Abstract
R loop, a transcription intermediate containing RNA:DNA hybrids and displaced single-stranded DNA (ssDNA), has emerged as a major source of genomic instability. RNaseH1, which cleaves the RNA in RNA:DNA hybrids, plays an important role in R loop suppression. Here, we show that replication protein A (RPA), a ssDNA-binding protein, interacts with RNaseH1 and colocalizes with both RNaseH1 and R loops in cells. In vitro, purified RPA directly enhances the association of RNaseH1 with RNA:DNA hybrids and stimulates the activity of RNaseH1 on R loops. An RPA binding-defective RNaseH1 mutant is not efficiently stimulated by RPA in vitro, fails to accumulate at R loops in cells, and loses the ability to suppress R loops and associated genomic instability. Thus, in addition to sensing DNA damage and replication stress, RPA is a sensor of R loops and a regulator of RNaseH1, extending the versatile role of RPA in suppression of genomic instability.
Introduction
Genomic instability arises from a variety of cellular processes in the genome, including DNA replication and transcription. R loop is a transcription intermediate resulting from stable RNA:DNA hybrids (Santos-Pereira and Aguilera, 2015; Skourti-Stathaki and Proudfoot, 2014; Sollier and Cimprich, 2015). A typical R loop contains an RNA:DNA hybrid and a displaced strand of single-stranded DNA (ssDNA). R loops have physiological functions. For example, the R loops in switch regions of the immunoglobulin locus promote class switch recombination (Yu et al., 2003). The R loops at promoters with high GC skew protect these regions from DNA methylation (Ginno et al., 2012), and the R loops at terminators of certain genes facilitate transcription termination (Sanz et al., 2016; Skourti-Stathaki et al., 2011, 2014). However, R loops are also associated with genomic instability, especially when their levels and distributions are aberrant (Santos-Pereira and Aguilera, 2015; Skourti-Stathaki and Proudfoot, 2014; Sollier and Cimprich, 2015). The displaced ssDNA in R loops is implicated in transcription-associated mutagenesis (Polak and Arndt, 2008). AID (activation-induced cytidine deaminase), which acts on R loops in the switch regions, could trigger chromosomal translocation (Chiarle et al., 2011). The collision between R loops and DNA replication forks gives rise to DNA double-strand breaks (DSBs) (Gan et al., 2011; Santos-Pereira and Aguilera, 2015; Tuduri et al., 2009; Wellinger et al., 2006). R loops are also processed into DSBs by endonucleases (Sollier et al., 2014). Consistent with their association with genomic instability, R loops are found at common fragile sites (Helmrich et al., 2011). The negative impact of R loops on genomic stability creates a demand for tight control of R loops in the genome.
Cells have evolved several mechanisms to down regulate R loops. The formation of R loops is suppressed by Topoisomerase I, which removes the negative supercoils behind RNA polymerases (Li et al., 2015; Tuduri et al., 2009). A number of factors involved in mRNA biogenesis, such as certain splicing factors and components of the RNA exosome complex, are important for antagonizing R loop formation (Huertas and Aguilera, 2003; Li and Manley, 2005; Paulsen et al., 2009; Stirling et al., 2012; Wahba et al., 2011). Many R loop suppressors travel with the transcription complex through binding to RNA polymerase II, pre-mRNA, or their associated factors. BRCA2 and FANCD2, which are required for R loop suppression, interact with an mRNA export factor and colocalize with RNA polymerase II, respectively (Bhatia et al., 2014; García-Rubio et al., 2015; Schwab et al., 2015). Once R loops are formed, they could be unwound by RNA:DNA helicases, such as SETX (Senataxin) and AQR (Aquarius) (De et al., 2015; Hatchi et al., 2015; Mischo et al., 2011; Skourti-Stathaki et al., 2011; Sollier et al., 2014). Additionally, the RNA in RNA:DNA hybrids can be degraded by RNaseH1 and RNaseH2 (Cerritelli and Crouch, 2009; Wahba et al., 2011). Both RNaseH1 and RNaseH2 contribute to the suppression of R loops in the genome (Arora et al., 2014; Chan et al., 2014; Groh and Gromak, 2014; El Hage et al., 2014; Helmrich et al., 2011; Lim et al., 2015), indicating non-redundant functions of these two enzymes. Overexpression of RNaseH1 is sufficient to reduce R loops and associated genomic instability (Paulsen et al., 2009; Stirling et al., 2012). Among the factors that process R loops, SETX and RNaseH2 may travel with replication forks (Alzu et al., 2012; Bubeck et al., 2011). The recruitments of SETX to R loops and RNA polymerase II require BRCA1 and SMN, respectively (Hatchi et al., 2015; Yanling Zhao et al., 2015). Overall, how R loop-processing enzymes recognize R loops and how they are regulated remains poorly understood.
Replication Protein A (RPA), a ssDNA-binding heterotrimeric complex, is crucial for both DNA replication and the DNA damage response (Maréchal and Zou, 2015). At stalled replication forks and sites of DNA damage, RPA functions as a key sensor of ssDNA to coordinate DNA damage signaling and DNA repair or recombination (Flynn and Zou, 2010; Maréchal and Zou, 2015; Zeman and Cimprich, 2014; Zou and Elledge, 2003). Interestingly, RPA is also detected in transcribed regions, suggesting the presence of ssDNA during transcription (Sikorski et al., 2011). RPA was shown to interact with AID, which may facilitate the action of AID during the transcription of immunoglobulin genes (Chaudhuri et al., 2004). In Arabidopsis, the ssDNA-binding protein AtNDX was shown to stabilize the R loops at a promoter (Sun et al., 2013). These tantalizing links among transcription, R loops, ssDNA and ssDNA-binding proteins raise the interesting question as to whether RPA is a sensor of R loops.
In this study, we show that RPA interacts with RNaseH1 and colocalizes with RNaseH1 at R loops in cells. In vitro, RPA directly stimulates the binding of RNaseH1 to RNA:DNA hybrids and enhances its activity on R loops. When the interaction of RNaseH1 with RPA is compromised, RNaseH1 is no longer efficiently stimulated by RPA in vitro, fails to accumulate at R loops in cells, and is unable to suppress R loops and associated genomic instability. Notably, RNaseH1 relies on its interaction with RPA to suppress R loops in multiple pathological or therapeutic contexts, including (1) loss of SETX (Moreira et al., 2004), (2) inhibition of mRNA splicing by pladienolide B (Kotake et al., 2007; Lagisetti et al., 2009; Yokoi et al., 2011), and (3) expression of the cancer-associated U2AF1 mutant (Graubert et al., 2011; Ilagan et al., 2015). These results reveal that RPA is not only important for sensing DNA replication stress but also R loops, highlighting the role of RPA as a master sensor of genomic stress arising from diverse sources.
Results
RPA interacts with RNaseH1
In a previous study, we have used RPA-coated ssDNA (RPA-ssDNA) as bait to identify proteins involved in the DNA damage response (Maréchal et al., 2014). In addition to a number of known and previously uncharacterized RPA-interacting proteins, we identified RNaseH1 as a potential interactor of RPA-ssDNA (Fig. S1A). Immunoprecipitation of GFP-tagged nuclear RNaseH1 (RNaseH1-GFP), but not GFP alone, captured endogenous RPA from cell extracts (Fig. 1A). Immunoprecipitation of SFB (S, Flag, and Biotin-binding peptide)-tagged RPA70, but not RPA32, also captured RNaseH1-GFP (Fig. 1B), suggesting that RNaseH1 interacts with the RPA complex through RPA70. Treatment of cell extracts with DNaseI or RNaseA did not disrupt the interaction between RPA and RNaseH1, suggesting that DNA and RNA are not essential for this interaction (Fig. S1B). Consistent with our results, the interaction between RPA and RNaseH1 was detected using a fragment of RNaseH1 as bait (Bhatia et al., 2014). In contrast to RNaseH1, no interaction was detected between RPA and the three subunits of RNaseH2 (data not shown). Given the known function of RNaseH1 in R loop suppression, the interaction between RPA and RNaseH1 raises the possibility that RPA is involved in the regulation of R loops.
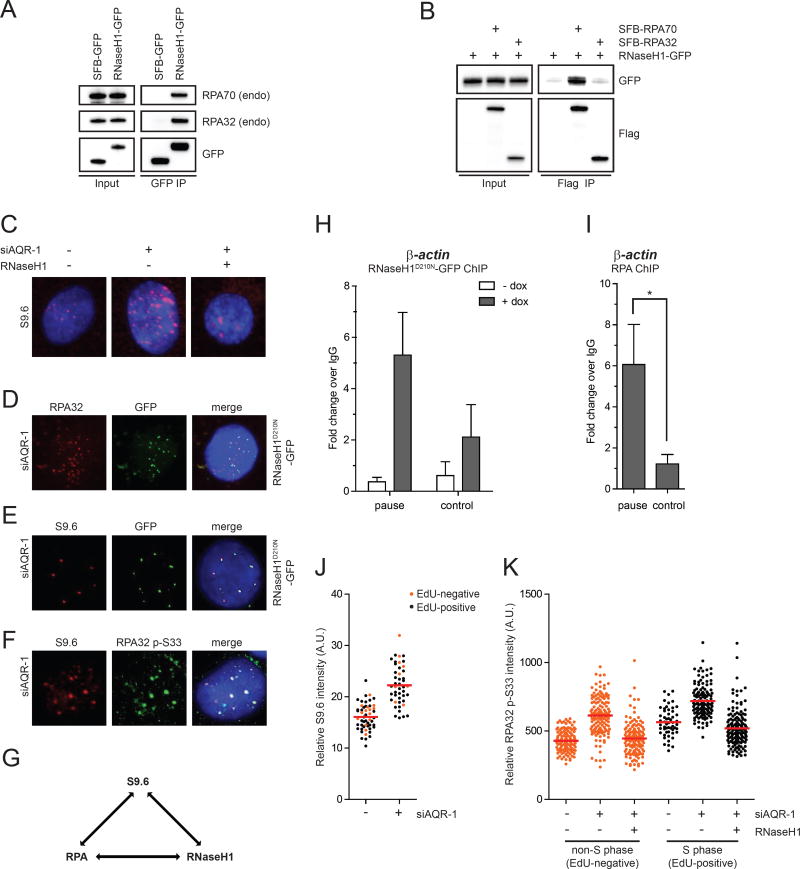
Fig. 1. RPA interacts with RNaseH1 and colocalizes with RNaseH1 and R loops.
(A) RNaseH1-GFP (the mitochondrial localization signal of RNaseH1 was removed) or SFB-GFP was expressed in HEK293T cells and immunoprecipitated using anti-GFP antibody. Endogenous RPA32 associated with RNaseH1-GFP was detected by Western blot. Cell lysates (1%) were loaded as input. (B) RNaseH1-GFP was expressed in HEK293T cells either alone or in combination with SFB-RPA70 or SFB-RPA32 and immunoprecipitated using anti-Flag antibody conjugated to agarose beads. Indicated tagged proteins were detected by Western blot. (C) HeLa-derived RNaseH1 inducible cells were transfected with control or AQR siRNA and cultured for 60 h. RNaseH1 expression was induced by doxycycline for 48 h. R loop levels in individual cells were analyzed using the S9.6 antibody. Representative images for each condition are shown. S9.6 intensity quantification and expression of RNaseH1-GFP are shown in Fig. S1E–F. (D-G) Pair-wise colocalization of RNaseH1D210N-GFP, endogenous RPA32 (or RPA32 p-S33), and R loops (S9.6 foci) were analyzed in AQR knockdown cells as indicated in D, E, and F. RNaseH1D210N-GFP was induced in D and E similar to C. A summary of the three-way colocalization is shown in G. (H-I) HeLa-derived RNaseH1D210N-GFP cells were induced with doxycycline for 48 h prior to fixation and fragmented. Chromatin was immunoprecipitated using anti-GFP (in H) and anti-RPA32 (in I). The association of RNaseH1D210N and RPA with the indicated loci was analyzed by ChIP-qPCR (n=3). Asterisks indicate p<0.05. (J) HeLa cells were transfected with control or AQR siRNA and cultured for 48 h. S-phase and non-S-phase cells were distinguished by 30-min EdU labeling. R loop levels in individual cells were analyzed with the S9.6 antibody (n=50). Red bars represent the mean S9.6 intensities of the indicated cell populations. (K) HeLa-derived cells were transfected with control or AQR siRNA and induced to express RNaseH1-GFP as indicated. S-phase and non-S-phase cells were distinguished by EdU labeling as in J. Levels of RPA32 p-S33 in individual cells were analyzed with the RPA p-S33 antibody (n>100). Red bars represent the mean RPA32 p-S33 intensities of the indicated cell populations. See also Fig. S1.
RPA colocalizes with RNaseH1 and R loops
To determine if RPA is present at R loops, we used the monoclonal antibody S9.6, which specifically recognizes RNA:DNA hybrids, to detect R loops in cells (Boguslawski et al., 1986). Using a previously described protocol (Wan et al., 2015), we detected nuclear foci in HeLa cells with the S9.6 antibody (Fig. 1C). Knockdown of AQR, a putative RNA:DNA helicase (Paulsen et al., 2009; Sollier et al., 2014), increased the foci detected by S9.6 (Fig. 1C). As reported, AQR knockdown also elevated the phosphorylation of KAP1 and H2AX, consistent with the R loop-associated genomic instability (Fig. S1C) (Sollier et al., 2014). To verify whether the foci detected by S9.6 are R loops, we generated a stable cell line that inducibly expresses RNaseH1 (Fig. S1D). Induction of RNaseH1 in cells suppressed the baseline S9.6 staining (Fig. S1E). Upon AQR knockdown, RNaseH1 induction also substantially reduced S9.6 foci (Fig. 1C, S1F). Furthermore, induction of RNaseH1 diminished the p-KAP1 in AQR knockdown cells (Fig. S1G). Together, these results confirm the induction of R loops by AQR knockdown, the suppression of R loops by RNaseH1, and the detection of R loops by the S9.6 antibody.
Next, we tested whether RPA and RNaseH1 are both present at R loops. Because wild-type RNaseH1 actively removes R loops (Fig. 1C), we used a catalytically inactive RNaseH1 mutant, RNaseH1D210N-GFP, to examine the localization of RNaseH1 (Wu et al., 2001). Consistent with the interaction of RNaseH1 with RPA, RNaseH1D210N colocalized with endogenous RPA32 in AQR knockdown cells (Fig. 1D). RNaseH1D210N also colocalized with S9.6 foci, confirming the presence of RNaseH1 at R loops (Fig. 1E). Finally, a phosphorylated form of RPA32 (RPA32 p-S33) was detected in the R loop foci in AQR knockdown cells (Fig. 1F). The phosphorylation of RPA32 at S33 is likely mediated by the ATR-ATRIP kinase complex, which directly interacts with RPA (Shiotani et al., 2013; Zou and Elledge, 2003). The three-way colocalization of RPA, RNaseH1, and R loops suggests that both RPA and RNaseH1 are present at R loops (Fig. 1G).
To corroborate the immunostaining experiments, we used chromatin immunoprecipitation (ChIP) to analyze the association of RNaseH1D210N and RPA with the termination pause sites of β-ACTIN and GEMIN7 genes, two loci known to form R loops (Hatchi et al., 2015; Skourti-Stathaki et al., 2011). In cells induced to express RNaseH1D210N, the RNaseH1 protein was enriched at both termination pause sites compared to nearby control loci (Fig. 1H, S1H). In addition, endogenous RPA was also specifically enriched at the pause sites (Fig. 1I, S1I). These results confirm the colocalization of RNaseH1 and RPA at two R loop-forming loci.
The colocalization of RPA and R loops suggests that RPA may recognize the ssDNA in R loops. Alternatively, RPA may be recruited by R loop-derived structures, such as those induced by the collision of R loops with replication forks. To determine if DNA replication is required for the localization of RPA to R loops, we analyzed the staining of RPA and R loops in S-phase (EdU-positive) and non-S-phase (EdU-negative) cells. Knockdown of AQR increased S9.6 staining in both S-phase and non-S-phase cells (Fig. 1J). AQR knockdown also elevated p-RPA staining in both S-phase and non-S-phase cells (Fig. 1K, S1J). Induction of RNaseH1 in AQR knockdown cells suppressed p-RPA staining, confirming that it arises from R loops (Fig. 1K). These results suggest that R loop formation is not restricted to S phase, and that RPA localizes to R loops independently of DNA replication. Nonetheless, the levels of p-RPA staining were higher in S-phase cells than in non-S-phase cells (Fig. 1K). It is possible that the collision of R loops with replication forks further induces ssDNA in R loops, stalled replication forks, or even DSBs, leading to elevated p-RPA staining. Alternatively, DNA replication may contribute to p-RPA staining independently of R loops. Thus, RPA is likely able to recognize the ssDNA in R loops independently of DNA replication, but its recruitment may be enhanced by R loop-derived structures during S phase.
RPA stimulates the activity of RNaseH1 on R loops
To investigate whether and how RPA regulates RNaseH1 at R loops, we purified both human RPA heterotrimer complex and RNaseH1 from E. coli and tested them in biochemical assays (Fig. S2A). Using a substrate that contains a 25-bp RNA:DNA hybrid and a 65-nt ssDNA overhang (R:D+ssDNA), we confirmed that wild-type human RNaseH1 (RNaseH1WT), like E. coli RNaseH1, was capable of cleaving the RNA in RNA:DNA hybrid efficiently (Fig. S2B, left panel). In contrast, the catalytically inactive RNaseH1 mutant, RNaseH1D210N, was unable to cleave the RNA in R:D+ssDNA (Fig. S2C). Furthermore, RNaseH1WT was not active on a substrate containing double-stranded DNA and a ssDNA overhang, confirming the specificity of RNaseH1 towards RNA:DNA hybrids (Fig. S2B, right panel). These experiments established an in vitro assay for human RNaseH1.
We next tested if RPA affects the activity of RNaseH1. RPA alone did not display any RNase activity on R:D+ssDNA (Fig. S2D). However, in the presence of RNaseH1, RPA stimulated the cleavage of substrate in a concentration-dependent manner (Fig. 2A). Additionally, RPA accelerated the cleavage of R:D+ssDNA by RNaseH1 in time courses (Fig. 2B). To test more rigorously if RPA stimulates RNaseH1 on R loops, we generated an “R loop-like” substrate that contains two arms of dsDNA (30-bp), a bubble of ssDNA (31-nt) in the middle, and an RNA:DNA hybrid (25-nt) in the bubble (this substrate is referred to as the R loop) (Fig. S2E). The predicted structural features of the R loop substrate were verified by mobility in native gels, heat denaturation and restriction digestion (Fig. S2E). Both human and E. coli RNaseH1, but not RPA, were able to cleave the RNA in the R loop substrate (Fig. S2F–G). Again, RPA stimulated the activity of human RNaseH1 on the R loop substrate in a concentration-dependent manner (Fig. 2C). Furthermore, RNaseH1 cleaved the R loop substrate more rapidly in the presence of RPA (Fig. 2D). These results demonstrate that RPA directly stimulates the activity of RNaseH1 on R loops in vitro.
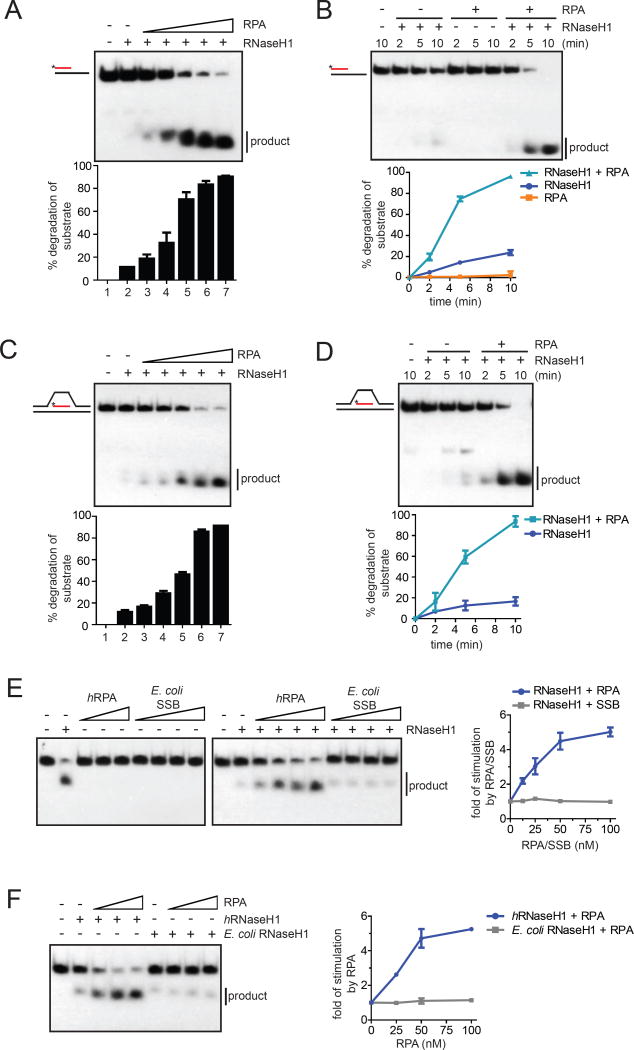
Fig. 2. RPA stimulates the activity of RNaseH1 on R loops.
(A) The R:D+ssDNA substrate with 32P-labeled RNA (25 nM) was incubated with RNaseH1 (2 nM) and increasing concentrations of RPA (0, 12.5, 25, 50, 100, 200 nM) for 5 min. The fractions of substrate cleaved by RNaseH1 were quantified. Data are presented as mean ± SD (n=3). (B) The R:D+ssDNA substrate (100 nM) was incubated with RNaseH1 (5 nM), RPA (100 nM), or both for the indicated amounts of time. Data are presented as mean ± SD (n=3). (C-D) In C, the R loop substrate (25 nM) was incubated with RNaseH1 (2 nM) and increasing concentrations of RPA (0, 12.5, 25, 50, 100, 200 nM) for 5 min. In D, the R loop substrate (100 nM) was incubated with RNaseH1 (2 nM) in the presence or absence of RPA (100 nM) for the indicated amounts of time. Data are presented as mean ± SD (n=3). (E) Left panel, RNA:DNA hybrid (25 nM) was incubated human RPA (0, 12.5*, 25, 50, 100 nM) or E. coli SSB (0, 12.5, 25, 50, 100 nM) in the presence or absence of human RNaseH1 for 5 min. *: only used in the presence of RNaseH1. Right panel, the cleavage of substrate was quantified, and the fold of simulation by RPA was determined. Data are presented as mean ± SD (n=3). (F) Left panel, RNA:DNA hybrid (25 nM) was incubated with human (2 nM) or E. coli RNaseH1 (0.05 nM) and increasing concentrations of human RPA (0, 25, 50, 100 nM). Right panel, the fold of stimulation by RPA was measured as in E. Data are presented as mean ± SD (n=3). See also Fig. S2.
To further assess the specificity of the stimulation of RNaseH1 by RPA, we compared the abilities of human RPA and E. coli SSB to stimulate human RNaseH1 (Fig. S2H). Both RPA and SSB bound ssDNA efficiently (Fig. S2I, see Fig. S3A). However, only RPA but not SSB was able to stimulate the activity of human RNaseH1 (Fig. 2E). Moreover, RPA only stimulated the activity of human RNaseH1, but not E. coli RNaseH1 (Fig. 2F). Thus, the stimulation of RNaseH1 by RPA is species-specific, lending further support to the specificity of this RPA function.
RPA promotes the association of RNaseH1 with RNA:DNA hybrids
RPA may stimulate RNaseH1 through several possible mechanisms. For example, RPA may recruit RNaseH1 to the ssDNA in R loops, thereby increasing the local concentration of RNaseH1. Indeed, only RPA but not RNaseH1 has the ability to bind ssDNA (Fig. S3A). However, even on a substrate consisting of RNA:DNA hybrid but not ssDNA (R:D), RPA still stimulated RNaseH1 in a concentration-dependent manner (Fig. 3A, S3B). RPA also accelerated the cleavage of R:D by RNaseH1 (Fig. 3B). Furthermore, although we captured RNaseH1 from nuclear extracts in our RPA-ssDNA pulldown (Fig. S1A), a tertiary complex of ssDNA, RPA, and RNaseH1 was barely detectable in EMSA (Fig. S3C). Thus, RPA is able to stimulate RNaseH1 on a substrate without ssDNA, and the tertiary complex of RPA, RNaseH1 and ssDNA appears to be unstable, suggesting that the main function of RPA in RNaseH1 stimulation is unlikely to bring RNaseH1 to the ssDNA exposed in R loops. These results do not exclude the possibility that RPA binds to the ssDNA generated during the action of RNaseH1 on RNA:DNA hybrids, which may allow RPA to stimulate the RNaseH1 already on substrates.
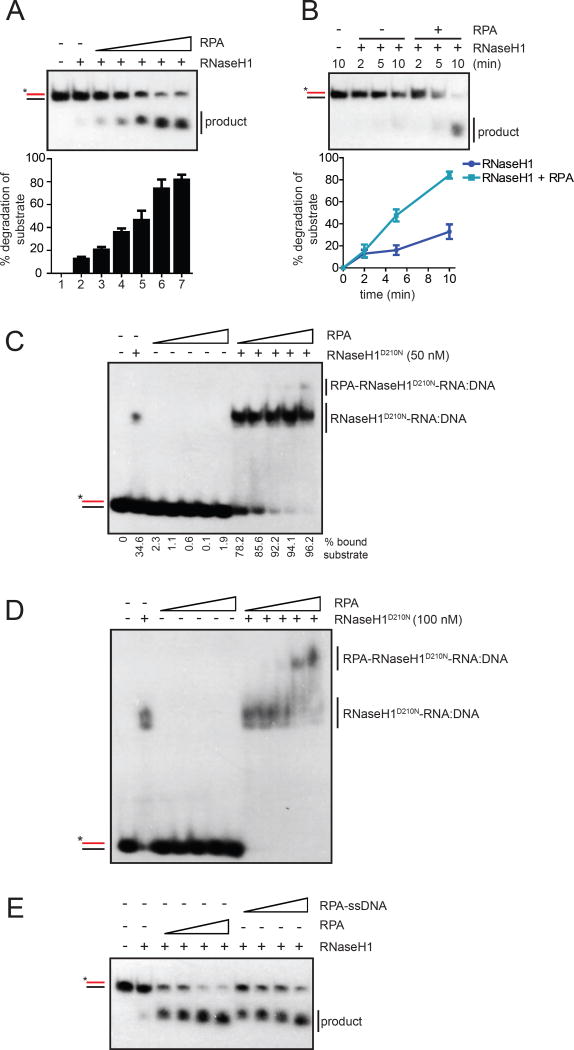
Fig. 3. RPA promotes association of RNaseH1 with RNA:DNA hybrids.
(A-B) In A, the R:D substrate (25 nM) was incubated with RNaseH1 (2 nM) and increasing concentrations of RPA (0, 6.25, 12.5, 25, 50, 100 nM) for 5 min. In B, the R:D substrate (100 nM) was incubated with RNaseH1 (5 nM) in the presence or absence of RPA (100 nM) for the indicated amounts of time. The cleavage of substrate was quantified. Data are presented as mean ± SD (n=3). (C) A 25-bp probe of RNA:DNA hybrid (25 nM) was labeled with 32P and incubated with increasing concentrations of RPA (0, 25, 50, 100, 200, 400 nM) in the presence or absence of RNaseH1D201N (50 nM). The RNaseH1D210N-RNA:DNA complex was detected on a native polyacrylamide gel. (D) The RNA:DNA probe (20 nM) was incubated with RNaseH1D210N (100 nM) and increasing concentrations of RPA (0, 50, 100, 200, 400, 800 nM). The tertiary RPA-RNaseH1D210N-RNA:DNA complex was detected on a native polyacrylamide gel. (E) Increasing concentration of RPA (12.5, 25, 50 and 100 nM) were incubated without or with 80-nt ssDNA (1, 2, 4 and 8 nM, respectively) for 5 min (to form fully covered RPA-ssDNA complex) and RNaseH1WT (2 nM) was incubated for 5 min. Then after R:D substrate (25 nM) was incubated for 5 min in Buffer A with 50 mM KCl. See also Fig. S3.
A second possible mechanism by which RPA stimulates RNaseH1 is to enhance the association of RNaseH1 with RNA:DNA hybrids. Consistent with this possibility, RPA stimulated formation of the RNaseH1-RNA:DNA complex in a concentration-dependent manner (Fig. 3C). In contrast to RPA, E. coli SSB failed to stimulate the binding of RNaseH1 to RNA:DNA hybrids (Fig. S3D). Additionally, while only RNaseH1 but not RPA was able to bind RNA:DNA hybrids in EMSA (Fig. S3E), the presence of both RNaseH1 and RPA led to formation of a tertiary complex (Fig. 3D). These results show that RPA directly interacts with RNaseH1 and promotes the association of RNaseH1 with RNA:DNA, providing a mechanistic explanation of how RPA stimulates RNaseH1.
If RPA binds to the displaced ssDNA in R loops, the binding of RPA to ssDNA may affect its ability to stimulate RNaseH1 either positively or negatively. We pre-incubated RPA with saturating amounts of ssDNA to form RPA-ssDNA, and then added it to RNaseH1 and the R:D substrate (Fig. 3E). Compared to free RPA, RPA-ssDNA displayed a similar activity in RNaseH1 stimulation. We also compared the stimulation of RNaseH1 by RPA on R:D, which contains no ssDNA, and R:D+ssDNA, which carries a ssDNA overhang (Fig. S3F). RPA simulated RNaseH1 on both substrates similarly regardless of the presence or absence of ssDNA. These results suggest that ssDNA-bound RPA is capable of stimulating RNaseH1, and that the specific activity of RPA in RNaseH1 stimulation is not significantly affected by ssDNA binding. While RPA does not need to bind ssDNA to stimulate RNaseH1, the binding of RPA to the ssDNA in R loops in cells may increase the local concentrations of RPA, allowing it to stimulate RNaseH1 more efficiently.
The basic ridge of RNaseH1 is critical for RPA binding but not catalytic activity
The human RNaseH1 contains an RNA:DNA hybrid-binding (HB) domain, an interconnecting domain, and a catalytic domain (Fig. 4A) (Cerritelli and Crouch, 2009). To map the region of RNaseH1 that interacts with RPA, we generated a series of GFP-tagged RNaseH1 fragments and tested their binding to RPA by coimmunoprecipitation. The interconnecting and catalytic domains of RNaseH1 were dispensable for RPA binding, whereas the HB domain was required (Fig. 4A–B). Crystal structure of the HB domain has revealed a positively charged surface adjacent to the RNA:DNA-binding pocket, which was termed basic ridge (Fig. 4C) (Nowotny et al., 2008). Alanine substitutions of several positively charged residues on this surface, including R32 and R33 (RR32AA), or R72 and K73 (RK72AA), did not affect RPA binding (Fig. 4D). However, when the R57 of RNaseH1 was singly mutated to alanine (RNaseH1R57A) or mutated in combination with RR32AA (RNaseH13RA), the binding of RNaseH1 to RPA was largely disrupted (Fig. 4D). The RNaseH1R57A mutant was previously shown to have intact abilities to bind and cleave RNA:DNA hybrids in vitro (Nowotny et al., 2008). Consistently, we found that purified RNaseH1R57A was slightly more active than RNaseH1WT in cleaving R loop and R:D substrates (Fig. S2A, Fig. S4A–B). Purified RNaseH13RA was also more active than RNaseH1WT on R:D (Fig. S4C–D). We conclude that R57A and 3RA mutations specifically disrupt the RPA binding of RNaseH1 without compromising its intrinsic activity on RNA:DNA hybrids.
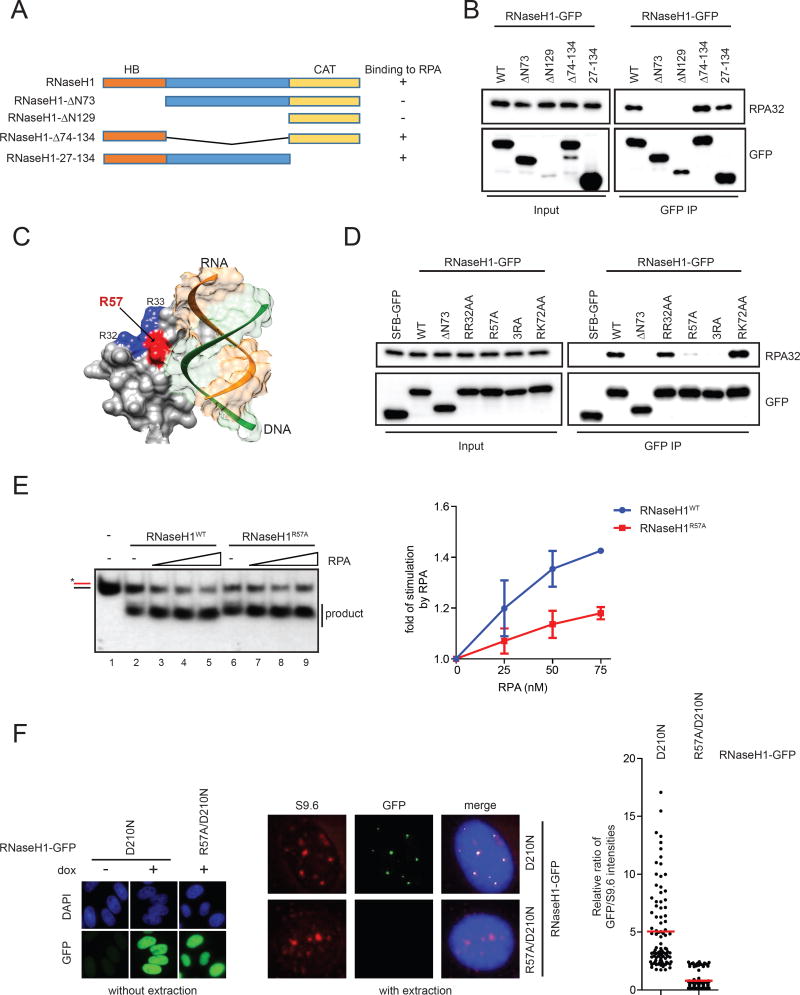
Fig. 4. RNaseH1R57A is compromised for RPA binding and RPA-mediated regulation.
(A) A schematic representation of the domain structure of the nuclear RNaseH1. A summary of the RNaseH1 fragments tested in this study and their abilities to interact with RPA is shown on the right. (B) GFP-tagged RNaseH1 and the indicated mutant derivatives were expressed in HEK293T cells and immunoprecipitated using anti-GFP antibody. GFP and RPA32 in the immunoprecipitates were analyzed by Western blot. Cell lysates (0.5%) were loaded as input. (C) A structural image of the hybrid domain of human RNaseH1 (Nowotny et al., 2008) with a modeled RNA:DNA hybrid substrate was generated using the UCSF Chimera software (PDB ID: 3BSU) (Pettersen et al., 2004). Positively charged residues R32 and R33 are colored in blue, and R57 is highlighted in red. (D) GFP-tagged RNaseH1 and the indicated mutants were expressed in HEK293T cells and tested for RPA binding as in B. Cell lysates (0.5%) were loaded as input. (E) The R:D substrate (25 nM) substrate was incubated with RNaseH1WT (2 nM) or RNaseH1R57A (1 nM) and increasing concentrations of RPA (0, 25, 50, 75 nM) for 5 min in Buffer A with 150 mM KCl. The cleavage of substrate was quantified, and the fold of stimulation by RPA was determined. Data are presented as mean ± SD (n=3). (F) Left panel, induction of RNaseH1D210N-GFP and RNaseH1R57A/D210N-GFP was confirmed using anti-GFP antibody. Cells were not extracted with triton, and soluble GFP proteins were detected. Middle panel, localizations of GFP-tagged RNaseH1 proteins and R loops were analyzed with GFP and S9.6 antibodies. Cells were extracted and fixed with methanol/acetic acid, and only chromatin-bound proteins were detected. Right panel, intensities of S9.6 and GFP foci were quantified in individual cells, and GFP/S9.6 ratios were determined (n=100). Red bars represent the mean GFP/S9.6 ratios of the indicated cell populations. See also Fig. S4.
RNaseH1R57A and RNaseH13RA are compromised for RPA-mediated stimulation in vitro
We next used the RNaseH1R57A and RNaseH13RA mutants to test if the interaction between RNaseH1 and RPA is required for the stimulation of RNaseH1 by RPA. Since RNaseH1R57A and RNaseH13RA mutants are more active than RNaseH1WT, we first normalized the amounts of RNaseH1WT, RNaseH1R57A, and RNaseH13RA to achieve similar cleavage of the R:D substrate (Fig. 4E, lanes 2 and 6; Fig. S4E, lanes 2 and 6). We then titrated RPA into the reactions and followed its effects on the cleavage of R:D by RNaseH1WT, RNaseH1R57A, and RNaseH13RA (Fig. 4E, S4E). At all concentrations tested, RPA stimulated RNaseH1WT more efficiently than RNaseH1R57A and RNaseH13RA. Moreover, we determined the RPA:RNaseH1 ratios in reactions, and compared the RPA stimulation of RNaseH1WT and RNaseH1R57A at the same ratios. Again, RPA stimulated RNaseH1WT more efficiently than RNaseH1R57A at the same RPA:RNaseH1 ratios (Fig. S4F). Consistent with the experiments using the R:D substrate, the stimulation of RNaseH13RA by RPA on the R loop substrate was also compromised (Fig. S4G). Furthermore, compared to that of RNaesH1WT, the binding of RNaseH1R57A to RNA:DNA hybrids was stimulated by RPA less efficiently (Fig. S4H). It should be noted that the stimulation of RNaseH1 by RPA was only partially reduced but not eliminated by R57A and 3RA mutations. Thus, although the basic ridge of RNaseH1 is required for RPA binding and contributes to RPA-mediated stimulation, another surface of RNaseH1 may transiently interact with RPA and be responsible for the remaining RPA-mediated stimulation when the basic ridge is disrupted.
RNaseH1R57A fails to associate with R loops in cells
Given that RPA promotes the association of RNaseH1 with RNA:DNA hybrids in vitro, we asked whether the interaction between RPA and RNaseH1 is important for the recognition of R loops by RNaseH1 in cells. We generated stable lines that express RNaseH1D210N-GFP and RNaseH1R57A/D210N-GFP at similar levels (Fig. S4I). Both RNaseH1D210N-GFP and RNaseH1R57A/D210N-GFP entered the nucleus efficiently (Fig. 4F, left panel). After removal of non-chromatin-bound GFP-tagged proteins by triton extraction, only RNaseH1D210N-GFP but not RNH1R57A/D210N-GFP colocalized with R loop foci in AQR knockdown cells (Fig. 4F, middle panel). When measured quantitatively, the signals of RNaseH1R57A/D210N-GFP in R loop foci were significantly lower than those of RNaseH1D210N-GFP (Fig. 4F, right panel). Thus, the RPA-RNaseH1 interaction mediated by the basic ridge is critical for RNaseH1 to recognize R loops in cells.
RNaseH1R57A fails to suppress R loops and associated genomic instability
To investigate the functional impact of RPA on RNaseH1 in cells, we generated a cell line that inducibly expresses RNaseH1WT-GFP and two other inducible lines expressing RNaseH1R57A-GFP (Fig. S5A–B). All of these cell lines expressed GFP-tagged RNaseH1 proteins at similar levels, and all the RNaseH1 variants entered the nucleus efficiently. As expected, induction of RNaseH1WT significantly reduced R loop and p-RPA foci in AQR knockdown cells (Fig. 5A, top panel). In marked contrast, the RNaseH1R57A mutant lost the ability to suppress foci of R loops and p-RPA in two independent cell lines (Fig. 5A, middle and bottom panels). When AQR knockdown cells were plotted in 2D according to their S9.6 and p-RPA staining, the majority of the cell population ended up in an upper right area (Fig. 5B). Expression of RNaseH1WT shifted the majority of cells leftward and downward in 2D plots, showing that RNaseH1WT suppressed R loops and p-RPA simultaneously in most of the cells. In contrast, induction of RNaseH1R57A in two independent cell lines failed to shift AQR knockdown cells significantly. These results show that the interaction with RPA is critical for RNaseH1 to suppress R loops in cells.
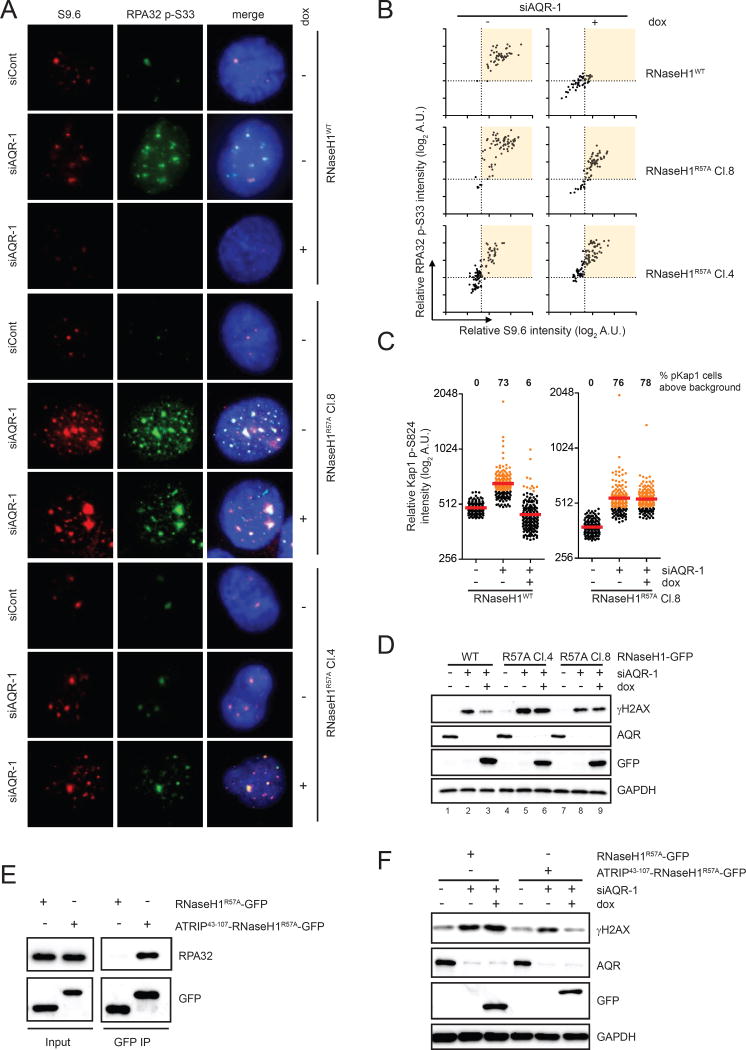
Fig. 5. RNaseH1R57A fails to suppress R loops and associated genomic instability.
(A-B) Inducible cell lines of RNaseH1WT-GFP or RNaseH1R57A-GFP were transfected with control or AQR siRNA and cultured for 60 h. GFP-tagged RNaseH1 proteins were induced by doxycycline for 48 h. Cells were analyzed by immunofluorescence using RPA32 p-S33 and S9.6 antibodies. Representative images are shown in A. Intensities of p-RPA32 and S9.6 staining were quantified in individual cells and plotted in 2-dimensional blots in B (n=80). Induction of RNaseH1WT and RNH1R57A was confirmed by immunofluorescence and Western blot in Fig. S5A and S5B. (C) Inducible cell lines of RNaseH1WT and RNH1R57A were transfected with control or AQR siRNA and induced by doxycycline for 48 h. Levels of Kap1 p-S824 in individual cells were analyzed by immunofluorescence (n>150). Red bars represent the mean p-Kap1 intensities of the indicated cell populations. AQR knockdown cells with p-Kap1 signals above the control cells are colored in orange, and the percentages of these cells in their respective cell populations were quantified. (D) Inducible cell lines of RNaseH1WT and RNH1R57A were treated as in C. Levels of γH2AX and other indicated proteins were analyzed by Western blot. (E) ATRIP43-107-RNaseH1R57A-GFP and RNaseH1R57A-GFP was expressed in HEK293T cells and immunoprecipitated using anti-GFP antibody. Endogenous RPA32 associated with RNaseH1-GFP was detected by Western blot. (F) Inducible cell lines of RNaseH1WT-GFP or RNaseH1R57A-GFP were treated as in D. Levels of γH2AX and other indicated proteins were analyzed by Western blot. See also Fig. S5.
If RPA binding is important for RNaseH1 to remove R loops, the RNaseH1R57A mutant is expected to be defective for suppressing R loop-induced genomic instability. In AQR knockdown cells, the signals of p-KAP staining were significantly elevated compared to control knockdown cells (Fig. 5C), indicating R loop-induced genomic instability. Expression of RNaseH1WT but not RNaseH1R57A in AQR knockdown cells effectively reduced p-KAP signals. Similarly, the levels of phosphorylated H2AX (γH2AX) were significantly increased by AQR knockdown (Fig. 5D). Again, in contrast to RNaseH1WT, RNaseH1R57A failed to suppress the R loop-induced γH2AX in two independent cell lines (Fig. 5D, lanes 3, 6, 9). Together, these results demonstrate that RPA plays a key role in regulating RNaseH1 for the suppression of R loop-induced genomic instability.
While the reduced ability of RNaseH1R57A to interact with RPA is likely responsible for its functional defects in cells, we wanted to exclude the possibility that the basic ridge of RNaseH1 is important for R loop suppression for reasons other than RPA binding. Fusion of the RPA-binding domain of ATRIP to the N terminus of RNaseH1R57A restored its interaction with RPA (Fig. 5E). Furthermore, the RPA-binding domain of ATRIP also largely restored the ability of RNaseH1R57A to suppress the γH2AX in AQR knockdown cells (Fig. 5F). These results strongly suggest that the defect of RNaseH1R57A in RPA binding is the responsible for its functional loss in cells.
A general role of the RPA-RNaseH1 interaction in R loop suppression
The importance of the RPA-RNaseH1 interaction for the suppression of R loops in AQR knockdown cells prompted us to investigate whether this interaction has a general role in R loop suppression in various pathological and therapeutic contexts. Ataxia with oculomotor apraxia type 2 (AOA2) has been linked to SETX mutations (Moreira et al., 2004). Similar to AQR knockdown, depletion of SETX by siRNA elevated levels of R loops and γH2AX (Fig. 6A–B). Expression of RNaseH1WT, but not RNaseH1R57A, suppressed the γH2AX in SETX knockdown cells (Fig. 6B), suggesting that the RPA-RNaseH1 interaction is required for suppressing the R loops arising from SETX loss.
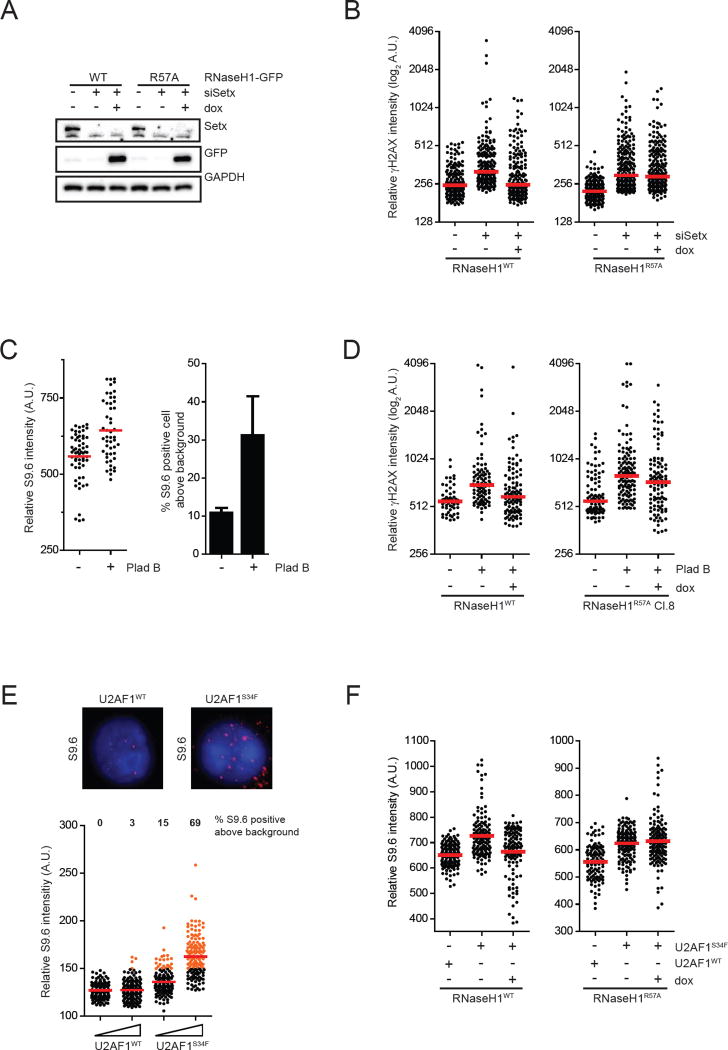
Fig. 6. RNaseH1R57A fails to suppress R loops in a variety of contexts.
(A-B) Inducible cell lines of RNaseH1WT-GFP or RNaseH1R57A-GFP were transfected with control or SETX siRNA and cultured for 60 h. GFP-tagged RNaseH1 proteins were induced by doxycycline for 48 h. In A, levels of indicated proteins were analyzed by Western blot. In B, levels of γH2AX in individual cells were analyzed by immunofluorescence (n>180). Red bars represent the median γH2AX intensities of the indicated cell populations. (C) HeLa cells were either treated with DMSO or 1nM Plad-B for 24 h. Left panel, intensities of S9.6 staining in individual cells were analyzed by immunofluorescence (n>50). Red bars represent the mean S9.6 intensities of the indicated cell populations. Percent of S9.6 positive cells were calculated and plotted on the right panel (n=3). (D) Inducible cell lines of RNaseH1WT-GFP or RNaseH1R57A-GFP were either treated with DMSO or Plad-B for 24h. RNaseH1-GFP proteins were expressed 24 h prior to and the whole duration of Plad-B treatment. Levels of γH2AX in individual cells were analyzed by immunofluorescence (n>60). Red bars represent the median γH2AX intensities of the indicated cell populations. (E) K562-derived cells stably expressing different levels of either U2AF1WT or U2AF1S34F were seeded for 48h prior to analysis by immunofluorescence using S9.6 antibody. Representative images are shown on the top panel. Bottom panels, intensities of S9.6 staining in individual cells were analyzed by immunofluorescence (n>130). Red bars represent the mean S9.6 intensities of the indicated cell populations. Protein expression levels of U2AF1WT and U2AF1S34F are shown in Fig. S6A. (F) Inducible cell lines of RNaseH1WT-GFP or RNaseH1R57A-GFP were stably expressing either U2AF1WT or U2AF1S34F for 60 h. RNaseH1-GFP proteins were expressed by addition of doxycycline. Intensities of S9.6 staining in individual cells from two experiments were combined and analyzed by immunofluorescence (n>100). Red bars represent the mean S9.6 intensities of the indicated cell populations. Protein expression levels of U2AF1WT and U2AF1S34F are shown in Fig. S6B. See also Fig. S6.
Given the link between R loops and defects in splicing factors, we also asked if pharmacological inhibition of mRNA splicing is sufficient to induce R loops. The antitumor drug pladienolide B (Plad-B) induces splicing defects, such as intron retention, by targeting the splicing factor SF3b (Kashyap et al., 2015; Kotake et al., 2007). Treatment of cells with Plad-B increased both S9.6 and γH2AX staining (Fig. 6C–D), suggesting that inhibition of splicing leads to R loop accumulation. Compared to RNaseH1WT, RNaseH1R57A suppressed the γH2AX in Plad-B-treated cells less efficiently (Fig. 6D), which suggests that the RPA-RNaseH1 interaction is important for suppressing the R loops resulting from splicing inhibition.
Finally, we tested if a prevalent splicing factor mutation in cancers induces R loops. The U2AF1-S34F mutation is frequently found in myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML), and it affects the alternative splicing of a number of genes (Graubert et al., 2011; Ilagan et al., 2015). Since U2AF1 mutations are always heterozygous and accumulate at codons S34 and Q157, the U2AF1 mutant proteins are believed to cause gain of function (Ilagan et al. 2015). When U2AF1WT and U2AF1S34F were expressed in K562 cells at similar levels, U2AF1S34F induced higher levels of S9.6 staining than U2AF1WT (Fig. 6E, S6A), suggesting that the splicing defects caused by U2AF1S34F lead to R loop accumulation. Similar observations were made in HeLa-derived cell lines expressing U2AF1WT and U2AF1S34F (Fig. S6B–C). Compared to RNaseH1WT, RNaseH1R57A displayed a reduced ability in suppressing R loops in U2AF1S34F-expressing cells (Fig. 6F), suggesting that the RPA-RNaseH1 interaction is important for suppressing R loops in this oncogenic context.
Collectively, the results above suggest that the interaction between RPA and RNaseH1 plays a general role in R loop suppression in various pathological and therapeutic contexts, thereby highlighting the critical functions of RPA as a general sensor of R loops and a regulator of RNaseH1.
Discussion
RPA as a regulator of RNaseH1
In this study, we show that RPA interacts with RNaseH1 and colocalizes with it at R loops in cells. Furthermore, RPA promotes the accumulation of RNaseH1 at R loops in cells and directly stimulates its activity on R loops in vitro. The ability of RPA to regulate RNaseH1 is not simply attributed to the binding of RPA to ssDNA because E. coli SSB lacks this activity (Fig. 2E). Importantly, RPA directly interacts with RNaseH1 and promotes the association of RNaseH1 with RNA:DNA hybrids (Fig. 3C–D), providing a mechanistic explanation of how RPA facilitates the recognition of R loops by RNaseH1 in cells. In addition, these results suggest that RPA may stimulate the activity of RNaseH1 by enabling the catalytic domain to engage RNA:DNA more efficiently.
Our results show that the interaction between RPA and RNaseH1 is important for the regulation of RNaseH1 by RPA. The R57A mutation in the basic ridge of RNaseH1 significantly weakened the interaction between RNaseH1 and RPA, and drastically reduced the localization of RNaseH1 to R loops in cells (Fig. 4F) (Nowotny et al., 2008). Nonetheless, disruption of the basic ridge by R57A or 3RA mutations only reduced but did not eliminate the stimulation by RPA in vitro (Fig. 4E, S4E). It is possible that RNaseH1 uses its basic ridge to engage RPA, which allows RPA to transiently interact with a second surface of RNaseH1 and stimulate its activity. When RPA is present at high concentrations in vitro, the role of basic ridge in engaging RPA may be partially alleviated. Notably, fusion of the RPA-binding domain of ATRIP with RNaseH1R57A largely restored its localization to R loops and its function in suppressing R loop-associated genomic instability (Fig. 5E–F), confirming that the reduced ability of RNaseH1R57A to interact with RPA is indeed responsible for the defects of this mutant in cells. A recent study showed that E. coli SSB also stimulates the E. coli RNaseH1 in vitro (Petzold et al., 2015), suggesting that the regulation of RNaseH1 by ssDNA-binding proteins is evolutionarily conserved.
RPA as a sensor of R loops
RPA is a crucial sensor of DNA replication stress and DNA damage in eukaryotic cells (Maréchal and Zou, 2015). At stalled replication forks and resected DSBs, RPA-ssDNA acts as a key platform to recruit the ATR kinase and its regulators and substrates. RPA also directly participates in DNA repair by interacting with ssDNA and a number of repair proteins. Like replication forks and repair intermediates, R loop is a ssDNA-containing structure. In this study, we present evidence that RPA is present at R loops in cells. The recognition of ssDNA in R loops by RPA is likely influenced by the length of ssDNA, duration of ssDNA exposure, and secondary DNA structures such as G quadruplexes (Maizels and Gray, 2013). The size and dynamics of R loops are variable at different sites in the genome, and in different physiological or pathological contexts (Sanz et al., 2016; Santos-Pereira and Aguilera, 2015; Skourti-Stathaki and Proudfoot, 2014; Sollier and Cimprich, 2015). Some R loops, such as those at class switch regions, are >1 kb long and stable (Yu et al., 2003). Non-denaturing bisulfite footprinting of several other genomic loci with R loops detected >600 bp of ssDNA (Ginno et al., 2012). These long and persistent R loops may be particularly detrimental to genomic stability, and also likely recognized by RPA.
How exactly RPA recognizes the ssDNA at R loops remains to be elucidated. R loop-induced p-RPA foci were detected in non-S-phase cells (Fig. 1K, S1J), suggesting that RPA can recognize R loops independently of DNA replication. The RPA-RNaseH1 interaction is required for the suppression of R loop-associated DNA damage (Fig. 5C–D), suggesting that RPA recognizes R loops before they are converted to DSBs. The most straightforward possibility is that RPA directly recognizes the displaced ssDNA in R loops. However, our results do not exclude the possibility that R loops need to be processed by certain repair or recombination factors to be recognized by RPA. Notably, R loop-induced p-RPA staining was higher in S-phase cells than in non-S-phase cells (Fig. 1K), suggesting that DNA replication contributes to the recognition of R loops by RPA. During S phase, the collision between replication forks and R loops may increase the ssDNA in R loops, and/or induce ssDNA at the forks stalled by R loops (Gan et al., 2011). Furthermore, the encounter of replication forks and transcription complexes may give rise to additional R loops (Helmrich et al., 2011). Such inductions of ssDNA at or near R loops may allow RPA to sense the increase of genomic stress during replication, helping remove R loops in front of replication forks.
A general role of RPA in R loop suppression
Our results suggest that RPA is important for R loop suppression in a variety of cellular contexts. For example, in cells lacking AQR or SETX, the RPA binding-defective RNaseH1R57A mutant is compromised for R loop suppression (Fig. 5A–B, Fig. 6B). We also present evidence that pharmacological inhibition of mRNA splicing and a cancer-associated splicing factor mutation induce R loop accumulation (Fig. 6C, 6E, S6C). These results raise the possibility that R loop formation may contribute to both cancer development and therapeutic responses in specific contexts. Again, the RPA-RNaseH1 interaction is important for R loop regulation in these contexts (Fig. 6D, 6F). It is tempting to speculate that RPA is a key sensor of R loops in addition to its role in sensing DNA replication stress and DNA damage. While our current data are limited to the interplays between RPA and RNaseH1, RPA may regulate additional factors involved in R loop metabolism. The induction of ssDNA by alterations in transcription or mRNA biogenesis may enable RPA to sense a much broader spectrum of genomic stress and suppress genomic instability in previously unappreciated contexts.
STAR⋆ METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Please direct any requests for further information or reagents to the lead contact, Professor Lee Zou (zou.lee@mgh.harvard.edu), Massachusetts General Hospital Cancer Center, Harvard medical School, Boston, MA 02129, USA.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
HeLa and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2mM Glutamine, and 1% penicillin/streptomycin. The transfection reagent Lipofectamine RNAiMAX (ThermoFisher Scientific) was used for siRNA transfection. Transfection of plasmid DNA to HEK293T cells was carried out by the calcium phosphate transfection method. The HeLa-derived cell lines that inducibly express GFP-tagged nuclear RNaseH1 or its mutant derivatives were generated by lentiviral infection and neomycin selection. All HeLa-derived cell lines were cultured in medium supplemented with G418 (600 μg/ml). RNaseH1-GFP expression was typically induced by doxycycline (200 ng/ml) for 48 h. Viruses expressing Flag-tagged U2AF1WT and U2AF1S34F were used to infect HeLa and HeLa-derived RNaseH1-GFP-expressing cell lines. The plasmids expressing U2AF1WT and U2AF1S34F contain an IRES-GFP, which is used to sort for infected cells.
Plasmids and siRNAs
The plasmid containing GFP-tagged RNaseH1 (the M27 form) under the control of a CMV promoter was kindly provided by Dr. Robert Crouch. The M27 form of RNaseH1 primarily localizes to the nucleus (Suzuki et al., 2010). Various RNaseH1 mutations were generated using the Infusion HD Cloning Kit (Clontech). For lentiviral expression, RNaseH1-GFP coding sequence was cloned into the pDONR221 Gateway vector and transferred into the pINDUCER20 vector for viral packaging. For recombinant protein expression, RNaseH1 coding sequence was cloned into the pDONR221 vector and transferred into a destination vector carrying a N-terminal 6xHis tag. The plasmids containing N-terminally GFP-tagged RNaseH2 subunits (A-C) were kindly provided by Dr. Min Ae Lee-Kirsch.
Silencer Select pre-designed siRNAs targeting Aquarius (AQR) [siAQR-1 (ID # s18725), siAQR-2 (ID # s18726), and siAQR-3 (ID # s18727)] were purchased from ThermoFisher Scientific and used at a final concentration of 4 nM.
METHOD DETAILS
Immunoprecipitation
HET293T cells were transfected with various plasmids (10–15 μg) with calcium phosphate and collected in 72 h. Cell pellets were lysed in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% Igapel-60, 1mM DTT, 1mM Na3VO4, 1x Protease inhibitors). SFB- or GFP-tagged proteins were immunoprecipitated using anti-FLAG or anti-GFP antibodies pre-conjugated with Protein G Dynal beads for 2 h. Subsequently, immunoprecipitates were washed three times with lysis buffer and once with lysis buffer containing 250 mM NaCl.
Immunofluorescence
For immunofluorescence using the S9.6 antibody alone or combinations of S9.6 with phospho-RPA32 pS33 or GFP antibodies, samples were prepared using a previously described protocol (Wan et al., 2015). Briefly, HeLa cells were trypsinized and pelleted in 15 ml Falcon tube (1,000 rpm, 5 min, 25°C). Media was aspirated down to approximately 300–500 μl, and cell pellets were resuspended. Pre-warmed 75 mM KCl solution at 37°C was added to cells in a drop-wise manner while the cells were agitated on a vortex at low speed. Cells were then incubated at 37°C for 12 min, 5–6 drops of freshly made, ice-cold methanol:acetic acid (3:1) were added to cells in a drop-wise manner while the cells were agitated. Cells were pelleted (1,000 rpm, 5 min, 25°C) and supernatants were aspirated down to 300–500 μl. Cells were resuspended in methanol:acetic acid (3:1, 5 ml), which was added in a drop-wise manner while the cells were agitated. Cells were fixed in methanol:acetic acid on ice for 20 min. Cells were washed once with methanol:acetic acid before being spotted onto slides. Slides were left to dry and immediately treated with blocking buffer (1x PBS, 5% BSA, 0.5% Triton X-100) for 1 h at room temperature, followed by incubation with primary antibody overnight at 4°C. Cells were washed three times with wash buffer (1x PBS + 0.1% Triton X-100) and subsequently incubated with appropriate secondary antibodies conjugated with fluorophores (Cy3 or Alexa-488) for 1 h at room temperature. After three washes with wash buffer, cells were stained with 4,6-diamidino-2-phenylindole (DAPI) and mounted using Vectorshield (Vector Laboratories).
For immunofluorescence using phospho-KAP1 pS428 antibody, cells were extracted with PBS containing 0.25% Triton X-100 prior to fixation with 3% paraformaldehyde/2% sucrose. Subsequently, cells were permeabilized with 1x PBS containing 0.5% Triton X-100, and treated in blocking buffer (1x PBS, 3% BSA, 0.05% Tween-20, and 10% milk) prior to primary antibody incubation for 2 h at room temperature. After the incubation in primary antibody, cells were washed three times with 1x PBS containing 0.05% Tween-20 and incubated with Cy3 conjugated anti-rabbit secondary antibody for 1 h. To visualize nuclei, cells were stained with DAPI after the final wash in PBS.
Immunofluorescence using phospho-RPA32 pS33 antibody alone was performed as described previously (Shiotani et al., 2013). To identify S-phase cell, cells were pulsed-labeled with 10 mM EdU for 30 min and processed with the Click-IT EdU Alexa Fluor 488 Imaging kit according to manufacturer’s manual (ThermoFisher Scientific). All images were captured using Nikon 90i microscope and analyzed using Image J software.
Chromatin Immunoprecipitation (ChIP)
Chromatin Immunoprecipitation was carried out as previously described (Giri et al., 2015) with modifications to the sonication protocol. Briefly, 5–6 million HeLa cells (in 10 cm plates) were fixed by addition of Formaldehyde (Sigma) to the culture medium to a final concentration of 1%. The crosslinking was carried out at room temperature for 10 min followed by quenching with glycine (final concentration: 0.125 M). Two washes with ice-cold PBS were then carried out followed by the following extraction steps. First extraction step (10 min at 4°C) was with Buffer 1 (50 mM Hepes/KOH pH 7.5; 140 mM NaCl; 1 mM EDTA; 10% Glycerol; 0.5% NP-40; 0.25% Triton) followed by the second extraction step with Buffer 2 (200 mM NaCl; 1mM EDTA; 0.5mM EGTA; 10 mM Tris pH 8). Nuclei were then pelleted by centrifugation, resuspended in SDS lysis buffer (1% SDS, 10mM EDTA, 50mM Tris pH 8.0) and subjected to sonication with Q800R2 sonicator (Qsonica) generating genomic DNA fragments with an average size of 200–600 bp. Chromatin (100 μg) was pre-cleared with protein G Dynabeads (Invitrogen) for 1 h at 4°C followed by immunoprecipitation (overnight at 4°C) with 5 µg of either GFP or RPA antibody. This was followed by pulldown with BSA-pretreated protein G Dynabeads (Invitrogen) and incubated for 2 h at 4°C. Beads were washed once with Low salt buffer (0.1% SDS; 1% Triton; 2 mM EDTA; 20 mM Tris pH 8; 150 mM NaCl), once with High salt buffer (0.1% SDS; 1% Triton; 2 mM EDTA; 20 mM Tris pH 8; 500 mM NaCl), once with LiCl wash buffer (10 mM Tris pH 8.0; 1% sodium deoxycholate; 1% NP- 40, 250 mM LiCl; 1 mM EDTA) and twice with TE . Elution of the beads (to recover immune complexes) was then carried out twice (10 min each) in TE + 1% SDS + 0.1% NaHCO3 at 65°C and followed by reverse crosslinking overnight at 65°C. The eluted DNA was then subjected to RNaseA treatment (10 µg/ml) for 1 h at 37°C followed by Proteinase K treatment (4 μl 0.5M EDTA, 8 μl 1M Tris pH 6.9, 2 μl Proteinase K 20 mg/ml) for 1 hour at 45°C. DNA isolation was then carried out by using QIAquick PCR purification kit (Qiagen), resuspended in elution buffer and q-PCR performed using Faststart Universal SYBR Green mix - Rox added (Roche) and analyzed on a LightCycler 480II machine (Roche). ChIP-qPCR results were analyzed and plotted as either fold change over IgG or percentage (%) of IP/input signal (% input). Significance (p-value) was calculated using Student’s t test.
Protein Purification
Various N-terminally 6xHis-tagged human RNaseH1 proteins (wild-type and mutants) were purified from E. Coli. Plasmids were transformed into BL21 derivative Rosetta (DE3) cells. Cells were grown at 37°C to OD600 = 0.8, and protein expression was induced by 0.3 mM IPTG at 30°C for 3 h. Cell pellets (8 g) were suspended in 40 ml of T-buffer (25 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 10% glycerol, 1 mM DTT, 0.1% Igepal-60) containing 1 mM PMSF, 500 mM KCl and protease inhibitor cocktail (Sigma). Cell lysates were sonicated (30 seconds on/off for 10 cycles), and clarified by centrifugation (16,000 × g for 60 min). The supernatants were diluted 5 times to reach the final concentration of 100 mM KCl and allowed to bind SP-Sepharose resin (10 ml) in T-buffer containing 100 mM KCl. The resin was washed with 50 ml T-buffer containing 100 mM KCl and eluted with 100 ml linear gradient from 100 to 800 mM KCl using FPLC. RNaseH1 was eluted at ∼450 mM KCl, the peak fractions were collected and added to Nickel-NTA resin (5 ml) with T-buffer containing 500 mM KCl and 10 mM Imidazole for 2 h. Resin was washed with 50 ml of T-buffer with 500 mM KCl and protein was eluted with 50 ml gradient of 10 to 250 mM Imidazole. RNaseH1 was eluted at ∼150 mM Imidazole. Peak fractions were pooled together, dialyzed against T-buffer containing 300 mM KCl and passed through Superose-12 column in T-buffer containing 300 mM KCl. RNaseH1 fractions were pooled and concentrated using Ultracel-30K centrifugal filters and stored at −80°C in small aliquots. RNaseH1 R57A, 3RA and D210N mutant proteins have been purified as WT purification. E. coli RNaseH1 was purchased from New England BioLabs.
Human RPA complex (containing the 70, 32 and 14 kDa subunits) was overexpressed in Rosetta (DE3) cells with 0.1 mM IPTG for 14 h at 16°C. The cell pellets (∼30 g) were resuspended in 150 ml T-buffer (25 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 10% glycerol, 1mM DTT, 0.1 % Igepal-60) with 1 mM PMSF, 100 mM KCl and protease inhibitor cocktail (Sigma), and lysed by sonication (1 min on/off, 12 cycles). Lysed solution was cleared by centrifugation (16,000 g for 1 h) and loaded on Affi-gel blue resin (15 ml) with T-buffer containing 100 mM KCl. Affi-gel blue resin was washed by 30 ml of T-buffer containing 100 mM KCl, followed by 60 ml of T-buffer containing 800 mM KCl. Protein was eluted from resin by a NaSCN gradient from 0.5 M to 2.5 M (100 ml). Eluted fractions containing RPA were pooled together and dialyzed against T-buffer with 50 mM KCl and 20% sucrose. After dialysis, sample was allowed to bind Hydroxyapatite beads in T-buffer with 50 mM KCl and 10 mM KH2PO4. Resin was washed with 80 ml of the same buffer and protein was eluted by 100 ml of gradient (10 to 300 mM KH2PO4). Eluted fraction containing RPA were collected and allowed to bind on a MonoQ column (1 ml). The column was washed with 10 ml of T-buffer with 50 mM KCl and eluted with a KCl gradient from 50 to 500 mM. Fractions containing purified RPA were collected and stored in – 80°C. E. coli SSB was purchased from Abcam.
RNA:DNA hybrid substrates
The list of oligonucleotide sequences used in this study is shown below. To prepare the substrates, the indicated RNA or DNA oligos were 5’-labeled with ATP-[γ32P] (PerkinElmer Life Sciences) using T4 polynucleotide kinase (New England BioLabs). Radiolabeled oligos were separated from free ATP-[γ32P] by passing through a 1 ml bio-spin column (Bio-Rad), containing Tris buffer. Radiolabeled oligos were then annealed to a complementary strand by heating to 95°C and slow cooling over a long period of time in buffer H (90 mM Tris-HCl pH 7.5, 10 mM MgCl2, 50 mM NaCl). Annealed substrates were separated on a 10% native PAGE in Tris acetate/ EDTA buffer at 4°C. The gel band corresponding to the annealed substrate was excised, purified, and finally eluted. The eluted substrates were further concentrated using a micro-concentrator. The concentrated substrates were ready to use in in vitro assays or stored at −20°C. For the R:D+ssDNA substrate, the RNA oligo was annealed with oligo 5. Oligos 3 and 5 were used to make double-stranded DNA with ssDNA overhang. For the R loop substrate, the RNA oligo was annealed with oligos 1 and 2. For the R:D substrate, the RNA oligo was annealed with oligo 4.
RNA:DNA hybrid and R loop resolution by RNaseH1 and RPA
Indicated concentration of RNaseH1 was pre-incubated with varying amounts of RPA on ice for 10 min in buffer A (25 mM Tris-HCl pH 7.5, 1 mM DTT, 5 mM MgCl2, 50 µg/ml BSA, 50 mM KCl), followed by the addition of indicated substrates in the reaction. The reactions were incubated at 30°C for indicated time and halted by addition of proteinase K (0.5 mg/ml), 5 mM EDTA and SDS (0.5% final concentration) for 10 min at 37°C. The reaction mixtures were resolved on 10% native polyacrylamide gel in 1X TBE buffer (45 mM Tris-borate, 1 mM EDTA, pH 8.0) at 4°C. Gels were dried, exposed on X-ray film scanned and analyzed with Image Lab (Bio-Rad).
Electrophoresis mobility shift assay (EMSA)
RNA:DNA hybrid substrates containing 5’-32P-labelled RNA were incubated with various amounts of RNaseH1 and/or RPA at 30°C for 15 min in buffer B (25 mM Tris-HCl pH 7.5, 1 mM DTT, 10 mM EDTA, 50 µg/ml BSA, 50 mM KCl). The resulting protein-substrate complexes were resolved on 6% polyacrylamide gels using 1X TBE buffer (Yadav et al., 2012). Gels were dried, exposed to X-ray film, and analyzed using Image Lab (Bio-Rad).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters are described in the Figures and the Figure Legends.
Supplementary Material
Table 1.
Primers for qRT-PCR of the ChIP experiment
| βActin Pause Fp | GGGACTATTTGGGGGTGTCT |
| βActin Pause Rp | TCCCATAGGTGAAGGCAAAG |
| βActin Control Fp (GFP ChIP) | CAGTGGTGTGGTGTGATCTTG |
| βActin Control Rp (GFP ChIP) | GGCAAAACCCTGTATCTGTGA |
| βActin Control Fp (RPA ChIP) | GATGCCACCACATCTGACTAA |
| βActin Control Rp (RPA ChIP) | GGGAGACAGACCCTGTTATATTG |
| Gemin7 Pause Fp | AGCTCACGCTGGTTCTTTCTT |
| Gemin7 Pause Rp | GAAATTCCAAAGGCGAGAGAC |
| Gemin7 Control Fp | GATTCTATTTGGGCCACCTATG |
| Gemin7 Control Rp | GGGAGGCATCTAAACCTCATC |
Table 2.
List of oligonucleotides used in vitro substrates
| RNA oligo | 5’GCAGCUGGCACGACAGGUAUGAAUC |
|---|---|
| Oligo 1 | 5’GCCAGGGACGAGGTGAACCTGCAGGTGGGCGGCTACTACTTAGATGTCATCCGAGGCTTATTGGTAGAATTCGGCAGCGTCATGCGACGGC |
| Oligo 2 | 5’GCCGTCGCATGACGCTGCCGAATTCTACCACGCGATTCATACCTGTCGTGCCAGCTGCTTTGCCCACCTGCAGGTTCACCTCGTCCCTGGC |
| Oligo 3 | 5’GCAGCTGGCACGACAGGTATGAATC |
| Oligo 4 | 5’GATTCATACCTGTCGTGCCAGCTGC |
| Oligo 5 | 5’GCAGTAGCATGACGCTGCTGAATTCTACCACGCTATGCTCTCGTCTAGGTTCACTCCGTCCCTGCGATTCATACCTGTCGTGCCAGCTGC |
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AQR | Bethyl | A302-547A–T |
| Flag | Sigma | F7425 |
| Anti-Flag M2 affinity gel | Sigma | A2220 |
| GAPDH | Millipore | ABS16 |
| γH2AX-pS139 | Cell Signaling | 9718S |
| γH2AX-pS139 | Millipore | 05–636 |
| GFP | Thermo Fisher Scientific | A11122 |
| Kap1-pS428 | Bethyl | A300-767A |
| Ku70 | GeneTex | GTX70271 |
| RPA32 | Thermo Fisher Scientific | MA1-26418 |
| RPA32-pS33 | Bethyl | A300-246A |
| S9.6 | Dr. Stephen Leppla; Antibodies Incorporated | Protein A purified from hybridoma S9.6 |
| SETX | Novus Biologicals | NB100-57542 |
| U2AF1 | Abcam | ab197591 |
| Chemicals, and Recombinant Proteins | ||
| T4 polynucleotide Kinase | NEB | M0201S |
| Proteinase K | Thermo Fischer | 25530049 |
| E.coli RNase H | NEB | M0297S |
| E. coli SSB | Abcam | ab123224 |
| BSA | NEB | B9000S |
| ATP gamma P32 | Perkin Elmer | NEG002250uc |
| IPTG | CalBiochem | 420322 |
| Ni++-NTA beads | Qiagen | 30230 |
| Affi-Gel Blue Gel | BioRad | 1537301 |
| SP Sepharose | GE Healthcare | 17–0729–01 |
| DNase I | Qiagen | 79254 |
| RNase A | ThermoFisher Scientific | 12091021 |
| Sequence-Based Reagents | ||
| Primers for ChIP | This Study | Table 1 |
| Oligos for in vitro substrates preparation | This study | Table 2 |
| Software and Algorithms | ||
| Image Lab | BioRad | http://www.bio-rad.com/en-us/product/image-lab-software |
| ImageJ | https://imagej-nih-gov.ezp-prod1.hul.harvard.edu/ij/index.html | |
| Prism 5 | http://www.graphpad.com/scientific-software/prism/ | |
Acknowledgments
We thank Drs. R. Crouch, M.A. Lee-Kirsch, and S. Leppla for reagents, Dr. A. Maréchal for characterizations of reagents, and members of the Zou and Dyson labs for helpful discussions. Molecular graphic image in Fig. 4C was produced using the UCSF Chimera package from the Computer Graphics Laboratory, University of California, San Francisco (supported by NIH P41 RR-01081). H.D.N. is partly supported by a postdoctoral fellowship from the Medical Discovery Fund. L.Z. is the James & Patricia Poitras Endowed Chair in Cancer Research, and was supported by a Jim & Ann Orr Massachusetts General Hospital Research Scholar Award. This work is supported by grants from the NIH (GM076388 and CA197779) to L.Z. and the Evans Foundation to T.G.
Footnotes
Author contributions
H.D.N., T.Y., and L.Z. designed the experiments. H.D.N. performed all cell-based experiments; T.Y. performed all biochemistry experiments; S.G. performed ChIP experiments. B.S. generated K562-derived U2AF1 expressing cells. T.A.G. and L.Z. supervised the experiments. H.D.N. and L.Z. wrote the manuscript with inputs from all authors.
References
- Alzu A, Bermejo R, Begnis M, Lucca C, Piccini D, Carotenuto W, Saponaro M, Brambati A, Cocito A, Foiani M, et al. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell. 2012;151:835–846. doi: 10.1016/j.cell.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, Azzalin CM. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014;5:5220. doi: 10.1038/ncomms6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia V, Barroso SI, García-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–365. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods. 1986;89:123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- Bubeck D, Reijns MAM, Graham SC, Astell KR, Jones EY, Jackson AP. PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res. 2011;39:3652–3666. doi: 10.1093/nar/gkq980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YA, Aristizabal MJ, Lu PYT, Luo Z, Hamza A, Kobor MS, Stirling PC, Hieter P. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014;10:1004288. doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho Y-J, Myers DR, Choi VW, Compagno M, Malkin DJ, et al. Genome-wide Translocation Sequencing Reveals Mechanisms of Chromosome Breaks and Rearrangements in B Cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De I, Bessonov S, Hofele R, dos Santos K, Will CL, Urlaub H, Lührmann R, Pena V. The RNA helicase Aquarius exhibits structural adaptations mediating its recruitment to spliceosomes. Nat. Struct. Mol. Biol. 2015;22:138–144. doi: 10.1038/nsmb.2951. [DOI] [PubMed] [Google Scholar]
- Flynn RL, Zou L. Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Crit. Rev. Biochem. Mol. Biol. 2010;45:266–275. doi: 10.3109/10409238.2010.488216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011;25:2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Rubio ML, Pérez-Calero C, Barroso SI, Tumini E, Herrera-Moyano E, Rosado IV, Aguilera A. The Fanconi Anemia Pathway Protects Genome Integrity from R-loops. PLoS Genet. 2015;11:1005674. doi: 10.1371/journal.pgen.1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri S, Aggarwal V, Pontis J, Shen Z, Chakraborty A, Khan A, Mizzen C, Prasanth KV, Ait-Si-Ali S, Ha T, et al. The preRC protein ORCA organizes heterochromatin by assembling histone H3 lysine 9 methyltransferases on chromatin. Elife. 2015;4 doi: 10.7554/eLife.06496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, Krysiak K, Harris CC, Koboldt DC, Larson DE, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet. 2011;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh M, Gromak N. Out of Balance: R-loops in Human Disease. PLoS Genet. 2014;10:1004630. doi: 10.1371/journal.pgen.1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, Webb S, Kerr A, Tollervey D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 2014;10:1004716. doi: 10.1371/journal.pgen.1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML, et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell. 2015;57:636–647. doi: 10.1016/j.molcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Ilagan JO, Ramakrishnan A, Hayes B, Murphy ME, Zebari AS, Bradley P, Bradley RK. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2015;25:14–26. doi: 10.1101/gr.181016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap MK, Kumar D, Villa R, La Clair JJ, Benner C, Sasik R, Jones H, Ghia EM, Rassenti LZ, Kipps TJ, et al. Targeting the spliceosome in chronic lymphocytic leukemia with the macrolides FD-895 and pladienolide-B. Haematologica. 2015;100:945–954. doi: 10.3324/haematol.2014.122069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- Lagisetti C, Pourpak A, Goronga T, Jiang Q, Cui X, Hyle J, Lahti JM, Morris SW, Webb TR. Synthetic mRNA Splicing Modulator Compounds with in Vivo Antitumor Activity. J. Med. Chem. 2009;52:6979–6990. doi: 10.1021/jm901215m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Li M, Pokharel S, Wang J-T, Xu X, Liu Y. RECQ5-dependent SUMOylation of DNA topoisomerase I prevents transcription-associated genome instability. Nat. Commun. 2015;6:6720. doi: 10.1038/ncomms7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YW, Sanz LA, Xu X, Hartono SR, Chédin F. Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi-Goutières syndrome. Elife. 2015;4 doi: 10.7554/eLife.08007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Zou L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015;25:9–23. doi: 10.1038/cr.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Li J-M, Ji XY, Wu C-S, Yazinski SA, Nguyen HD, Liu S, Jiménez AE, Jin J, Zou L. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol. Cell. 2014;53:235–246. doi: 10.1016/j.molcel.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Gómez-González B, Grzechnik P, Rondón AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira M-C, Klur S, Watanabe M, Németh AH, Ber ILe, Moniz J-C, Tranchant C, Aubourg P, Tazir M, Schöls L, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat. Genet. 2004;36:225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- Nowotny M, Cerritelli SM, Ghirlando R, Gaidamakov SA, Crouch RJ, Yang W. Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD. EMBO J. 2008;27:1172–1181. doi: 10.1038/emboj.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee M-C, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Petzold C, Marceau AH, Miller KH, Marqusee S, Keck JL. Interaction with Single-stranded DNA-binding Protein Stimulates Escherichia coli Ribonuclease HI Enzymatic Activity. J. Biol. Chem. 2015;290:14626–14636. doi: 10.1074/jbc.M115.655134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Arndt PF. Transcription induces strand-specific mutations at the 5’ end of human genes. Genome Res. 2008;18:1216–1223. doi: 10.1101/gr.076570.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015;16:583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- Sanz LA, Hartono SR, Lim YW, Steyaert S, Rajpurkar A, Ginno PA, Xu X, Chédin F. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol. Cell. 2016;63:167–178. doi: 10.1016/j.molcel.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, Liang C-C, Cohn MA, Gibbons RJ, Deans AJ, Niedzwiedz W. The Fanconi Anemia Pathway Maintains Genome Stability by Coordinating Replication and Transcription. Mol. Cell. 2015;60:351–361. doi: 10.1016/j.molcel.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani B, Nguyen HD, Håkansson P, Maréchal A, Tse A, Tahara H, Zou L. Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1. Cell Rep. 2013;3:1651–1662. doi: 10.1016/j.celrep.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski TW, Ficarro SB, Holik J, Kim T, Rando OJ, Marto JA, Buratowski S. Sub1 and RPA associate with RNA polymerase II at different stages of transcription. Mol. Cell. 2011;44:397–409. doi: 10.1016/j.molcel.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28:1384–1396. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014;516:436–439. doi: 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, Cimprich KA. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015;25:514–522. doi: 10.1016/j.tcb.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, Stork CT, García-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. Transcription-Coupled Nucleotide Excision Repair Factors Promote R-Loop-Induced Genome Instability. Mol. Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Chan YA, Minaker SW, Aristizabal MJ, Barrett I, Sipahimalani P, Kobor MS, Hieter P. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 2012;26:163–175. doi: 10.1101/gad.179721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. R-Loop Stabilization Represses Antisense Transcription at the Arabidopsis FLC Locus. Science (80-. ) 2013;340:619–621. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Holmes JB, Cerritelli SM, Sakhuja K, Minczuk M, Holt IJ, Crouch RJ. An upstream open reading frame and the context of the two AUG codons affect the abundance of mitochondrial and nuclear RNase H1. Mol. Cell. Biol. 2010;30:5123–5134. doi: 10.1128/MCB.00619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuduri S, Crabbé L, Conti C, Tourrière H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Zheng X, Chen H, Guo Y, Jiang H, He X, Zhu X, Zheng Y. Splicing function of mitotic regulators links R-loop-mediated DNA damage to tumor cell killing. J. Cell Biol. 2015;209:235–246. doi: 10.1083/jcb.201409073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RE, Prado F, Aguilera A. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol. Cell. Biol. 2006;26:3327–3334. doi: 10.1128/MCB.26.8.3327-3334.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Lima WF, Crooke ST. Investigating the structure of human RNase H1 by site-directed mutagenesis. J. Biol. Chem. 2001;276:23547–23553. doi: 10.1074/jbc.M009676200. [DOI] [PubMed] [Google Scholar]
- Yadav T, Carrasco B, Myers AR, George NP, Keck JL, Alonso JC. Genetic recombination in Bacillus subtilis: a division of labor between two single-strand DNA-binding proteins. Nucleic Acids Res. 2012;40:5546–5559. doi: 10.1093/nar/gks173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanling Zhao D, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, Zhong G, Liu K, Li W, Moffat J, et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature. 2015;529:48–53. doi: 10.1038/nature16469. [DOI] [PubMed] [Google Scholar]
- Yokoi A, Kotake Y, Takahashi K, Kadowaki T, Matsumoto Y, Minoshima Y, Sugi NH, Sagane K, Hamaguchi M, Iwata M, et al. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J. 2011;278:4870–4880. doi: 10.1111/j.1742-4658.2011.08387.x. [DOI] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh C-L, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat. Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.