Abstract
MicroRNAs (miRNAs, miRs) are short, non-coding molecules engaged in normal functioning of eukaryotic cells, as negative regulators of gene expression. Since the first discovery of miRNA in the early 1990s, hundreds of different miRNAs and their targets have been identified. A growing number of studies have aimed to search for microRNAs which have a key role in the regulation of insulin signaling and metabolic homeostasis. Recent evidence indicates that dysregulation of miRNA expression is involved in the development of various diseases, including type 2 diabetes mellitus (T2DM), obesity and cardiovascular diseases. This review summarizes the biogenesis of miRNAs and their role in pancreatic β cell biology, insulin signaling and metabolism. We also discuss recent findings of miRNAs associated with metabolic disorders and vascular diabetic complications, their diagnosis and therapeutic value. The PubMed database and published reference lists were searched for articles published between 1990 and 2016 using the following keywords: miRNA, miRNA and pancreas; miRNA and insulin; miRNA and type 2 diabetes mellitus, miRNA and obesity, and miRNA and microvascular or macrovascular diabetic complication. This review indicates that miRNA functioning is significantly different in metabolic diseases than in the normal condition.
Keywords: microRNA, insulin signaling, type 2 diabetes, obesity, vascular diabetic complications
Introduction
In recent years, diseases associated with disturbances in glucose and lipid metabolism, including obesity, diabetes and metabolic syndrome, have become a global health problem. The increasing prevalence of these metabolic disorders is associated with unhealthy lifestyles characterized by limited physical activity and excessive caloric intake. Recent evidence has shown that lifestyle is associated with an inherited epigenetic pattern, which then affects gene expression and the activity of proteins involved in the development of metabolic disease. One of the most crucial components of this pattern is the microRNA (miRNA). These non-coding single-stranded RNAs ranging from 19 to 25 nucleotides in length are involved in the post-transcriptional regulation of gene expression [1, 2]. It is estimated that miRNAs occupy 3% of the human genome and regulate the expression of 30–50% of all protein-coding genes [3, 4]. They are engaged in multiple signaling pathways in both animals and plants, as well as some viruses. miRNAs have been identified to regulate cell proliferation, necrosis, apoptosis, migration and differentiation [2]. The first miRNA, discovered in 1993 by Lee et al. in Caenorhabditis elegans, was found to be involved in normal temporal control of diverse postembryonic developmental events. They isolated the lin-4 gene, which did not produce an mRNA encoding a protein, but produced two short non-coding RNAs. One of them, a 22-nucleotide RNA, contained sequences partially complementary to a repeated sequence element in the 3′ untranslated region (UTR) of lin-14 mRNA. Lee et al. proposed that this complementarity was responsible for the inhibition of the translation of the lin-14 mRNA into the LIN-14 protein [5]. However, the full understanding of how lin-4 controls the larval development of C. elegans was recognized in the early 2000s [6–8]. Since then, miRNAs as the negative regulators of expression of various genes have been identified in many biological processes.
The plausible role of miRNA in fat metabolism was for the first time suggested by Brennecke et al. in Drosophila during tissue growth [9]. Nevertheless, the first study which clearly showed the involvement of miRNAs in the control of metabolic processes in higher mammals was performed by Morin et al. [10]. They showed that metabolic changes during periods of euthermia and hibernation in ground squirrel organs are under the control of several miRNAs, including miR-29a, miR-223, miR-195, miR-486 and miR-378. miR-29a targets mRNAs encoding peroxisome proliferator-activated receptor-gamma coactivator (PGC)-1α and glucose-6-phosphatase-a (G6Pase-a), directly involved in glucose metabolism, whereas miR-223 targets forkhead box protein O1 (FOXO1), a crucial transcription factor of metabolic regulation by insulin signaling. Up-regulated miR-29a coincided with a decreased level of PGC-1α and G6Pase-a and reduced hepatic glucose production. miR-195 is a fatty acid synthase (FAS) regulator, a main enzyme of the fatty acid synthesis pathway. The hibernating liver tissues of I. tridecemlineatus showed overexpression of miR-195 and reduced FAS protein levels. The liver tissue of hibernating squirrels showed a significantly down-regulated insulin growth factor (IGF) pathway, whose components were found to be under the control of miR-486. The expression of miR-486 in liver tissue was also increased. Decreased expression of miR-378 was observed in hibernating skeletal muscle tissue samples. It was demonstrated that miR-378 expression might be associated with expression of MyoD, a key transcription factor of skeletal muscle myogenesis [11]. Interestingly, recent evidence has demonstrated that biochemical adaptation found in hibernating animals is also frequently deregulated in the insulin resistance state of human type 2 diabetes [12]. Emerging studies are focused on investigation of microRNAs that are relevant to diabetic complications [13].
This review provides an overview of microRNAs engaged in insulin signaling and related glucose and fat metabolism as key players in the pathogenesis, diagnosis, and treatment of metabolic diseases.
Methodology
The PubMed database and published reference lists were searched for articles published between 1990 and 2014 in September 2014 with an update in August 2016. To obtain information needed for this review the following keywords were used: miRNA, miRNA and pancreas; miRNA and insulin; miRNA and type 2 diabetes mellitus, miRNA and obesity, and miRNA and microvascular or macrovascular diabetes complication.
Biogenesis of miRNAs
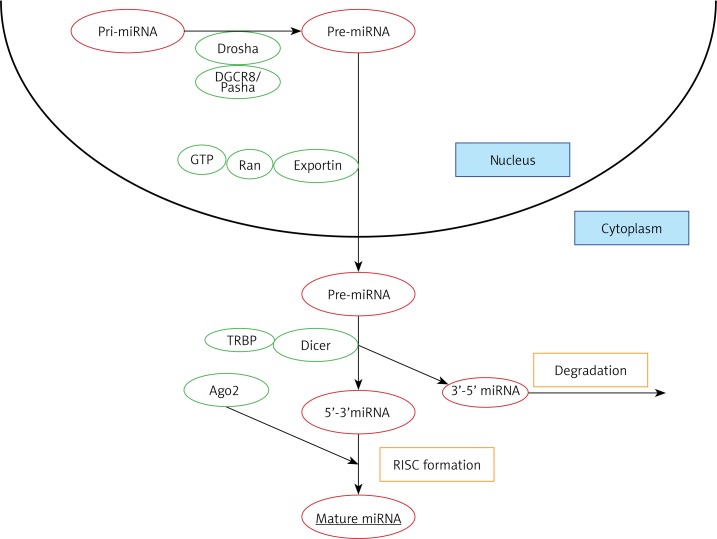
Most miRNAs are formed in canonical pathways comprising several consecutive steps (Figure 1). In the initial stage, RNA forms a long precursor transcript containing stem-loop structures, which is then processed by Drosha, an RNA-se III enzyme, and the DGCR8/Pasha protein. This processing stage leads to the formation of a 70-nucleotide pre-miRNA with a hairpin structure. This pre-miRNA is then transported to the cytoplasm by Exportin-5 using energy from GTP bound to Ran protein and further processed by Dicer, another ribonuclease III-activity enzyme associated with TRBP protein (TAR RNA binding protein in mammals, TRBP) to form a 22-nucleotide RNA duplex. This two-stranded RNA duplex is then separated into two independent strands, one being attached to the RISC complex (RNA-induced silencing complex) containing Argonaute protein, thus being capable of silencing complementary mRNAs. Another strand is usually degraded [14, 15]. Whether the target mRNA will be degraded or its translation merely inhibited depends on the degree of miRNA-mRNA complementarity: the target mRNA will be fully degraded if it is fully complementary to the formed miRNA molecule, otherwise it will only be inhibited to some degree. In both cases, translation will be arrested [16]. Therefore, identification of the specific miRNAs targeting the genes engaged in key metabolic processes may improve our understanding of metabolic disorders and influence their diagnosis and therapy.
Figure 1.
The biogenesis of miRNA. Initially, RNA forms a long precursor transcript containing stem-loop structures (pri-miRNA), which is then processed by Drosha, an RNase III enzyme, and the DGCR8/Pasha protein. This processing leads to the formation of a 70-nucleotide pre-miRNA with a hairpin structure (pre-miRNA). pre-miRNA is then transported to the cytoplasm by Exportin-5 using energy from GTP bound to Ran protein. In the cytoplasm pre-miRNA is processed by Dicer-TRBP complex to form a 22-nucleotide RNA duplex. This two-stranded RNA duplex is separated into two independent strands, one being attached to the RISC complex containing Argonaute protein, thus being capable of silencing complementary mRNAs. The second strand is usually degraded. The formed complex interacts with complementary sequences, thus influencing the gene expression
TRBP – TAR RNA binding protein in mammals, RISC – RNA-induced silencing complex.
The role of microRNAs in β-cell biology
Several miRNAs have been found to be engaged in pancreatic development. Specific miRNAs (miR-124a2, miR-375) were demonstrated to be involved in the regulation of the differentiation of β-cells and formation of pancreatic islets [17, 18]. Microarray analysis of the human fetal pancreatic samples revealed variable expression of 212 miRNAs during developmental processes [19]. In the pancreatic fetal samples from 10 to 22 weeks of gestational age, Rosero et al. identified a group of miRNA with increased expression, containing 4 miRNAs, a group of 35 miRNAs with decreased expression, and a group of miRNAs wherein the level of expression was stable, comprising 173 miRNAs. Furthermore, the authors searched for target genes for miRNAs exhibiting differential expression in the pancreatic samples, based on a previous genome-wide microarray analysis by Sarkar et al. [20]. It was found that one miRNA may target several genes and, conversely, one gene could be targeted by different miRNAs. Importantly, one of the miRNAs’ target, NEUROD1, which is a transcription factor involved in the transition of progenitor cells of the pancreas into endocrine cells, has been identified to be targeted by 5 different miRNAs: miR-17-5p, miR-18a, miR-92, miR-103 and miR-494. Other miRNAs have been correlated with genes involved in endoderm formation or differentiation and maturation of β-cells. This dual role has been attributed to miR-342.
Furthermore, it was demonstrated that micro-RNAs play crucial role in the survival of β-cells in the course of diabetes. Belgardt et al. [21] detected an increase in the level of miR-200 in the pancreatic tissue of diabetic mice. Experimental over-expression of miR-200 was associated with the apoptosis of β-cells and subsequent death of an animal. The authors found that miR-200 regulated apoptosis and stress-response pathways, mainly via the control of expression of Dnajc3 and Xiap – a β-cell chaperon protein and a caspase inhibitor, respectively. Furthermore, it has been observed that miR-200 also regulated the expression of Trp53, which led to the formation of a specific pro-apoptotic signature of gene expression, characteristic for the pancreatic tissue of diabetic mice, evoking a cascade of events resulting in apoptosis [21]. It is speculated that microRNAs may participate in the autoimmunological destruction of β-cells, which is fundamental for type 1 diabetes (T1D) development. Hezova et al. found that regulatory T cells isolated from T1D patients had significantly higher expression of miRNA-146a and lower expression of eight specific miRNAs (20b, 31, 99a, 100, 125b, 151, 335, and 365) as compared to healthy controls. However, the molecular role of these microRNAs has not been recognized in diabetes [22]. A summary of microRNAs involved in β-cell biology is presented in Table I.
Table I.
The role of different microRNAs and their target genes in the development and functioning of the pancreas
| miRNA | Target gene | Function | Reference |
|---|---|---|---|
| miR-124a, miR-342 | Foxa2 | Differentiation of β-cells | [17] |
| miR-375 | Pdx1, NeuroD1 | Formation of pancreatic islets, proliferation and growth | [18] |
| miR-382, miR-432, miR-182 | Isl1 | Embryogenesis of pancreatic islets | [20] |
| miR-17-5p, miR-18a, miR-92, miR-103, miR-494 | NeuroD1 | Transition of pancreatic progenitor cells into endocrine cells | [20] |
| miR-99b, miR-345, miR-494 | Insm1 | Regulation of β-cell development | [20] |
| miR-187, miR-130b, miR-301 | Pax6 | Embryonic development | [20] |
| miR-200 | p58IPK, Xiap, Trp53 | β-cell apoptosis | [21] |
Insulin signaling and microRNAs
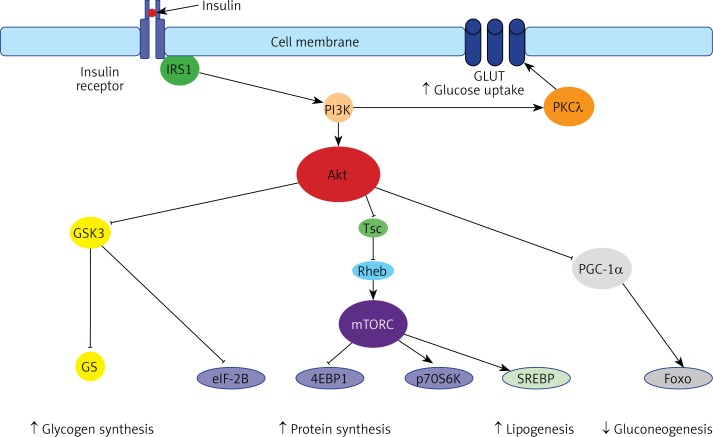
A growing body of evidence shows that miRNAs participate in the physiological processes associated with insulin signaling and energy homeostasis. The signaling cascade initiated by insulin results in transcriptional suppression of gluconeogenesis and the activation of lipogenesis (Figure 2). Insulin binds to the insulin receptor that belongs to the tyrosine kinase family and induces the phosphorylation of insulin-receptor substrates (IRS). Then, IRS recruits phosphoinositide-3-kinase (PI3K), which phosphorylates phosphatidylinositol [4, 5] bisphosphate (PIP2) to produce phosphatidylinositol [3–5] triphosphate (PIP3). PIP3, as a second messenger, recruits kinase protein kinase B (Akt) and 3-phosphoinositide-dependent protein kinase-1 (PDK1) to the cell membrane. PDK1 phosphorylates Akt. Active Akt phosphorylates downstream targets engaged in cell metabolism and growth, including glycogen synthase kinase 3 (GSK3), forkhead box protein O1 (Foxo1), and tuberous sclerosis 2 (TSC2).
Figure 2.
The insulin pathway. The insulin-mediated effect on metabolism is composed of (1) the stimulation of glycogen synthesis (via Akt-mediated inhibition of glycogen synthase kinase-3, GSK3 and glycogen synthase, GS), (2) the stimulation of protein synthesis (via Akt-mediated: inhibition of GSK3 and eIF-2B, as well as stimulation of mammalian target for rapamycin complex, mTORC, which inhibits 4EBP1 and activates p70S6K), (3) the stimulation of lipogenesis (via Akt/mTORC-mediated stimulation of sterol-regulated element binding protein, SREBP), the inhibition of gluconeogenesis (via Akt-mediated inhibition of peroxisome proliferator-activated receptor gamma coactivator 1-a, PCG-1a, which coactivates forkhead box O1 (Foxo1) transcription factor), and protein kinase C
GSK3, a serine/threonine protein kinase, phosphorylates and inhibits its downstream targets, including glycogen synthase and the eukaryotic initiation factor 2B (eIF2B). In the insulin stimulated pathway, Akt phosphorylates and inhibits GSK3, leading to the dephosphorylation of glycogen synthase and eIF2B and stimulation of glycogen and protein synthesis [23].
FOXO1 is a transcription factor that in an unphosphorylated state increases the rate of transcription of glucose 6-phosphatase (G6P), phosphoenolpyruvate carboxykinase, and microsomal triglyceride transfer protein (MTP) and thereby controls glucose and triglyceride metabolism [24]. However, when Foxo1 is phosphorylated by Akt, it is translocated and degraded in the cytoplasm, and in consequence the rate of hepatic glucose production decreases.
TSC2 phosphorylated by Akt ceases to inhibit the Ras homolog enriched in brain (Rheb) protein that is required for activation of mammalian target of rapamycin (mTOR) activation [25]. The mTOR functions in two distinct multiprotein complexes, TORC1 and TORC2. Both complexes are important regulators of cell growth. mTORC1, consisting of mTOR, raptor, and mLST8, plays a crucial role in the regulation of protein anabolism, nucleotide biosynthesis, lipogenesis, glycolysis, and autophagy. mTORC2, composed of mTOR, rictor, SIN1, and mLST8, regulates lipogenesis, glucose metabolism, the actin cytoskeleton, and apoptosis [26]. Among key downstream targets of mTORC involved in glucose and lipid metabolism are glucose transporters, rate-limiting glycolytic enzymes, and sterol regulatory element binding protein-1c (SREBP-1c) [25, 27].
It is documented that the PI3K/Akt/mTOR pathway plays a significant role in metabolic diseases such as diabetes and cancer development [28]. Genetic murine models provided evidence that the mTOR pathway is crucial for pancreatic β-cell proliferation. Constitutive activation of mTORC1 promoted an increase of pancreatic β-cell number and size, and a decrease of blood glucose levels and hyperinsulinemia. Conversely, the chronic inhibition of mTORC1 and/or mTORC2 reduced β-cell mass and function, evoked severe insulin resistance and glucose intolerance, and promoted hepatic glucose production [28].
Wang et al. found that miR-7a is expressed in adult pancreatic islets and targets multiple components of the mTOR signaling pathway. They demonstrated that the inhibition of miR-7 activated mTOR signaling and promoted adult β-cell proliferation in both mouse and human primary islets, suggesting that miR-7 is a negative regulator of adult β-cell proliferation [29].
Karolina et al. demonstrated that the insulin pathway is under control of microRNAs. The circulating level of miR-144 was significantly increased in patients with diabetes. miR-144 was found to target insulin receptor substrate 1 (IRS1) [30]. Bork-Jensen et al. identified downregulated miR-15b and miR-16 in twins with overt type 2 diabetes, and they suggested that they regulate the key insulin signaling components IRS1 and PI3KR1 [31].
Under physiological conditions, insulin secreted into the blood acts on the peripheral tissues, what causes glucose uptake by various isoforms of glucose transporters (GLUTs). Nonetheless, diabetes is accompanied by insulin resistance, the inability of tissues to respond to insulin action. In this case, the mechanism depicted in Figure 2 is disturbed. Several miRNAs have been found to be differentially expressed in tissues taken from T2DM rats exhibiting an insulin-resistant profile: for example, miR-222 and miR-27a were found to be upregulated in adipose tissue, and miR-125a, miR-195 and miR-103 were upregulated in the liver, while miR-10b was significantly downregulated in muscle tissue [32, 33].
The diverse expression profile demonstrated by the level of miRNAs in various tissues may potentially be associated with the development of insulin resistance, and this condition has been strongly linked to dyslipidemia and obesity. Although insulin inhibits lipolysis under physiological conditions, thus promoting the storage of triglycerides in adipocytes, these tissues become resistant to insulin in obese individuals and enter a hyper-lipolytic state. Non-esterified fatty acids (NEFA) undergo greater mobilization, leading to their accumulation in plasma; this is accompanied by ectopic deposition of triglycerides in the muscles and the liver, impairing their metabolism and leading to overproduction of glucose. In addition, lipids accumulate in non-adipose tissue, including the β-cell mass, resulting in functional impairments and apoptosis. As a consequence, persistently elevated glucose levels demonstrate toxic effects with serious, metabolism-impairing consequences.
The oxidation of fatty acids in the liver results in the enhancement of oxidative stress, defined as an imbalance between the amount of reactive oxygen species (ROS) and the ability of the organism to detoxify ROS or to repair oxidative damage. Oxidative stress is also promoted by excessive production of pro-inflammatory cytokines in hypertrophic adipose tissue (Figure 3). Adipose tissue serves as an endocrine organ, secreting a number of hormones and cytokines, including adiponectin, leptin and resistin, which are crucial for glucose and lipid metabolism. When adipose tissue becomes hypertrophic and insulin resistant, its endocrine function is altered, resulting in abnormal levels of adipocyte-derived cytokines being observed [34–36]. Several miRNAs are believed to be involved in the process of adipogenesis, including miR-143 and miR-103. Both miRNAs were found to significantly accelerate the process of adipocyte differentiation, thus further increasing triglyceride accumulation (Figure 3). Importantly, an association has been established between the level of miR-143 and vital glucose-metabolism-related genes, including GLUT and the peroxisome proliferator-activated receptor γ2 (PPARγ2) [37, 38]. Ahn et al. found that miR-146b is involved in the regulation of sirtuin-1 (SIRT1) expression in the adipose tissues of diet-induced and genetically obese mice [39]. SIRT1 was found to be involved in lipid and glucose metabolism, inflammation, and adipogenesis [40]. SIRT1 deacetylates and affects activity of specific transcription factors, including Foxo1, PGC1α, and NF-κB [41–43]. Picard et al. demonstrated that SIRT1 inhibits adipogenesis by docking with PPARγ cofactors, nuclear receptor co-repressor (NCoR) and silencing mediator of retinoid and thyroid hormone receptors (SMRT) [44]. Significant upregulation of miR-146b and decrease of SIRT1 were found in obese mice. Ahn et al. demonstrated that miR-146b downregulated SIRT1 expression, leading to a decrease of Foxo1 deacetylation. In consequence, Foxo1-mediated inhibition of PPARγ transcription was diminished and adipocyte differentiation was stimulated. A knockdown of miR-146b by LNA-miR-146b antagomir evoked an increase of SIRT1, and a reduction of body weight and adiposity [39].
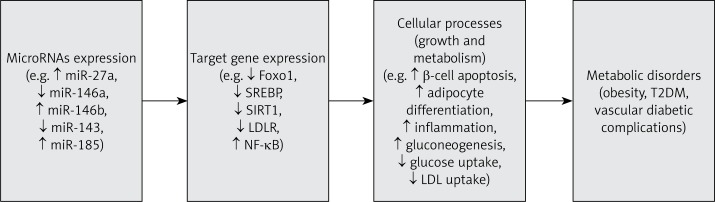
Figure 3.
The dysregulated microRNAs and metabolic disorders. Dysregulation of microRNA expression leads to significant changes in the expression of their target genes. The consequences of these changes are disturbances in cellular processes, including growth and metabolism, which result in the development of obesity, diabetes and its complications
A comprehensive set of experiments by Fu et al. showed that miR-26a is a key regulator of hepatic metabolism and insulin signaling [45]. An examination of liver samples collected from patients undergoing abdominal surgery demonstrated lower levels of miR-26a expression in overweight individuals in comparison with their lean counterparts. Fu et al. also reported a decreased level of miR-26a in obese mice and that inducible transgenic overexpression of miR-26a in mice fed with a high-fat diet decreased insulin resistance, gluconeogenesis and fatty acid synthesis. In contrast, the silencing of miR-26a in mice fed with a conventional diet impaired insulin sensitivity, enhanced glucose production, and increased fatty acid synthesis [45].
Kornfeld et al. note that miR-802, which targets Hnf1b (hepatocyte nuclear factor-1-β, TCF-2), plays an important role in hepatic glucose metabolism and insulin signaling [46]. Increased expression of miR-802 was found in the liver tissue of obese mice and humans. Using transgenic mice, the authors demonstrated that overexpression of miR-802 evoked the silencing of Hnf1b in the liver, resulting in impaired glucose tolerance, attenuated insulin sensitivity and elevated gluconeogenesis. Conversely, the silencing of miR-802 expression was associated with hepatic Hnf1b protein expression, as well as improved glucose tolerance and insulin action [46].
In a study performed on transgenic mice with inducible expression of the Let-7 family, Frost and Olson found that the Let-7 family of microRNAs regulates the glucose metabolism in multiple organs [47]. Mice overexpressing Let-7 were smaller and had decreased fat mass, and both global and pancreas-specific overexpression of Let-7 in mice resulted in impaired glucose tolerance and reduced glucose-induced pancreatic insulin secretion. They also note that global silencing of the Let-7 family in mice fed with a high-fat diet improved glucose tolerance by the reduction of insulin resistance in liver and muscle. Additionally, silencing of the Let-7 family in diet-induced obese mice increased lean and muscle mass, but not fat mass, and prevented ectopic fat deposition in the liver [47].
Finally, Trajkovski et al. reported the expression of miR-103 and miR-107 to be increased in the liver of obese mice and humans with insulin resistance and hepatic steatosis [48]. Global silencing of miR-103 and miR-107 increased insulin signaling in both liver and adipose tissue in obese (ob/ob) and diet-induced obese (DOI) mice. However, they demonstrated that hepatic silencing of miR-103 and miR-107 was not sufficient to reverse the metabolic abnormalities in both the obese ob/ob and DOI mice, but in adipocytes, the silencing of miR-103 and miR-107 improved insulin sensitivity. In contrast, miR-103 or miR-107 gain-of-function was sufficient to induce impaired glucose homeostasis in both hepatic and adipocyte tissue. Both miR-103 and miR-107 were found to target caveolin-1 (Cav1), which is a critical regulator of the insulin receptor. They demonstrated that silencing of miR-103 and miR-107 in adipocytes of ob/ob mice increased Cav1 expression, leading to the activation of insulin signaling and enhanced insulin-stimulated glucose uptake [48]. A summary of the dysregulation of microRNA expression leading to metabolic disorders is presented in Figure 3.
miRNAs as potential biomarkers of metabolic disorders
Interestingly, some miRNAs have been identified as potential biomarkers for distinguishing obesity from T2DM [49, 50]. In an analysis of patient sera, Wu et al. found the expression of the deregulated miRNAs, including miR-152, miR-17, miR-138, miR-593, miR-150, miR-205, miR-376a, miR-432-5p and miR-500a, to differ between a group of 25 healthy control subjects and groups of 25 patients with T2DM, 25 obese (OB) and 25 OB + T2DM patients. An analysis of the area under the curve (AUC) values based on receiver operating characteristic curves for each miRNA revealed that miR-152, miR-17, miR-593 and miR-138 alone can be used to distinguish patients with OB from those with T2DM, OB + T2DM or healthy subjects [49]. Similarly, Pescador et al. performed an analysis of miRNA expression profiles in serum samples taken from 13 T2DM, 20 OB, 16 OB + T2DM and 20 healthy control subjects. They identified miR-138, miR-15b and miR-376a as potential predictive biomarkers in obesity, and found that miR-138 and miR-376a can be used to distinguish OB from healthy controls, T2DM, and OB + T2DM. Moreover, the combination of miR-503 and miR-138 can differentiate T2DM from OB + T2DM patients [50].
A study by Klöting et al. investigated whether miRNA expression is fat-deposit specific, and whether it is associated with obesity and the glucose metabolism [51]. Paired samples of abdominal subcutaneous (SC) and intra-abdominal omental adipose tissue were obtained from nine individuals with normal glucose tolerance (NGT) and from six newly-diagnosed individuals with T2DM. An expression analysis of 155 miRNAs revealed that 106 miRNAs were detectable in both omental and SC adipose tissue samples, but none were exclusively expressed in either type of fat deposit. However, the expression pattern differed between the NGT and T2DM patients: for omental fat, significantly higher expression of miR-17-5p, miR-132 and miR-134 was observed among the NGT subjects than among those with T2DM, whereas the opposite was seen for miR-181a. In the case of SC fat, the expression of miR-27a, miR-30e, miR-140, miR-155 and miR-210 was significantly higher, and expression of miR-147 and miR-197 lower, in the NGT than the T2DM group. The analysis of the pattern of miRNA expression and metabolic parameters revealed that omental fat area was negatively associated with the expression of miR-17-5p and miR-132. In SC fat, miR-34a and miR-145 expression significantly correlated with BMI, miR-145 expression significantly correlated with HDL cholesterol level and 2-hour oral glucose tolerance test (OGTT) glucose level, miR-197 expression correlated with fasting plasma glucose and glucose infusion rate during the clamp, and miR-95 expression was related to serum concentrations of adiponectin, CrP and IL-6. Both types of fat deposit demonstrated a significant association between miRNA-95 expression and adipocyte size. The number of macrophages infiltrating the fat depot was found to be significantly associated with miRNA-132 expression for omental fat, and miRNA-26b and miRNA-155 expression for SC fat [51].
Obesity and T2DM are known to be characterized by low-grade chronic inflammation caused by increased secretion of pro-inflammatory cytokines and adipokines by the macrophages and adipocytes present in adipose tissue. BaldeÓn et al. examined the expression of miR-146a and miR-155a, key regulators of inflammation, in serum samples from 56 Ecuadorian T2DM patients and 40 non-diabetic controls, to determine whether they would be associated with glucose control, dyslipidemia, obesity and the serum level of a number of inflammatory mediators: TNF-α, IL-1β, IL-6, NFG, HGF, PAI, resistin, CCL2, adiponectin, leptin, IL-8, and CCL4. It was found that the serum level of miR-146a, but not miR-155, was significantly lower in T2DM patients than the non-diabetic controls. A positive correlation was found between miR-146a and serum PAI level, and the serum miR-155 level correlated significantly with the levels of leptin and IL-8 [52]. Similarly, in a study of 23 morbidly obese, 30 obese, and 30 normal or overweight (non-obese) subjects by Kilic et al., the levels of miR-143 and miR-223, two other key regulators of adipocyte differentiation and inflammation, were found to be significantly lower in blood from the morbidly obese and obese patients than non-obese subjects. However, no correlation was found between these miRNAs and HOMA-IR, BMI, WHR or hsCRP levels [53].
To identify prediabetes miRNA biomarkers, PÁrrizas et al. compared the sera isolated from 29 control subjects with those of 22 pre-diabetic individuals demonstrating impaired fasting glucose, 21 pre-diabetic subjects with impaired glucose tolerance and 20 newly diagnosed T2DM patients. It was found that the pre-diabetic phenotype could be identified based on miR-192 and miR-193b level, and that the amounts of the two miRNAs correlated with glucose level, HOMA-IR, triglyceride level and fatty liver index. A similar experiment compared the expression of serum miR-192 and miR-193b between 12 control and six pre-diabetic individuals (3 impaired fasting glucose and 3 impaired glucose tolerance patients) after a 16-week exercise program, as well as a comparison of high-fat diet (HFD)-induced glucose-intolerant obese mice and controls after a 5-week intervention consisting of caloric restriction and exercise. After the former intervention, serum miR-192 and miR-193b expression significantly decreased in pre-diabetic patients, reaching the levels in the control subjects. The authors note that the 5-week intervention caused a significant decrease in serum miR-192 and miR-193b in glucose-intolerant HFD mice, and this was associated with normalization of body weight, glucose tolerance and triglyceride level [54].
Taken together, although microRNAs correlate with some metabolic parameters, their use in the diagnosis of T2DM and obesity requires further clinical studies and experimental validation. It is because we are at the beginning of our way to understand how microRNAs regulate glucose and fat metabolism in various tissues and their involvement in the pathogenesis of metabolic disorders (Table II).
Table II.
miRNAs, their targets and related disorders
| miR | Target gene | Target tissue | Pathway/function | Disorder | References |
|---|---|---|---|---|---|
| miR-7a | mTOR pathway components (p70S6K, eIF4E) | Pancreatic islets | mTOR pathway/β-cell proliferation | Diabetes | [29] |
| miR-144 | IRS1 | Blood | Insulin pathway | T2DM | [30] |
| miR-15b miR-16 | IRS1, PI3KRI | Skeletal muscle | Insulin pathway | T2DM | [31] |
| miR-143 | GLUT, PPARγ2 | Adipose tissue | Insulin pathway | Obesity | [37, 38] |
| miR-146b | SIRT1 | Adipose tissue | Deacetylation of Foxo1, PGC1α, NF-κB | Obesity | [39] |
| miR-802 | Hnf1b (TCF2) | Liver | Insulin pathway | Obesity | [46] |
| miR-103, miR-107 | Caveolin-1 (Cav1) | Adipose tissue, liver | Insulin pathway | Obesity | [48] |
| miR-152 | PTEN, ROCK1, PPARγC1A | Serum | Insulin pathway, Wnt pathway, adipocytokine pathway | Obesity | [49] |
| miR-17 | SPRY4, JAK1, CCND1 | Serum | Jak-STAT pathway | Obesity | [49] |
| miR-138 | SHP | Serum | PPARγ2 pathway | Obesity | [49, 50] |
| Nerogenin3 (NGN3) | Serum | Obesity | [50] | ||
| miR-376a | p85α, PIK3R1 | Serum | Apoptotic pathway | Obesity | [50] |
| miR-192 | Smad-interacting protein 1 (SIP1) | Kidney | Collagen deposition | Diabetic nephropathy | [60] |
| miR-200b | VEGF | Retina | Angiogenesis pathway | Diabetic retinopathy | [61] |
| miR-146a | NF-κB | Sciatic neural tissue | Inflammatory pathway | Diabetic neuropathy | [64] |
| miR-24 | HMGB1 | Vascular smooth muscle cells | Proinflammatory signaling pathway | Diabetic atherosclerosis | [68] |
| miR-27a | LDLR | Blood | Cholesterol homeostasis | Diabetic atherosclerosis | [68] |
| miR-185 | SERBP-2 | Hepatocytes | Cholesterol biosynthesis, LDL uptake | Diabetic atherosclerosis | [70] |
miRNAs in vascular diabetic complications
The combination of long-term diabetes and uncontrolled blood glucose levels significantly increases the risk of serious life-threatening microvascular complications, such as nephropathy, retinopathy and neuropathy, as well as macrovascular complications such as heart attack, stroke and insufficient blood flow to the legs. A growing body of evidence shows that the development of diabetic vascular diseases is associated with excessive proliferation and migration of vascular smooth muscle cells exposed to chronic hyperglycemia [55].
Recent evidence indicates that several miRNAs may be involved in the development of diabetic angiopathies. Efforts have been made to determine whether specific miRNAs may serve as a diagnostic tool that could help elucidate the risk for the development of such complications in the vascular system of diabetic subjects. By using a diabetic rat model, it has been shown that several miRNAs (miR-29 family, miR-24) are differentially expressed during the course of diabetes [56, 57], while Zampetaki et al. report that alterations in the expression of several miRNAs (miR-15a, miR-28-3p, miR-126, miR-223, miR-320) preceded the onset of diabetes [58]. Furthermore, as this specific “miR-diabetic signature” was shown to be accompanied by subclinical artery disease, the authors suggest that this change may be a potential marker for identifying diabetic patients with a high risk of angiopathy.
Currently, among vascular diabetic complications, diabetic nephropathy has been associated with the largest number of studied miRNAs (miR-21, miR-93, miR-377, miR-216a, miR-192, miR-29), with miR-192 being the most intensively studied in this condition [59]. Kato et al. report a link between increased expression of miR-192, reduced expression of Smad-interacting protein 1 (SIP1), a transcription factor engaged in the regulation of collagen gene expression, and enhanced deposition of collagen in diabetic rat and mouse models [60]. In vitro studies showed that increased miR-630 expression promoted cell proliferation, apoptosis, migration and invasion of renal cancer cells [61].
It is well documented that the development of diabetic retinopathy (DR) is associated with increased expression of vascular endothelial growth factor (VEGF). As VEGF mRNA is a known potential target for miR-200b, McArthur et al. examined its potential role in the development of DR. They found that diabetic rats demonstrated decreased expression of retinal miR-200b accompanied by over-expression of VEGF [62]. Furthermore, endothelial cells transfected with a mimic of miR-200b exhibited a decline in VEGF level. These findings may serve as the initial steps in the development of miRNA-based therapy for DR.
To date, little is known about whether micro-RNAs are involved in the development of diabetic neuropathy. Wang et al. identified miR-1 and miR-133a as potential regulators of peripheral nerve homeostasis [63]. A recent study performed on diabetic rats identified a regulatory pathway, inclusive miR-146-a and NF-κB, that may be involved in the development of diabetic neuropathy [64]. Yousefzadeh et al. observed that a 2-month intervention of uncontrolled diabetes was associated with significantly increased levels of miR-146a, NF-κB and inflammation-related mediators (TNF-α, IL-6, IL-1β), but reduced expression of TRAF6 (TNF-receptor-associated factor 6) in the sciatic neural tissue of rats.
Regarding macrovascular diabetes-induced complications, recently, Yang et al. summarized the data on cardiac-specific microRNAs that are involved in myocardial fibrosis of the left atrium. miR-1, -21, -25, -29, -29a, -132, -133, -208, and -590 are able to target genes associated with fibrosis and connexin involved in the pathophysiology of atherosclerosis [65]. miR-133, a molecule exhibiting high cardiac expression, was found to be involved in the regulation of muscular tissue formation [59]. Increased expression of miR-133 was associated with conduction disturbances in the heart [66]. However, down-regulation of miR-133 has been associated with cardiac hypertrophy [67], a process found to be mediated by genes and proteins involved in heart formation: RhoA, a GDP-GTP exchange protein, Cdc42, a signal transduction kinase, hypertrophy-related proteins, and Nelf-A/WHSC2, a nuclear factor engaged in cardiogenesis, all of which are regulated by miR-133.
Yang et al. provided evidence that miR-24 was implicated in diabetes-associated atherosclerosis. Vascular smooth muscle cells (VSMC) cultured in high glucose concentration (30 mM) showed a decreased expression level of miR-24 and an increase of TNF-α and IL-6 production. MiR-24 targets high mobility group box-1 (HMGB1), which is involved in proinflammatory signaling. They demonstrated that upregulation of miR-24 reduced proliferation and migration of VSMC as a result of suppressing HMGB1/NF-κB associated production of proinflammatory cytokines [68]. miR-27a was found to decrease the expression of low-density lipoprotein receptor (LDLR), responsible for LDL removal from the circulation, by Alvarez et al. [69]. They showed that insulin induced an increase in the levels of miR-27a and a related decrease in LDLR level in HepG2 cells. However, knockdown of miR-27a evoked an increase in the level of LDLR [69]. These results suggest that miR-27a may be a therapeutic target for the prevention of atherosclerosis, especially in patients with metabolic syndrome and type 2 diabetes patients, who were found to have an increased circulating 27a level [70]. Yang et al. found that atherosclerosis may be associated with overexpression of miR-185. They showed that miR-185 repressed the sterol response element binding protein (SREBP)-2 transcription in HepG2 cells. SREBP2 is a transcription factor that induces the expression of genes involved in de novo cholesterol biosynthesis and low-density lipoprotein (LDL) uptake. Moreover, they detected that mice fed with a high-fat diet and individuals with high cholesterol had a significantly increased miR-185 level [71] (Table II).
microRNAs as a target of therapy
Growing in vivo evidence indicates that manipulation of miRNA levels may provide beneficial therapeutic effects. Recently, several strategies have been developed to inhibit or mimic therapeutic miRNA [72]. The most widely used approach to inhibit miRNA is by using anti-miRs. As modified antisense oligonucleotides, anti-miRs reduce the expression of endogenous miRNA, which results in an increase of target gene expression. In order to increase the in vivo stability of anti-miRs, chemical modifications have been used, including 2′-O-methyl group-modified oligonucleotides and LNA-modified oligonucleotides [73]. To re-express the miRNA of interest, synthetic RNA duplexes or tissue-specific viral vectors delivering target miRNA have been employed [74]. MicroRNAs as a target of therapy have already been tested for use in cancer and heart failure therapy, with the most advanced, miR-122, currently in phase 2 clinical trials to treat hepatitis C virus [75].
Regarding metabolic disease, several micro-RNAs have been proposed as candidates for potential therapy. miR-33 is known to be engaged in the regulation of cholesterol biosynthesis. Mice treated with anti-miR-33 oligonucleotides exhibited increased cholesterol efflux and elevated HDL cholesterol levels [76, 77]. Further research confirmed that anti-miR-33 treatment may be atheroprotective, as mice with established atherosclerotic plaques showed lower plaque size and lipid content, elevated plaque stability, and decreased expression of inflammation-related genes following anti-miR therapy [78]. Interestingly, it is suggested that anti-miR-33 treatment as an adjunct to statin therapy may be beneficial in reducing primary and secondary cardiovascular events [79]. Furthermore, treatment targeted at miR-103/107 may improve glucose homeostasis and insulin sensitivity. It was found that the level of this miRNA is elevated in obese mice. Experimental silencing of miR-103/107 with an antisense oligonucleotide was found to elevate the level of caveolin-1, resulting in a reduction of adipocyte size, stabilization of the insulin receptor, and elevated insulin signaling and glucose uptake [48].
It is also worth noting that microRNAs are regulated by various environmental factors, including physical activity and different components of the diet. Results presented in a systematic review performed by Momtazi et al. suggested that curcumin modulated the expression of several miRs in brain, ocular, renal, and liver diseases and therefore may protect against the onset of noncancerous diseases associated with these organs [80].
Conclusions and future remarks
A number of studies have indicated that microRNAs are involved in the control of metabolic processes in various tissues, mainly by the regulation of genes encoding proteins responsible for metabolism. Dysregulation of microRNAs was found to be involved in the development of obesity, T2DM, and diabetic complications.
It was established that expression of micro-RNAs may be influenced by the modification of lifestyle. Several microRNAs were found to be correlated with metabolic parameters, and their levels were found to change after exercise intervention. Further elucidation of the involvement of specific miRNAs in the pathogenesis of metabolic disorders can result in the identification of potential therapeutic targets. Attempts are underway to incorporate specific miRNAs in diagnostic procedures. Moreover, as microRNAs may play a role in the development of life-threatening diabetic complications, further research may well prove valuable in reducing the risk of their occurrence.
Acknowledgments
This paper was partially supported by a grant from the Medical University of Lodz (no. 503/0-077-09/503-01-002), grant no. 2015/17/B/NZ7/03019 from the Polish National Science Centre and grant from Polish Society of Metabolic Disease.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.O’Driscoll L. The emerging world of microRNA. Anticancer Res. 2006;26:4271–8. [PubMed] [Google Scholar]
- 4.Huppi K, Volfovsky N, Mackiewicz M, et al. MicroRNAs and genomic instability. Sem Cancer Biol. 2007;17:65–73. doi: 10.1016/j.semcancer.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 6.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 7.Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev. 1996;10:3041–50. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- 8.Reinhart BJ, Ruvkun G. Isoform-specific mutations in the Caenorhabditis elegans heterochronic gene lin-14 affect stage-specific patterning. Genetics. 2001;157:199–209. doi: 10.1093/genetics/157.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 10.Morin P, Jr, Dubuc A, Storey KB. Differential expression of microRNA species in organs of hibernating ground squirrels: a role in translational suppression during torpor. Biochim Biophys Acta. 2008;1779:628–33. doi: 10.1016/j.bbagrm.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Gagan J, Dey BK, Layer R, Yan Z, Dutta A. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J Biol Chem. 2011;286:19431–8. doi: 10.1074/jbc.M111.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu CW, Biggar KK, Storey KB. Biochemical adaptations of mammalian hibernation: exploring squirrels as a perspective model for naturally induced reversible insulin resistance. Braz J Med Biol Res. 2013;46:1–13. doi: 10.1590/1414-431X20122388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kantharidis P, Wang B, Carew RM, Lan HY. Diabetes complications: the microRNA perspective. Diabetes. 2011;60:1832–7. doi: 10.2337/db11-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–32. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian micro-RNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baroukh N, Ravier MA, Loder MK, et al. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J Biol Chem. 2007;282:19575–88. doi: 10.1074/jbc.M611841200. [DOI] [PubMed] [Google Scholar]
- 18.Keller DM, Clark EA, Goodman RH. Regulation of micro-RNA-375 by cAMP in pancreatic beta-cells. Mol Endocrinol. 2012;26:989–99. doi: 10.1210/me.2011-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosero S, Bravo-Egana V, Jiang Z, et al. MicroRNA signature of the human developing pancreas. BMC Genomics. 2010;11:509–18. doi: 10.1186/1471-2164-11-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkar SA, Kobberup S, Wong R, et al. Global gene expression profiling and histochemical analysis of the developing human fetal pancreas. Diabetologia. 2008;51:285–97. doi: 10.1007/s00125-007-0880-0. [DOI] [PubMed] [Google Scholar]
- 21.Belgardt BF, Ahmed K, Spranger M, et al. The micro-RNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med. 2015;21:619–27. doi: 10.1038/nm.3862. [DOI] [PubMed] [Google Scholar]
- 22.Hezova R, Slaby O, Faltejskova P, et al. microRNA-342, microRNA-191 and microRNA-510 are differentially expressed in T regulatory cells of type 1 diabetic patients. Cell Immunol. 2010;260:70–4. doi: 10.1016/j.cellimm.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 23.Cohen P, Frame S. The renaissance of GSK3. Nature Rev Mol Cell Biol. 2001;2:769–76. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 24.Kamagate A, Qu S, Perdomo G, et al. FoxO1 mediates insulin-dependent regulation of hepatic VLDL production in mice. J Clin Invest. 2008;118:2347–64. doi: 10.1172/JCI32914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X, So JS, Park JG, Lee AH. Transcriptional control of hepatic lipid metabolism by SREBP and ChREBP. Semin Liver Dis. 2013;33:301–11. doi: 10.1055/s-0033-1358523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Betz C, Hall MN. Where is mTOR and what is it doing there? JCB. 2013;203:563–74. doi: 10.1083/jcb.201306041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finlay DK, Rosenzweig E, Sinclair LV, et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med. 2012;209:2441–53. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Liu J, Liu C, Naji A, Stoffers DA. MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic beta-cells. Diabetes. 2013;62:887–95. doi: 10.2337/db12-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karolina DS, Armugam A, Tavintharan S, et al. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS One. 2011;6:e22839. doi: 10.1371/journal.pone.0022839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bork-Jensen J, Scheele C, Christophersen DV, et al. Glucose tolerance is associated with differential expression of microRNAs in skeletal muscle: results from studies of twins with and without type 2 diabetes. Diabetologia. 2015;58:363–73. doi: 10.1007/s00125-014-3434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrera BM, Lockstone HE, Taylor JM, et al. MicroRNA-125a is over-expressed in insulin target tissues in a spontaneous rat model of type 2 diabetes. BMC Med Genom. 2009;2:54–65. doi: 10.1186/1755-8794-2-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrera BM, Lockstone HE, Taylor JM, et al. Global microRNAexpression profiles in insulin target tissues in a spontaneous rat model of type 2 diabetes. Diabetologia. 2010;53:1099–109. doi: 10.1007/s00125-010-1667-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis GF, Carpentier A, Adeli K, Giacca A. Disordered fat storage and mobilization in the pathogenesis of insulin resistance and type 2 diabetes. Endocr Rev. 2002;23:201–29. doi: 10.1210/edrv.23.2.0461. [DOI] [PubMed] [Google Scholar]
- 35.Raz I, Eldor R, Cernea S, Shafrir E. Diabetes: insulin resistance and derangements in lipid metabolism. Cure through intervention in fat transport and storage. Diabetes Metab Res Rev. 2005;21:3–14. doi: 10.1002/dmrr.493. [DOI] [PubMed] [Google Scholar]
- 36.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–53. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–5. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 38.Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050–7. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahn J, Lee H, Jung CH, Jeon TI, Ha TY. MicroRNA-146b promotes adipogenesis by suppressing the SIRT1-FOXO1 cascade. EMBO Mol Med. 2013;5:1602–12. doi: 10.1002/emmm.201302647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silva JP, Wahlestedt C. Role of sirtuin 1 in metabolic regulation. Drug Discov Today. 2010;15:781–91. doi: 10.1016/j.drudis.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Daitoku H, Hatta M, Matsuzaki H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:10042–7. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–60. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 43.Stafeev IS, Menshikov MY, Tsokolaeva ZI, Shestakova MV, Parfyonova YV. Molecular mechanisms of latent inflammation in metabolic syndrome. Possible role of sirtuins and peroxisome proliferator-activated receptor type gamma. Biochemistry (Mosc) 2015;80:1217–26. doi: 10.1134/S0006297915100028. [DOI] [PubMed] [Google Scholar]
- 44.Picard F, Kurtev M, Chung N, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–6. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu X, Dong B, Tian Y, et al. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J Clin Invest. 2015;125:2497–509. doi: 10.1172/JCI75438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kornfeld JW, Baitzel C, Könner AC, et al. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature. 2013;494:111–5. doi: 10.1038/nature11793. [DOI] [PubMed] [Google Scholar]
- 47.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A. 2011;108:21075–80. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trajkovski M, Hausser J, Soutschek J, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–53. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 49.Wu L, Dai X, Zhan J, et al. Profiling peripheral microRNAs in obesity and type 2 diabetes mellitus. APMIS. 2015;123:580–5. doi: 10.1111/apm.12389. [DOI] [PubMed] [Google Scholar]
- 50.Pescador N, Pérez-Barba M, Ibarra JM, CorbatÓn A, MartÍnez-Larrad MT, Serrano-RÍos M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS One. 2013;8:e77251. doi: 10.1371/journal.pone.0077251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klöting N, Berthold S, Kovacs P, et al. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS One. 2009;4:e4699. doi: 10.1371/journal.pone.0004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.BaldeÓn RL, Weigelt K, de Wit H, et al. Decreased serum level of miR-146a as sign of chronic inflammation in type 2 diabetic patients. PLoS One. 2014;9:e115209. doi: 10.1371/journal.pone.0115209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kilic ID, Dodurga Y, Uludag B, et al. MicroRNA-143 and -223 in obesity. Gene. 2015;560:140–2. doi: 10.1016/j.gene.2015.01.048. [DOI] [PubMed] [Google Scholar]
- 54.PÁrrizas M, Brugnara L, Esteban Y, et al. Circulating miR-192 and miR-193b are markers of prediabetes and are modulated by an exercise intervention. J Clin Endocrinol Metab. 2015;100:E407–15. doi: 10.1210/jc.2014-2574. [DOI] [PubMed] [Google Scholar]
- 55.Wu WY, Yan H, Wang XB, et al. Sodium tanshinone IIA silate inhibits high glucose-induced vascular smooth muscle cell proliferation and migration through activation of AMP-activated protein kinase. PLoS One. 2014;9:e94957. doi: 10.1371/journal.pone.0094957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol Endocrinol. 2007;21:2785–94. doi: 10.1210/me.2007-0167. [DOI] [PubMed] [Google Scholar]
- 57.Huang B, Qin W, Zhao B, et al. MicroRNA expression profiling in diabetic GK rat model. Acta Biochim Biophys Sin (Shanghai) 2009;41:472–7. doi: 10.1093/abbs/gmp035. [DOI] [PubMed] [Google Scholar]
- 58.Zampetaki A, Kiechl S, Drozdov I, et al. Plasma micro-RNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–7. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 59.Kantharidis P, Wang B, Carew RM, Lan HY. Diabetes complications: the microRNA perspective. Diabetes. 2011;60:1832–7. doi: 10.2337/db11-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kato M, Zhang J, Wang M, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA. 2007;104:3432–7. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao JJ, Chen PJ, Duan RQ, Li KJ, Wang YZ, Li Y. miR-630 functions as a tumor oncogene in renal cell carcinoma. Arch Med Sci. 2016;12:473–8. doi: 10.5114/aoms.2016.59918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McArthur K, Feng B, Wu Y, Chen S, Chakrabarti S. MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes. 2011;60:1314–23. doi: 10.2337/db10-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Q, Wang Y, Minto AW, et al. MicroRNA-377 is upregulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 2008;22:4126–35. doi: 10.1096/fj.08-112326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yousefzadeh N, Alipour MR, Soufi FG. Deregulation of NF-kappaB-miR-146a negative feedback loop may be involved in the pathogenesis of diabetic neuropathy. J Physiol Biochem. 2015;71:51–8. doi: 10.1007/s13105-014-0378-4. [DOI] [PubMed] [Google Scholar]
- 65.Yang H, Wu C, Xiao Y, Zhou S. Connexin and fibrosis related microRNAs in complex fractionated atrial electrograms. Arch Med Sci. 2015;11:679–82. doi: 10.5114/aoms.2015.52375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiao J, Luo X, Lin H, et al. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J Biol Chem. 2007;282:12363–7. doi: 10.1074/jbc.C700015200. [DOI] [PubMed] [Google Scholar]
- 67.Carè A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–8. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 68.Yang J, Chen L, Ding J, et al. MicroRNA-24 inhibits high glucose-induced vascular smooth muscle cell proliferation and migration by targeting HMGB1. Gene. 2016;586:268–73. doi: 10.1016/j.gene.2016.04.027. [DOI] [PubMed] [Google Scholar]
- 69.Alvarez ML, Khosroheidari M, Eddy E, Done SC. Micro-RNA-27a decreases the level and efficiency of the LDL receptor and contributes to the dysregulation of cholesterol homeostasis. Atherosclerosis. 2015;242:595–604. doi: 10.1016/j.atherosclerosis.2015.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karolina DS, Tavintharan S, Armugam A, et al. Circulating miRNA profiles in patients with metabolic syndrome. J Clin Endocrinol Metab. 2012;97:E2271–6. doi: 10.1210/jc.2012-1996. [DOI] [PubMed] [Google Scholar]
- 71.Yang M, Liu W, Pellicane C, et al. Identification of miR-185 as a regulator of de novo cholesterol biosynthesis and low density lipoprotein uptake. J Lipid Res. 2014;55:226–38. doi: 10.1194/jlr.M041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Rooij E. The art of microRNA research. Circ Res. 2011;108:219–34. doi: 10.1161/CIRCRESAHA.110.227496. [DOI] [PubMed] [Google Scholar]
- 73.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–33. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–80. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 75.Bader AG, Lammers P. The therapeutic potential of microRNAs. Discovery technology. Available at: http://www.mirnarx.com/pdfs/The%20Therapeutic%20Potential%20of%20microRNAs.pdf.
- 76.Najafi-Shoushtari SH, Kristo F, Li Y, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–9. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marquart TJ, Wu J, Lusis AJ, BaldÁn Á. Anti-miR-33 therapy does not alter the progression of atherosclerosis in low-density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2013;33:455–8. doi: 10.1161/ATVBAHA.112.300639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rayner KJ, SuÁrez Y, DÁvalos A. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–3. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sahebkar A, Maffioli P, Banach M, Derosa G. Micro-RNA-33 inhibition: a potential adjunct to statin therapy? Curr Vasc Pharmacol. 2016;14:321–2. doi: 10.2174/1570161114999160513150737. [DOI] [PubMed] [Google Scholar]
- 80.Momtazi AA, Derosa G, Maffioli P, Banach M, Sahebkar A. Role of microRNAs in the therapeutic effects of curcumin in non-cancer diseases. Mol Diagn Ther. 2016;20:335–45. doi: 10.1007/s40291-016-0202-7. [DOI] [PubMed] [Google Scholar]