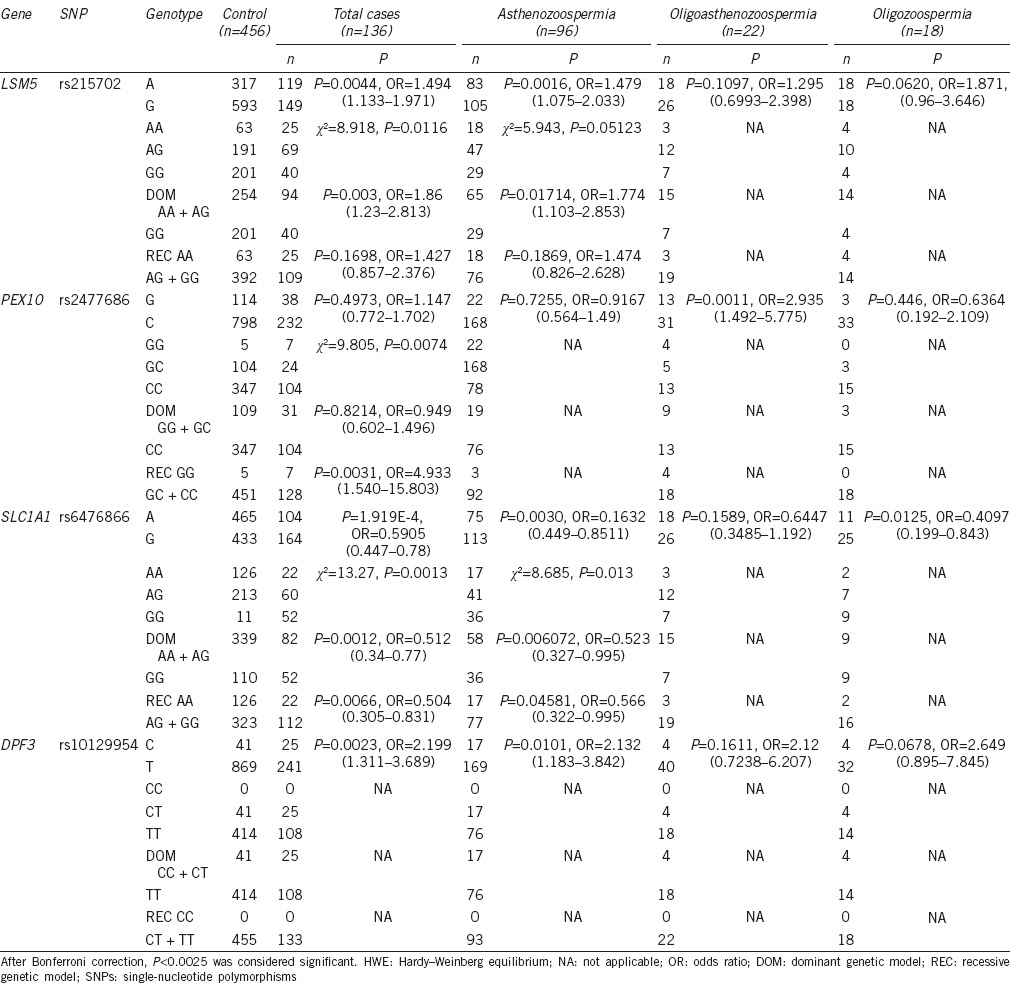
In this study, we performed comparisons of allele and genotype frequencies of 20 SNPs and applied dominant and recessive genetic model analysis between case and control groups (Table 3 and Supplementary Table 1 (117.2KB, pdf) ). With the exception of SNPs rs3749474, there were no deviations from HWE observed within control group for all studied SNPs. After Bonferroni correction, we found that rs6476866 and rs10129954 were strongly associated with idiopathic male infertility when the total group of subfertile men was analyzed. rs6476866 located in SLC1A1 gene showed the greatest association with idiopathic male infertility (P = 1.919E-4, OR = 0.5905, 95% CI: 0.447–0.78). There was a notable difference in the genotype distribution between case and control groups (P = 0.0013), which was also observed in analysis using the dominant genetic model (AA + AG vs GG; P = 0.0012, OR = 0.512, 95% CI: 0.340–0.770). rs10129954 in DPF3 gene displayed a higher minor allele C frequency in subfertile group (P = 0.0023, OR = 2.199, 95% CI: 1.311–3.689). Strong associations were also observed between rs215702 and idiopathic asthenozoospermia (P = 0.0016, OR = 1.479, 95% CI: 1.075–2.033) and between rs2477686 and oligoasthenozoospermia (P = 0.0011, OR = 2.935, 95% CI: 1.492–5.775; Table 3).
Table 3.
SNPs significantly associated with idiopathic male infertility
Allelic and genotypic frequencies of 20 SNPs in cases and controls
Correlations between semen parameters and SNP genotypes
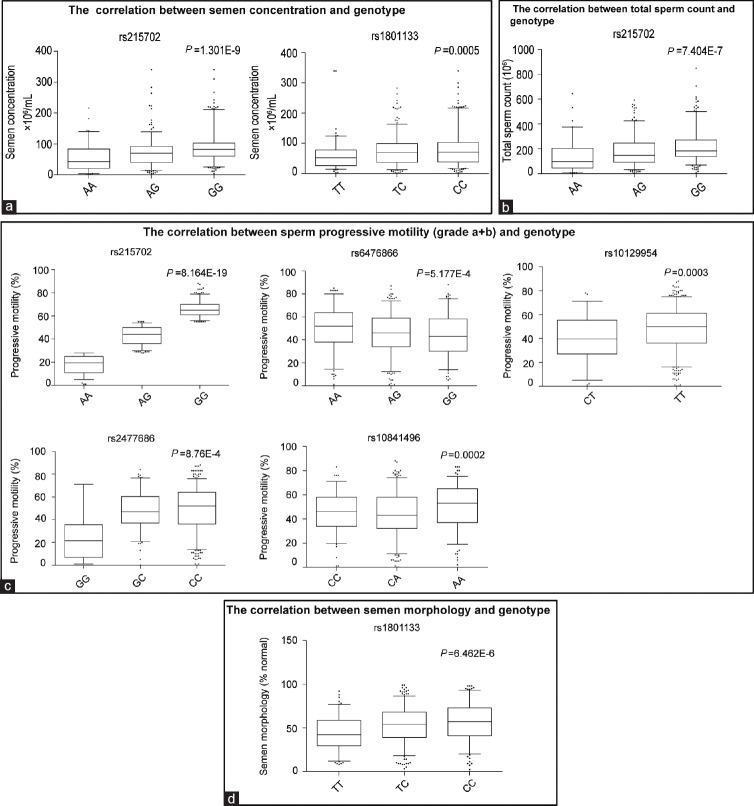
We compared the equality of means of clinical semen parameters across genotypes (Supplementary Table 2 (38.4KB, pdf) ) and found that sperm concentration was significantly different among different genotypes of two SNPs (rs215702 and rs1801133) (P < 6.25E-4). The total sperm number per ejaculate was similar among genotypes of most SNPs except for rs215702, which showed a significant correlation with total sperm count (P = 7.404E-7). For rs215702, the total sperm number per ejaculate was much higher in subjects carrying GG genotype compared to those with AA genotype. In addition, rs215702, rs10129954, rs10841496, rs6476866, and rs2477686 were highly correlated with sperm progressive motility (P < 6.25E–4), and rs1801133 was significantly correlated with semen morphology (Figure 1a–1d and Supplementary Table 2 (38.4KB, pdf) ).
Figure 1.
Box-and-whisker plots of different semen parameters across different genotypes in Chinese men. (a) Semen concentration, (b) total sperm count per ejaculate, (c) sperm progressive motility (Grade a + b), and (d) semen morphology. Box-and-whiskers extend to the maximum and minimum values; the horizontal lines within boxes show the means for each genotype group. P values were calculated by linear regression across genotypes. After Bonferroni correction, P < 0.000625 was considered statistically significant.
Different semen parameters across different genotypes in Chinese men
DISCUSSION
Of the 20 SNPs we examined, we found that rs6476866 and rs10129954 were strongly associated with idiopathic male infertility. rs6476866 in SLC1A1 showed the highest association with idiopathic male infertility, and when analyzed in stratification subgroups, rs6476866 also showed a slight association with asthenozoospermia. When we analyzed semen parameters from different genotypes, we found that semen progressive motility was much greater in AA carriers compared to other genotypes (P = 5.177E-4), and that semen concentration and total sperm count were also very high in AA carriers (P = 0.003 and P = 0.014, respectively). SLC1A1 is a neuronal glutamate transporter gene encoding a member of neuronal and epithelial high-affinity glutamate transporters,12 which are critical for terminating the postsynaptic action of glutamate and regulating extrasynaptic glutamate levels by rapidly removing released glutamate from the synaptic cleft.13 The role of the SLC1A1 gene in semen quality and spermatogenesis is still unclear. In one GWAS, rs6476866 was significantly associated with oligozoospermia.4 Several studies have also indicated that polymorphisms of the SLC1A1 gene may be associated with obsessive-compulsive disorder.12,13 Based on our results, we speculate that neuropsychiatric factors may affect normal spermatogenesis and semen quality as several lines of evidence have suggested an association between mental illness and sperm quality and reduced chance of pregnancy.14 Evaluating the role of SLC1A1 in semen quality and male reproduction will be helpful for clinical diagnosis in future.
Our results showed that the allele distribution of rs10129954 in DPF3 gene was significantly different between subfertile and fertile men. We also found that semen progressive motility (Grade a + b) was greatly reduced in CT heterozygotes (P = 0.0003) and the concentration of semen in CT heterozygotes was also low (P = 0.0319). DPF3 encodes a member of the D4 protein family of zinc finger proteins which is implicated as an epigenetic factor specifically binding acetylated and methylated lysine residues of histone 3 and histone 4 to provide easy access of transcription factors to DNA.15,16 DPF3-regulated histone modification plays a key role in preparing mature sperm DNA for early embryogenesis. Recently, studies indicated that the epigenetic factors such as histone modification, DNA methylation, and spermatozoal RNA transcripts in sperm play a potential role in spermatogenesis and embryogenesis; for example, in chromatin packaging, most of the histones in the sperm head are replaced with protamines, which are crucial for sperm motility and protect DNA from the hostile environment in the female reproductive tract.17 It was reported that sperm epigenetic abnormalities may be associated with male infertility, poor embryogenesis, and other diseases.17 Our results indicated that rs10129954 in an intron of the DPF3 gene was correlated with both poor motility and sperm count. Kosova et al.6,16 found rs10129954 associated with total motile count, beat frequency and linearity, and sperm morphology in men from Chicago. This polymorphism locus may thus affect histone modifications by altering the splicing of dpf3, which may influence the expression or normal function of dpf3 and eventually reduce semen motility and spermatogenesis. However, this hypothesis and its mechanism need to be evaluated in future studies.
In this study, rs215702 in LSM5 showed a positive association with idiopathic asthenozoospermia. AA genotype carriers showed correlations with decreased sperm concentration (P = 1.301E-9), sperm number per ejaculate (P = 7.404E-7), and semen progressive motility (P = 8.164E-19). Our results thus suggest that rs215702 may play an important role in spermatogenesis and sperm quality; however, the role of LSM5 gene in male infertility or reproduction remains to be clarified and may be an interesting loci for further study.
A report by Hu et al. indicated that rs2477686, rs1207821, rs10842262, and rs6080550 were risk loci for NOA in Chinese men.3 In this study, we detected an association of rs12097821, rs2477686, and rs6080550 with idiopathic male infertility. We also observed that rs2477686 was significantly associated with oligoasthenozoospermia and that sperm progressive motility in rs2477686 GG homozygote men was greatly reduced compared with that of CC carriers (P = 8.76E-4). The nearest genes to rs2477686 in genome are PEX10 and MMEL1, which have been previously reported to have functional roles in male infertility in Drosophila and mouse, respectively.18,19 Although the sample size of oligoasthenozoospermia subjects was rather small in our study, from the results of Hu et al. and ours, we suggest that PEX10 and MMEL1 may be risk genes for human idiopathic male infertility, and a greater number of oligoasthenozoospermic patients will be required to validate this hypothesis. Additional genetic association and functional studies are needed to reveal the roles of those two genes on spermatogenesis and sperm motility.
Another GWAS on NOA in Chinese men found variants of HLA regions were significantly associated with NOA (rs3129878 in HLA-DRA and rs498422 in C6orf10 and BTNL2). In this study, the association of these two SNPs with idiopathic male infertility was not significant. It is possible that these inflammation-related genes may not be involved in sperm quality.
Although other SNPs analyzed in this study were not significantly associated with male infertility, several SNPs including rs1801133 in MTHFR and rs10841496 in PED3A were correlated with genotype and semen qualities. The correlation of rs1801133 in MTHFR gene with male infertility has been studied in Asia, Europe, and America.20,21,22,23,24 Some of these studies have reported that rs1801133 may be a risk factor for male infertility, but other studies have not reproduced this result. Folate deficiency is considered to be a risk factor for male infertility, and methylenetetrahydrofolate reductase (MTHFR) is one of the key enzymes in the folate metabolic pathway which may play an important role in spermatogenesis.24 The rs1801133 site located in the coding region of the MTHFR gene substitutes an alanine for a valine residue (Ala 222 Val) which may reduce enzyme activity.24,25 It was reported that MTHFR enzyme activity is 35% lower in rs1801133 heterozygotes (CT) and 70% lower in mutant homozygotes (TT) compared to wild-type homozygotes (CC).24 In this study, rs1801133 TT carriers had lower sperm concentrations (P = 4.836E-4) and altered semen morphologies (P = 3.439E-4) compared to CC genotype carriers, similar to results of other studies.20,26,27,28 Therefore, we conclude that MTHFR polymorphism may be associated with spermatogenesis in Chinese men.
rs10841496 (A > C) is another locus correlated with semen qualities. The CC carriers in our study showed a reduction in semen progressive motility compared to AA carriers (P = 0.0002). rs10841496 is located in the 5’ untranslated region of the PDE3A gene, which has been suggested to play an important role in sperm motility, capacitation, and the acrosome reaction.29 rs10841496 was significantly associated with azoospermia in previous GWAS.4 Although the role of PED3A in male reproduction has not been described, the enzyme is crucial for oocyte maturation. Our results suggest that rs10841496 is a locus correlated with Chinese male idiopathic infertility and indicate that further study on the function of this gene and the association of other genetic polymorphisms within the phosphodiesterase genes and male infertility is warranted.
CONCLUSION
Twenty potential SNPs were evaluated for their association with idiopathic male infertility in Chinese Han men. rs6476866 in SLC1A1 and rs10129954 in DPF3 showed strong associations with Chinese idiopathic male infertility whereas rs215702 in LSM5 was significantly associated with asthenozoospermia and rs2477686 was significantly associated with oligoasthenozoospermia. In addition, we found six SNPs highly correlated with semen qualities (rs215702 in LSM5, rs6476866 in SLC1A1, rs10129954 in DPF3, rs1801133 in MTHFR, rs2477686 in PEX10, and rs10841496 in PED3A). Our results suggest that idiopathic male infertility in different ethnic groups may share the same mechanism or pathway. Cohort expansion and further mechanistic studies on the role of genetic factors that influence spermatogenesis and sperm progressive motility are necessary in future.
AUTHOR CONTRIBUTIONS
SYL carried out the genetic studies and participated in the clinical andrological examinations. CJZ conducted the clinical diagnosis and andrological examinations. HYP participated in the clinical andrological examinations and sample collection. HS performed the statistical analysis together with SYL. KQL, XQH, and KH participated in DNA extraction. JYC participated in the design of this study. ZQY participated in the design and coordination of this study and helped draft the manuscript. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declared no competing financial interests.
ACKNOWLEDGMENTS
We would like to thank all the volunteers who participated in this study, and we also thank the staff of the Reproductive Medical Research Centre of People's Hospital of Shiyan in Hubei Province. This work was supported by the Chinese National High Technology Research and Development Program (Grant Number: 2012AA021802).
Supplementary information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Poongothai J, Gopenath TS, Manonayaki S. Genetics of human male infertility. Singapore Med J. 2009;50:336–47. [PubMed] [Google Scholar]
- 2.Ferlin A, Foresta C. New genetic markers for male infertility. Curr Opin Obstet Gynecol. 2014;26:193–8. doi: 10.1097/GCO.0000000000000061. [DOI] [PubMed] [Google Scholar]
- 3.Hu Z, Xia Y, Guo X, Dai J, Li H, et al. A genome-wide association study in Chinese men identifies three risk loci for non-obstructive azoospermia. Nat Genet. 2012;44:183–6. doi: 10.1038/ng.1040. [DOI] [PubMed] [Google Scholar]
- 4.Aston KI, Carrell DT. Genome-wide study of single-nucleotide polymorphisms associated with azoospermia and severe oligozoospermia. J Androl. 2009;30:711–25. doi: 10.2164/jandrol.109.007971. [DOI] [PubMed] [Google Scholar]
- 5.Aston KI, Krausz C, Laface I, Ruiz-Castane E, Carrell DT. Evaluation of 172 candidate polymorphisms for association with oligozoospermia or azoospermia in a large cohort of men of European descent. Hum Reprod. 2010;25:1383–97. doi: 10.1093/humrep/deq081. [DOI] [PubMed] [Google Scholar]
- 6.Kosova G, Scott NM, Niederberger C, Prins GS, Ober C. Genome-wide association study identifies candidate genes for male fertility traits in humans. Am J Hum Genet. 2012;90:950–61. doi: 10.1016/j.ajhg.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou S, Li Z, Wang Y, Chen T, Song P, et al. Association study between polymorphisms of PRMT6, PEX10, SOX5, and nonobstructive azoospermia in the Han Chinese population. Biol Reprod. 2014;90:96. doi: 10.1095/biolreprod.113.116541. [DOI] [PubMed] [Google Scholar]
- 8.Sato Y, Jinam T, Iwamoto T, Yamauchi A, Imoto I, et al. Replication study and meta-analysis of human nonobstructive azoospermia in Japanese populations. Biol Reprod. 2013;88:87. doi: 10.1095/biolreprod.112.106377. [DOI] [PubMed] [Google Scholar]
- 9.Zhao H, Xu J, Zhang H, Sun J, Sun Y, et al. A genome-wide association study reveals that variants within the HLA region are associated with risk for nonobstructive azoospermia. Am J Hum Genet. 2012;90:900–6. doi: 10.1016/j.ajhg.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu M, Qin Y, Qu J, Lu C, Wang Y, et al. Evaluation of five candidate genes from GWAS for association with oligozoospermia in a Han Chinese population. PLoS One. 2013;8:e80374. doi: 10.1371/journal.pone.0080374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteves SC, Zini A, Aziz N, Alvarez JG, Sabanegh ES, Jr, et al. Critical appraisal of World Health Organization's new reference values for human semen characteristics and effect on diagnosis and treatment of subfertile men. Urology. 2012;79:16–22. doi: 10.1016/j.urology.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Stewart SE, Mayerfeld C, Arnold PD, Crane JR, O’Dushlaine C, et al. Meta-analysis of association between obsessive-compulsive disorder and the 3’ region of neuronal glutamate transporter gene SLC1A1. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:367–79. doi: 10.1002/ajmg.b.32137. [DOI] [PubMed] [Google Scholar]
- 13.Wu H, Wang X, Xiao Z, Yu S, Zhu L, et al. Association between SLC1A1 gene and early-onset OCD in the Han Chinese population: a case-control study. J Mol Neurosci. 2013;50:353–9. doi: 10.1007/s12031-013-9995-6. [DOI] [PubMed] [Google Scholar]
- 14.Dooley M, Dineen T, Sarma K, Nolan A. The psychological impact of infertility and fertility treatment on the male partner. Hum Fertil (Camb) 2014;3:1–7. doi: 10.3109/14647273.2014.942390. [DOI] [PubMed] [Google Scholar]
- 15.Ninkina NN, Mertsalov IB, Kulikova DA, Alimova-Kost MV, Simonova OB, et al. Cerd4, third member of the d4 gene family: expression and organization of genomic locus. Mamm Genome. 2001;12:862–6. doi: 10.1007/s00335-001-3039-1. [DOI] [PubMed] [Google Scholar]
- 16.Kosova G, Hotaling JM, Ohlander S, Niederberger C, Prins GS, et al. Variants in DPF3 and DSCAML1 are associated with sperm morphology. J Assist Reprod Genet. 2014;31:131–7. doi: 10.1007/s10815-013-0140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gannon JR, Emery BR, Jenkins TG, Carrell DT. The sperm epigenome: implications for the embryo. Adv Exp Med Biol. 2014;791:53–66. doi: 10.1007/978-1-4614-7783-9_4. [DOI] [PubMed] [Google Scholar]
- 18.Carpentier M, Guillemette C, Bailey JL, Boileau G, Jeannotte L, et al. Reduced fertility in male mice deficient in the zinc metallopeptidase NL1. Mol Cell Biol. 2004;24:4428–37. doi: 10.1128/MCB.24.10.4428-4437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Liu Z, Huang X. Drosophila models of peroxisomal biogenesis disorder: peroxins are required for spermatogenesis and very-long-chain fatty acid metabolism. Hum Mol Genet. 2010;19:494–505. doi: 10.1093/hmg/ddp518. [DOI] [PubMed] [Google Scholar]
- 20.Mfady DS, Sadiq MF, Khabour OF, Fararjeh AS, Abu-Awad A, et al. Associations of variants in MTHFR and MTRR genes with male infertility in the Jordanian population. Gene. 2014;536:40–4. doi: 10.1016/j.gene.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Wei B, Xu Z, Ruan J, Zhu M, Jin K, et al. MTHFR 677C>T and 1298A>C polymorphisms and male infertility risk: a meta-analysis. Mol Biol Rep. 2012;39:1997–2002. doi: 10.1007/s11033-011-0946-4. [DOI] [PubMed] [Google Scholar]
- 22.Gupta N, Gupta S, Dama M, David A, Khanna G, et al. Strong association of 677 C>T substitution in the MTHFR gene with male infertility - A study on an indian population and a meta-analysis. PLoS One. 2011;6:e22277. doi: 10.1371/journal.pone.0022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu W, Shen O, Qin Y, Lu J, Niu X, et al. Methylenetetrahydrofolate reductase C677T polymorphism and the risk of male infertility: a meta-analysis. Int J Androl. 2012;35:18–24. doi: 10.1111/j.1365-2605.2011.01147.x. [DOI] [PubMed] [Google Scholar]
- 24.Naqvi H, Hussain SR, Ahmad MK, Mahdi F, Jaiswar SP, et al. Role of 677C-->T polymorphism a single substitution in methylenetetrahydrofolate reductase (MTHFR) gene in North Indian infertile men. Mol Biol Rep. 2014;41:573–9. doi: 10.1007/s11033-013-2894-7. [DOI] [PubMed] [Google Scholar]
- 25.Paracchini V, Garte S, Taioli E. MTHFR C677T polymorphism, GSTM1 deletion and male infertility: a possible suggestion of a gene-gene interaction? Biomarkers. 2006;11:53–60. doi: 10.1080/13547500500442050. [DOI] [PubMed] [Google Scholar]
- 26.Safarinejad MR, Shafiei N, Safarinejad S. Relationship between genetic polymorphisms of methylenetetrahydrofolate reductase (C677T, A1298C, and G1793A) as risk factors for idiopathic male infertility. Reprod Sci. 2011;18:304–15. doi: 10.1177/1933719110385135. [DOI] [PubMed] [Google Scholar]
- 27.Gava MM, Chagas Ede O, Bianco B, Christofolini DM, Pompeo AC, et al. Methylenetetrahydrofolate reductase polymorphisms are related to male infertility in Brazilian men. Genet Test Mol Biomarkers. 2011;15:153–7. doi: 10.1089/gtmb.2010.0128. [DOI] [PubMed] [Google Scholar]
- 28.Montjean D, Benkhalifa M, Dessolle L, Cohen-Bacrie P, Belloc S, et al. Polymorphisms in MTHFR and MTRR genes associated with blood plasma homocysteine concentration and sperm counts. Fertil Steril. 2011;95:635–40. doi: 10.1016/j.fertnstert.2010.08.054. [DOI] [PubMed] [Google Scholar]
- 29.Leclerc P, de Lamirande E, Gagnon C. Cyclic adenosine 3’,5’monophosphate-dependent regulation of protein tyrosine phosphorylation in relation to human sperm capacitation and motility. Biol Reprod. 1996;55:684–92. doi: 10.1095/biolreprod55.3.684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Allelic and genotypic frequencies of 20 SNPs in cases and controls
Different semen parameters across different genotypes in Chinese men