Abstract
The watershed-hypothesis of schizophrenia asserts that over 200 different mutations dysregulate distinct pathways that converge on an unspecified common mechanism(s) that controls disease ontogeny. Consistent with this hypothesis, our RNA-sequencing of neuron committed cells (NCCs) differentiated from established iPSCs of 4 schizophrenia patients and 4 control subjects uncovered a dysregulated transcriptome of 1349 mRNAs common to all patients. Data reveal global dysregulation of developmental genome, deconstruction of coordinated mRNA, and mRNA networks, and the formation of aberrant, new coordinated mRNA networks indicating a concerted action of the responsible factor(s). Sequencing of miRNA transcriptomes demonstrated an overexpression of 16 miRNAs and deconstruction of coordinated miRNA–mRNA networks in schizophrenia NCCs. ChiPseq revealed that the nuclear (n) form of FGFR1, a pan-ontogenic regulator, is overexpressed in schizophrenia NCCs and overtargets dysregulated mRNA and miRNA genes. The nFGFR1 targeted 54% of all human gene promoters and 84.4% of schizophrenia dysregulated genes. The upregulated genes reside within major developmental pathways that control neurogenesis and neuron formation, whereas downregulated genes are involved in oligodendrogenesis. Our results indicate (i) an early (preneuronal) genomic etiology of schizophrenia, (ii) dysregulated genes and new coordinated gene networks are common to unrelated cases of schizophrenia, (iii) gene dysregulations are accompanied by increased nFGFR1-genome interactions, and (iv) modeling of increased nFGFR1 by an overexpression of a nFGFR1 lead to up or downregulation of selected genes as observed in schizophrenia NCCs. Together our results designate nFGFR1 signaling as a potential common dysregulated mechanism in investigated patients and potential therapeutic target in schizophrenia.
Keywords: Schizophrenia, developmental hypothesis, genome regulation, miRNA Fibroblast Growth Factor Receptor 1
Introduction
Schizophrenia is one of the most debilitating mental illnesses worldwide (Hanzawa et al., 2013), with a lifetime prevalence of about 1.5 – 2% (Saha et al., 2005), and current treatments are only partially effective (Blanchard et al., 2011; Rummel-Kluge et al., 2012). Although the primary onset of schizophrenia is during adolescence to young adulthood, schizophrenia is commonly thought to be a neurodevelopmental disorder(Fatemi and Folsom, 2009; Rehn and Rees, 2005). According to the neurodevelopmental hypothesis, both genetic and environmental factors broadly affect the brain development and thus contribute to the etiology of schizophrenia. (Kneeland and Fatemi, 2013). The early developmental origin of schizophrenia is supported by the changes in brain morphology, which most likely develop in utero during the late first and early second trimester (Kneeland and Fatemi, 2013). Studies of such changes have revealed improperly clustered neurons in layers II, III and V of the cortex (Arnold et al., 1997), differences in a number of nonpyramidal neurons in CA2 and in hippocampal shape (Benes et al., 1998), hypoplastic dopamine neurons, and cerebellar atrophy (Akbarian et al., 1993; Bogerts et al., 1983; Connor et al., 2004; Schiller et al., 2006). Additionally, disorganization of the white matter tract has been observed in schizophrenia (Davis et al., 2003), suggesting that the disease affects the development and function of not only neurons, but also oligodendrocytes. The wide spread of brain malformations are thought to underlie the variety of clinical findings: positive symptoms (delusions and hallucinations), negative symptoms (affective flattening, amotivation and anhedonia) (Blanchard et al., 2011; Foussias et al., 2011), and cognitive symptoms (disorganized speech and cognitive deficits) (DSM 4th edition). Abnormal development during the first trimester is also consistent with minor physical anomalies associated with schizophrenia (Lloyd et al., 2008).
Current evidence points to schizophrenia as a familial disorder with a complex mode of inheritance and variable expression (Sullivan et al., 2003). Due to the polygenetic nature of schizophrenia and its complexity, it has been difficult to dissect out the underlying genetic mechanisms. Schizophrenia linkage studies are generally characterized by a lack of highly significant and consistently reproducible results (Need et al., 2009). The advent of high-throughput sequencing allowed researchers to look at hundreds of thousands of single nucleotide polymorphisms (SNPs) simultaneously. To date, over 600 SNPs have been found to have statistically significant associations with schizophrenia (GWAS Catalog)(Welter et al., 2014). However, like the linkage studies, GWAS have been inconsistent, even with respect to the top few genes identified. A common theme in the investigation of copy-number variants in schizophrenia is an enrichment of those that are rare (< 1% minor allele frequency) and large (>100kb) CNVs (International Schizophrenia, 2008; Malhotra et al., 2011; Walsh et al., 2008), and those that occur de novo (Kirov et al., 2012; Malhotra et al., 2011; Xu et al., 2008). Even the strongest genetic predictors of schizophrenia appear in only 1–2% of schizophrenia cases (International Schizophrenia, 2008; Stefansson et al., 2008; Xu et al., 2008). Hence, the genetic causes of schizophrenia appear to be a multiplicity of rare risk alleles and schizophrenia has been defined as s a common, rare-variant disease.
In 2006, Cannon and Keller proposed a watershed hypothesis (Cannon and Keller, 2006) to explain how so many different mutations throughout the genome give rise to a common disease like schizophrenia. According to this hypothesis, individual mutations dysregulate distinct biological pathways that in turn converge on a common ontogenic pathway(s). Dysregulation of such common pathway leads to developmental malformations, which increase the risk of the disease. However, the nature of these pathways and their organization have not been understood.
The candidate pathway investigated in present study is one recently described as pan-ontogenic, Integrative Nuclear FGFR1 Signaling (INFS) (Stachowiak, 2011; Stachowiak et al., 2015; Stachowiak et al., 2007), which integrates signals from diverse pathways in which the schizophrenia-linked mutations have been found. Genetic experiments have positioned the fgfr1 gene at the top of the gene regulatory network that governs gastrulation and subsequent development of the major body axis, nervous system, muscles, and bones, by affecting downstream mRNAs that control the cell cycle, pluripotency and differentiation (Chioni and Grose, 2012; Ciruna and Rossant, 2001; Ciruna et al., 1997; Dequeant and Pourquie, 2008; Partanen et al., 1998), as well as microRNAs (miRNAs) (Bobbs et al., 2012; Stuhlmiller and Garcia-Castro, 2012). This regulation is executed by a single protein, the nuclear isoform of FGFR1 (nFGFR1), which operates at the interface of genomic and epigenomic information, integrates signals from diverse developmental pathways and provides feedback regulation of those pathways. (Chioni and Grose, 2012; Fang et al., 2005; Han et al., 2015; Stachowiak and Stachowiak, 2016; Terranova et al., 2015).
Evidence from RNA-seq and ChIP-seq analyses has delineated global and direct gene programing by nFGFR1 and its partner CREB Binding Protein (CBP), which guide pluripotent embryonic stem cells (ESCs) towards development into multipotent neural progenitor cells (NPCs) and towards further differentiation(Stachowiak and Stachowiak, 2016; Terranova et al., 2015). nFGFR1 cooperates with a multitude of transcription factors (TFs), including RXR, RAR, and orphan nuclear receptors, and targets thousands of genes that encode mRNAs or miRNAs in top ontogenic networks. nFGFR1 binds to the promoters of ancient proto-oncogenes and tumor suppressor genes, in addition to those encoding metazoan morphogens that delineate the body axis, and that construct the nervous system as well as the mesodermal and endodermal tissues (Stachowiak and Stachowiak, 2016; Terranova et al., 2015). The discovery of pan-ontogenic gene programming by INFS expands our understanding of ontogeny, and could also help to understand developmental pathologies such as schizophrenia and autism spectrum disorders.
Jungerius reported that single nucleotide point mutations in the fgfr1 gene are linked to schizophrenia (Jungerius et al., 2008). Furthermore, manipulation of FGFR1 function in dopaminergic neurons of transgenic mice led to developmental brain malformation and behavioral changes that the mimic the positive, negative and cognitive deficits observed in humans (Stachowiak et al., 2013a). Although a number of mouse models of schizophrenia exist, it is unclear how accurately effects on the mouse genome recapitulate human disease. A robust human-based system is needed to study schizophrenia and to understand the pathways and mechanisms involved. Postmortem analysis of brain tissues from schizophrenia patients is affected by lifetime events such as substance abuse, drug treatment, and fluctuations in brain pH caused by hypoxia or nutritional deficiency (Deep-Soboslay et al., 2011). Moreover, a body of evidence for the early in utero origination of schizophrenia indicates that elucidation of the genomic-developmental etiology of this disease will require a focus on the early stages of neuro-ontogenesis. To this end, human induced pluripotent stem cells (iPSCs) are invaluable. In 2011, two laboratories reported successful development of iPSCs from schizophrenic patients (Brennand et al., 2011; Chiang et al., 2011). Brennand et al. had developed iPSCs from four patients diagnosed with schizophrenia or its schizo-affective variant, and from control non-schizophrenic subjects (Brennand et al., 2011). These iPSCs were differentiated into mature neurons and their gene expression was analyzed using an mRNA microarray. 596 genes were differentially expressed in neurons derived from schizophrenic patients versus controls, reflecting the phenotypic differences between the differentiated neuronal populations (Brennand et al., 2011). Previous work has shown that schizophrenia hiPSC-derived NPCs have aberrant migration (Brennand et al., 2014b) and cellular polarity (Yoon et al., 2014), perturbed WNT signaling (Srikanth et al., 2015; Topol et al., 2015b), increased oxidative stress (Brennand et al., 2014b; Paulsen et al., 2011; Robicsek et al., 2013) and altered responses to environmental stressors (Hashimoto-Torii et al., 2014); while schizophrenia hiPSC-derived neurons exhibit decreased neurite number (Brennand et al., 2011), reduced synaptic maturation (Brennand et al., 2011; Robicsek et al., 2013; Wen et al., 2014; Yu et al., 2014) and synaptic activity (Wen et al., 2014; Yu et al., 2014), and blunted activity-dependent response (Roussos et al., 2016). Extensive immunocytochemical validation of the NPC lines used in the present study showed they are positive for diverse NPC markers including nestin, Sox2, vimentin, Pax6 and TBR2(Brennand et al., 2015). Furthermore, comparisons of the NPC microarray gene expression profiles to AllenBbrain Span Atlas of the developing human brain using Spearmen Rank Correlation Analysis, showed the highest similarility to the first trimester human fetal brain tissue.
To investigate the early neural developmental origin of schizophrenia, we used the same established iPSCs from four schizophrenia and four control individuals as described in (Brennand et al., 2011). The iPSCs were developed into NPCs, which were then briefly (2 days) stimulated to initiate differentiation, producing neuron-committed cells (NCCs). We tested the hypothesis that the NCCs from different patients share transcriptional dysregulation of one or more developmental gene programs, and that INFS is a common cause of those changes. To this end, we performed RNA-seq and nFGFR1 ChIP-seq experiments. Our results reveal that different schizophrenia cases have common dysregulated transcriptomes as well as newly formed coordinated gene networks. The gene dysregulation is accompanied by increased interactions of nFGFR1 with the mRNA and miRNA genes, and can be modeled in human ESCs by transfection of a nuclear variant of FGFR1. These results indicate that schizophrenia is programmed early (before neurons develop), and designate INFS as a one common dysregulated mechanism of the Cannon and Keller model of schizophrenia and a prospective target for therapeutic interventions.
Experimental/Materials and Methods
Plasmids and transfections
Plasmids expressing FGFR1 constructs: FGFR1 (TK-)-deleted tyrosine kinase domain, FGFR1 (SP-/NLS)-signal peptide replaced with the nuclear localization signal (NLS) from the SV40 large T antigen, and FGFR1 (SP-/NLS) (TK-) were described in (Peng et al., 2001; Peng et al., 2002). All transfections were carried out using Fugene 6, per manufacturer’s instructions. hESC-derived NPCs were transfected using 6 μg of either control β-gal, FGFR1(SP-/NLS) or FGFR1(SP-/NLS)(TK-) per well in a 6-well plate. Three wells were combined into a single sample.
IPSCs and ESCs culture and differentiation
Human induced pluripotent stem cell lines (Brennand et al., 2011) (Table 1) were differentiated into NPCs as described previously (Brennand et al., 2011). To initiate neuronal development, NPCs were cultured in neuronal differentiating media for 2 days (DMEM/F12, 1× N2, 1× B27-RA, 20 ng ml−1 BDNF (Peprotech), 20 ng ml−1 GDNF (Peprotech), 1 mM dibutyryl-cyclic AMP (Sigma), and 200 nM ascorbic acid (Sigma). After two days of treatment, the neuron comitted cells (NCCs) were harvested for RNAseq and ChiP-seq analyses. RNAseq analysis of mRNA transcriptomes were performed on established and characterzed iPSC lines from all 4 patient and 4 control individuals (Brennand et al., 2011) (Table 1). Further information about genotype and CNV analysis can be found in previous publication(Brennand et al., 2011). Due to financial constrains, RNAseq of miRNA transcriptomes were performed on 3 control and 3 schziophrenia lines.
Table 1. Description of clinical information available for control and SZ patients.
The information in the table is from (Topol et al., 2015a; Topol et al., 2016; Xu et al., 2016). Fibroblasts from all repository patients and controls have been recently genotyped by PsychChip and exome sequencing(Brennand et al., 2011). CNV analysis was published in (Brennand et al., 2011).
| Patient ID | Coriell ID | Sex | Ethnicity | Age at Biopsy (years) | Age of Onset (years) | Phenotype | Hospitalizations? | Family History |
|---|---|---|---|---|---|---|---|---|
| C1 | BJ | M | Causasian | 0 | - | - | - | unknown |
| C2 | GM03440 | M | Causasian | 20 | - | - | - | unknown |
| C4 | GM04506 | F | Causasian | 20 | - | - | - | unknown |
| C6 | AG09429 | F | Causasian | 25 | - | - | - | unknown |
| P1 | GM02038 | M | Causasian | 22 | 6 | suicide | ? | unknown |
| P2 | GM01792 | M | Causasian Jewish / Scandanavian | 26 | unknown | episodes of agitation, delusions of persecution, and fear of assassination; at age four mild features of pervasive developmental disorder | ? | father and sister affected; brother autistic at age four |
| P3 | GM01835 | F | Causasian Jewish | 27 | unknown | drug abuse; schizo-affective disorder | Yes | father and brother affected |
| P4 | GM02497 | M | Causasian Jewish | 23 | 15 | paralogical thinking, affective shielding, splitting of affect from content, and suspiciousness | Yes | affected father, anorexic /schizoid sister |
hESC line, H9, from WiCell Research Institute (Madison, WI), was used for these experiments under the approval of the Committee for Stem Cell Research Oversight at SUNY-Buffalo. hESCs were differentiated into NPCs utilizing the same protocol as for iPSCs. In brief, hESCs were transferred to a non-adherent plate (Corning). Embryoid bodies were grown in suspension in N2 medium (DMEM/F12-GlutaMAX (Invitrogen), 1× N2 (Invitrogen). After seven days, embryoid bodies were plated in N2 medium with 1 μg ml−1 laminin (Invitrogen) onto polyornithine (PORN)/laminin-coated plates. Visible rosettes formed within one week and were detached using Neural Rosette Selection reagent (Stem Cell). Rosettes were cultured in NPC medium (DMEM/F12, 1x N2, 1x B27-RA (Invitrogen), 1 μg ml−1 laminin and 20 ng ml−1 FGF2). NPCs were then transferred from polyornithine/laminin-coated plates onto matrigel-coated plates and were maintained in NPC media. NPCs were differentiated into NCCs with neuronal differentiating media for 2 days.
Antibody verification
The nuclear presence of FGFR1 was demonstrated in several laboratories in non-transformed cells (Maher, 1996; Reilly and Maher, 2001), cancer cell lines (Bryant and Stow, 2005; Peng et al., 2001; Stachowiak et al., 2003; Stachowiak et al., 1996a, b), stem cells and in the rat and mouse brain (Clarke et al., 2001; Gonzalez et al., 1995) (Fang et al., 2005; Leadbeater et al., 2006; Stachowiak et al., 2003) using an array of antibodies that target different FGFR1 epitopes. Furthermore, transfected FGFR1-EGFP in live cells was detected using native fluorescence and FGFR1-Flag using αFlag. Gene targeting by FGFR1 was shown by EMSA and ChiP assays using diverse FGFR1 antibodies: SC121, Abcam ab10646, FGFR1McAb6, Abcam FGFR1 Mab; (SC 121G) (Baron et al., 2012b; Chioni and Grose, 2012; Fang et al., 2005; Lee et al., 2013; Lee et al., 2012; Peng et al., 2002). FGFR1-DNA interactions were not detected in cells that do not express endogenous FGFR1 and were restored by transfection of FGFR1 (Fang et al., 2005). Gene binding by FGFR1-flag was verified with αFlag (Lee et al., 2012). Dynamic transcription-dependent interactions of FGFR1-EGFP with chromatin was shown in live cells by confocal microscopy and FRAP (Dunham-Ems et al., 2009). In the present study, we used ChiP- and ChiPseq-validated αFGFR1, Abcam ab10646 (Lee et al., 2013; Lee et al., 2012; Narla et al., 2013; Terranova et al., 2015). Detection of nFGFR1 by this antibody is prevented by siRNA knock down of FGFR1 mRNA (Coleman et al., 2014). The ChiP validated Notch1 antibodies were obtained as a gift from Dr. Jon C. Aster (Brigham and Women’s Hospital, Boston, MA) and used as previously in ChIPseq assays (Wang et al., 2011).
ChIP-Seq
ChIP was performed with 250 μg of chromatin DNA and 4 μg of FGFR1 or Notch1 antibodies using the Invitrogen MAGnify kits, according to manufacturer’s instructions and as previously described (Terranova et al., 2015), with slight modifications. Genomic DNA was precipitated with ethanol, treated with RNase A and proteinase K, and purified using the Qiagen PCR purification kit. ChiPseq analyses required a major expansion of the CNC cultures. Cells from twelve 35 mm plates were pooled together and the immunoprecipitated chromatin was further processed using the Tru-seq ChIP Sample Preparation Kit. The purified library DNA was captured on an Illumina flowcell for cluster generation and sequenced on an Illumina Hi-seq 2500, following the manufacturer’s protocols.
RNA-seq and RT-qPCR
RNAseq was performed as previously described (Terranova et al., 2015). RNA was extracted using Trizol or the Qiagen RNA extraction kit, according to manufacturer’s instructions. RNA was prepared using the Tru-Seq RNA kit and purified library cDNA was captured on an Illumina flowcell for cluster generation and sequenced on an Illumina Hi-seq 2500, following the manufacturer’s protocols. For independent mRNA assays, cDNA synthesis was carried out using 1 μg of RNA and the iScript cDNA Synthesis Kit (Bio-Rad; Hercules CA). One tenth of the synthesized cDNA was used as the template for real time-quantitative PCR. 25 μl real-time PCR reactions were performed on the BioRad MyiQ Cycler with iQ SYBR Green Supermix (Bio-Rad). RT-qPCR using the amplification cycles: initial denaturation for 8:30 min at 95C, followed by 35x cycle 2 (denaturation for 15 s at 95C and annealing for 1 min at 60C). Melt curve data collection was enabled by decreasing the set point temperature after cycle two by 0.58C. The specificity of amplicons was confirmed by generating the melt curve profile of all amplified products. Levels of individual mRNAs were analyzed relative to GADPH mRNA (Terranova et al., 2015).
ChIP-seq and RNA-seq data processing
For ChIP-seq, raw RASTQ reads were aligned to the homo sapien genome build hg19 (UCSC) using bowtie2. All peaks were called using Model-based Analysis of ChIP-Seq(MACS) version 2.0 using a q-value cutoff of 0.01(Zhang et al., 2008). MACS2 uses a dynamic Poisson distribution to calculate peaks.
RNA-seq results were analyzed using the tuxedo pipeline and its statistics (Trapnell et al., 2012). Raw FASTQ reads were mapped to the homo sapiens genome build hg19 (UCSC) using tophat 2. These mapped reads were assembled into transcripts and gene expression levels using Cufflinks version 2.0.2. Transcript assemblies were merged together using Cuffmerge and Cuffdiff was used to determine significant differences in gene expression (FPKM) between control and schizophrenia cell lines. Heatmaps of differentially expressed genes were generated using R.
Raw data of next- generation sequencing data can be found at GEO: ChIP-seq GSE92873, RNA-seq GSE92874, miRNA-seq GSE92875
Results
Schizophrenia patients share a common pattern of gene dysregulation
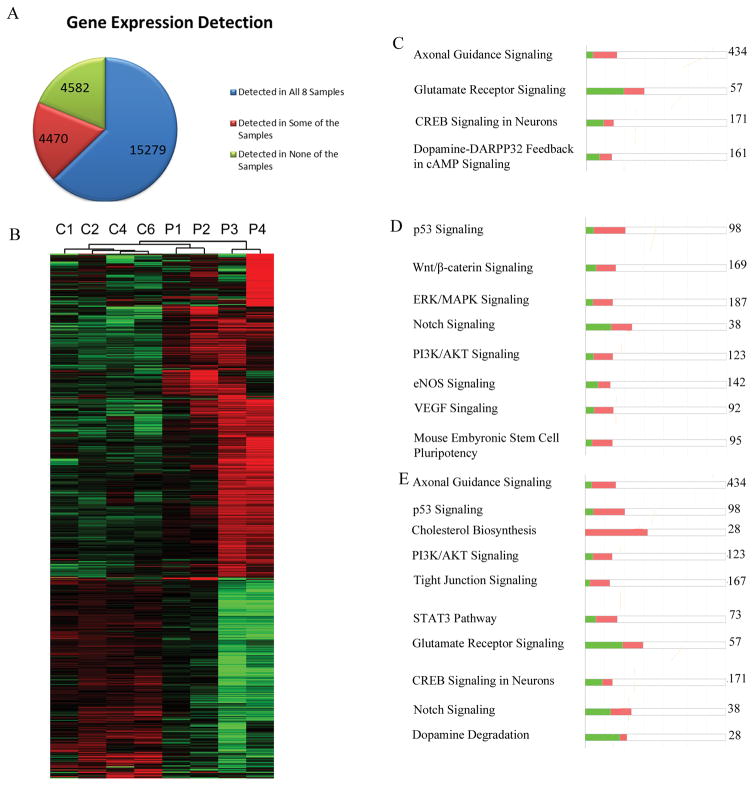
To identify all genes for which the mRNA was expressed in NCC lines from both controls and patients, we performed RNA-seq using an Illumina HiSeq2500 instrument. Raw data were analyzed using the Tuxedo pipeline (Bowtie2 -> Tophat2 -> Cufflinks2) and aligned to the UCSC genome hg19 build. The expression levels of over 24,000 genes were assessed using the Tuxedo pipeline. We found 15,279 genes expressed at detectable levels (63%) in all eight samples, whereas 4,582 genes (19%) were not detectable in any of the eight samples. The remaining 4,470 (18%) were (Figure 1A) not detected in all eight samples and were eliminated from further analysis due to their near-threshold expression. Out of the 15,279 genes, 1,349 showed expression levels that differed significantly between the schizophrenia and control NCC lines (FC ≥ −/+1.5 and q value ≥ 0.05). Among these dysregulated genes, 839 were upregulated, and 510 were downregulated in schizophrenia NCC (Figure 1B).
Figure 1.
RNAseq of control and schizophrenia NCCs. A) Distribution of gene expression across 8 samples: 4 control and 4 schizophrenia NCC lines. 15,279 expressed genes (mRNAs) were detected in all 8 samples. 4470 expressed genes were detected in some but not all samples. 4582 annotated genes were not detected in any sample. B) Heatmap of 1349 genes that were dysregulated in all 4 schizophrenia NCCs samples (FC ≥ −/+1.5 and q value ≥ 0.05). Among these, 839 were upregulated and 510 were downregulated. Raw expression data were log transformed and then centered to the median of all 8 samples. Red indicates higher value than median, green indicates lower value than median. C–E) IPA analysis of dysregulated genes. The number on the right represents the total number of genes in the pathway. The bar height represents the percentage of genes in the pathway that were found to be dysregulated. Green represents downregulated genes. C) Neuronal pathways, D) developmental pathways, and E) pathways that are predominantly up- or downregulated.
We used Gene Ontology (GO) to identify functions of the dysregulated genes. In overrepresentation analysis, the 1349 dysregulated genes were tested for the likelihood of belonging to a specific biological category and were then compared to a randomly selected group from among all human genes. This analysis revealed that many neuronal GO categories were overrepresented (Table S1) including genes involved in neural crest development, synaptic plasticity, learning, memory, and synapse organization. Several GO groups related to glial development were also overrepresented, including those involved in the processes of myelination, axon ensheathment, glial cell differentiation, and oligodendrocyte differentiation. Lastly, GO groups related to general ontogeny, including regulators of endothelial cell differentiation, artery development, collagen metabolic process, and mesenchyme differentiation, were also overrepresented among the dysregulated genes (Table S1).
Next, pathway analysis was performed using Reactome, an open-source, manually curated and peer-reviewed database of pathways and biological processes (Haw et al., 2011). This analysis estimated gene overrepresentation within specific pathways, taking into account gene interactions. Due to the limitations of manual curation and incomplete knowledge of genes in pathways, out of the initial 1349 differentially regulated genes, only 886 were mapped to Reactome. The identified pathways clustered around the terms: neuronal systems, developmental biology, and extracellular organization (Table S2). One hub was centered around the neurotransmitter release cycle and included pathways related to the release cycles for dopamine, serotonin, norepinephrine, and glutamate neurotransmitters (Fig. S1). L1cam signaling, which plays a role in axonal growth (Pollerberg et al., 2013), was also significantly overrepresented (Fig. S2), as were Notch Signaling, various extracellular matrix pathways, and transcriptional regulation by TP-53 (Table S2).
Further analysis was performed using Ingenuity pathway analysis (IPA), a proprietary curation of pathways by Qiagen. Analysis by IPA of the 1349 commonly dysregulated genes n schizophrenia NCC revealed multiple neuronal pathways, such as axonal guidance (Figure S3), glutamate receptor signaling (Figure S4), CREB signaling in neurons, and dopamine degradation (Table S3, Figure 1C). Number of the IPA-identified neuronal development pathways were overrepresented consistent with the Reactome analysis. These included Notch signaling (Figure S5), Wnt/β-catenin signaling (Figure S6), and pluripotency regulation (Table S3, Figure 1D). Other affected pathways represented major general signaling pathways, such as PI3K/AKT signaling, eNOS signaling, and VEGF signaling (Figure 1D), as shown in Table S3. Together, these analyses revealed that the dysregulation of gene expression in NCCs derived from patients with schizophrenia was centered on neuronal genes as well as other developmental genes.
To further characterize the observed gene dysregulation, we performed separate analyses of genes that were upregulated and genes that were downregulated in schizophrenia NCCs. Genes involved in glial differentiation and axon ensheathment, were present only in the downregulated category (Table S4) while, neuronal ontologies such as axonogenesis, neurotransmitter transport, and learning were overrepresented in the upregulated group (Table S5). IPA analysis indicated that in many pathways, genes were up- and downregulated, but in certain pathways, the majority of dysregulated genes leaned more in one direction or the other. For example, the majority of dysregulated genes in glutamate receptor signaling, CREB signaling in neurons, Notch signaling, and dopamine degradation were downregulated, whereas most of those involved in axon guidance, p53 signaling, cholesterol biosynthesis, PI3K/AKT signaling, tight-junction signaling, and STAT3 pathway signaling were upregulated (Figure 1E). Analysis of pathways through Reactome verified segregation of distinct pathways between the up- and downregulated categories. The upregulated genes were involved in neurotransmitter release, axon guidance, (TP53-dependent) transcription of cell cycle genes, and development (Table S6); whereas downregulated genes were involved in cell junction organization, cell-cell junctions, neurotransmitter receptor binding, and cell-cell communication (Table S7).
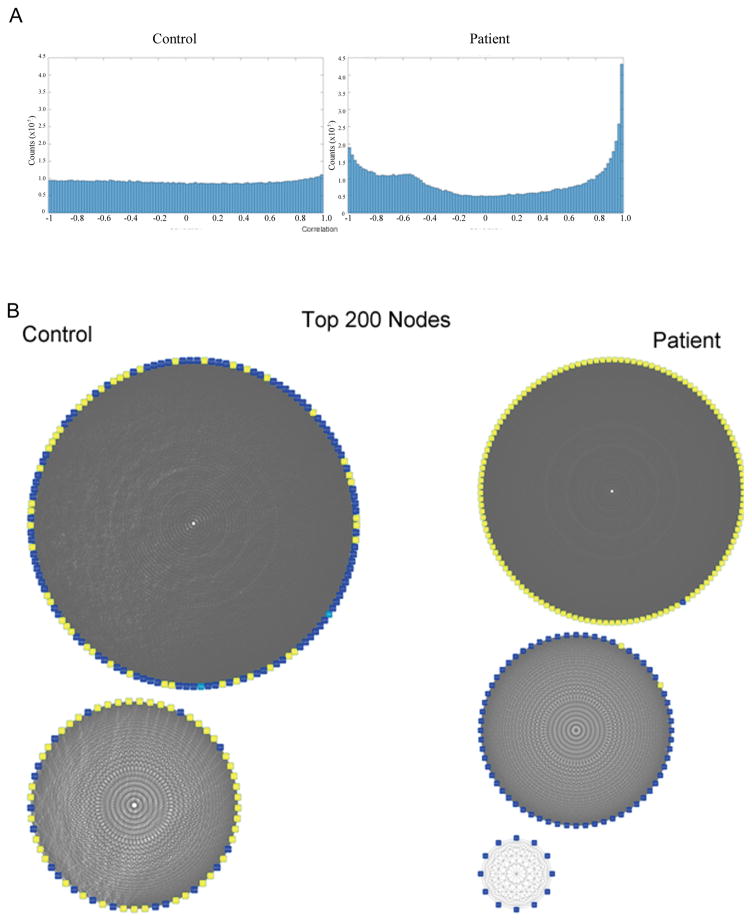
To identify genes that were co-regulated, we performed a pairwise correlation network analysis. First, we correlated the expression of all 1349 dysregulated genes in pairs (Figure 2A), for 909,226 correlations. In control cells (n=4), the distribution of correlations was flat, whereas in patients cells (n=4) peaks were present at the edges of both positive (the same direction) correlations and negative (opposite direction) correlations. Next, we implemented a cutoff of 0.9 to identify and focus on genes for which expression was highly correlated. The 200 most connected genes from control and patient networks were analyzed using circular network graphs. A common feature of both networks was that the top 200 genes were highly interconnected (Figure 2B). However, the sets of most connected genes in control and patient cells were different. The top 200 genes interconnected in controls were largely unconnected in patients and vice versa (Supplemental Figure 7). These results suggest that whereas the control networks were disrupted in the patient NCCs, a new network of connected genes formed in their place. Control networks included both up- and downregulated genes. In the schizophrenia networks, however, the up- and downregulated genes clearly segregated into separate networks indicate a concerted up- and down gene dysregulation.
Figure 2.
A) Histogram of pairwise correlations. Correlation was performed using 4 control and 4 patient NCCs samples. A flat distribution of correlation is observed in controls, while in patients an increase in the number of positively and negatively correlated genes was observed. B) Top 200 nodes (genes whose expression is highly correlated with that of multiple other genes) in control and in patient NCCs were identified. Yellow represents upregulated genes, and blue represents downregulated genes. Grey lines link pairs of genes whose correlation is greater than 0.9. In the control set, two separate networks are observed and each contains both upregulated and downregulated genes. In the patient set the upregulated and downregulated genes form two separate networks.
Common dysregulated miRNAs in schizophrenia NCCs
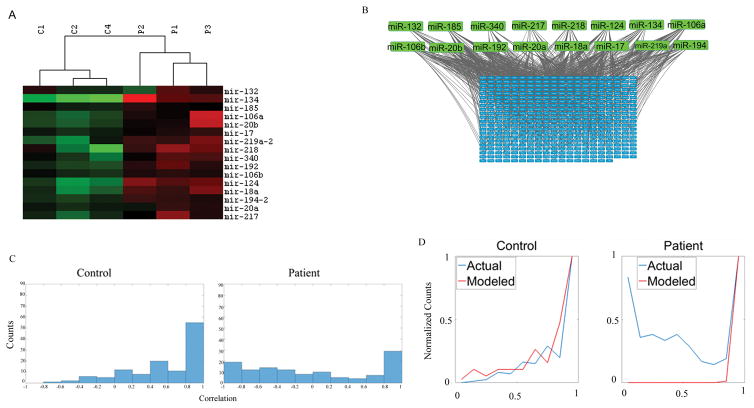
Factors that could elicit a concerted dysregulation of transcriptome included miRNAs, which influence overlapping gene sets in a coordinated fashion. miRNAs influence gene expression at the transcription level, by promoting mRNA degradation and by inhibiting mRNA translation (Bartel, 2009; Younger and Corey, 2011). Earlier studies have shown that some miRNAs may be dysregulated in the brains of schizophrenia patients (de Bartolomeis et al., 2015; Hill et al., 2014). We performed small-RNA seq (Terranova et al., 2015) to identify and to measure the miRNA levels in NCC lines from the three controls (C1, C2, C4) and three schizophrenic patients (P1, P2, P3). The expression of 1391 miRNAs was quantified utilizing the CLC Genomics Workbench and the most up-to-date miRBASE. Among these, 440 miRNAs were expressed in all six samples, whereas 479 miRNAs were not detected in any of the samples. The remaining 472 miRNAs were detected only in some samples at the threshold levels of detection and, therefore, were eliminated from further analysis. Out of 440 miRNAs expressed in all six samples, 16 were differentially expressed in patients as compared to controls (Fig 3A). All 16 miRNAs were upregulated, but to different degrees (1.5- to >70-fold). In contrast, in control cells the expression levels of these individual miRNAs were similar. TargetScan and MirTarBase analyses predicted that the overexpressed miRNAs may interact with >400 dysregulated mRNAs, in a largely overlapping manner as illustrated on Figure 3B.
Figure 3.
A) NCCs from three control subjects and three patients were analyzed. Heatmap shows 16 dysregulated miRNA genes (all upregulated) in patient NCCs. Raw expression data were log transformed, and then centered to the median of all 6 samples. Red indicates higher value than median, green indicates, lower value than median. B) 16 dysregulated miRNAs (Green) target 440 dysregulated mRNA (Blue). Each gray line indicates a connection between an miRNA and an mRNA. C) In pairwise correlation of dysregulated miRNA genes a high correlation is observed in control NCCs (n=3), but not in patient NCCs (n=3). D) Modeled versus actual miRNA-mRNA correlations. Red line shows correlations for miRNAs predicted based on correlations of their target mRNAs as in (Huttenhower et al., 2009). Blue line shows measured correlations between miRNAs and their target mRNAs. In control NCCs the predicted and actual correlations are similar. In patients, the patterns of predicted and actual correlations differ markedly.
Given the complexity of these individual miRNA-mRNA interactions, we analyzed relations between the networks of miRNAs and their target mRNA. First, we preformed pairwise correlations of the 16 miRNAs (Figure 3C) which revealed a high degree of positive correlation in control NCCs. This high correlation was consistent with the model in which different miRNAs controlled shared mRNA targets. In schizophrenia NCCs, the correlation among 16 miRNAs was lost. Subsequently, we modeled the miRNA correlations based on the correlations among their targeted mRNAs by constructing combined miRNA-mRNA networks using protocol of Huttenhower et. al. (Huttenhower et al., 2009). In control NCCs, the predicted and actual miRNA networks were similar (Fig. 3D). In contrast, in the schizophrenia NCCs, the predicted miRNA networks differed markedly from the observed networks. This result indicated a disruption of the miRNA-mRNA network in the schizophrenia NCCs.
nFGFR1 binds throughout the genome
Another candidate factor that could potentially contribute to the concerted transcriptome dysregulation in schizophrenia is panontogenic INFS. To identify sites of the genome targeted by nFGFR1, we performed ChIP-seq of both control and patient samples. Initially, we performed a detailed ChiP-seq analysis of a single control line, female C4, and a single patient line, female P3 (Table 1 in Experimental Methods/Materials), using previously tested, ChiP and ChiPseq validated anti-FGFR1 antibody (Terranova et al., 2015). IP and input samples were sequenced on Illumina HiSeq2500. Raw data were aligned to the UCSC hg19 human genome using the Bowtie2 tool. Peaks were identified with MACS2 at a q-value ≤ 0.01. The input sequences were used as the background from which peaks were called. This ensured that our peak identification was not influenced by a biased fragmentation of the genome. DNA sequences located at the center of the specific peak are expected to be more conserved than the random DNA sequences away from the binding site. This was verified in both our control and patient conservation plots using the Conservation Plot feature of Cistrome. (Supplemental figure 8A).
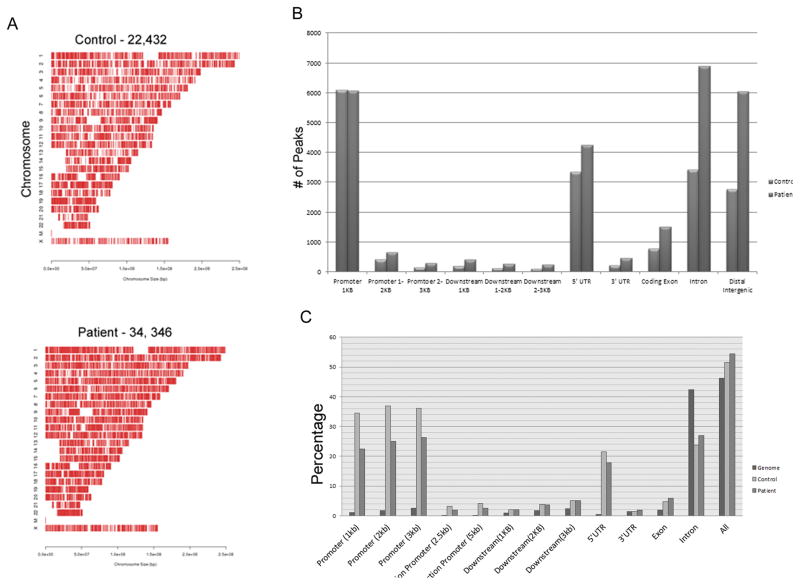
In the control sample, 22,432 peaks of FGFR1 binding were identified. More peaks were found in the patient sample (34, 346; Figure 4A). In both the control and patient samples, peaks were distributed throughout the genome and across all chromosomes. In both control and patient DNA, the highest concentrations of FGFR1 peaks were on chromosomes 17, 19, and 22, and the lowest were on chromosome 13. However, this enrichment was not observed when the values were normalized to gene density. Thus, the chromosomal differences in nFGFR1 binding reflect the distribution of genes across the chromosomes.
Figure 4.
A) Distribution of nFGFR1 peaks throughout the genomes of NCCs from a female control (C4) and a female patient (P3). nFGFR1 binds to all chromosomes. The binding profiles are similar for both control and patient genomes. B) Distributions of nFGFR1 peaks in genomes of control and patient NCCss. In patients, a two-fold increase is observed in introns, distal intergenic regions, downstream (0–3kb), and distal promoters (1–3kb). C) Enrichment of nFGFR1 peaks in genomes of control and patient NCCs within the promoters and bidirectional promoters, and downstream of the TSS. No enrichment is observed in introns or intergenic regions.
Next, we analyzed the distribution of nFGFR1 peaks within various genomic regions. The most upstream transcription start sites (TSS) in individual genes were identified and used as landmarks for promoters and 5′UTRs. Overall, in both control and patient genomes, the peaks were located primarily within the promoters (±1kb), 5′ UTRs, introns, and distal intergenic regions. Distribution of overall genomic length was compared to that of FGFR1 peaks to identify potential sites of enrichment in certain genomic regions. We observed a large enrichment of peaks in the promoters (>20 Fold) and 5′UTRs (>5fold), and a small enrichment (>2fold) in downstream DNA (Figure 4B). In contrast, peaks were not enriched in introns and intergenic regions. The distribution of nFGFR1 peaks was similar in both the control and patient samples.
To identify the locations of the additional ~12,000 peaks that were present only in schizophrenia NCCs, we tabulated the number of peaks in each genome location (Figure 4C). A two-fold increase was found in distal promoters (1–3kb from TSS) and downstream promoters (0–3kb). Interestingly, a majority of peaks unique to patient samples were distributed within introns and distal intergenic regions.
Binding of nFGFR1 to active genes
To determine how nFGFR1 binding relates to gene expression, we first identified the set of peaks at the proximal promoters (+/− 1kb of the TSS) of all of the genes in the human genome. For this analysis, all TSS within a gene that could potentially be used to produce alternative transcripts were taken into account. For the proximal promoters of all the annotated human genes (24,331), nFGFR1 bound to 13,182 in control and to 15,324 genes in patient cells. nFGFR1 also bound to the distal promoters (1 to 3kb from TSS) of 377 genes in control cells and 672 genes in patient cells. In majority of those genes (>80%) nFGFR1 bound also to the proximal promoter suggesting its cooperative regulation. Approximately 90% of genes that were expressed in all eight samples had promoters targeted by nFGFR1 (Figure 5A). Thus, in both control and schizophrenia NCC nFGFR1 primarily associated with the promoters of active genes.
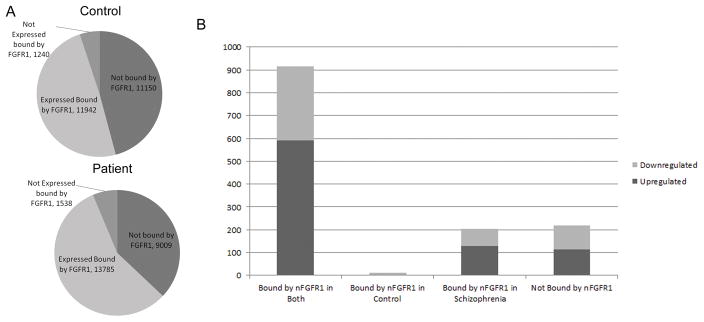
Figure 5.
A) nFGFR1 binds to 54% of all annotated human genes in control (C4) NCCs and to 63% in schizophrenia patient (P3) NCCs. B) nFGFR1 binds to 915 dysregulated genes under both conditions, and to an additional 203 genes in schizophrenia NCCs only. In total, nFGFR1 binds to 84% of the genes that are dysregulated in schizophrenia NCCs. Only 14 genes are targeted by nFGFR1 in control but not schizophrenia NCCs.
Among the 1,349 genes dysregulated in schizophrenia NCCs, nFGFR1 bound the proximal promoters of 1,124 genes in patient cells and 929 genes in control cells (Fig. 5B). Likewise, the number of dysregulated genes in which nFGFR1 bound to distal promoters was greater in schizophrenia (96 genes) than in control cells (63 genes). In both control and patient cells 915 genes were bound by nFGFR1 (Figure 5C). Of these, 592 were upregulated, and 323 downregulated, in patient samples. 203 genes (128 upregulated and 75 downregulated) were bound by nFGFR1 only in patient cells. Only 12 genes were bound by nFGFR1 exclusively in control cells. Those genes were found upregulated (3 genes) and downregulated (9 genes) in patient cells (Figure 5C). Finally, 219 genes were not bound by nFGFR1. Among these, 115 were upregulated and 104 were downregulated in patient cells. In summary, the majority of dysregulated genes have promoters targeted by nFGFR1 and the number of genes bound nFGFR1 was increased in schizophrenia NCCs
Categories of dysregulated genes targeted by nFGFR1 were identified using GO, IPA, and Reactome. Due to their small number, the 14-dysregulated genes that bound nFGFR1 only in control cells (listed in Table S8) were not analyzed. As found for all 1,349 dysregulated genes, the 915 genes in which nFGFR1 was bound in both control and schizophrenia NCCs overrepresented the pathways involved in axon guidance, neurotransmitter release, and glial-cell differentiation (GO, Table S9). Pathway analysis using Reactome revealed that nFGFR1 targeted the pathways including neurotransmitter release, axon guidance, TP53-regulated cell cycle genes, and L1Cam signaling (Table S10). IPA verified and identified pathways such as axonal guidance, PI3K/AKT signaling, Notch signaling, and Wnt/β-catenin signaling. Overall, the 915 genes bound by nFGFR1 were distributed among pathways that overlap with those selected by analysis of all dysregulated genes (Table S11).
Second, we analyzed the 203 genes bound in patient cells but not in controls. GO terms for neuron development, axon guidance, extracellular matrix, and others were overrepresented in this set (Table S12). Reactome pathway analysis revealed a spread across various functions, including Notch signaling, RHO GTPase pathways, p75 NTR axonogenesis pathway, and MHC Class 1 Antigen processing and presentation (Table S13). In this case, IPA also revealed axonal guidance signaling, tight junction signaling, urea cycle, along with others. The 203 genes, which were targeted by nFGFR1 only in schizophrenia cells, were engaged in the same functions as the remaining genes targeted by nFGFR1 in both control and patient cells (Table S14).
In addition to the nFGFR1 binding gene sets, we also analyzed the 211 dysregulated genes to which nFGFR1 did not bind. Overrepresented GO terms included synaptic transmission and neuron projection development (Table S15). Reactome analysis showed that overrepresented pathways involved in hemostasis, metabolism, lipid digestion, and the regulation of cell death. These categories were not overrepresented in the set of the 1,124 nFGFR1-targeted genes (Table S16). In addition, IPA identified glutamate receptor signaling, calcium signaling, and CREB signaling in neurons among these nFGFR1 no-binding genes (Table S17). However, neither IPA nor Reactome identified key developmental pathways, such Wnt signaling or Notch signaling, among the genes that did not bind nFGFR1. Thus, categories of dysregulated genes that were targeted by nFGFR1 differed from those that did not bind nFGFR1.
The relationship between gene dysregulation and nFGFR1 binding was analyzed further in 915 genes targeted by nFGFR1 in both control and patient cells. To compare nFGFR1 binding at the same sites of control and patient genomes we used MACS2 analysis to determine the nFGFR1 binding score. The score reflects the abundance of nFGFR1 at a particular genomic locus in a cell population. The binding was compared only when the centers of peaks in control and patient cells were within 50 base pairs of each other. This analysis showed that 15,451 peaks had a stronger binding (higher binding score) in patients than in control cells. Out of the 915 genes that bound nFGFR1 in both samples, 828 showed stronger nFGFR1 binding in patient cells. The average binding profile of nFGFR1 over the genome showed a sharp increase in nFGFR1 binding score near the promoter (Figure 6A). This binding was stronger in schizophrenia NCCs than in control NCCs. These differences were verified in a separate ChiP-seq experiment on a pair of male control and male patient NCCs (Figure S8B).
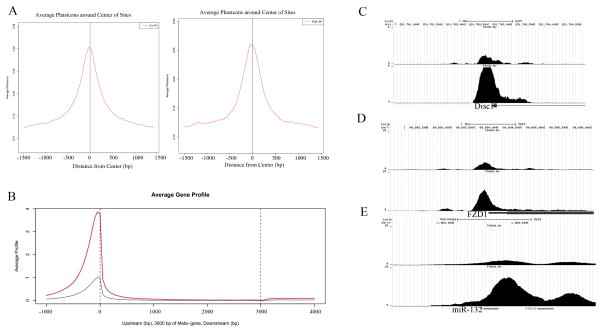
Figure 6.
A) Conservation plot of sequences of FGFR1 peaks in NCCs from a female control (C4) and female patient (P3). The expected increase in conservation was observed near the center of the nFGFR1 peak. B) Strength of nFGFR1 binding across a modeled average gene in NCCs from female C4 (blue) and male P3 (red). C) UCSC genome browser views of nFGFR1 binding for Disc1, FZD1, and Sox3. Tag distribution of nFGFR1 – increased binding is observed in patient (P3) compared to control (C4).
Total of 15,451 peaks had a higher binding score in patient than in control cells. Examples of the loci (Disc1, FZD1, Sox3 TH, and WNT genes) with increased nFGFR1 binding in schizophrenia NCCs are shown on Figure 6B and Figure S8C. Relatively few peaks, 102 located on 24 genes, had binding scores higher in control cells than in schizophrenia NCCs. Only three of these genes were dysregulated, GAS7 was downregulated, and HIF3A and TFAP2C upregulated.
nFGFR1-targted motifs
Although nFGFR1 lacks a DNA binding domain, it can bind to promoters indirectly through CBP, which interacts with diverse TFs. Analysis of Motif Enrichment (AME) program identified number of overrepresented sequences targeted by nFGFR1. nFGFR1 bound at canonical target sequences of RAR, RXR, Nurr, and CREB, all of which were shown interact with nFGFR1 (Baron et al., 2012b; Lee et al., 2013; Lee et al., 2012; Peng et al., 2002; Terranova et al., 2015). In addition, we identified motifs for various TFs, including homeodomain (HOX) family, Krüppel-like factor (KLF), sex determining region-Y box (Sox), and octamer-binding transcription factor (OCT). Importantly, in patient NCCs, but not in control cells, nFGFR1 bound at the motif of the chromatin organizing CTCF protein. Similarly, motifs for Olig1 and Olig2 were targeted by nFGFR1 only in patient cells. Next, we determined if specific motifs were overrepresented only in the dysregulated genes. We found that members of the HOX, high mobility box family (which include SOXs) and zinc finger proteins (ZNF354C) were overrepresented in the nFGFR1 peaks on dysregulated genes relative to all nFGFR1 peaks. Also, number of nFGFR1-targeted motifs (i.e., Esrrg, Mybl2, MAFB, HoxB5, and NR2C2) were enriched among the upregulated genes. Downregulated genes were enriched for the FGFR1-targeted Olig1, Sox2, Creb3l1, Pax6, and PDX1 motifs (Figure S9).
Co-targeting of gene promoters by Notch1 and nFGFR1
Several of the nFGFR1-targeted motifs are known to bind an important developmental TF – Notch1 (Wang et al., 2011). We performed ChiP-seq with a Notch1 Ab to determine whether nFGFR1 may be targeting any part of the genome in schizophrenia NCCs together with Notch1. Hence, we identified genomic locations in which binding of nFGFR1 and Notch1 overlapped by at least 1 base pair (bp) in the same ChiP-seq sample. Conservation of Notch1 binding is plotted on Figure S10A. The total number of Notch1 peaks was 21,508, and nearly half of these (9,664) colocalized with nFGFR1 peaks (Figure S10B). Even though we found no general enrichment for Notch peaks at the promoters or 5′UTR, the overlapping nFGFR1/Notch-1 peaks were enriched in these regions (Figure S10C). In contrast, the Notch1-only peaks were enriched in the intergenic part of genome (Figure S10C, D). Thus, Notch1 may regulate gene promoters together with nFGFR1 but act independently elsewhere. Among 1,349 genes dysregulated in schizophrenia NCCs, <20 had promoter sites that were targeted only by Notch1. In contrast, nearly half (560) of the dysregulated genes had promoter sites cotargeted by Notch1 and nFGFR1 (Figure S10E).The numbers of those genes that were upregulated (57%) and number of genes which were downregulated (63%) were similar. Thus, nFGFR1 could act together with Notch-1 to affect gene expression in schizophrenia NCCs. The examples of genes which are regulated by nFGFR1 (Chris PLOS) and are targeted by both the nFGFR1 and Notch1 include PAX3, IRX3, ID3 (Figure S11).
nFGFR1-targeting miRNAs
Among 440 miRNAs expressed by control and schizophrenia NCCs, the promoter sites of 68 (15%) were bound by nFGFR1 (Table S18). Among 16 miRNAs that were dysregulated in schizophrenia NCCs, 5 (miR-132, miR-17, miR-2V, miR-124 and miR-219a) were targeted by nFGFR1. For miR-132 (Figure 6C) and other miRNAs genes (not shown), the strength of nFGFR1 binding was increased in schizophrenia NCCs.
Role of nFGFR1 in dysregulation
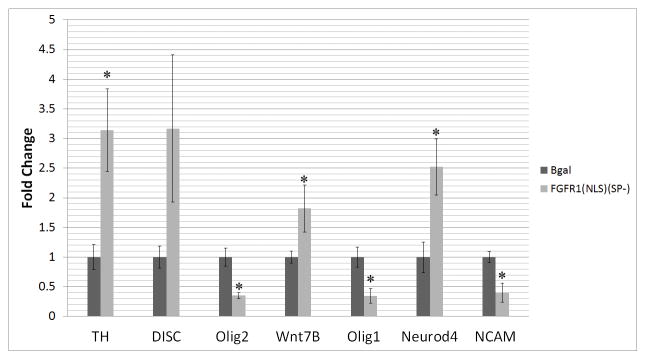
Previous work showed that an increase in nFGFR1 can lead to changes in the transcriptional activity of diverse genes (Baron et al., 2012a; Baron et al., 2012b; Lee et al., 2013; Lee et al., 2012; Stachowiak et al., 2013b; Terranova et al., 2015). Here, in schizophrenia NCCs, we observed an increase in nFGFR1 binding accompanied by altered gene activities. Increased nFGFR1 binding to NuRE, RARE, nBRE, CRE, AP1, NfkB, and Ebox were previously shown to activate transcription, whereas binding to Smad targeted site, inhibited transcription (Terranova et al., 2015). Also, nFGFR1 activated the promoters of neural genes βIII- tubulin, doublecortin (Lee et al., 2013; Lee et al., 2012; Narla et al., 2013), Pax, Id3, Cdx1, IRX3, and Hox A, but inhibited promoters of pluripotency Klf4, STAT3, and Sox2 genes (Terranova et al., 2015). To inquire further if the increased nFGFR1 binding plays a role in genome-wide dysregulation of gene expression in schizophrenia cells, we transfected FGFR1(SP-)(NLS) or a control β-galactosidase expressing vector into hESC-derived NCCs, which were differentiated into a neuronal lineage as described for iPSCs. qPCR of selected genes, Olig2, Disc1, TH, Wnt 7B, Neurod4, NCAM, and Olig1 showed that transfection of constitutive active nuclear FGFR1(SP-)(NLS) resulted in a change in mRNA expression (Figure 7). Olig1, Olig2, and NCAM mRNA were downregulated to a similar to that observed in patient cells. mRNA of TH, WNt7B, and Neurod4 was upregulated by FGFR1(SP-)(NLS), similar to the upregulation found in the schizophrenia NCCs. The NCAM is one of several genes targted by both R1 and Notch1.
Figure 7.
Overexpression of constitutively active nuclear FGFR1(NLS/SP-) affects expression of selected schizophrenia dysregulated genes. Human ESCs were stimulated to differentiate into NPCs and transfected with constitutively active nuclear FGFR1(NLS/SP-) or control β-galactosidase (β-gal). Transfected NPCs were induced to commit to a neuronal lineage (NCCs) within 2 days of treatment, and specific mRNAs were analyzed by RT-qPCR. FGFR1(NLS/SP-) transfection upregulates TH, Wnt7B, and Neurod4 mRNAs and downregulates Olig2, Olig1, and NCAM mRNAs.
Discussion
The present investigation into iPSCs neural progeny supports the watershed hypothesis of schizophrenia, revealing early developmental, common dysregulation of the genome, in four patients with diverse genetic backgrounds and different schizophrenia-linked copy-number variants. The common 1349 dysregulated genes displayed varying degrees of dysregulation in the individual patients, including the siblings. Variations in the expression of these individual genes were not observed in unrelated control subjects, and thus, may represent different forms of the disease. The gene dysregulation in schizophrenia was observed also in the mature neuronal populations differentiated from the same four control and four schizophrenia patients iPSCs as used in the present studies (Brennand et al., 2011). Indeed, these earlier studies concluded that neuronal populations develop differently from schizophrenia than from control iPSCs and that the synaptic connectivity is altered among the schizophrenia neurons. Many of the changes in gene expression observed in mature neurons could reflect differences in the types of neurons that were generated from the patient and control IPSCs (Brennand et al., 2015; Brennand and Gage, 2011; Brennand et al., 2014a; Brennand et al., 2011). To identify the genomic mechanisms that lead to altered neuronal and brain development and thus underlie the etiology of schizophrenia, we focused on early neural development, i.e., the transition from NPCs into the committed neuronal stage, NCCs.
Gene ontology and pathway analyses (both Reactome and IPA) revealed that many of the dysregulated NCC genes are involved in pathways that govern the early stages of neural development, e.g., Wnt/Beta-Catenin signaling, Notch signaling, ERK/MAPK signaling, PI3K/AKT signaling, and growth hormone signaling. The dysregulated genes reside also in pathways that guide axonal guidance and myelination, glial differentiation, as well as signaling by dopamine and glutamate receptors and CREB signaling in neurons. This type of changes may forecast malfunctions in maturing and in mature neuronal systems.
Our recent studies have shown that nFGFR1 interacts with the same genes which were affected in the schizophrenia NCCs pathways also in the mouse genome (Stachowiak and Stachowiak, 2016; Terranova et al., 2015). Like in the mouse genome, in both control and schizophrenia NCCs, the nFGFR1 peaks are enriched at the promoters (both distal and proximal) and 5′ UTRs (Terranova et al., 2015), and are found primarily on expressed genes. Of all annotated human genes (24,331), 54% (13,182 genes) were bound by nFGFR1 in control and patients NCCs. However, among the 1324 dysregulated genes, nFGFR1 bound to 84.4% (1,118) dysregulated genes. The majority of nFGFR1 peaks were in the same location in control and patient NCCs.
While many pathways had both up- and downregulated genes, in several neurodevelopmental pathways, the number of upregulated genes was higher than that of downregulated genes in schizophrenia NCCs. The upregulated genes are involved in the differentiation and maturation of neurons and in axonal guidance, whereas the downregulated genes are involved in glial differentiation and myelination. An in-depth analysis of selected pathways revealed a concerted augmentation of the pro-neuronal developmental mechanisms and an attenuation of pro-glial/anti-neuronal signaling. For instance, within the pro-neuronal Wnt pathway, the schizophrenia NCCs displayed a concerted activation of genes that enhance Wnt signals and repression of genes that inhibit the Wnt signals (Figure S6). In contrast, genes within the Notch1 pathway, which inhibits neuronal development but promotes gliogenesis, displayed a concerted downregulation (Figure S5). A similar coordinated downregulation was found in schizophrenia NCCs genes that mediate glutamatergic transmission (Figure S4). The latter suggests that the glutamate synaptic insufficiency is programmed during early neural development. Together, these findings imply that in the context of schizophrenia, neural progenitor cells may exit from the cell cycle prematurely and differentiate into neurons, and that the development of glia, specifically oligodendroglia, is suppressed. These gene dysregulation could therefore account for the excessive numbers of small neurons often found in schizophrenia brains upon post-mortem examination (Akbarian et al., 1993; Connor et al., 2004; Schiller et al., 2006), and for the abnormal myelination observed by MRI and PET imaging (Palaniyappan et al., 2013).
Many of the genes dysregulated in schizophrenia NCCs are involved in general ontogenic pathways. Their malfunction could impact neural development as well potentially make it vulnerable to schizophrenia-linked environmental factors(Hamlyn et al., 2013). The early neuronal committed cells used in our study, still express number of non-neural genes, some of which were dysregulated in patients’ NCCs. Changes in the activities of genes involved in angiogenesis, development of muscles and other tissues may be predictive of non-neural developmental changes, more frequently observed in the schizophrenia compared to the control population. The well-known examples include curved fifth digit, asymmetrical ears, hypertelorism, etc. (Compton and Walker, 2009). Programing development of the schizophrenia iPSCs to non-neural tissues may further elaborate our findings,
Within the dysregulated transcriptome of schizophrenia NCCs, we observed a marked rise in the number of genes displaying a highly correlated expression. The analysis of the 200 most connected genes revealed a unique set of genes whose expression was highly correlated in control NCCs and different sets of genes that were highly correlated in the schizophrenia NCCs. Within the patient network, the upregulated genes correlated almost exclusively with other upregulated genes, while the downregulated correlated with the downregulated genes. Such separation was not observed within the control network in which up and downregulated genes displayed strong regulatory connection.
The control networks, which became disrupted in patient NCCs, included genes involved in neural development and cell motility both of which are affected in schizophrenia (Fatemi and Folsom, 2009). The newly formed highly correlated networks in schizophrenia NCCs included genes involved in the biology of the extracellular matrix, consistent with the proposed role of brain extracellular matrix in the pathophysiology of schizophrenia (Berretta, 2012). In addition, analysis of all 15,279 genes expressed by schizophrenia NCCs revealed increased positive correlation among genes that participate in brain development, cell motility, differentiation, synaptogenesis as well as glutamatergic transmission, all implicated in the etiology of schizophrenia (Figure S12).
The formation of the coordinated changes in gene activities and the separate up- and down-networks in schizophrenia NCCs suggested a concerted action of a global gene activator/inhibitor. Our present investigation points to two types of global gene regulators that may be involved: miRNAs and nFGFR1. In schizophrenia NCCs, we found a concerted dysregulation of 19 miRNAs, all of which were overexpressed, albeit to a different extent. Within this group, mir-132 (Miller et al., 2012), mir-134 (Moreau et al., 2011; Santarelli et al., 2011), mir-218 (Perkins et al., 2007), and mir-17 (Shi et al., 2012) had previously been implicated in schizophrenia (Miller et al., 2012; Santarelli et al., 2011). Postmortem human studies revealed that miRNAs are upregulated in schizophrenia brain accompanied by an increase in the expression of DICER and other miRNA processing genes (Santarelli et al., 2011). In schizophrenia NCCs, we found no changes in the expression of miRNA processing genes. Therefore, the upregulation of the 16 specific miRNAs may not be due to generalized changes in miRNA processing. In control NCCs, the expression of these 16 miRNAs showed high level of correlation consistent with the largely shared population of their 440 targeted mRNAs (Figure 4B). This highly correlated network of miRNAs can be correctly modeled based on their target mRNAs expression. In contrast, in schizophrenia NCCs, the expression of 16 miRNAs was not correlated and was disconnected from their highly correlated target mRNA network (Fig 3D). We hypothesize that separation of miRNAs and a malfunctioning mRNA gene regulator that takes over the control of mRNA genes, causing their hyper-correlation, could enforce mRNAs networks. Another possibility is suggested by the study of Schmiedel (Schmiedel et al., 2015), who showed that miRNAs may be more effective in controlling mRNAs expressed at low versus high levels (Schmiedel et al., 2015). Thus, the efficacy of miRNAs may change as their targeted mRNA become dysregulated in schizophrenia NCCs. Further studies, including manipulation of individual miRNAs, and new computational techniques that analyze global genome miRNA-mRNA interactions as a function of their levels, could to shed light on the mechanisms of the disintegration of joint miRNA-mRNA network in schizophrenia NCCs.
A novel mechanism that could elicit global genomic dysregulation as observed in schizophrenia NCCs is the pan-ontogenic INFS. Similar as found in mouse ESCs and NCCs (Terranova et al., 2015), nFGFR1 binds throughout all chromosomes in human NCCs. Like in the mouse genome, in both control and schizophrenia NCCs, the nFGFR1 peaks are enriched at the promoters (both distal and proximal) and 5′ UTRs (Terranova et al., 2015), and are found primarily (90%) on expressed genes. Of all annotated human genes (24,331), 54% (13,182 genes) were bound by nFGFR1 in control and patient NCCs. Among 1118 nFGFR1 targeted dysregulated genes, nFGFR1 bound to 915 genes, in both the control and patient genomes, and to 203 genes in only the patient genome. The majority of nFGFR1 peaks were in the same location in control and patient NCCs. However, the total number of peaks was higher in patient NCCs, and the overall binding to the nFGFR1 promoter stronger in schizophrenia NCCs than in control cells. Thus, the increase in nFGFR1 binding and de-novo targeting of gene promoters by nFGFR1 in schizophrenia cells may contribute to the observed gene dysregulation. Analysis by qPCR confirmed that the increase in nFGFR1 in the nucleus is sufficient to increase expression of the exemplary neuronal genes, TH, WNT7B, and Neurod4, as well as to suppress the expression of the oligodendrocytic genes, Olig1, Olig2 and NCAM, thus modeling the changes observed in patient cells. Furthermore, nFGFR1 targeted motifs, many of which were the same in the mouse genome (Terranova et al., 2015), are effectively regulated by nFGFR1. For example, increased binding of nFGFR1 to DNA motifs targeted by Nur (NuRE, NBRE), RAR and RXR (RARE), CREB (CRE), myc (Ebox), AP1 and NfκB promotes transcription by RNA Polymerase II. In contrast nFGFR1 binding to the Smad site inhibits transcription (Stachowiak et al., 2015; Terranova et al., 2015). Therefore, the generalized increase in nFGFR1 binding could lead to global changes in gene activities as observed in schizophrenia cells.
Our investigation has revealed a close relationship between the nFGFR1 and the Notch1 systems the significance of which will require further investigation. Among genes whose expression is dysregulated in schizophrenia NCCs, Notch pathway genes showed a concerted suppression (Figure S10), and several of those genes (i.e., NOTCH2, DTX4, HES5, MAML2, HEY2, DLL3, and JAG11) were directly targeted by nFGFR1 (Table S2, S11). Also, many of the nR1 regulated genes (IRX3, ID3, PAX3(Terranova et al., 2015) and NCAM present study) ) are targeted by both nR1 and Notch (Figure S11). Thus R1 may control expression of genes involed in Notch synthesis and signaling as well as interact with the Notch1 targted genes. The global interactions between the nFGFR1 and Notch systems could play an important role in the etiology of schizophrenia and thus should be addressed by future experiments.
In addition to the nFGFR1 peaks that were associated with the proximal promoters, we observed changes in the number of peaks in the distal promoters and the intergenic regions in schizophrenia NCCs. The role of distal intergenic binding by nFGFR1 is under investigation. It is possible that these peaks play a role in chromatin organization. Motif analysis revealed that in the patient NCC genome, nFGFR1 targets DNA motifs that are bound by CTCF, a chromatin insulator and an organizer of topologically associating domains (Ciabrelli and Cavalli, 2015). Therefore, the binding of FGFR1 to CTCF in these distal regions may be related to global gene dysregulation over longer distances in developing schizophrenia neurons.
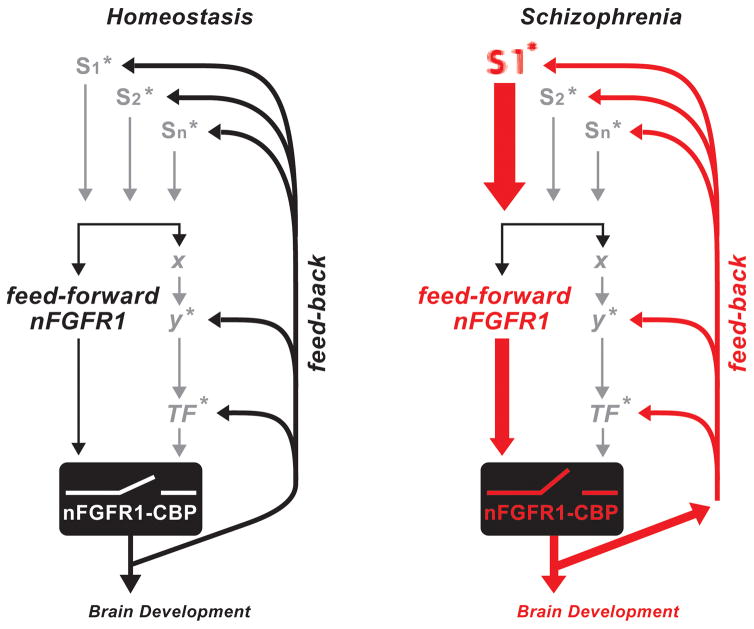
In conclusion, our detailed investigation of global transcriptomes in four different schizophrenia patients and four unrelated control subjects lends strong support to the watershed model of the polygenic, rare variant disease, schizophrenia. Our investigation designates pan-ontogenic INFS as a potential central, converging mechanism, which receives aberrant signals from schizophrenia genes and elicits genome dysregulation in differentiating neural progenitor cells (Fig. 8).
Figure 8.
Disruption of INFS in schizophrenia. “Feed-Forward-and-Gate” signaling by INFS during development(Fang et al., 2005; Stachowiak et al., 2015). Neurogenic signals generated by diverse extracellular stimuli (S: neurotransmitters, hormones, growth factors, cell contact receptors) are propagated through signaling pathways (SiP; cAMP, Ca++/PKC, MAPK, etc.) to sequence-specific transcription factors (TF: CREB, AP1, NfkB, Smads, Klf4, Stat3, RXR/RAR, etc.). In parallel, newly synthesized nFGFR1 translocates into the nucleus and “feeds forward” (F-F) developmental signals directly to CREB binding protein (CBP), an essential transcriptional co-activator and gene-gating factor. The coupled activation of TFs and CBP by nFGFR1 allows genes to respond to developmental signals in a coordinated fashion. In addition, INFS reinforces or turns off the input signals via a feedback loop (Stachowiak et al., 2013a). * marks signaling pathways in which schizophrenia-linked genes have been found, including cAMP, G-protein, PKC, MAPK, NfkB, CREB, RXR, and Nurr1 pathways (Stachowiak et al., 2013a). In schizophrenia and other neurodevelopmental diseases, mutations of these individual genes, including “weak” copy variations, could deregulate this auto-regulated genomic circuit (red lines) and thus lead to broad molecular and developmental dysfunction.
Overactive INFS may dysregulate multitudes of common, nFGFR1-targeted developmental pathways in patients with different schizophrenia-linked mutations. Given that nFGFR1 is both sufficient and essential for neuronal programing of stem and neural progenitor cells (transfected constitutively active nuclear (FGFR1(SP-/NLS) induces neuronal differentiation in ESC and NPC, while the dominant negative (FGFR1(SP-/NLS)(TK-) prevents retinoic acid, cAMP, NGF or BMP4 neuronal programing (Fang et al., 2005; Lee et al., 2013; Lee et al., 2012; Stachowiak et al., 2003)), treatments that target the overactive INFS may reduce developmental progression of brain malformations and emerge as a potential strategy for treating schizophrenia and other neurodevelopmental disorders, including autism. Our findings warrant further INFS-focused investigation of the diverse groups of patients.
Supplementary Material
Acknowledgments
We thank Dr. Michael Buck (NextGen Sequencing and Expression Analysis Core, SUNY, Buffalo), Drs. Barbara Birkaya and Scott Doyle and graduate students Brandon Decker, David Freedman and Seerat Elahi for critical discussion of this work. This work was supported by grants from New York State Department of Health (NYSTEM C026415, C026714), National Science Foundation (CBET-1555720) and Patrick P. Lee Foundation to MKS. KB was supported by grants from NIH (R01 MH101454 and R01 MH106056), and the New York Stem Cell Foundation.
Footnotes
Competing Financial Interests: authors declare no competing financial interests.
References
- Akbarian S, Bunney WE, Jr, Potkin SG, Wigal SB, Hagman JO, Sandman CA, Jones EG. Altered distribution of nicotinamide-adenine dinucleotide phosphate-diaphorase cells in frontal lobe of schizophrenics implies disturbances of cortical development. Archives of general psychiatry. 1993;50(3):169–177. doi: 10.1001/archpsyc.1993.01820150007001. [DOI] [PubMed] [Google Scholar]
- Arnold SE, Ruscheinsky DD, Han LY. Further evidence of abnormal cytoarchitecture of the entorhinal cortex in schizophrenia using spatial point pattern analyses. Biological psychiatry. 1997;42(8):639–647. doi: 10.1016/s0006-3223(97)00142-x. [DOI] [PubMed] [Google Scholar]
- Baron O, Foerthmann B, Lee YW, Terranova C, Ratzka A, Stachowiak EK, Grothe C, Claus P, Stachowiak MK. Cooperation of nuclear FGFR1 and Nurr1 offers a new interactive mechanism in postmitotic development of mesencephalic dopaminergic neurons. The Journal of biological chemistry. 2012a doi: 10.1074/jbc.M112.347831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron O, Forthmann B, Lee YW, Terranova C, Ratzka A, Stachowiak EK, Grothe C, Claus P, Stachowiak MK. Cooperation of nuclear fibroblast growth factor receptor 1 and nurr1 offers new interactive mechanism in postmitotic development of mesencephalic dopaminergic neurons. The Journal of biological chemistry. 2012b;287(24):19827–19840. doi: 10.1074/jbc.M112.347831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, Kwok EW, Vincent SL, Todtenkopf MS. A reduction of nonpyramidal cells in sector CA2 of schizophrenics and manic depressives. Biological psychiatry. 1998;44(2):88–97. doi: 10.1016/s0006-3223(98)00138-3. [DOI] [PubMed] [Google Scholar]
- Berretta S. Extracellular matrix abnormalities in schizophrenia. Neuropharmacology. 2012;62(3):1584–1597. doi: 10.1016/j.neuropharm.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard JJ, Kring AM, Horan WP, Gur R. Toward the next generation of negative symptom assessments: the collaboration to advance negative symptom assessment in schizophrenia. Schizophrenia bulletin. 2011;37(2):291–299. doi: 10.1093/schbul/sbq104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobbs AS, Saarela AV, Yatskievych TA, Antin PB. Fibroblast growth factor (FGF) signaling during gastrulation negatively modulates the abundance of microRNAs that regulate proteins required for cell migration and embryo patterning. The Journal of biological chemistry. 2012;287(46):38505–38514. doi: 10.1074/jbc.M112.400598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerts B, Hantsch J, Herzer M. A morphometric study of the dopamine-containing cell groups in the mesencephalon of normals, Parkinson patients, and schizophrenics. Biological psychiatry. 1983;18(9):951–969. [PubMed] [Google Scholar]
- Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, Beaumont KG, Kim HJ, Topol A, Ladran I, Abdelrahim M, Matikainen-Ankney B, Chao SH, Mrksich M, Rakic P, Fang G, Zhang B, Yates JR, 3rd, Gage FH. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Molecular psychiatry. 2015;20(3):361–368. doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, Gage FH. Concise review: the promise of human induced pluripotent stem cell-based studies of schizophrenia. Stem cells. 2011;29(12):1915–1922. doi: 10.1002/stem.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, Landek-Salgado MA, Sawa A. Modeling heterogeneous patients with a clinical diagnosis of schizophrenia with induced pluripotent stem cells. Biological psychiatry. 2014a;75(12):936–944. doi: 10.1016/j.biopsych.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, Silvas J, Kim Y, Tran N, Simone A, Ladran I, Beaumont K, Kim H-J, Mrksich M, Fang G, Zhang B, Yates JR, 3rd, Gage FH. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Molecular psychiatry. 2014b doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, McCarthy S, Sebat J, Gage FH. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473(7346):221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DM, Stow JL. Nuclear translocation of cell-surface receptors: lessons from fibroblast growth factor. Traffic. 2005;6(10):947–954. doi: 10.1111/j.1600-0854.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- Cannon TD, Keller MC. Endophenotypes in the genetic analyses of mental disorders. Annual review of clinical psychology. 2006;2:267–290. doi: 10.1146/annurev.clinpsy.2.022305.095232. [DOI] [PubMed] [Google Scholar]
- Chiang CH, Su Y, Wen Z, Yoritomo N, Ross CA, Margolis RL, Song H, Ming GL. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Molecular psychiatry. 2011;16(4):358–360. doi: 10.1038/mp.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chioni AM, Grose R. FGFR1 cleavage and nuclear translocation regulates breast cancer cell behavior. J Cell Biol. 2012;197(6):801–817. doi: 10.1083/jcb.201108077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciabrelli F, Cavalli G. Chromatin-driven behavior of topologically associating domains. J Mol Biol. 2015;427(3):608–625. doi: 10.1016/j.jmb.2014.09.013. [DOI] [PubMed] [Google Scholar]
- Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1(1):37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Ciruna BG, Schwartz L, Harpal K, Yamaguchi TP, Rossant J. Chimeric analysis of fibroblast growth factor receptor-1 (Fgfr1) function: a role for FGFR1 in morphogenetic movement through the primitive streak. Development. 1997;124(14):2829–2841. doi: 10.1242/dev.124.14.2829. [DOI] [PubMed] [Google Scholar]
- Clarke WE, Berry M, Smith C, Kent A, Logan A. Coordination of fibroblast growth factor receptor 1 (FGFR1) and fibroblast growth factor-2 (FGF-2) trafficking to nuclei of reactive astrocytes around cerebral lesions in adult rats. Molecular and cellular neurosciences. 2001;17(1):17–30. doi: 10.1006/mcne.2000.0920. [DOI] [PubMed] [Google Scholar]
- Coleman SJ, Chioni AM, Ghallab M, Anderson RK, Lemoine NR, Kocher HM, Grose RP. Nuclear translocation of FGFR1 and FGF2 in pancreatic stellate cells facilitates pancreatic cancer cell invasion. EMBO molecular medicine. 2014;6(4):467–481. doi: 10.1002/emmm.201302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton MT, Walker EF. Physical manifestations of neurodevelopmental disruption: are minor physical anomalies part of the syndrome of schizophrenia? Schizophrenia bulletin. 2009;35(2):425–436. doi: 10.1093/schbul/sbn151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor SE, Ng V, McDonald C, Schulze K, Morgan K, Dazzan P, Murray RM. A study of hippocampal shape anomaly in schizophrenia and in families multiply affected by schizophrenia or bipolar disorder. Neuroradiology. 2004;46(7):523–534. doi: 10.1007/s00234-004-1224-0. [DOI] [PubMed] [Google Scholar]
- Davis KL, Stewart DG, Friedman JI, Buchsbaum M, Harvey PD, Hof PR, Buxbaum J, Haroutunian V. White matter changes in schizophrenia: evidence for myelin-related dysfunction. Archives of general psychiatry. 2003;60(5):443–456. doi: 10.1001/archpsyc.60.5.443. [DOI] [PubMed] [Google Scholar]
- de Bartolomeis A, Iasevoli F, Tomasetti C, Buonaguro EF. MicroRNAs in Schizophrenia: Implications for Synaptic Plasticity and Dopamine-Glutamate Interaction at the Postsynaptic Density. New Avenues for Antipsychotic Treatment Under a Theranostic Perspective. Molecular neurobiology. 2015;52(3):1771–1790. doi: 10.1007/s12035-014-8962-8. [DOI] [PubMed] [Google Scholar]
- Deep-Soboslay A, Benes FM, Haroutunian V, Ellis JK, Kleinman JE, Hyde TM. Psychiatric brain banking: three perspectives on current trends and future directions. Biological psychiatry. 2011;69(2):104–112. doi: 10.1016/j.biopsych.2010.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequeant ML, Pourquie O. Segmental patterning of the vertebrate embryonic axis. Nature reviews Genetics. 2008;9(5):370–382. doi: 10.1038/nrg2320. [DOI] [PubMed] [Google Scholar]
- Dunham-Ems SM, Lee YW, Stachowiak EK, Pudavar H, Claus P, Prasad PN, Stachowiak MK. Fibroblast growth factor receptor-1 (FGFR1) nuclear dynamics reveal a novel mechanism in transcription control. Molecular biology of the cell. 2009;20(9):2401–2412. doi: 10.1091/mbc.E08-06-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Stachowiak EK, Dunham-Ems SM, Klejbor I, Stachowiak MK. Control of CREB-binding protein signaling by nuclear fibroblast growth factor receptor-1: a novel mechanism of gene regulation. The Journal of biological chemistry. 2005;280(31):28451–28462. doi: 10.1074/jbc.M504400200. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Folsom TD. The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophrenia bulletin. 2009;35(3):528–548. doi: 10.1093/schbul/sbn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foussias G, Mann S, Zakzanis KK, van Reekum R, Agid O, Remington G. Prediction of longitudinal functional outcomes in schizophrenia: the impact of baseline motivational deficits. Schizophrenia research. 2011;132(1):24–27. doi: 10.1016/j.schres.2011.06.026. [DOI] [PubMed] [Google Scholar]
- Gonzalez AM, Berry M, Maher PA, Logan A, Baird A. A comprehensive analysis of the distribution of FGF-2 and FGFR1 in the rat brain. Brain Res. 1995;701(1–2):201–226. doi: 10.1016/0006-8993(95)01002-x. [DOI] [PubMed] [Google Scholar]
- Hamlyn J, Duhig M, McGrath J, Scott J. Modifiable risk factors for schizophrenia and autism--shared risk factors impacting on brain development. Neurobiology of disease. 2013;53:3–9. doi: 10.1016/j.nbd.2012.10.023. [DOI] [PubMed] [Google Scholar]
- Han X, Xiao Z, Quarles LD. Membrane and integrative nuclear fibroblastic growth factor receptor (FGFR) regulation of FGF-23. J Biol Chem. 2015;290(33):20101. doi: 10.1074/jbc.A114.609230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa S, Bae JK, Bae YJ, Chae MH, Tanaka H, Nakane H, Ohta Y, Zhao X, Iizuka H, Nakane Y. Psychological impact on caregivers traumatized by the violent behavior of a family member with schizophrenia. Asian journal of psychiatry. 2013;6(1):46–51. doi: 10.1016/j.ajp.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Torii K, Torii M, Fujimoto M, Nakai A, El Fatimy R, Mezger V, Ju MJ, Ishii S, Chao SH, Brennand KJ, Gage FH, Rakic P. Roles of heat shock factor 1 in neuronal response to fetal environmental risks and its relevance to brain disorders. Neuron. 2014;82(3):560–572. doi: 10.1016/j.neuron.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haw RA, Croft D, Yung CK, Ndegwa N, D’Eustachio P, Hermjakob H, Stein LD. The Reactome BioMart. Database : the journal of biological databases and curation 2011. 2011:bar031. doi: 10.1093/database/bar031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MJ, Donocik JG, Nuamah RA, Mein CA, Sainz-Fuertes R, Bray NJ. Transcriptional consequences of schizophrenia candidate miR-137 manipulation in human neural progenitor cells. Schizophrenia research. 2014;153(1–3):225–230. doi: 10.1016/j.schres.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Haley EM, Hibbs MA, Dumeaux V, Barrett DR, Coller HA, Troyanskaya OG. Exploring the human genome with functional maps. Genome Res. 2009;19(6):1093–1106. doi: 10.1101/gr.082214.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia C. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455(7210):237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungerius BJ, Hoogendoorn ML, Bakker SC, Van’t Slot R, Bardoel AF, Ophoff RA, Wijmenga C, Kahn RS, Sinke RJ. An association screen of myelin-related genes implicates the chromosome 22q11 PIK4CA gene in schizophrenia. Mol Psychiatry. 2008;13(11):1060–1068. doi: 10.1038/sj.mp.4002080. [DOI] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, Grozeva D, Fjodorova M, Wollerton R, Rees E, Nikolov I, van de Lagemaat LN, Bayes A, Fernandez E, Olason PI, Bottcher Y, Komiyama NH, Collins MO, Choudhary J, Stefansson K, Stefansson H, Grant SG, Purcell S, Sklar P, O’Donovan MC, Owen MJ. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Molecular psychiatry. 2012;17(2):142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneeland RE, Fatemi SH. Viral infection, inflammation and schizophrenia. Progress in neuro-psychopharmacology & biological psychiatry. 2013;42:35–48. doi: 10.1016/j.pnpbp.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadbeater WE, Gonzalez AM, Logaras N, Berry M, Turnbull JE, Logan A. Intracellular trafficking in neurones and glia of fibroblast growth factor-2, fibroblast growth factor receptor 1 and heparan sulphate proteoglycans in the injured adult rat cerebral cortex. J Neurochem. 2006;96(4):1189–1200. doi: 10.1111/j.1471-4159.2005.03632.x. [DOI] [PubMed] [Google Scholar]
- Lee YW, Stachowiak EK, Birkaya B, Terranova C, Capacchietti M, Claus P, Aletta JM, Stachowiak MK. NGF-induced cell differentiation and gene activation is mediated by integrative nuclear FGFR1 signaling (INFS) PloS one. 2013;8(7):e68931. doi: 10.1371/journal.pone.0068931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YW, Terranova C, Birkaya B, Narla S, Kehoe D, Parikh A, Dong S, Ratzka A, Brinkmann H, Aletta JM, Tzanakakis ES, Stachowiak EK, Claus P, Stachowiak MK. A novel nuclear FGF Receptor-1 partnership with retinoid and Nur receptors during developmental gene programming of embryonic stem cells. J Cell Biochem. 2012;113(9):2920–2936. doi: 10.1002/jcb.24170. [DOI] [PubMed] [Google Scholar]
- Lloyd T, Dazzan P, Dean K, Park SB, Fearon P, Doody GA, Tarrant J, Morgan KD, Morgan C, Hutchinson G, Leff J, Harrison G, Murray RM, Jones PB. Minor physical anomalies in patients with first-episode psychosis: their frequency and diagnostic specificity. Psychological medicine. 2008;38(1):71–77. doi: 10.1017/S0033291707001158. [DOI] [PubMed] [Google Scholar]
- Maher PA. Nuclear Translocation of fibroblast growth factor (FGF) receptors in response to FGF-2. The Journal of cell biology. 1996;134(2):529–536. doi: 10.1083/jcb.134.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, McCarthy S, Michaelson JJ, Vacic V, Burdick KE, Yoon S, Cichon S, Corvin A, Gary S, Gershon ES, Gill M, Karayiorgou M, Kelsoe JR, Krastoshevsky O, Krause V, Leibenluft E, Levy DL, Makarov V, Bhandari A, Malhotra AK, McMahon FJ, Nothen MM, Potash JB, Rietschel M, Schulze TG, Sebat J. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72(6):951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, Willoughby D, Kenny PJ, Elsworth JD, Lawrence MS, Roth RH, Edbauer D, Kleiman RJ, Wahlestedt C. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci U S A. 2012;109(8):3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biological psychiatry. 2011;69(2):188–193. doi: 10.1016/j.biopsych.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla ST, Klejbor I, Birkaya B, Lee YW, Morys J, Stachowiak EK, Prokop D, Bencherif M, Stachowiak MK. Activation of developmental nuclear fibroblast growth factor receptor 1 signaling and neurogenesis in adult brain by alpha7 nicotinic receptor agonist. Stem cells translational medicine. 2013;2(10):776–788. doi: 10.5966/sctm.2012-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KL, Hunt P, Ge D, Heinzen EL, Maia JM, Shianna KV, Weale ME, Cherkas LF, Clement G, Spector TD, Gibson G, Goldstein DB. A genome-wide study of common SNPs and CNVs in cognitive performance in the CANTAB. Hum Mol Genet. 2009;18(23):4650–4661. doi: 10.1093/hmg/ddp413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniyappan L, Al-Radaideh A, Mougin O, Gowland P, Liddle PF. Combined white matter imaging suggests myelination defects in visual processing regions in schizophrenia. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2013;38(9):1808–1815. doi: 10.1038/npp.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partanen J, Schwartz L, Rossant J. Opposite phenotypes of hypomorphic and Y766 phosphorylation site mutations reveal a function for Fgfr1 in anteroposterior patterning of mouse embryos. Genes & development. 1998;12(15):2332–2344. doi: 10.1101/gad.12.15.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen BD, Maciel RD, Galina A, da Silveira MS, Souza CD, Drummond H, Pozzato EN, Junior HS, Chicaybam L, Massuda R, Setti-Perdigao P, Bonamino M, Belmonte-de-Abreu PS, Castro NG, Brentani H, Rehen SK. Altered oxygen metabolism associated to neurogenesis of induced pluripotent stem cells derived from a schizophrenic patient. Cell Transplant. 2011 doi: 10.3727/096368911X600957. [DOI] [PubMed] [Google Scholar]
- Peng H, Moffett J, Myers J, Fang X, Stachowiak EK, Maher P, Kratz E, Hines J, Fluharty SJ, Mizukoshi E, Bloom DC, Stachowiak MK. Novel nuclear signaling pathway mediates activation of fibroblast growth factor-2 gene by type 1 and type 2 angiotensin II receptors. Molecular biology of the cell. 2001;12(2):449–462. doi: 10.1091/mbc.12.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Myers J, Fang X, Stachowiak EK, Maher PA, Martins GG, Popescu G, Berezney R, Stachowiak MK. Integrative nuclear FGFR1 signaling (INFS) pathway mediates activation of the tyrosine hydroxylase gene by angiotensin II, depolarization and protein kinase C. J Neurochem. 2002;81(3):506–524. doi: 10.1046/j.1471-4159.2002.00833.x. [DOI] [PubMed] [Google Scholar]
- Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome biology. 2007;8(2):R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollerberg GE, Thelen K, Theiss MO, Hochlehnert BC. The role of cell adhesion molecules for navigating axons: density matters. Mechanisms of development. 2013;130(6–8):359–372. doi: 10.1016/j.mod.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Rehn AE, Rees SM. Investigating the neurodevelopmental hypothesis of schizophrenia. Clinical and experimental pharmacology & physiology. 2005;32(9):687–696. doi: 10.1111/j.1440-1681.2005.04257.x. [DOI] [PubMed] [Google Scholar]
- Reilly JF, Maher PA. Importin beta-mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation. J Cell Biol. 2001;152(6):1307–1312. doi: 10.1083/jcb.152.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robicsek O, Karry R, Petit I, Salman-Kesner N, Muller FJ, Klein E, Aberdam D, Ben-Shachar D. Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Mol Psychiatry. 2013 doi: 10.1038/mp.2013.67. [DOI] [PubMed] [Google Scholar]
- Roussos P, Guennewig B, Kaczorowski DC, Barry G, Brennand KJ. Activity-Dependent Changes in Gene Expression in Schizophrenia Human-Induced Pluripotent Stem Cell Neurons. JAMA Psychiatry. 2016;73(11):1180–1188. doi: 10.1001/jamapsychiatry.2016.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rummel-Kluge C, Komossa K, Schwarz S, Hunger H, Schmid F, Kissling W, Davis JM, Leucht S. Second-generation antipsychotic drugs and extrapyramidal side effects: a systematic review and meta-analysis of head-to-head comparisons. Schizophrenia bulletin. 2012;38(1):167–177. doi: 10.1093/schbul/sbq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Chant D, Welham J, McGrath J. A systematic review of the prevalence of schizophrenia. PLoS medicine. 2005;2(5):e141. doi: 10.1371/journal.pmed.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biological psychiatry. 2011;69(2):180–187. doi: 10.1016/j.biopsych.2010.09.030. [DOI] [PubMed] [Google Scholar]
- Schiller D, Zuckerman L, Weiner I. Abnormally persistent latent inhibition induced by lesions to the nucleus accumbens core, basolateral amygdala and orbitofrontal cortex is reversed by clozapine but not by haloperidol. Journal of psychiatric research. 2006;40(2):167–177. doi: 10.1016/j.jpsychires.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Schmiedel JM, Klemm SL, Zheng Y, Sahay A, Bluthgen N, Marks DS, van Oudenaarden A. Gene expression. MicroRNA control of protein expression noise. Science. 2015;348(6230):128–132. doi: 10.1126/science.aaa1738. [DOI] [PubMed] [Google Scholar]
- Shi W, Du J, Qi Y, Liang G, Wang T, Li S, Xie S, Zeshan B, Xiao Z. Aberrant expression of serum miRNAs in schizophrenia. Journal of psychiatric research. 2012;46(2):198–204. doi: 10.1016/j.jpsychires.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Srikanth P, Han K, Callahan DG, Makovkina E, Muratore CR, Lalli MA, Zhou H, Boyd JD, Kosik KS, Selkoe DJ, Young-Pearse TL. Genomic DISC1 Disruption in hiPSCs Alters Wnt Signaling and Neural Cell Fate. Cell reports. 2015;12(9):1414–1429. doi: 10.1016/j.celrep.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak EK, Fang X, Myers J, Dunham S, Stachowiak MK. cAMP-induced differentiation of human neuronal progenitor cells is mediated by nuclear fibroblast growth factor receptor-1 (FGFR1) J Neurochem. 2003;84(6):1296–1312. doi: 10.1046/j.1471-4159.2003.01624.x. [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Birkaya B, Aletta JM, Narla ST, Benson CA, Decker B, Stachowiak EK. Nuclear FGF Receptor-1 and CREB Binding Protein: An Integrative Signaling Module. Journal of cellular physiology. 2015;230(5):989–1002. doi: 10.1002/jcp.24879. [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Kucinski A, Curl R, Syposs C, Yang Y, Narla S, Terranova C, Prokop D, Klejbor I, Bencherif M, Birkaya B, Corso T, Parikh A, Tzanakakis ES, Wersinger S, Stachowiak EK. Schizophrenia: a neurodevelopmental disorder--integrative genomic hypothesis and therapeutic implications from a transgenic mouse model. Schizophrenia research. 2013a;143(2–3):367–376. doi: 10.1016/j.schres.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Kucinski A, Curl R, Syposs C, Yang Y, Narla S, Terranova C, Prokop D, Klejbor I, Bencherif M, Birkaya B, Corso T, Parikh A, Tzanakakis ES, Wersinger S, Stachowiak EK. Schizophrenia: a neurodevelopmental disorder - integrative genomic hypothesis and therapeutic implications from a transgenic mouse model. Schizophrenia research. 2013b;143(2–3):367–376. doi: 10.1016/j.schres.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Maher PA, Joy A, Mordechai E, Stachowiak EK. Nuclear accumulation of fibroblast growth factor receptors is regulated by multiple signals in adrenal medullary cells. Molecular biology of the cell. 1996a;7(8):1299–1317. doi: 10.1091/mbc.7.8.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak MK, Maher PA, Joy A, Mordechai E, Stachowiak EK. Nuclear localization of functional FGF receptor 1 in human astrocytes suggests a novel mechanism for growth factor action. Brain research Molecular brain research. 1996b;38(1):161–165. doi: 10.1016/0169-328x(96)00010-1. [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Maher PA, Stachowiak EK. Integrative nuclear signaling in cell development--a role for FGF receptor-1. DNA and cell biology. 2007;26(12):811–826. doi: 10.1089/dna.2007.0664. [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Stachowiak EK. Evidence-Based Theory for Integrated Genome Regulation of Ontogeny-An Unprecedented Role of Nuclear FGFR1 Signaling. Journal of cellular physiology. 2016;231(6):1199–1218. doi: 10.1002/jcp.25298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak MK, Stachowiak EK, Aletta JM, Tzanakakis ES. A common integrative nuclear signaling module for stem cell development. In: Stachowiak MK, EST, editors. Stem Cells from mechanisms to technologies. World Scientific; 2011. pp. 87–132. [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di Forti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St Clair D, Stefansson K Group. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455(7210):232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhlmiller TJ, Garcia-Castro MI. FGF/MAPK signaling is required in the gastrula epiblast for avian neural crest induction. Development. 2012;139(2):289–300. doi: 10.1242/dev.070276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Archives of general psychiatry. 2003;60(12):1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- Terranova C, Narla ST, Lee YW, Bard J, Parikh A, Stachowiak EK, Tzanakakis ES, Buck MJ, Birkaya B, Stachowiak MK. Global Developmental Gene Programing Involves a Nuclear Form of Fibroblast Growth Factor Receptor-1 (FGFR1) PloS one. 2015;10(4):e0123380. doi: 10.1371/journal.pone.0123380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol A, English JA, Flaherty E, Rajarajan P, Hartley BJ, Gupta S, Desland F, Zhu S, Goff T, Friedman L, Rapoport J, Felsenfeld D, Cagney G, Mackay-Sim A, Savas JN, Aronow B, Fang G, Zhang B, Cotter D, Brennand KJ. Increased abundance of translation machinery in stem cell-derived neural progenitor cells from four schizophrenia patients. Transl Psychiatry. 2015a;5:e662. doi: 10.1038/tp.2015.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol A, Zhu S, Hartley BJ, English J, Hauberg ME, Tran N, Rittenhouse CA, Simone A, Ruderfer DM, Johnson J, Readhead B, Hadas Y, Gochman PA, Wang YC, Shah H, Cagney G, Rapoport J, Gage FH, Dudley JT, Sklar P, Mattheisen M, Cotter D, Fang G, Brennand KJ. Dysregulation of miRNA-9 in a Subset of Schizophrenia Patient-Derived Neural Progenitor Cells. Cell reports. 2016;15(5):1024–1036. doi: 10.1016/j.celrep.2016.03.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol A, Zhu S, Tran N, Simone A, Fang G, Brennand KJ. Altered WNT Signaling in Human Induced Pluripotent Stem Cell Neural Progenitor Cells Derived from Four Schizophrenia Patients. Biol Psychiatry. 2015b;78(6):e29–34. doi: 10.1016/j.biopsych.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320(5875):539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, Pear WS, Schug J, Blacklow SC, Arnett KL, Bernstein BE, Kieff E, Aster JC. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci U S A. 2011;108(36):14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, Kim NS, Yoon KJ, Shin J, Zhang C, Makri G, Nauen D, Yu H, Guzman E, Chiang CH, Yoritomo N, Kaibuchi K, Zou J, Christian KM, Cheng L, Ross CA, Margolis RL, Chen G, Kosik KS, Song H, Ming GL. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014 doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nature genetics. 2008;40(7):880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Xu J, Hartley BJ, Kurup P, Phillips A, Topol A, Xu M, Ononenyi C, Foscue E, Ho SM, Baguley TD, Carty N, Barros CS, Muller U, Gupta S, Gochman P, Rapoport J, Ellman JA, Pittenger C, Aronow B, Nairn AC, Nestor MW, Lombroso PJ, Brennand KJ. Inhibition of STEP61 ameliorates deficits in mouse and hiPSC-based schizophrenia models. Molecular psychiatry. 2016 doi: 10.1038/mp.2016.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Nguyen HN, Ursini G, Zhang F, Kim NS, Wen Z, Makri G, Nauen D, Shin JH, Park Y, Chung R, Pekle E, Zhang C, Towe M, Hussaini SM, Lee Y, Rujescu D, St Clair D, Kleinman JE, Hyde TM, Krauss G, Christian KM, Rapoport JL, Weinberger DR, Song H, Ming GL. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell. 2014;15(1):79–91. doi: 10.1016/j.stem.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger ST, Corey DR. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic acids research. 2011;39(13):5682–5691. doi: 10.1093/nar/gkr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DX, Di Giorgio FP, Yao J, Marchetto MC, Brennand K, Wright R, Mei A, McHenry L, Lisuk D, Grasmick JM, Silberman P, Silberman G, Jappelli R, Gage FH. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Reports. 2014;2(3):295–310. doi: 10.1016/j.stemcr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.