Abstract
Aquilaria malaccensis Lam, commonly known as Agarwood, is a highly valuable species used in production of agar oil from its infected wood, which is utilized in pharmaceutical and perfumery industry. Agar oil formation in agarwood takes years through the natural process which is induced by natural or artificial injury or microbial infection. The role of soil fungi and bacteria in artificial induction is still an unexplored area. In the present study, we isolated the fungal and bacterial community residing inside the stem of A. malaccensis tree and circumventing soil, samples collected from 21 different sites of the north-eastern state Assam of India and explored their potential in induction of Agarospirol (2-(6,10-Dimethylspiro[4,5]dec-6-en-2-yl)-2-propanol) production by artificially infecting the trees with these microorganisms. A total 340 fungi and 131 bacteria were isolated from 50 stem samples, and 188 fungi and 148 bacteria were isolated from 50 soil samples. Highest Shannon (H′ = 2.43) and Fisher (α = 5.57) diversity index was observed in the stem isolates. The dominant fungal genus was Trichoderma in stem with Pi value of 0.18; while in soil, Aspergillus showed dominance with Pi value 0.73. In bacteria, Bacillus genera showed dominance in both stem and soil samples with Pi = 0.62 and 0.51, respectively. Forty fungal and bacterial isolates were used to assess their potential to induce formation of agarwood in A. malaccensis by artificial infection method. Gas chromatography mass spectroscopy (GC-MS) analysis confirmed development of Agarwood by the presence of Agarospirol compound in samples collected after 3 months of the artificial infection. Only 31% of bacterial and 23% of fungal isolates showed their ability in production of Agarospirol by artificial infection method. Bacteria Pantoea dispersa and fungi Penicillium polonicum showed the highest production in comparison to other isolates.
Keywords: endophytes, diversity, artificial infection, Pantoea dispera, agarwood, agarospirol, GC-MS
Introduction
Aquilaria malaccensis is an oleoresin-producing tree which occurs in Malaysia, Myanmar, Sumatra, Borneo Island, Philippines, Bangladesh, Vietnam, China, and India (Gibson, 1977; Hai et al., 1999). The oleoresin produced by the tree is a highly valuable component of incense, perfumes, drugs, stimulants, cardiac tonics, and carminatives (Mengling et al., 2005). Further, it is used as an active ingredient in traditional medicines of China and has also been included in different pharmaceutical products of coughs, acroparalysis, asthma, and anti-histamine (Kim et al., 1997; Bhuiyan and Bhuiyan, 2008). The dark wood of Aquilaria tree having oleoresin is known as agarwood, which emits a pleasant smell on burning and is widely used as incense in Islamic, Buddhist, and Hindu ceremonies. The essential oil extracted from agarwood also has antimicrobial properties (Chen et al., 2011). Commercial exploitation and uncontrolled cutting is leading to a decline in number of these trees. Eight species of Aquilaria are listed in Appendix II of the Convention on International Trade in Endangered Species (CITES) of wild fauna and flora (CITES, 1994) and are categorized as vulnerable by the International Union for the Conservation of Nature (IUCN, 2009).
Agarwood is the oleoresin containing part of Aquilaria tree which is formed after natural and artificial injury and tree produced oleoresin to prevent or recover the injury in response to plant defense mechanism (Zhang et al., 2012). Naturally the wound is produced by microbial invasion, gnawing of insects, lightning strikes and heavy winds (Xu et al., 2013; Zhang et al., 2014). As a defense response to these outbreaks, normal heartwood converts into dark agarwood. Firstly, Bhattacharyya in 1952 reported role of endophytic fungi in inducing agarwood production in the tree trunk. Later many scientists isolated several endophytes from the agarwood tree (Chhipa et al., 2017). Mohamed et al. (2010) showed the presence of Curvularia, Cunninghamella, Trichoderma, and Fusarium species in the agarwood fungal community in Malaysia. Similarly, Tian et al. (2013) reported the presence of Phomopsis, Botryosphaeria, Cylindrocladium, and Colletotrichum gloeosporioides species in wounded Aquilaria tree; and the presence of Alternaria, Mycosphaerella, Phoma, Ramichloridium, and Sagenomella species in the non-resinous tree internally. Bhore et al. (2013) demonstrated the presence of endophytic bacteria and reported Bacillus pumilus to be a dominating species among 18 different types of bacteria. In another study, Huang et al. (2015) reported the presence of distinct bacterial community in agarwood and non-agarwood plant of A. sinensis. It has been reported that local tree environment and wound microclimate also affect the succession pattern of the fungal population (Mohamed et al., 2014). The community of bacterial endophytes are also affected by the formation of agarwood and on metabolic changes in the plant (Huang et al., 2015). It is important to compare the microbial community structure and their association with the artificially induced agarwood formation at different locations for determining responsible microbes in artificial infection.
The natural development of agarwood takes 25–30 years and the yield is less, thus it unable to meet the demand of the growing market (Zhang et al., 2010). The increasing commercial demand has led to forced development of artificial infection methods. Different methods, such as physical, chemical, and biological or combinations have been reported for artificially inducing resinous oil in Aquilaria tree (Ito et al., 2005; Pojanagaroon and Kaewrak, 2005; Gong and Guo, 2009; Okudera and Ito, 2009; Kumeta and Ito, 2010; Novriyanti et al., 2010; Chen et al., 2011; Wei et al., 2012; Liu et al., 2013). The physical methods include severe bark removal, inflicting nail and axe wounds, making hole with screws, inflicting wounds with chisels, partial-trunk pruning, burning-chisel-drilling, pin-hole, and agar wit are recently reported (Pojanagaroon and Kaewrak, 2005; Liu et al., 2013; Tian et al., 2013). The biological methods include fungi-based inoculation, introduced by Tunstall (Gibson, 1977). Few fungal isolates, such as Melanotus flavolivens, Phytium sp., Penicillium sp., Diplodia sp., Botryodyplodis sp., Lasiodiplodis sp., and Fusarium sp. A2, have been used for artificial infection (Gong and Guo, 2009; Zhang et al., 2014). The role of bacteria in artificial inoculation is less explored in comparison to fungi. More research is required to look at microbial strain or consortia that can develop high yield of agarwood by artificial infection. In the present study, microbial diversity residing in A. malaccensis tree and surrounding soil of A. malaccensis growing in north-eastern region of India was explored and their potential for inducing agarwood production by artificial inoculation method was assessed.
Materials and methods
Sample collection and isolation of microbes
Fifty healthy trees from 21 different sites (Figure S1) in Assam, India were identified by the State Forest Research Institute, Itanagar. Stem and soil samples were collected from these sites and transported to the lab in an icebox. Isolation of microbes was done within 48 h of the collection. List of collected samples is provided at Table S1. The isolation of stem endophytes was done using surface sterilization according to the method established in our laboratory (Kumar et al., 2011) and the soil microbes were isolated by serial dilution method according to protocol of Kasana et al. (2008). Grouping of these isolates was done on the basis of morphological similarities and agar block of axenic cultures were stored in 15% glycerol stock at −70°C for long-time preservation.
Extraction of DNA, PCR, and sequencing
The genomic DNA of pure fungal isolates was extracted using DNeasy Plant mini kit (Qiagen GmbH, Hilden Cat. No 69104) by following the manufacture's instruction. Bacterial DNA was isolated using the CTAB method (Nishiguchi et al., 2002). The extracted DNA was used for PCR amplification by primer ITS1 and ITS4 according to Kumar et al. (2011) for fungi and 518F and 1492 R universal primers for bacterial sequence. The Polymerase chain reaction was achieved in 25 μL of reaction mixture, which contained 2.5 μL of 10× PCR Buffer with 15 mM MgCl2 (Applied Biosystem, India), 0.5 μL of dNTP mix (10 mM, Applied Biosystem), 2.5 μL of ITS1, ITS4 for fungi, and 518F and 1492 R primers for bacteria (10 picomole/μL), 1 μL of DNA template, and 0.5 μL of Ampli Teg Gold (5 U/μL). The ITS1, ITS4, 518F, and 1492 R primers were synthesized from Merck nucleotide synthesis services, Bengaluru, India, according ITS1 5′TCCGTAGGTGAACCTGCGG3′, ITS4 5′TCCTCCGCTTATTGATATGC3′, 518F 5′-CCAGCAGCCGCGGTAAT-3′, and 1492R 5′-GGYTACCTTGTTACGACTT-3′ sequence. The Polymerase Chain Reaction was done on Veriti Thermal Cycler (Applied Biosystem) using following programmes: initial denaturation at 94°C for 2 min; 30 cycles of denaturation, annealing, and elongation at 94°C for 1 minute; 57°C for 1.30 min, and 72°C for 2 min followed by final elongation at 72°C for 4 min. The negative control was also run using sterile water. The amplified product was checked on 1.5% agarose gel by gel electrophoresis. The amplified product was sequenced by Merck sequencing services, Bengaluru, India.
Identification of microbes and phylogenetic evaluation
The sequences were aligned and trimmed by DNA Baser 4.2 software on the basis of quality read values. The identification of microbes was done on the basis of similarity of amplified sequence with NCBI database using Basic Local Alignment Search Tool (nBLAST) of the US National Centre for Biotechnology Information (NCBI), SILVA and UNITE databases. The sequences were submitted to NCBI database. The phylogenetic tree was developed using Neighbor-Joining and Maximum Likelihood method by MEGA 6.0 software using sequence alignment by Clustal W pairwise sequence Alignment tool of the EMBL Nucleotide Sequence Database. The evolutionary distances were computed using the Maximum Composite Likelihood method.
Evaluation of microbial diversity
The microbial diversity in the stem and surrounding soil of A. malaccensis was determined by the Shannon diversity index, Simpson's index, and Fisher Alpha index. Species richness of the isolated microbes was estimated according to the Menhinick's index (Dmn) by the following equation (Whittaker, 1972):
| (1) |
Where “s” represents the number of different type of species in a sample and “N” represents the total number of isolates in a given sample. Simultaneously, dominance of class was also determined by Camargo's index (1/Dmn), where “Dmn” denotes species richness. A species was explained as dominant if Pi > 1/ Dmn, where Pi represents the relative abundance of a species, i explains the total number of competing species in the community (Camargo, 1992). The diversity was also evaluated to understand the distribution of organisms as randomly, aggregated, or uniformly distributed. Shannon diversity index (H′), Simpson's index, and Fisher Alpha index were calculated by SPADE programme version 3.1 (Chao and Shen, 2010). The heat map analysis was also done for soil and stem isolates using online tool Heatmapper1 (Babicki et al., 2016). The pairwise heat map was generated by average linkage calculation using Euclidean distance measurement method.
Agarwood formation by artificial infection method
Forty microbial isolates selected on the basis of their dominance were screened for their potential to induce agarwood formation by artificial infecting the wood with these micobes. For this, fungal mycelium was grown on PDA media and 5 days old fungal plugs were transferred to 100 ml of potato dextrose broth. The cultures were incubated at 25°C for 7 days. While, bacterial cultures were grown in 100 ml of nutrient broth medium and incubated for 48 h at 37°C. The grown fungal biomass was collected by filtration with Whatman filter paper no 1 and bacterial biomass with centrifugation at 8,000 rpm for 10 min and lyophilized to get powder form. The lyophilized sample was transported in sterile falcon tubes at experimental site. The inoculum was developed in 10 ml of 2% glucose solution. The artificial infection was done in A. malaccensis tree in Dergaon, Assam using syringe method. Artificial infection was introduced in 4–5 years old A. malaccensis trees. Holes were made in a zigzag manner with a drill machine (5 mm diameter bit size) in the trunk of the tree. The initial hole was made 20 cm above from the ground and next wound was drilled at 10 cm interval and the depth of the drill was 1.5 cm. Around 1 ml of homogenized microbial culture was inoculated in each hole using sterile syringe and hole was covered with para film. Each strain was infected in replicates. A sterilized medium was used as a syringe control.
Gas chromatography–mass spectroscopy analysis
The wood samples (~2–3 g) were collected 3 months after inducing artificial infection by drilling the wood around the inoculated hole. The collected of infected wood dust sample (1 g) were extracted in ethyl acetate by the reflux method. Uninoculated wood dust (wood control) and media control wood dust (syringe control) were also treated in similar manner to compare chemical composition with treated wood dust samples. The extract was concentrated in rotary evaporator. The concentrated extract was dissolved in 1 mL of dichloromethane and analyzed by Gas Chromatography-Mass Spectroscopy (GC-MS) (7890A/5975C, Agilent, California, United States) using an DB-WAX capillary column (30 m × 0.25 mm; film thickness 0.25 μm) and using Helium (purity > 99.999%) as the carrier gas with the constant flow rate of 1.0 mL/min. Around 1 μL of injection volume was used. The temperature of injector part was 230°C and oven temperature programing was used. The initial oven temperature was maintained at 80°C for 1 minute, then increased to 150°C @ 10°C/min in 7 min followed by temperature increase to 250°C @ 5°C/min till 22.5 min. The Mass Spectroscopic system was operated in EI mode (70 eV). Mass of the compounds was analyzed in the range of m/z 50–500 amu.
Data analysis
Unscrambler version 10 (CAMO, USA) was used to perform Principal Component Analysis (PCA) and statistical analysis.
Results
Identification of fungi and bacteria and their distribution
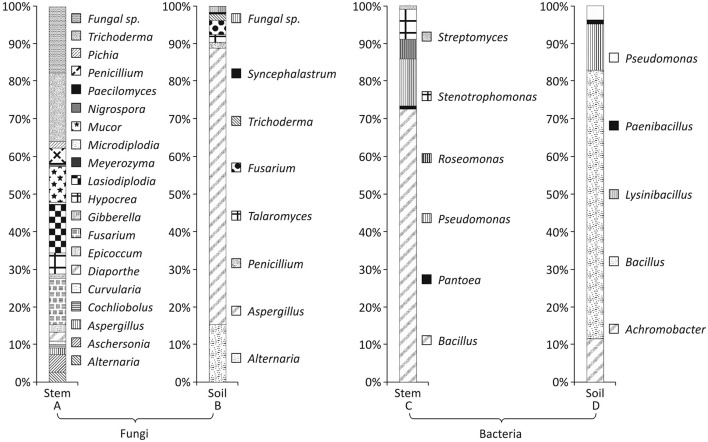
In all 340 fungi and 131 bacteria were isolated from 50 stem samples, and 188 fungi and 148 bacteria were isolated from 50 soil samples. These isolates were grouped on the basis of similarity of their external morphology and microscopic examination and representative isolates were used for molecular identification by 16s rRNA region amplification using 518F and 1492 R primers for bacteria and ITS region amplification using ITS1 and ITS4 primers for fungi and sequenced using Merck sequencing services, Bengaluru, India. Distribution of fungi and bacteria up to class level is given in Figure 1 and up to genus level in Figure 2. The list of identified strains and their accession number along with % similarity with the best match with different sequence databases is given in supplementary information Table S2. The identification based on ITS sequence of fungal strains showed maximum diversity in the stem samples (Table 3). It was observed that in the stem five fungal classes viz. Eurotiomycetes, Dothideomycetes, Zygomycetes, Saccharomycetes, and Sordariomycetes represented fungal community, while in the soil samples only four out of the five classes were present. Saccharomycetes were not isolated from stem samples. In the case of bacteria Bacilli class was dominant in the stem and soil samples.Actionbacteria and Alpha Proteobacteria members were obtained only from the stem and Beta Proteobacteria only from the soil samples.
Figure 1.
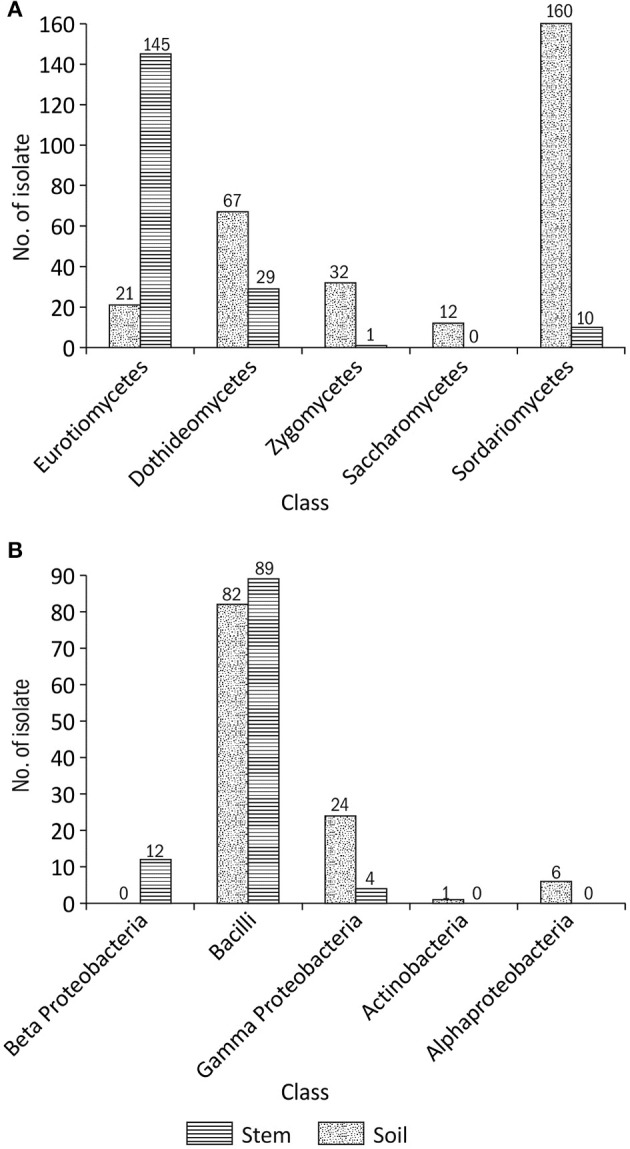
Diversity of fungal (A) and bacterial (B) isolates in the stem and soil samples of Aquilaria malaccensis.
Figure 2.
Distribution of fungi (A,B) and bacteria (C,D) in stem and soil samples at genus level.
Phylogenetic and microbial diversity analysis
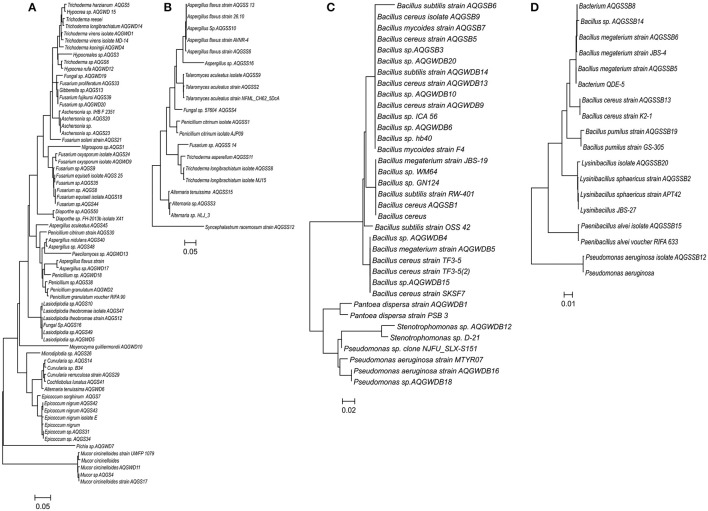
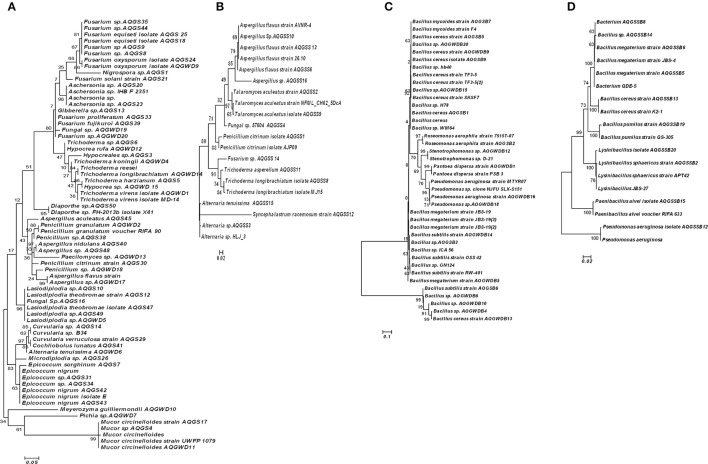
The phylogenetic relation between different morphotypes isolated from stem and soil samples were measured by MEGA version 6.06 using the Neighbor-Joining (N-J) and Maximum Likelihood (M-L) method (Figures 3, 4). Both type of method showed similar type of fungal and bacterial clusters. But in stem fungal isolate, Meyerozyma guilliermondii AQGWD10 showed less distant from Lasiodiplodia sp. cluster in N-J method while, in M-L method M. guilliermondii showed close to Pichia sp. (Figure 3A). In the case of soil fungal, soil and stem bacterial isolates cluster pattern was similar in both N-J and M-L method. In stem, 19 genera from 11 fungal families were identified using sequence similarity with NCBI database. In the stem, Hypocreaceae family was most dominant contributing 23.8% to richness while Hypocrea and Trichoderma genera, Botryospaeriaceae and Nectriaceae were second most commonly occurring family with 13.2 and 13.5% richness, having Microdiplodia, Lasiodiplodia, Fusarium, and Gibberella genera (Table 1). Pleosporace and Trichocomaceae family contributed only 6.5 and 6.2% richness, but contained 4 and 3 types of genera, including Cochliobolus, Curvularia, Alterneria, and Epicoocum in Pleosporace family and Aspergillus, Penicillium, and Paecilomyces in Trichocomaceae family. Around 14.1% of unidentified fungal sp. also contributed in fungal diversity. Likewise, in soil samples Trichocomaceae family showed the highest with 77.1% richness and members of Syncephalastraceae family was only isolated from soil samples.
Figure 3.
Phylogenetic trees based on Neighbor-Joining method of the r DNA and ITS sequence of different fungi from stem (A) and soil (B) and bacterial isolates from stem of Aquilaria malaccensis (C) and soil (D). 1000 replicates were calculated in the bootstrap.
Figure 4.
Phylogenetic trees based on Maximum likelihood method of the SSU ribosomal sequence of different fungi from stem (A) and soil (B) and bacterial isolates from stem of Aquilaria malaccensis (C) and soil (D).
Table 1.
Family dominance of fungus and bacteria isolated from stems and soil samples of A. malaccensis.
| S. No. | Fungal family | Richness (%) | Bacterial family | Richness (%) | ||
|---|---|---|---|---|---|---|
| Stem | Soil | Stem | Soil | |||
| 1 | Botryosphaeriaceae | 13.20 | 0.0 | Acetobacteraceae | 4.6 | 0.0 |
| 2 | Clavicipitaceae | 04.70 | 0.0 | Alcaligenaceae | 0.0 | 8.11 |
| 3 | Debaryomycetaceae | 00.30 | 0.0 | Bacillaceae | 62.6 | 59.5 |
| 4 | Diaporthaceae | 02.40 | 0.0 | Enterobacteriaceae | 0.8 | 0.0 |
| 5 | Fungal sp | 17.60 | 1.6 | Paenibacillaceae | 0.0 | 0.68 |
| 6 | Hypocreaceae | 23.80 | 1.6 | Pseudomonadaceae | 10.7 | 2.7 |
| 7 | Mucoraceae | 09.40 | 0.0 | Xanthomonadaceae | 6.9 | 0.0 |
| 8 | Nectriaceae | 13.50 | 3.7 | |||
| 9 | Pleosporaceae | 06.50 | 15.4 | |||
| 10 | Saccharomycetaceae | 01.80 | 0.0 | |||
| 11 | Syncephalastraceae | 00.00 | 0.5 | |||
| 12 | Trichocomaceae | 06.20 | 77.1 | |||
| 13 | Trichosphaeriaceae | 00.60 | 0.0 | |||
The phylogenetic relation of these isolates is given in the Figures 3, 4. In the case of bacteria, Bacillaceae is the dominant family in the stem and soil with 62.6 and 59.5% richness, respectively. Various bacterial families were isolated only in stem samples. Xanthomonadaceae showed 10.7% dominance, followed by Acetobacteraceae (4.6%) and Enterobacteriaceae (0.8%), while Alcaligenaceae and Paenibacillaceae showed presence only in the soil samples with 8.11% and 0.7% richness. Debaryomycetaceae, Saccharomycetaceae, Diaporthaceae, Clavicipitaceae, and Trichosphaeriaceae fungal families were exclusively isolated from stem, while Syncephalastraceae from soil samples. Similarly in bacteria, Enterobacteriaceae, Acetobacteraceae, and Xanthomonadaceae family was exclusively isolated from stem.
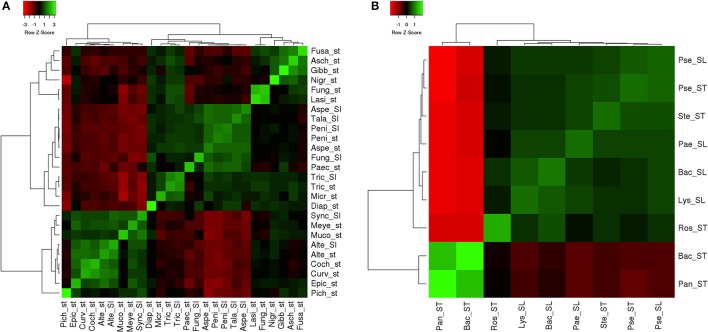
The fungal diversity of A. malaccensis in the North Eastern part of India was measured using Shannon diversity index (H′), Simpson index (D), and Fisher alpha index (α). Species richness in each source was measured by the Menhinick's diversity index (Dmn). The diversity index for all samples were measured by SPADES software and presented in Table 2. It was observed that stem were rich in fungal species (Dmn = 0.034) followed by soil (Dmn = 0.02). A similar pattern was measured in bacteria also (stem = 0.03 and soil = 0.02). Source-specific fungal dominance by Camargo's index was the highest measured in soil (47.00) followed by stem (29.57) and bacterial dominance also observed the highest in soil (49.33) than stem (37.43). The dominant genus was Tricoderma in stem with their Pi = 0.18, while in soil Aspergillus showed dominance with Pi = 0.73. The next dominance genus was Lasiodiplodia (Pi = 0.13) and Fusarium (Pi = 0.12) in stem and Alternaria (Pi = 0.15) in soil. Rests of fungal species were less dominant <0.09. In bacteria, Bacillus genera showed dominance in both root and soil samples with Pi = 0.62 and 0.51, respectively. Pseudomonas was dominant in the stem and Lysinibacillus in soil with Pi = 0.10 and 0.09, respectively. The Pielou's evenness index showed the distribution of fungal and bacterial species evenly in soil (J′ = 0.18 and 0.25) and stem (J′ = 0.42 and 0.25). The Maximum Shannon diversity index was observed for stem fungal isolates (H′ = 2.43) accompanied by soil bacteria (H′ = 1.25), while the Simpson diversity index was the highest in soil bacteria (H′ = 2. 84) followed by stem bacteria in comparison to fungi. Fisher alpha (α) was measured the highest in fungal stem (α = 5.57), followed by fungal soil (α = 1.70) samples. The similarity of fungal and bacterial isolates from stem and soil were done using heat map expression. It is observed that stem and bacterial isolates showed less variation at genera level and creating a cluster in heat map (Figure 5).
Table 2.
Diversity indexes of fungi and bacteria in the different samples.
| Diversity index | Fungi | Bacteria | ||
|---|---|---|---|---|
| Stem | Soil | Stem | Soil | |
| Shannon (H') | 2.43 | 0.95 | 1.20 | 1.30 |
| Simpson index (D) | 0.10 | 0.56 | 0.42 | 0.40 |
| Simpson Diversity index (1-D) | 0.90 | 0.44 | 2.35 | 2.80 |
| Fisher (α) | 5.57 | 1.70 | 1.58 | 1.30 |
| Menhinick's Diversity (Dmn) | 0.034 | 0.02 | 0.03 | 0.02 |
| Pielou's Evenness index (J') | 0.42 | 0.18 | 0.25 | 0.25 |
| Camargo's index (1/ Dmn) | 29.57 | 47.0 | 37.43 | 49.33 |
Figure 5.
Heat map analysis of (A) fungi and (B) bacterial isolates from stem and soil samples. St, stem isolate; Sl, Soil isolate; Pich, Pichia; Epic, Epicoccum; Curv, Curvularia; Coch, Cochliobolus; Alte, Alternaria; Muco, Mucor; Meye, Meyerozyma; Sync, Syncephalastrum; Diap, Diaporthe; Micr, Microdiplodia; Tric, Trichoderma; Pace, Paecilomyces; Fung, Fungal; Aspe, Aspergillus; Peni, Penicillium; Tala, Talaromyces; Lasi, Lasiodiplodia; Nigr, Nigrospora; Gibb, Gibberella; Asch, Aschersonia; Fusa, Fusarium; Pan, Pantoea; Bac, Bacillus; Ros, Roseomonas; Lys, Lysinibacillus; Pae, Paenibacillus; Ste, Stenotrophomonas; Pse, Pseudomonas.
Induction of artificial infection
Total 21 fungal and 19 bacterial strains isolated from infectious stem and surrounding soil were screened to assess their potential for induction of artificial infection in A. malaccensis tree. It was observed that the color of wood changed from white to brown/black after 3 months of infection in infected trees (Figure 6). Wood dust was harvested after 3months of inoculation and subjected to GC-MS analysis for confirmation of artificial infection on the basis of production of compounds presented in agarwood oil. In our previous study on comparison of GC-MS profile of infected and uninfected woods, we identified agarospirol, benzyl acetone and anisyl acetone as marker compounds for infection therefore, these compounds were selected as indicator for agrawood formation. Presence of agarospirol compound in the harvested wood confirmed the initiation of infection in the tree (Figure S2 and Table 3). Other details such as percentage of benzyl acetone and anisyl acetone are provided in Table S3.
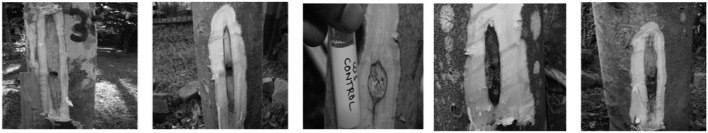
Figure 6.
Artificial infection in different Aquilaria malaccensis trees after 3 months of inoculation of fungal and bacterial strain: Dark brownish color of wood showing the initiation of artificial infection.
Table 3.
Agarospirol content and infection length obtained with different fungal and bacterial strains 3 months after the artificial inoculation in the A. malaccensis trees.
| S. No. | Species | Type of organism | Source | Agarospirol*a,b,c (2-(6,10-Dimethylspiro[4,5]dec-6-en-2-yl)-2-propanol) | Infection |
|---|---|---|---|---|---|
| Content (%) | Length (cm) | ||||
| 1 | Pantoea dispersa AQGWDB1 | Bacteria | Stem | 3.77 | 1.26 |
| 2 | Penicillium polonicum AQGGR1.1 | Fungi | Stem | 3.33 | 2.60 |
| 3 | Syncephalastrum racemosum AQGSS 12 | Fungi | Soil | 1.34 | 2.60 |
| 4 | Penicillium aethiopicum AQGGR1.2 | Fungi | Stem | 0.67 | 3.00 |
| 5 | Trichoderma asperellum AQGSS 11 | Fungi | Soil | 0.20 | 3.00 |
| 6 | Bacillus cereus AQGWDB9 | Bacteria | Stem | 0.09 | 3.45 |
| 7 | Penicillium polonicum AQGGR1.1 | Fungi | Stem | 0.08 | 2.60 |
| 8 | (Unidentified) AQGSSB23 | Bacteria | Soil | 0.06 | 2.00 |
| 9 | Bacillus anthricus strain AQGWDB2 | Bacteria | Stem | 0.05 | 2.91 |
| 10 | Rhizopus oryzae AQGGR1.3 | Fungi | Stem | 0.05 | 2.00 |
| 11 | (Unidentified) AQGWDB7 | Bacteria | Stem | 0.05 | 3.33 |
| 12 | Bacillus megaterium AQGWDB 3 | Bacteria | Stem | 0.03 | 2.35 |
| 13 | Wood control | Control | 0.00 | 0.00 |
Variance comparison at significant level: 0.05.
Fungi vs. control- P-value < 0.001.
Bacteria vs. control -P-value < 0.001.
Bacteria vs. Fungi -P-value = 0.0057231.
It was noted that in comparison to uninfected wood and experimental control (syringe and wood controls) agarospirol was detected only in artificially infected wood samples. It is reported that the presence of agarospirol is one of the responsible compound for fragrance in agarwood (Kalra and Kaushik, 2017). Highest agarospirol content was measured in wood infected by Pantoea dispersa (3.77%), followed by fungi Penicillium polonicum (3.33%), Syncephalastrum racemosum, Penicillium aethiopicum, and Trichoderma asperellum (Table 3). The infection length was measured in the range of 2.00 to 11.83 cm. Strain Paenibacillus alvei AQGSSB15 showed the highest infection up to 11.83 cm in length. Although, agarospirol could not be detected in this sample, however, it showed presence of benzyl acetone confirming early stage of infection (Table S3). Highest agarospirol was measured in the sample infected by Pantoea dispersa AQGWDB1 and length of infection was found only up to 1.26 cm. Therefore, no relation was observed between infection length and agarospirol production. Based on the agarospirol formation, 31% of bacterial and 23% of fungal isolates showed their ability to produce agarospirol in agarwood through artificial infection (Table 3). Production of agarospirol was significantly higher with bacterial inoculum than the fungal inoculum with P-value 0.0057 (Figure S3). Principal component analysis (PCA) also showed two distinct clusters of bacteria and fungi (Figures S4, S5).
Discussion
Fungi and bacteria identification and their distribution
Endophytes play an important role in plant physiology. Most of the endophytes are helpful in host plant growth, stress tolerance, and explained as the microorganisms that are not detrimental to the plant (Oses et al., 2008; Huang et al., 2009; Nimnoi et al., 2010). However, some had no beneficial impact and showed latent pathogenicity in plant species (Pojanagaroon and Kaewrak, 2005). Latent pathogenicity of endophytes could be an economical source of local community and Aquilaria is a live example in this regard, which produces oleoresin after infection and enhances the value of tree in market (Barden et al., 2000; Pojanagaroon and Kaewrak, 2005; Chhipa et al., 2017). Various studies have been done on isolation of endophytes from Aquilaria species, such as Malyasian A. malaccensis (Mohamed et al., 2010; Bhore et al., 2013); Chinese A. sinensis (Gong and Guo, 2009; Tian et al., 2013); Thai A. crassna (Nimnoi et al., 2010); A. agallocha (Tamuli et al., 2000), and Indian A. malaccensis (Bhore et al., 2013). However, very few reports are available on the development of artificial infection by using these endophytes (Mitra and Gogol, 2001; Mohamed et al., 2014). In the present study endophytic microbes of Aquilaria tree, and soil microbes isolated from circumventing soils of Aquilaria tree were successfully utilized for induction of agarospirol formation through artificial inoculation,. Trichoderma virens was found as the most abundant fungi in stem followed by Lasiodiplodia theobromae, Lasiodiplodia sp., and Fusarium equiseti (4.9%). While Mohamed et al. (2010) observed a reverse pattern with Fusarium sp. as the most dominant and Trichoderma sp. as the least dominant in infected and similar fungal strains in non-infected samples of A. malaccensis in Malaysia region and suggested that these fungi might have little role in resin formation. Similarly, Colletotrichum, Botryosphaeria, Phomopsis, Gloeosporioides, and Cylindrocladium species are reported in wounded Aquilaria tree, while Alternaria, Mycosphaerella, Ramichloridium, Sagenomella, and Phoma species is in non-resinous tree of A. sinensis in China (Tian et al., 2013). While in the case of bacteria dominance of Bacillus, genus was observed in both soil and stem samples. Pantoea, Roseomonas, Stenotrophomonas, and Streptomyces genera were only obtained from stem samples, while Achromobacter, Lysinibacillus, and Paenibacillus were only observed in soil samples. Presence of endophytic actinomycetes including Streptomyces, Nonomuraea, Actinomadura, Pesudonocardi, and Nocardia genera is also reported in A. crassna (Nimnoi et al., 2010). Similarly, Bhore et al. (2013) demonstrated dominance of Bacillus pumilus (36.4%) among 18 different bacterial species in Aquilaria sp. of Malaysia region.
Phylogenetic and microbial diversity analysis
In the case of the Aquilaria plant, microbial diversity and dominance could be species and region specific. Kusari et al. (2012) observed that 70% of isolates in soil and plant part are common. It suggests that the plant and endophytes association developed after overcoming many physical and chemical barriers and particular fungi established as endophytes in a particular niche or localized in the tissue in a systemic manner (Hyde and Soytong, 2008). Subsequently, pathogenicity is also affected by the surrounding environment as endophytic to host plant in one ecosystem can be pathogenic in another ecological niche. It is reported that endophytic and pathogenic lifestyle are inter convertible due to environmental, chemical, and molecular elicitors (Schulz et al., 2002; Eaton et al., 2011).
In the present study, higher diversity of microbial isolates has been observed in the stem than the soil samples. The Shannon and Fisher alpha diversity index in the stem measured the highest value showing diversity and abundance of endophytes in stem comparison to soil samples. The results are similar to previously reported by Kumar and Hyde (2004) in Tripterigium wilfordii and Gond et al. (2012) in Nyctanthes arbortristis showed the highest Shannon index in stem.
Induction of artificial infection
The development of artificial infection method using endophytic and soil microorganisms will provide significant impact on agarwood production. In present study, 31% of bacterial and 23% of fungal isolates showed positive response in agarwood formation by production of agarospirol compound within 3 months of infection. To the best of our knowledge no previous study has reported agarospirol production by artificial infection in 3 months. Zhang et al. (2012) reported agarwood production in 6 months after infection, while Lin et al. (2010) after 12 months. The maximum amount of Agarospirol was induced by Pantoea dispersa AQGWDB1 followed by Penicillium polonicum AQGGR1.1. Previously, various researchers have used fungi in Agarwood formation (Chhipa et al., 2017). However, this is the first report on use of bacteria in artificial infection and agarospirol production (Bose, 1934; Bhattacharyya et al., 1952; Jalaluddin, 1977; Venkataramanan et al., 1985; Beniwal, 1989; Tamuli et al., 2000; Mitra and Gogol, 2001). Further, Studies on artificial infection have been reported by Aspergillus sp., Lasiodiplodia sp., Fusarium sp., Penicillium sp., and Trichoderma sp. (Yunita, 2009; Akter et al., 2013). Recently, Mohamed et al. (2014) also reported artificial Agarwood formation in young A. malaccensis tree by fungal inoculation. Similarly, Lin et al. (2010) also reported that fungus Melanotus flavolivence is also capable of inducing Agarwood formation after 6 months of infection. It has been reported that the accumulation of plant secondary metabolites can be the result of microbial stimulation. Cui et al. (2013) also demonstrated the production of resin-containing organic volatile fatty acid in response to fungal attack. This process is called “tylosis.” The resin increases the density and changes the color of the wood from pale to dark brown or black. In present study, it was observed that 41% of stem isolates showed their potential in development of agarospirol while 13.6% soil isolates could induce agarwood infection, confirming the role of endophytes in agarwood formation. This study is extremely beneficial for local tribes of north-eastern part of India where Agarwood is treated as an economic resource (Lin et al., 2010). Identification of such microbes that can induce agarwood production at short time exposure and validation at field levels has lot of potentials for future research.
Conclusion
Endophytes are helpful in plant physiology by producing different compounds, which assist in plant growth, stress tolerance, and plant immunity. Isolation and identification of such microbes are an important aspect for biotechnological applications. In present study, we could isolate total 528 fungi and 279 bacteria from Aquilaria stem and surrounding soil samples collected from different sites of Assam. Bacilli were found most dominant class in the bacteria and Sordariomycetes in fungi. In stem, Hypocreaceae was measured most dominant fungal family with 23.8% richness while Bacileaceae was dominant bacterial family with 62.6 and 59.5% richness in both stem and soil isolates. Maximum Shannon diversity index was measured in stem fungal isolates (H′ = 2.43). Agarospirol production was induced successfully using bacteria Pantoea dispersa (3.77%) and fungi Penicillium polonicum (3.33%) within 3 months only, after inducing the infection. The agarospirol is one of the responsible compounds for fragrance in Agarwood. The use of such microbes in artificial production of agarospirol could be source of economic development of villagers of North Eastern part of India.
Author contributions
Conception and designed the experiments: HC and NK. Performed the experiments: HC. Analyzed the data: HC and NK. Contributed reagents/materials/analysis tools: HC and NK. Wrote and enriched the literature: HC and NK. Corrected the manuscript: NK.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are highly thankful to Mr. Surjit Bohra, Golaghat, Assam and R. K. Mohsang Society, Jairampur, Arunachal Pradesh for providing Aquilaria trees for experimental purpose. We are highly thankful to Dr. L. R. Bhuyan, SFRI, Itanagar, for identification of the Aquilaria malaccensis tree.
Funding. This work has been supported by the Department of Biotechnology, Government of India, Research Grant no BT/PR1505/NDB/38/223/2011.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01286/full#supplementary-material
Site map of the districts selected for sample collection.
GC-MS profile of oil extracted from artificial infected wood dust sample inoculated with Pantoea dispersa AQGWDB1 (A) and Penicillium aethiopicum AQGGR1.2 (B) showing the presence of the agarospirol compound in gas chromatogram mass spectrum of Agarospirol compound (RT 35.72) (C).
F-test of significance for agarospirol content variation between fungal infected and control wood (A), bacterial infected and control wood (B) and fungal and bacterial infected wood (C).
Bi plot of GC data of bacterial and fungal infected wood samples collected 3 months after artificial inoculation  Bacteria
Bacteria  Fungi.
Fungi.
PCA plot of GC data of bacterial and fungal infected wood samples collected 3 months after artificial inoculation  Bacteria
Bacteria  Fungi.
Fungi.
List of sample collected from different districts of Assam for microbial isolation.
Identification of fungal and bacterial isolates on the basis of sequence similarity with different sequence data base.
Per cent composition of compounds in oleoresin extract identified by gas chromatography mass spectroscopy.
References
- Akter S., Islam M. T., Zulkefeli M., Khan S. I. (2013). Agarwood production-a multidisciplinary field to be explored in Bangladesh. Int. J. Pharm. Life Sci. 2, 22–32. 10.3329/ijpls.v2i1.15132 [DOI] [Google Scholar]
- Babicki S., Arndt D., Marcu A., Liang Y., Grant J. R., Maciejewski A., et al. (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44, W147–W153. 10.1093/nar/gkw419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barden A., Awang A., Mulliken T., Song M. (2000). Heart of the Matter Agarwood Use and Trade and CITES Implementation for Aquilaria Malaccensis. Cambridge: Traffic International. [Google Scholar]
- Beniwal B. S. (1989). Silvical characteristics of Aquilaria agallocha Roxb. Indian Forester 115, 17–21. [Google Scholar]
- Bhattacharyya B., Datta A., Baruah H. K. (1952). On the formation and development of agaru in Aquilaria agallocha. Sci. Cult. 18, 240–243. [Google Scholar]
- Bhore S. J., Preveena J., Kandasamy K. I. (2013). Isolation and identification of bacterial endophytes from pharmaceutical agarwood-producing Aquilaria species. Pharmacognosy Res. 5, 134–137. 10.4103/0974-8490.110545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan M. N., Bhuiyan M. N. J. (2008). Analysis of essential oil of eaglewood tree (Aquilaria agallocha Roxb.) by gas chromatography mass spectrometry. Bangladesh J. Pharmacol. 4, 24–28. 10.3329/bjp.v4i1.851 [DOI] [Google Scholar]
- Bose S. R. (1934). The nature of agar formation. Sci. Cult. 4, 89–91 [Google Scholar]
- Camargo J. A. (1992). Temporal and spatial variations in dominance, diversity and biotic indices along a limestone stream receiving a trout farm effluent. Water Air Soil Pollut. 63, 343–359. 10.1007/BF00475501 [DOI] [Google Scholar]
- Chao A., Shen T. J. (2010). Program SPADE (Species Prediction and Diversity Estimation). Program and User's Guide. Available online at: http://chao.stat.nthu.edu.tw
- Chen H., Yang Y., Xue J., Wei J., Zhang Z., Chen H.f. (2011). Comparison of compositions and antimicrobial activities of essential oils from chemically stimulated agarwood, wild agarwood and healthy Aquilaria sinensis (Lour.) Gilg trees. Molecules 16, 4884–4896. 10.3390/molecules16064884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhipa H., Chowdhary K., Kaushik N. (2017). Artificial production of agarwood oil in Aquilaria sp. by fungi: a review. Phytochem. Rev. [Epub ahead of print]. 10.1007/s11101-017-9492-6 [DOI] [Google Scholar]
- CITES (1994). Resolution of the conference of the parties, in Ninth Meeting of the Conference of the Parties, Fort Lauderdale, FL. [Google Scholar]
- Cui J., Wang C., Guo S., Yang L., Xiao P., Wang M. (2013). Evaluation of fungus-induced agilawood from Aquilaria sinensis in China. Symbiosis 60, 37–44. 10.1007/s13199-013-0237-z [DOI] [Google Scholar]
- Eaton C. J., Cox M. P., Scott B. (2011). What triggers grass endophytes to switch from mutualism to pathogenism? Plant Sci. 180, 190–195. 10.1016/j.plantsci.2010.10.002 [DOI] [PubMed] [Google Scholar]
- Gibson I. A. (1977). The role of fungi in the origin of oleoresin deposits (agaru) in the wood of Aquilaria agallocha Roxb. Bano Biggyan Patrika 6, 16–26. [Google Scholar]
- Gond S. K., Mishra A., Sharma V. K., Verma S. K., Kumar J., Kharwar R. N., et al. (2012). Diversity and antimicrobial activity of endophytic fungi isolated from nyctanthes arbor-tristis, a well-known medicinal plant of India. Mycoscience 53, 113–121. 10.1007/S10267-011-0146-Z [DOI] [Google Scholar]
- Gong L., Guo S. (2009). Endophytic fungi from Dracaena cambodiana and Aquilaria sinensis and their antimicrobial activity. Afr. J. Biotechnol. 8, 731–736. Available online at: https://www.ajol.info/index.php/ajb/article/view/59937/48209 [Google Scholar]
- Hai L. E., Shyun C. Y., Yusoff A. M. (1999). Early survival and growth in field trials of Aquilaria malaccensis (karas) and Azadirachta excelsa (sentang). J. Trop. For. Sci. 11, 852–854. [Google Scholar]
- Huang W. Y., Cai Y. Z., Surveswaran S., Hyde K. D., Corke H., Sun M. (2009). Molecular phylogenetic identification of endophytic fungi isolated from three Artemisia species. Fungal Divers. 36, 69–88. Available online at: www.fungaldiversity.org/fdp/sfdp/FD36-5.pdf [Google Scholar]
- Huang Y. L., Kuang Z. Y., Song M. W., Zhang R. (2015). Community structure and difference of endophtic bacteria in Aquilaria sinensis with and without agarwood. Zhongguo Zhong Yao Za Zhi 40, 63–67. Available online at: http://europepmc.org/abstract/med/25993789 [PubMed] [Google Scholar]
- Hyde K. D., Soytong K. (2008). The fungal endophyte dilemma. Fungal Divers. 33, 163–173. Available online at: www.fungaldiversity.org/fdp/sfdp/33-9.pdf [Google Scholar]
- Ito M., Okimoto K. I., Yagura T., Honda G., Kiuchi F., Shimada Y. (2005). Induction of sesquiterpenoid production by methyl jasmonate in Aquilaria sinensis cell suspension culture. J. Essent. Oil Res. 17, 175–180. 10.1080/10412905.2005.9698867 [DOI] [Google Scholar]
- IUCN (2009). Asian Regional Workshop (Conservation & Sustainable Management of Trees, Vietnam) 1998. Aquilaria malaccensis, in IUCN 2009, IUCN Red List of Threatened Species, Version 2009.2.2. (Hanoi: ). Available online at: http://www.iucnredlist.org/details/32056/0 [Google Scholar]
- Jalaluddin M. A. (1977). Useful pathological condition of wood. Econ. Bot. 31, 222–224. 10.1007/BF02866592 [DOI] [Google Scholar]
- Kalra R., Kaushik N. (2017). A review of chemistry, quality and analysis of infected agarwood tree (Aquilaria sp.). Phytochem. Rev. [Epub ahead of print]. 10.1007/s11101-017-9518-0 [DOI] [Google Scholar]
- Kasana R. C., Salwan R., Dhar H., Dutt S., Gulati A. (2008). A rapid and easy method for the detection of microbial cellulases on agar plates using Gram's iodine. Curr. Microbiol. 57, 503–507. 10.1007/s00284-008-9276-8 [DOI] [PubMed] [Google Scholar]
- Kim Y. C., Lee E. H., Lee Y. M., Kim H. K., Song B. K., Lee E. J., et al. (1997). Effect of the aqueous extract of Aquilaria agallocha stems on the immediate hypersensitivity reactions. J. Ethnopharmacol. 58, 318. 10.1016/S0378-8741(97)00075-5 [DOI] [PubMed] [Google Scholar]
- Kumar D. S., Hyde K. D. (2004). Biodiversity and tissue-recurrence of endophytic fungi in Tripterygium wilfordii. Fungal Divers. 17, 69–90. Available online at: www.fungaldiversity.org/fdp/sfdp/17-6.pdf [Google Scholar]
- Kumar S., Kaushik N., Edrada-Ebel R., Ebel R., Proksch P. (2011). Isolation, characterization, and bioactivity of endophytic fungi of Tylophora indica. World J. Microbiol. Biotechnol. 27, 571–577. 10.1007/s11274-010-0492-6 [DOI] [Google Scholar]
- Kumeta Y., Ito M. (2010). Characterization of δ-guaiene synthases from cultured cells of Aquilaria, responsible for the formation of the sesquiterpenes in agarwood. Plant Physiol. 154, 1998–2007. 10.1104/pp.110.161828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusari S., Hertweck C., Spiteller M. (2012). Chemical ecology of endophytic fungi: origins of secondary metabolites. Chem. Biol. 19, 792–798. 10.1016/j.chembiol.2012.06.004 [DOI] [PubMed] [Google Scholar]
- Lin F., Mei W. L., Wu J., Dai H. F. (2010). GC-MS analysis of volatile constituents from Chinese eaglewood produced by artificial methods. Zhong Yao Cai 33, 222–225. [PubMed] [Google Scholar]
- Liu Y., Chen H., Yang Y., Zhang Z., Wei J., Meng H., et al. (2013). Whole-tree agarwood-inducing technique: an efficient novel technique for producing high-quality agarwood in cultivated Aquilaria sinensis trees. Molecules 18, 3086–3106. 10.3390/molecules18033086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengling H. E., Shu-yuan Q. I., Lan-juan H. U. (2005). Rapid in vitro propagation of medicinally important Aquilaria agallocha. J. Zhejiang Univ. Sci. B 6, 849–852. 10.1631/jzus.2005.B0849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra J., Gogol P. (2001). Fungi associated with the diseased wood (agarwood/agaru) of Aquilaria agallocha Roxb. grown in Assam, in Proceedings of Seminar on Scope and Dimension of Agar Plantation in NE Region of New Delhi, India, eds Ahmed M., Gogoi P., Ahmed G. U. (Hojai: AATMA; ), 61–69. [Google Scholar]
- Mohamed R., Jong P. L., Kamziah A. K. (2014). Fungal inoculation induces agarwood in young Aquilaria malaccensis trees in the nursery. J. Forestry Res. 25, 201–204. 10.1007/s11676-013-0395-0 [DOI] [Google Scholar]
- Mohamed R., Jong P. L., Zali M. S. (2010). Fungal diversity in wounded stems of Aquilaria malaccensis. Fungal Divers. 43, 67–74. 10.1007/s13225-010-0039-z [DOI] [Google Scholar]
- Nimnoi P., Pongsilp N., Lumyong S. (2010). Endophytic actinomycetes isolated from Aquilaria crassna Pierre ex Lec and screening of plant growth promoters production. World J. Microbiol. Biotechnol. 26, 193–203. 10.1007/s11274-009-0159-3 [DOI] [Google Scholar]
- Nishiguchi M. K., Doukakis P., Egan M., Kizirian D., Phillips A., Prendini L., et al. (2002). DNA isolation procedures, in Techniques in Molecular Systematics and Evolution, eds DeSalle R., Giribet G., Wheeler W. (Basel: Springer Science & Business Media; ), 249–287. [Google Scholar]
- Novriyanti E., Santosa E., Syafii W., Turjaman M., Sitepu I. R. (2010). Anti-fungal activity of wood extract of Aquilaria crassna pierre ex lecomte against agarwood-inducing fungi, Fusarium solani. Indones. J. Forestry Res. 7, 155–165. 10.20886/ijfr.2010.7.2.155-165 [DOI] [Google Scholar]
- Okudera Y., Ito M. (2009). Production of agarwood fragrant constituents in Aquilaria calli and cell suspension cultures. Plant Biotechnol. 26, 307–315. 10.5511/plantbiotechnology.26.307 [DOI] [Google Scholar]
- Oses R., Valenzuela S., Freer J., Sanfuentes E., Rodriguez J. (2008). Fungal endophytes in xylem of healthy Chilean trees and their possible role in early wood decay. Fungal Divers. 33, 77–86. Available online at: www.fungaldiversity.org/fdp/sfdp/33-4.pdf [Google Scholar]
- Pojanagaroon S., Kaewrak C. (2005). Mechanical methods to stimulate aloes wood formation in Aquilaria crassna Pierre ex H. Lec.(Kritsana) trees, in III WOCMAP Congress on Medicinal and Aromatic Plants-Volume 2: Conservation, Cultivation and Sustainable Use of Medicinal, eds Jatisatienr A., Paratasilpin T., Elliott S., Anusarnsunthorn V., Wedge D., Craker L. E., Gardner Z. E. (Chiang Mai: International Society for Horticultural Science; ) 161–166. [Google Scholar]
- Schulz B., Boyle C., Draeger S., Römmert A. K., Krohn K. (2002). Endophytic fungi: a source of novel biologically active secondary metabolites. Mycol. Res. 106, 996–1004. 10.1017/S0953756202006342 [DOI] [Google Scholar]
- Tamuli P., Boruah P., Nath S. C., Samanta R. (2000). Fungi from diseased agarwood tree (Aquilaria agallocha Roxb.): two new records. Adv. Forestry Res. India 22, 182–187. Available online at: https://www.cabdirect.org/cabdirect/abstract/20023060621 [Google Scholar]
- Tian J. J., Gao X. X., Zhang W. M., Wang L., Qu L. H. (2013). Molecular identification of endophytic fungi from Aquilaria sinensis and artificial agarwood induced by pinholes-infusion technique. Afr. J. Biotechnol. 12, 3115–3131. 10.5897/AJB11.3159 [DOI] [Google Scholar]
- Venkataramanan M. N., Borthakur R., Singh H. D. (1985). Occurrence of endotrophic myccorhizzal fungus in agarwood plant Aquilaria agallocha Roxb. Curr. Sci. 54, 928. [Google Scholar]
- Wei J. H., Zhang Z., Yang M. H., Gao Z. H., Chen W. P., Feng J. D., et al. (2012). Tambac Inducer and Preparation Method. CN101731282B.
- Whittaker R. H. (1972). Evolution and measurement of species diversity. Taxonomy 21, 213–251. 10.2307/1218190 [DOI] [Google Scholar]
- Xu Y., Zhang Z., Wang M., Wei J., Chen H., Gao Z., et al. (2013). Identification of genes related to agarwood formation: transcriptome analysis of healthy and wounded tissues of Aquilaria sinensis. BMC Genomics 14:227. 10.1186/1471-2164-14-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunita L. (2009). Effectivity of Acremonium sp. and Methyl Jasmonate to Improve Agarwood Quality of Aquilaria Microcarpa. IPB Scientific Repository, Bogor Agricultural University; Available online at: http://repository.ipb.ac.id/handle/123456789/14803 [Google Scholar]
- Zhang X. L., Liu Y. Y., Wei J. H., Yang Y., Zhang Z., Huang J. Q., et al. (2012). Production of high-quality agarwood in Aquilaria sinensis trees via whole-tree agarwood-induction technology. Chin. Chem. Lett. 23, 727–730. 10.1016/j.cclet.2012.04.019 [DOI] [Google Scholar]
- Zhang Z., Yang Y., Meng H., Sui C. H., Wei J. H., Chen H. Q. (2010). Advances in studies on mechanism of agarwood formation in Aquilaria sinensis and its hypothesis of agarwood formation induced by defense response. Chin. Trad. Herbal Drugs 41, 156–159. Available online at: http://en.cnki.com.cn/Article_en/CJFDTOTAL-ZCYO201001045.htm [Google Scholar]
- Zhang Z., Zhang X., Yang Y., Wei J. H., Meng H., Meng Z. H., et al. (2014). Hydrogen peroxide induces vessel occlusions and stimulates sesquiterpenes accumulation in stems of Aquilaria sinensis. Plant Growth Regul. 72, 81–87. 10.1007/s10725-013-9838-z [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Site map of the districts selected for sample collection.
GC-MS profile of oil extracted from artificial infected wood dust sample inoculated with Pantoea dispersa AQGWDB1 (A) and Penicillium aethiopicum AQGGR1.2 (B) showing the presence of the agarospirol compound in gas chromatogram mass spectrum of Agarospirol compound (RT 35.72) (C).
F-test of significance for agarospirol content variation between fungal infected and control wood (A), bacterial infected and control wood (B) and fungal and bacterial infected wood (C).
Bi plot of GC data of bacterial and fungal infected wood samples collected 3 months after artificial inoculation  Bacteria
Bacteria  Fungi.
Fungi.
PCA plot of GC data of bacterial and fungal infected wood samples collected 3 months after artificial inoculation  Bacteria
Bacteria  Fungi.
Fungi.
List of sample collected from different districts of Assam for microbial isolation.
Identification of fungal and bacterial isolates on the basis of sequence similarity with different sequence data base.
Per cent composition of compounds in oleoresin extract identified by gas chromatography mass spectroscopy.