Abstract
Angiogenesis, the formation of new blood capillaries from pre-existing vessels, is a hallmark of cancer. Thus far, strategies for reducing tumor angiogenesis have focused on inhibiting pro-angiogenic factors, and less is known about the therapeutic effects of mimicking the actions of angiogenesis inhibitors. Thrombospondin-1 (TSP1) is an important endogenous inhibitor of angiogenesis that has been investigated as an anti-angiogenic agent. TSP1 impedes the growth of new blood vessels in many ways, including crosstalk with pro-angiogenic factors. Owing to the complexity of TSP1 signaling, a predictive systems biology model would provide quantitative understanding of the angiogenic balance in tumor tissue. Therefore, we have developed a molecular-detailed, mechanistic model of TSP1 and vascular endothelial growth factor (VEGF), a promoter of angiogenesis, in breast tumor tissue. The model predicts the distribution of the angiogenic factors in tumor tissue, revealing that TSP1 is primarily in an inactive, cleaved form owing to the action of proteases, rather than bound to its cellular receptors or to VEGF. The model also predicts the effects of enhancing TSP1’s interactions with its receptors and with VEGF. To provide additional predictions that can guide the development of new anti-angiogenic drugs, we simulate administration of exogenous TSP1 mimetics that bind specific targets. The model predicts that the CD47-binding TSP1 mimetic markedly decreases the ratio of receptor-bound VEGF to receptor-bound TSP1, in favor of anti-angiogenesis. Thus, we have established a model that provides a quantitative framework to study the response to TSP1 mimetics.
Introduction
A hallmark of cancer is angiogenesis, the formation of new blood capillaries from pre-existing vessels. This process enables oxygen and nutrients from the surrounding microenvironment to reach the tumor. In fact, angiogenesis promotes cancer development, invasion, and metastasis. For these reasons, angiogenesis has become a prominent target for cancer drugs.1
Therapies aimed at inhibiting angiogenesis (‘anti-angiogenic therapies’) target many aspects in the process of new blood vessel growth, with a focus on inhibiting pro-angiogenic factors.2 Anti-angiogenic therapeutics that target signaling mediated by the vascular endothelial growth factor-A (VEGF), a potent promoter of angiogenesis, are approved for treatment of various cancer types.3 These agents include drugs that bind to VEGF and prevent it from binding to and activating its receptors, as well as tyrosine kinase inhibitors that impede activation of VEGF receptors intracellularly. These treatment strategies, however, have not been successful in all cancer types. In fact, antibody therapy targeting VEGF is no longer approved for breast cancer treatment. In addition, many tumors, including breast tumors, become resistant to anti-VEGF or other anti-angiogenic treatments.4 Numerous preclinical studies show that targeting a single factor within the angiogenesis signaling network is insufficient to arrest tumor growth and vascularization since tumors may ‘escape’ treatment by utilizing alternative pathways.5 Thus, there is a critical need to better understand the effects of these pro- and anti-angiogenic pathways in order to develop effective treatment strategies, including multi-modal therapies that can address the issue of drug resistance.6,7
Both pro- and anti-angiogenic factors determine the extent of vascularization8 and the response to anti-angiogenic therapy.9 Therefore, another means of increasing the efficacy of anti-angiogenic treatment may be to mimic the action of inhibitors of angiogenesis, while simultaneously inhibiting the promoters. For example, in a preclinical model of pancreatic cancer, altering the balance between pro- and anti-angiogenic factors was shown to modulate tumor growth.10
To this end, several anti-angiogenic factors have been identified as potential cancer therapeutics. Thrombospondins (TSPs) are a family of multi-domain, calcium-binding glycoproteins that are highly expressed during development.11 Of the five TSPs, thrombospondin-1 (TSP1) is the most studied, was the first endogenous anti-angiogenic factor identified,12 and has been investigated for anti-angiogenic therapy. TSP1 acts to impede the growth of new blood vessels in multiple ways. First, TSP1 influences growth factor availability. It can bind to VEGF and other pro-angiogenic factors to reduce intracellular signaling through their receptors and to clear the pro-angiogenic growth factors from the cell via the low-density lipoprotein receptor-related protein 1 (LRP1). TSP1 also inhibits the activation of matrix metalloprotease-9 (MMP9),13 which among its many functions, is able to cleave VEGF.14 In addition to altering growth factor availability, TSP1 inhibits angiogenesis by binding to and activating its own receptors. TSP1 signaling through the CD36 receptor results in reduced cell survival and activation of apoptosis pathways via caspase-3.13 TSP1-mediated activation of the CD47 receptors antagonizes nitric oxide signaling via endothelial cell-derived nitric oxide synthase,15 which is important in cell migration and proliferation.16 CD47 also couples to the VEGF receptor R2 to inhibit VEGF-mediated activation.17 In addition, TSP1 binds to β1 integrins, which further antagonizes VEGF signaling.13
Interestingly, TSP1 has been shown to elicit both pro- and anti-angiogenic effects, depending on the microenvironment.18 This effect is not fully understood; thus, the complex TSP1 interactome19 and its context-dependent role suggest that a predictive systems biology model would greatly aid in the optimization of TSP1-based therapeutics.
Quantitative models of angiogenesis provide insight into the fundamental mechanisms of neovascularization. For example, systems biology models are useful in optimizing anti-angiogenic treatment strategies and identifying prognostic biomarkers,20,21 thereby complementing preclinical and clinical studies. We have previously developed and applied mechanistic, systems biology models to examine the effects of drug and tumor properties on the response to anti-VEGF agents22 and to predict drug pharmacokinetics.23 The relative distribution of the angiogenic factors in their free and receptor-bound forms and the number of pro- and anti-angiogenic complexes, are key quantities that influence downstream signaling to modulate angiogenesis. These quantities are difficult to measure and enumerate experimentally; however, systems biology models enable their quantitative prediction.
Here we present a novel computational model that incorporates the molecular interactions between TSP1, VEGF, and their respective receptors in breast cancer tumor tissue. Our systems-level model predicts the concentrations of these factors and their distributions in breast tumor tissue. The model also predicts the effects of altering TSP1’s binding rates on various quantities that characterize the angiogenic state of the tumor. Finally, we apply the model to simulate the effects of TSP1 mimetics. We provide a quantitative framework to understand the distribution of angiogenic factors and to investigate the effects of anti-angiogenic strategies that mimic the actions of TSP1.
Results
Model construction
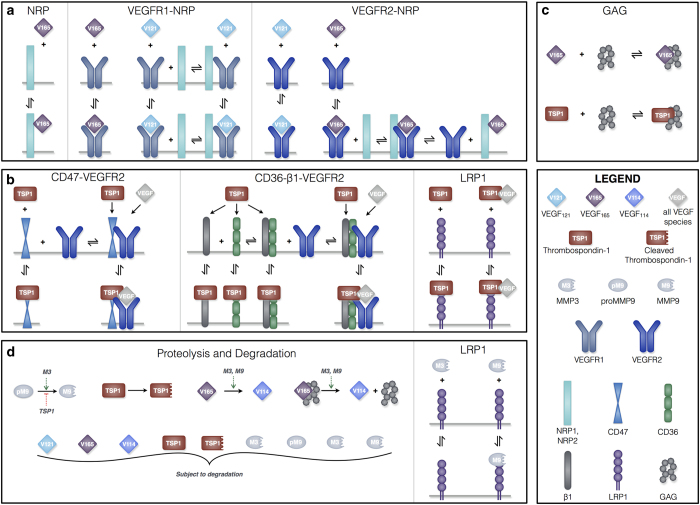
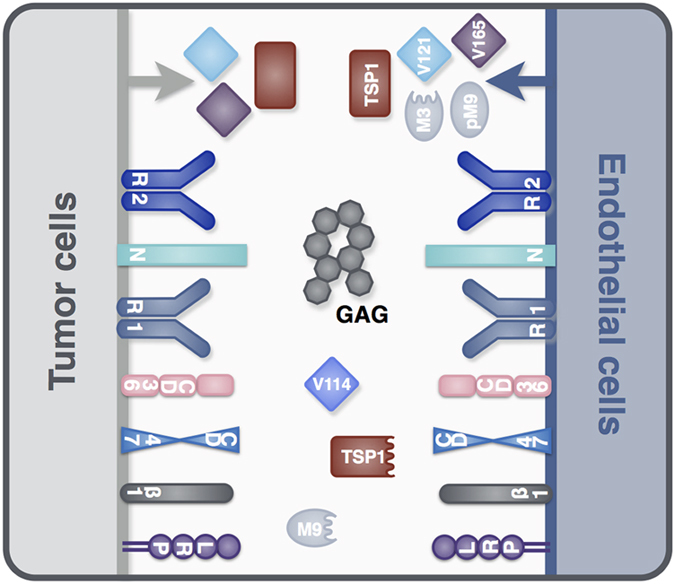
We have constructed a compartment model of TSP1, VEGF, and their receptors in tumor tissue (Figure 1). The molecular interactions are described below and illustrated in Figure 2, and the kinetic rates that govern the interactions are provided in the Materials and Methods section. VEGF and TSP1 are both secreted by endothelial and tumor cells. As in our previous work, we consider two main VEGF isoforms, VEGF121 and VEGF165. A third isoform (VEGF114) is produced following cleavage of VEGF165 by proteases. We also include receptors VEGFR-1 and -2 (R1 and R2, respectively), as well as the neuropilin co-receptors NRP-1 and -2 (N1 and N2, respectively), as shown in Figure 2a. TSP1 interacts with its receptors, CD36, CD47, LRP1, and αxβ1 integrins (β1, representing several species), illustrated in Figure 2b. All receptors are present on endothelial and tumor cells, at specific densities.
Figure 1.
Compartment model of TSP1 and VEGF. The model is comprised of a single compartment representing a solid breast tumor and involves 18 molecule types. The soluble species TSP1, VEGF121, VEGF165, proMMP9, and MMP3 are secreted by endothelial cells. Tumor cells secrete VEGF121 and VEGF165. Other soluble species can be formed, including cleaved TSP1, active MMP9, and VEGF114. All soluble species are subject to degradation. TSP1 receptors (CD36, CD47, β1, and LRP1) and VEGF receptors (R1, R2) and co-receptors (N1, N2) are localized on endothelial and tumor cells. Glycosaminoglycan (GAG) chains are present in the interstitial space, representing the extracellular matrix and endothelial and parenchymal cell basement membranes.
Figure 2.
Schematic of TSP1 and VEGF interactions. The compartment model includes: (a) molecular interactions of two VEGF isoforms (VEGF121 and VEGF165), receptors (R1 and R2), and co-receptors (N1 and N2); (b) interactions between TSP1, its receptors (CD36, CD47, β1, and LRP1), and VEGF; (c) sequestration of VEGF165 and TSP1 by GAG chains in the ECM and the cellular basement membranes; and (d) activation of proMMP9 via cleavage by MMP3, proteolysis of VEGF165 to form VEGF114 by active MMPs, cleavage of TSP1, and degradation of all soluble species. The receptor coupling and multi-protein complexes indicate the large amount of crosstalk between the pro- and anti-angiogenic interaction networks.
Receptors can be internalized by the cell and recycled back to the surface, and we assume that for each type of receptor the total number (ligated plus unbound) is conserved. In addition, receptors (whether ligated or unbound) can associate with one another, forming multi-protein complexes. Once bound to a cell surface receptor, TSP1 and VEGF can be removed from the tumor interstitial space via receptor-mediated internalization. TSP1 and VEGF bind to one another, other soluble factors, or heparan sulfate proteoglycans (termed glycosaminoglycan, GAG, chains) present in the ECM and basement membranes (Figure 2c). The growth factors can be degraded via proteolysis. In addition, TSP1 is subject to cleavage via multiple factors,24 generating an inactive TSP1 species.
The model includes three other soluble factors only secreted by endothelial cells: two active proteases, MMP3 and MMP9, and the inactive form of MMP9, proMMP9 (Figure 2d). MMP3 is able to activate MMP9, and both proteases cleave soluble and matrix-bound VEGF165 to form VEGF114, which is unable to bind to VEGF receptors.14 TSP1 decreases the protease action of MMP9 by binding to MMP3 and preventing it from activating proMMP9.13 The MMPs can be degraded or removed from the tissue via LRP1-mediated internalization.25
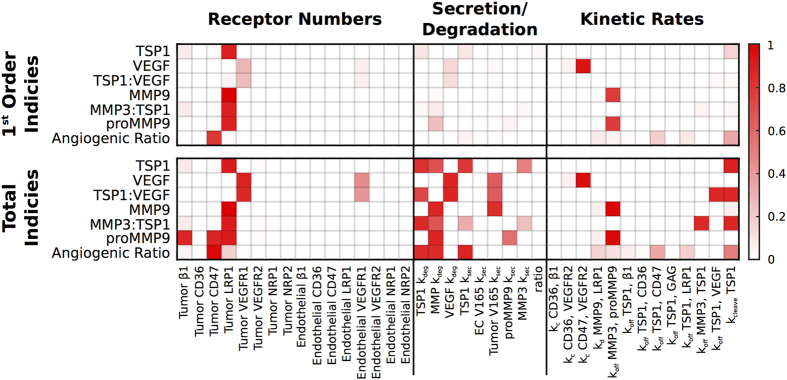
Global sensitivity analysis
We investigated the sensitivity of several model outputs to variations in three sets of parameters: receptor numbers; secretion and degradation rates; and kinetic parameters (Figure 3). We selected these parameters because there is uncertainty in their values owing to a lack of quantitative data available in the literature. We use the eFAST analysis (see Materials and Methods) to quantify how sensitive the steady-state concentrations of TSP1, VEGF, proMMP9, MMP9, the TSP1-VEGF and MMP3-TSP1 complexes, and the ratio of receptor-bound VEGF to receptor-bound TSP1 (termed the ‘angiogenic ratio’) are to those parameters.
Figure 3.
Sensitivity indices of model parameters. The extended Fourier Amplitude Sensitivity Test (eFAST) was used to estimate the variance in the model output with respect with variance in individual model inputs (first-order indices) and covariances in combinations of model inputs (total indices). The eFAST method was used to investigate the sensitivity to receptor numbers, secretion and degradation parameters, and kinetic parameters. The ‘ratio’ refers to the ratio of TSP1 secretion by the endothelial cells compared to the tumor cells.
This global sensitivity analysis reveals that the densities of CD47, LRP1, R1 and R2 on tumor cells influence the model outputs both alone and in combination with other receptor numbers. These results quantify the importance of receptor coupling. LRP1 density particularly affects the predicted concentrations of several species in the model, including TSP1 and the MMP species. This is because TSP1 binding to LRP1 leads to internalization, removing TSP1 from the tissue and influencing its availability to bind to other species. Significantly and non-intuitively, the angiogenic ratio is particularly influenced by the density of CD47 receptors, indicating that this species may have an important role in controlling the extent of angiogenesis.
The sensitivity analysis revealed that the degradation and production rates are more influential in combination, rather than alone, indicated by the higher total indices compared with the first indices. TSP1 is sensitive to its own production and degradation rates, as well as the degradation rate of the MMPs. The concentrations of the other soluble species are more sensitive to only their own production and degradation rates. Interestingly, the concentration of MMP9 is predicted to be sensitive to VEGF secretion. This is because both MMP9 and the TSP1-bound VEGF isoforms bind to and are internalized by LRP1. Thus, VEGF influences MMP9’s removal from the tissue. Such results are not immediately intuitive; therefore, the analysis provides valuable insight into the system.
Finally, we quantified the effects of variance in the kinetic rates. Several of the model outputs are more sensitive to variations in combinations of the kinetic parameters than individual kinetic rates. TSP1 tissue concentration is most sensitive to its cleavage rate. This is to be expected, as the rate at which TSP1 is cleaved is large compared with the other kinetic rates (see Materials and Methods). The concentrations of active and inactive MMP9 are particularly sensitive to the binding rate of MMP3 and proMMP9, and we set their values based on published studies.
The global sensitivity analysis identified the parameters to which the model outputs are most sensitive. Because of their impact on the model outputs, it is especially important to have high confidence in the values of these parameters. We use the model to predict the concentrations of the angiogenic factors, to investigate the effects of varying the rates at which TSP1 binds to certain species, and to simulate the response to TSP1 mimetics. For all of the model predictions, we present the mean and standard deviation of 5,000 Monte Carlo (MC) results. The MC analysis generates a range of model predictions by varying specific model parameters shown by the sensitivity analysis to significantly influence the outputs. This analysis enables interpretation of the robustness of the model predictions.
Predicted concentrations and tissue distributions
The model generates novel predictions for the absolute concentration of the angiogenic species in the tissue interstitial space, including unbound ligands and receptor-ligand complexes. We report the average concentrations for 5,000 MC simulations, along with the s.d.
The concentration of soluble TSP1 is predicted to be 2.3±0.06 nmol/l, including 185±5 pmol/l unbound and 2.2±0.06 nmol/l cleaved TSP1 (Table 1). The concentration of cleaved TSP1 is predicted to be several fold higher than unbound TSP1 (12-fold, s.d.=8×10−5); thus, a large proportion of soluble TSP1 is in an inactive form. In comparison, the concentration of total soluble VEGF is 178±8 pmol/l, including 165±5, 8±0.6, and 5±6 pmol/l of VEGF121, VEGF165, and VEGF114, respectively. Therefore, nearly all of soluble VEGF, 97%±3%, is in an active form (VEGF121 or VEGF165).
Table 1. Experimental and predicted steady-state concentrations.
| Predicted value mean±s.d. | Measured value | Cancer type and References | |
|---|---|---|---|
| VEGF (pmol/l)a | 178±8 | 8–389 | Multiple, compiled in ref. 22 |
|
TSP1 | |||
| Total (pmol) | 61±4 | 52–64b | Breast34 Hepatocellular carcinoma35 |
| Unbound (pmol/l)a | 185±5 | c | |
| Cleaved (nmol/l)a | 2.2±0.06 | c | |
|
MMP3 | |||
| Total (pmol) | 162±0.02 | 160b | Oral squamous cell carcinoma33 |
| Unbound (nmol/l)a | 9.0±5×10−3 | c | |
|
MMP9 | |||
| Total (pmol) | 161±0.1 | 160b | Oral squamous cell carcinoma33 |
| Active (% of total) | 3%±8×10−4% | 8% | Oral squamous cell carcinoma33 |
| Active (pmol/l)a | 295±7 | c | |
Refers to unbound protein in the available interstitial space.
The model predicts the extracellular protein concentrations, whereas experimental data reflect the total amount (extracellular and intracellular), as the tissue is homogenized prior to protein quantification. We assume that 50% of the total protein amount is localized to the extracellular space. This assumption can easily be changed, given more experimental data.
Model prediction, no data available.
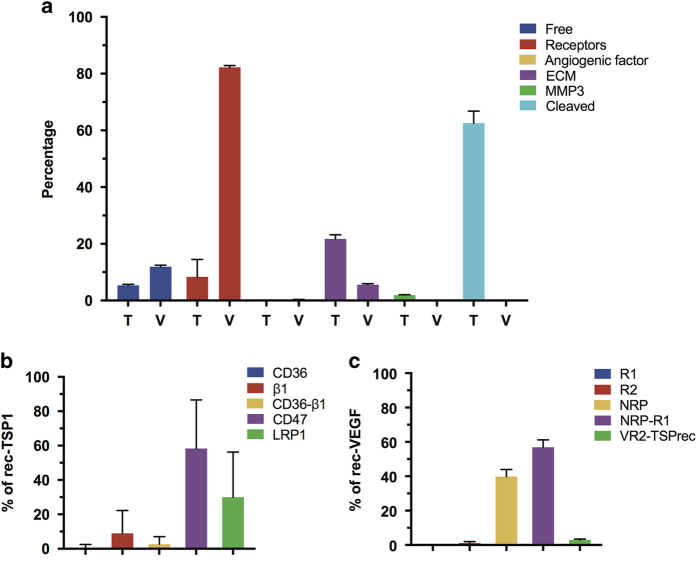
The model also predicts that pro- and anti-angiogenic ligands have significantly different soluble and bound distributions in tumor tissue (Figure 4a). We found that most TSP1 in the tumor tissue is either in the cleaved form, due to proteolysis, or is bound to the extracellular matrix (63±4% and 22±1%, respectively). Approximately 5±0.4% of TSP1 is unbound, while receptor- and VEGF-bound TSP1 comprise 8±6% and 0.1±0.04%, respectively, of total TSP1 in the tumor. In comparison, VEGF largely exists in complexes with its receptors (82±0.6% of total VEGF in the tumor). Unbound and ECM-bound VEGF comprise 12±0.5% and 6±0.4% of total VEGF, respectively. The predicted VEGF distribution is nearly the same as that predicted by our previous model of VEGF transport and kinetics in human cancer patients,23 indicating that the presence of TSP1 does not influence VEGF binding in tumor tissue.
Figure 4.
Distribution TSP1 and VEGF in tumor. (a) The predicted percentage of TSP1 (‘T’) and VEGF (‘V’) bound to different factors in the tissue, and the predicted percentage of receptor-bound forms of the angiogenic ligands: (b) bound TSP1 distribution; (c) bound VEGF distribution.
The model quantifies the distribution of receptor-bound ligands (Figure 4b,c). TSP1-CD47 interactions are predicted to comprise 58±28% of receptor-bound TSP1. These complexes include species that antagonize pro-angiogenic signaling mediated by VEGF, either by directly modulating nitric oxide signaling (TSP1 bound to CD47 alone) or through the formation of inhibitory complexes (TSP-CD47 complexes involving R2 and VEGF). Another significant percentage of receptor-bound TSP1 is complexed with LRP1 (30±26%). Binding of TSP1 to this receptor leads to internalization and endocytosis and serves as a means of removing TSP1 from the extracellular space. In contrast, more than half of the receptor-bound VEGF complexes are pro-angiogenic (56±4%). These include VEGF165-R2-N1 and VEGF121-R1-N1 complexes, which promote angiogenesis signaling. In addition, the range of model predictions for the distribution of VEGF receptors is tighter than the TSP1 receptor distribution.
To summarize these results, the model predicts that nearly all unbound VEGF is in one of its active forms, and receptor-bound VEGF is participating in pro-angiogenic complexes. Although the majority of receptor-bound TSP1 is promoting anti-angiogenic signaling, receptor-bound TSP1 comprises only a small percentage of total TSP1. A large percentage of TSP1 is in an inactive form. Altogether, these predictions indicate that the tumor is significantly shifted towards a pro-angiogenic state.
Effects of enhancing TSP1 binding rates
Using the model, we are able to quantify the contribution of specific TSP1-mediated anti-angiogenic mechanisms on particular quantities that characterize the angiogenic state of the tissue. Specifically, we investigated the effects of varying the binding of TSP1 with its receptors, as well as VEGF. We decreased the dissociation rates for β1, CD36, CD47, LRP1, and MMP3, individually, to make TSP1 bind 100-fold more tightly to each partner. It is possible, in principle, to evolve and select for TSP1 variants with desired kinetic properties.26 Thus, our simulations provide important predictions as to the effect of TSP1 mimetics with altered binding kinetics compared with the endogenous protein.
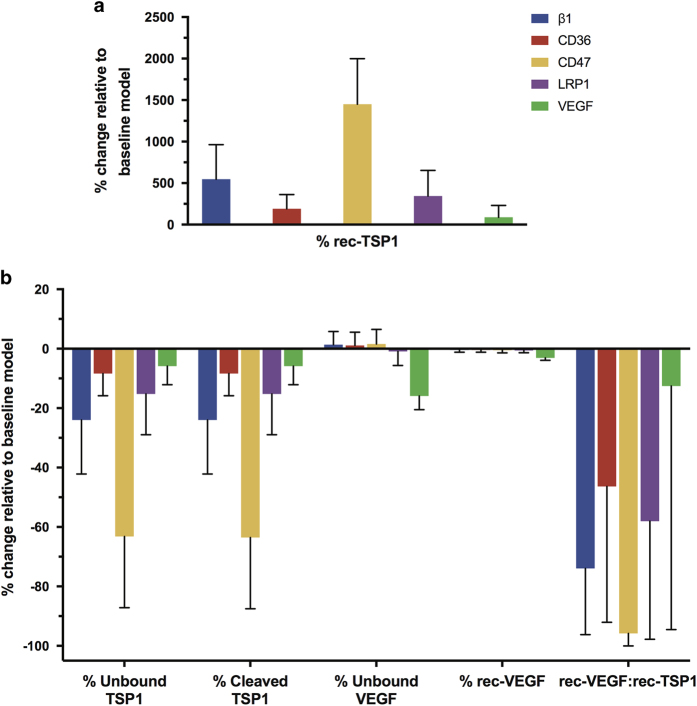
Increasing the affinity of TSP1 to its binding partners is predicted to affect the percentage of receptor-bound TSP1 (Figure 5a, ‘% rec-TSP1’). As expected, when TSP1 binds more tightly to receptors β1, CD36, CD47, or LRP1, more TSP1 is in the receptor-bound form. However, the increase in receptor-bound TSP1 varies to different degrees depending on the particular interaction strengthened. Allowing TSP1 to bind more tightly to CD47 has the largest impact on the percentage of receptor-bound TSP1 (an increase of more than 14-fold). Strengthening the association between TSP1 and β1 or LRP1 increases receptor-bound TSP1 by 1.9- and 5.5-fold, respectively. The different affinities and numbers of receptors contribute to these results. However, these results are highly variable, depending on the receptor coupling rates and TSP1 density values. Interestingly, the effect on receptor-bound TSP1 does not vary linearly with the affinity or receptor density; therefore, the model provides novel and quantitative predictions regarding the effects of enhancing TSP1’s interactions with its receptors.
Figure 5.
Effect of enhancing TSP1 affinity for its binding partners. The model predicts the effect on various quantities that characterize the angiogenic state of the tumor tissue in response to individually decreasing the dissociation rates between TSP1 and β1, CD36, CD47, LRP1, and MMP3 by 100-fold: (a) percentage change in total receptor-bound TSP1; (b) percentage change in the relative amounts of unbound and cleaved TSP1, unbound and receptor-bound VEGF, and the ratio of receptor-bound VEGF to receptor-bound TSP1.
Upon altering the affinity, we also used the model to predict the percent change in the relative amounts of unbound and cleaved TSP1, as well as the relative amounts of unbound and receptor-bound VEGF, and the ratio of receptor-bound VEGF to receptor-bound TSP1 (Figure 5b). Having more receptor-bound than unbound TSP1 may be a desired outcome of anti-angiogenic therapy. Therefore, we sought to understand whether enhancing the interactions between TSP1 and the selected binding partners could reduce the percentage of unbound TSP1. The model predicts that increasing the affinity to CD47 has the largest effect on the amount of unbound TSP1 (Figure 5b, ‘% Unbound TSP1’). Although this result varies across the 5,000 MC simulations, overall, increasing the binding affinity for TSP1 to CD47 decreases unbound TSP1 by 64±24%). Increasing the affinity of TSP1 to β1, CD36, LRP1, or VEGF reduces unbound TSP1 between 6 and 24%.
Reducing cleaved TSP1 is also of interest, as a significant proportion of TSP1 is in this inactive form (63±4%; Figure 4a), preventing TSP1 from exerting its anti-angiogenic actions. The model predicts that allowing TSP1 to bind more tightly to any of the five selected binding partners decreases the percentage of cleaved TSP1, indicating that the species are able to displace TSP1 from proteases and protect it from cleavage.
Altering the affinity of TSP1 to its binding partners also has a slight influence on the distribution of VEGF. The model predicts that altering TSP1 binding kinetics with VEGF can reduce unbound VEGF and shift the balance toward an anti-angiogenic state. Overall, increasing the association of TSP1 with VEGF has the largest effect on the distribution of VEGF in the tumor tissue (Figure 5b, ‘% Unbound VEGF’, ‘% rec-VEGF’). The effect of a tighter association with VEGF influences the relative amount of unbound VEGF the most (a decrease of 16±5%). Interestingly, the relative amount of receptor-bound VEGF is insensitive to changes in TSP1’s affinity to its binding partners. Only increasing the affinity of TSP1 to VEGF is able to slightly reduce the percentage of ligated VEGF (decreasing it by 3±0.8%).
Finally, the model predicts the effects of TSP1’s affinity on the ratio of receptor-bound VEGF to receptor-bound TSP1 (Figure 5b, ‘rec-VEGF:rec-TSP1’). This quantity provides an indication of TSP1’s anti-angiogenic effect, leading to reduced VEGF signaling and increased TSP1-mediated apoptosis. For the baseline model, the rec-VEGF:rec-TSP1 ratio is 7.8:1, indicating a state in which the signaling complexes that promote new blood vessel formation greatly outnumber those that oppose angiogenesis. We used the model to predict how this angiogenic ratio is affected by the affinity of TSP1 to selected binding partners.
Increasing the affinity of TSP1 to β1 or LRP1 decreases the rec-VEGF:rec-TSP1 ratio in the tumor tissue by 74±22% and 58±40%, respectively. Increasing the affinity of TSP1 to CD47 has the largest effect, decreasing the ratio by more than 96±4%. Allowing TSP1 to bind more tightly to VEGF also decreases the ratio. This is because TSP1, even when bound to VEGF, can bind to LRP1, increasing the number of anti-angiogenic receptor complexes, despite having a limited effect on the percentage of free or receptor-bound VEGF. Interestingly, the effects of tighter binding between TSP1 and CD47 are significantly less variable across the MC simulations, as compared to a tighter association between TSP1 and β1, CD36, or LRP1, or VEGF.
Effects of TSP1 mimetics
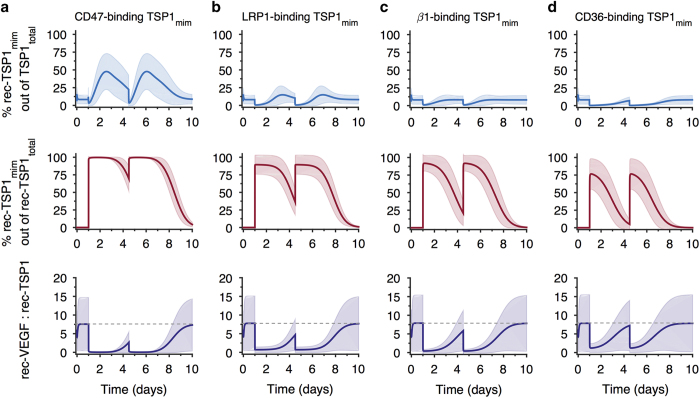
Building on the simulations to investigate the effect of increasing the affinity of TSP1 to its binding partners, we apply the model to predict the effects of administering TSP1 mimetics that bind to one of TSP1’s native receptors. Given the large size of endogenous TSP1 (~450 kDa), it is not possible to directly administer the protein as a therapeutic agent. Thus, short peptide sequences have been derived to specifically target TSP1 receptors and inhibit angiogenesis.27 Therapeutic use of peptide mimetics is enabled by delivery strategies that provide a slow release of the peptides, such as nanoparticle delivery or administration of the peptides within a biomaterial scaffold.28 We utilized the model to investigate the effects of CD47-, LRP1-, β1-, or CD36-binding TSP1 mimetics (‘TSP1mim’). We simulate twice-weekly dosing within an engineered biomaterial.29 The system is first allowed to reach steady state (~10 h), which is the same state described above, in terms of the concentrations and distributions of the soluble species. Two bolus doses of 10 μg of the therapeutic are given at days 1 and 3.5. The release of TSP1 peptides from the delivery vehicle follows an exponential decay with rate constant 2.8×10−5 s−1. The peptide mimetics are also subject to degradation, as are all soluble factors, with a half-life of 60 min.13 For these simulations, the mimetics bind to a particular receptor with the same affinity as endogenous TSP1, and cannot be cleaved. The model predicts the drug’s effects on the distribution of TSP1 in the tissue, the relative amounts of receptor-bound TSP1, and the rec-VEGF:rec-TSP1 ratio, quantities that are difficult to measure in the clinic or in preclinical models. Below, we report the mean predicted result from 5,000 MC simulations, and Figure 6 shows the mean±s.d.
Figure 6.
Effect of TSP1 mimetics. The model predicts the effect of TSP1 mimetics that specifically bind to: (a) CD47; (b) LRP1; (c) β1; and (d) CD36. Top panels: the percentage of total TSP1 (TSP1endogenous+TSP1mim) that is receptor-bound. Middle panels: the percentage of receptor-bound TSP1mim relative to rec-TSP1total in tumor tissue. Bottom panels: the ratio of rec-VEGF to rec-TSP1, including endogenous TSP1 and the TSP1 mimetic. The gray dashed line indicates the ratio for the baseline model without TSP1mim.
The relative amount of receptor-bound TSP1mim and the time over which the receptors remain bound to TSP1mim vary depending on the particular receptor that the mimetic is binding. We calculated the percentage of total TSP1 (‘TSP1total’, i.e., endogenous TSP1+TSP1mim) that is receptor-bound, as shown in Figure 6, top row. This is the predicted amount relative to all of the TSP1 and TSP1mim in the tissue (i.e., TSP1 in all of its various forms, including all of the binding partners previously shown in Figure 4a).
The CD47-binding mimetic is predicted to most significantly increase the relative amount of ligated receptors, and its effects last for the longest time. For this mimetic, the percentage of receptor-bound TSP1total peaks at 48% of TSP1total in the tissue ~1.5 days after the therapeutic is administered (Figure 6a, top panel). The maximum receptor-bound TSP1total for the LRP1-, β1-, and CD36-binding mimetics are 15%, 8%, and 7%, respectively, (Figures 6b–d, top panels, blue lines).
We next consider whether the receptor-bound TSP1mim complexes remain bound before a second dose of the therapeutic. The middle row of Figure 6 shows the percentage of receptor-bound TSP1mim relative to receptor-bound TSPtotal. After the first dose of the CD47-binding mimetic, the percentage of rec-TSP1mim remains above 69% until the next dose is given (Figure 6a, middle panels). In comparison, the percentage of rec-TSP1mim falls to 36%, 20%, and 5% for the LRP1-, β1-, and CD36-binding mimetics, respectively (Figures 6b–d, middle panels).
Lastly, the model predicts that the TSP1 mimetics are able to significantly shift the ratio of receptor-bound VEGF to receptor-bound TSP1total in favor of anti-angiogenesis. Following each dose of a particular TSP1 mimetic, the ratio of rec-VEGF:rec-TSP1total varies compared to the baseline value of 7.8:1. For the CD47-binding mimetic, the ratio shifts to 1:38 (Figure 6a, bottom panel), indicating that the number of ligated TSP1 receptors significantly outnumbers the number of ligated VEGF receptors. The ratio of rec-VEGF:rec-TSP1total is 1:1.2 and 1:2.4 for the LRP1- and β1-binding mimetics (Figure 6b,c, bottom panels). Interestingly, a TSP1 mimetic that selectively binds CD36 shifts the balance in favor of anti-angiogenesis (the ratio becomes 1.2:1), but does not make receptor-TSP1total complexes outnumber receptor-VEGF complexes as the other mimetics do.
Discussion and Conclusion
We have developed a molecular-detailed model of the interactions between two important angiogenic factors: TSP1 and VEGF. The biophysically realistic model represents the network of biochemical interactions between these molecular species, their receptors, and relevant soluble factors. The molecular interactions are governed by kinetic parameters taken from experimental measurements, where available. To our knowledge, this is the first model that explicitly accounts for these important factors in the context of tumor angiogenesis.
The model captures experimental observations, both qualitatively and quantitatively. However, there are limitations that can be addressed in future work. Given the limited amount of quantitative data, we used measurements from tumor types other than breast cancer. In addition, we have assumed a generic mechanism for TSP1 cleavage, without specifying the dynamics of the species that mediate TSP1 proteolysis. We have neglected soluble receptors sVEGFR-1 and -2, as there are limited data with which to specify their secretion rates, particularly for sVEGFR-2. We focus on the extracellular interactions between the angiogenic ligands and their receptors and do not include intracellular signaling pathways or downstream effects, which could be added to predict the rates of cell proliferation, apoptosis, and vascular sprouting.30
We performed a global sensitivity analysis to quantify how individual and combinations of model parameters influence the steady-state concentrations of the soluble species. The global sensitivity analysis showed that despite not having specific measurements for all of the molecular interactions included in the model, for most parameters, our assumed values do not significantly influence the model predictions. In cases where the model parameters were shown to be important based on the sensitivity analysis, we were able to estimate the values using existing data. We utilize results from the global sensitivity analysis to determine which parameters to vary in Monte Carlo (MC) simulations. The MC analysis shows that most of the model predictions have low s.d., indicating that the model generates robust quantitative predictions.
The model predicts the distributions of TSP1 and VEGF among their binding partners in tumor tissue. The species are predicted to have very different relative amounts that are unbound or receptor-bound. Specifically, a large percentage of VEGF (82%) is ligated to its receptors, while receptor-bound TSP1 only comprises 8% of total TSP1. In addition, the ratio of receptor-bound VEGF to receptor-bound TSP1 is predicted to strongly favor angiogenesis (7.8:1). Altogether, these results show that the angiogenic balance is significantly tipped towards a pro-angiogenic state. The model provides a quantitative and mechanistic description as to how this pro-angiogenic state occurs. Specifically, the predictions provide insight into why the absolute concentration of TSP1 can be significantly greater than VEGF, either in plasma or tumor tissue,9 but may not be sufficient to inhibit angiogenesis.
We have used the model to explore the effects of altering the interactions between TSP1 and its binding partners. It is possible, in principle, to select TSP1 variants with higher affinity to specific binding partners through directed evolution via random mutagenesis or exon shuffling.26 Therefore, these simulations represent more than simply changing the model parameters. Rather, they provide important quantitative predictions regarding the effect of TSP1 mimetics with altered binding kinetics compared with the endogenous protein.
The model predicts that increasing the rates at which TSP1 binds to CD47 has a global effect to alter the angiogenic balance in the tumor tissue. Specifically, enhancing TSP1’s binding interactions with CD47 decreases the relative amount of unbound TSP1 and protects TSP1 from cleavage. In addition, the formation of more TSP1-CD47 complexes can shift the angiogenic ratio (rec-VEGF:rec-TSP1) in favor of anti-angiogenic complexes, compared to the baseline model. Interestingly, it has been postulated that the ratio of VEGF to TSP1 correlates with response to chemotherapy and overall survival.31 Our model suggests that the amount of the receptor-bound form of the angiogenic factors may also be an important quantity to consider.
Of the strategies to enhance TSP1’s anti-angiogenic action, only increasing the affinity of TSP1 for VEGF can lead to a reduction in the relative amount of ligated VEGF (i.e., fewer pro-angiogenic complexes between VEGF and its receptors). Thus, TSP1’s ability to act as a sink for VEGF has the downstream effect of explicitly reducing the number of pro-angiogenic signaling complexes.
We applied the model to investigate the response to TSP1 mimetics. The model predicts the distribution of total TSP1 (TSP1endogenous+TSP1mim) in the tissue following two doses of a mimetic that specifically targets one of TSP1’s receptors. The CD47-binding mimetic significantly increases the relative amount of receptor-bound total TSP1, compared with the other mimetics. In addition, a majority of the CD47 receptors are predicted to remain ligated to the TSP1 mimetic longer than what occurs with the other receptor-binding mimetics. Our results provide quantitative and mechanistic support for the development of TSP1 mimetics that specifically bind CD47.27 Interestingly, compared with targeting CD47, CD36-binding peptides have a longer history of development in vitro, in preclinical studies, and in clinical trials; however, these drugs have only achieved limited benefits to patients.27 Our model predicts that very few CD36 receptors are being ligated following administration of the CD36-binding mimetic and the effects of this mimetic are short-lived. These predictions provide some hypotheses regarding the disappointing results of targeting CD36.
In summary, our molecular-detailed model quantifies specific molecular interactions that determine the balance between two opposing angiogenic factors. The model provides mechanistic and quantitative explanations of the effects of altering the kinetics of TSP1 binding and the response to TSP1 mimetics. Due to the highly complex and interconnected nature of the reaction network, it is difficult to predict these effects a priori. In addition, although high-throughput quantitative approaches to study ligand-receptor interactions are becoming possible,32 experimental quantification of the formation of these complexes and the effects of disrupting the interactions is challenging. Therefore, the model provides relevant and useful insight into the effects of TSP1 binding. The model is a much-needed quantitative framework that can be used to study the response to anti-angiogenic therapies and generate testable hypotheses regarding optimal TSP1 mimetics.
Materials and Methods
Computational model
Tumor tissue
The tumor tissue is parameterized as a breast tumor 4 cm in diameter. We assume the tissue to be a spatially averaged compartment and do not consider concentration gradients of the molecular species, as the timescale for the biochemical interactions is slower than that of diffusion. The tissue is comprised of tumor cells, endothelial cells representing the blood vessels, and interstitial space. The interstitial space consists of the interstitial fluid, the space available to the soluble species, and the solid phase, which consists of the extracellular matrix and basement membranes surrounding tumor and endothelial cells. Geometric parameters (Supplementary Table S1) characterize the compartment and enable conversion of the concentrations from units used in the model (moles per cm3 tissue) to more standard units (pmol/l). Although the model represents a spatially averaged compartment, we still denote the density of receptors on specific cell types (tumor or endothelial cells). In addition, we specify the origin of the soluble species from the specific cell types. This provides a molecularly detailed model and enables investigation of the extent of ligation for receptors on distinct cell surfaces and other mechanistic and molecular-level questions related to the angiogenic balance.
Model parameters
The values for the VEGF secretion rates, receptor densities, and kinetic parameters and the concentrations of ECM binding sites are the same as those used in our previous models.22,23 Below, we present the parameters involving TSP1 and its receptors. We explore the effects of varying these parameters in the global sensitivity analysis (see Results).
Production of soluble species
The rates at which TSP1, MMP3, and proMMP9 are secreted are not readily available in published literature. Therefore, these rates must be set in order to match experimental measurements of the proteins’ concentrations in tumor tissue.33–35 We manually changed the secretion rates for TSP1, MMP3, and proMMP9 in combination such that the total concentration of each species given by the model matches the experimentally measured concentrations.
To our knowledge, there is no quantitative data on the relative TSP1 secretion rates by endothelial and tumor cells. Therefore, we assume that the ratio of endothelial cell secretion compared to tumor cell secretion is 10:1, the opposite of the estimated ratio at which endothelial cells and tumor cells secrete VEGF.23 Thus, we fixed the secretion ratio and only tuned a single secretion rate for TSP1.
The values of the secretion rates required for the predicted total protein concentrations (bound and unbound) to match experimental measurements are given in Table 1. The data used to calibrate the TSP1 and MMP secretion rates come from multiple tumor types, including breast cancer, hepatocellular carcinoma, and oral squamous cell carcinoma. In addition, the tuned secretion rates represent one possible combination that enables the predicted steady-state concentration to match experimental data. Therefore, these parameters are used in the sensitivity analysis.
Degradation of soluble species
We set the degradation rate constants to be ln(2)/t1/2, where t1/2 is the species’ half-life (Supplementary Table S2). As in our previous models, we use a half-life of 60 min for VEGF. TSP1’s half-life is nearly two-fold shorter, at 35 min.36 The half-life of MMP species is reported to be on the order of 10 min.37
Receptor density
There is a paucity of quantitative data for the number of TSP1 receptors on endothelial and tumor cells. We use information from the Human Protein Atlas,38 which provides qualitative levels of protein levels in tumor tissue. The database shows that CD47 and β1 receptors are highly expressed in many tumor types, while LRP1 and CD36 are expressed at moderate and low levels, respectively. Using this qualitative information, we assume the number of receptors on tumor cells to be: 10,000 for CD47 and β1, 5,000 for LRP1 and 2,500 for CD36 (Supplementary Table S3). The numbers of receptors on the endothelial cell within the tumor tissue are set to be half of that for tumor cells, assuming equal distribution on the luminal and abluminal surfaces.
Molecular interactions
The kinetic rate parameters are given in Supplementary Table S4 and described below. The Kd values of TSP1 to CD36, CD47, and LRP1 are 230, 10, and 12 nmol/l, respectively.39–41 It has been shown that TSP1 binds less tightly to β1 integrins compared with CD47.39 Therefore, we assume the affinity of TSP1 binding β1 to be 100 nmol/l (10-fold less tight binding compared with TSP1-CD47); to our knowledge, there are no measurements of the kinetics of that interaction. The Kd value for TSP1 binding to VEGF is 10 nmol/l,42 and we assume the affinity of TSP1 is the same for each of the three VEGF isoforms. The coupling rate for CD36 and β1 is assumed to be the same as the R2-NRP coupling rate (3.1×1013 (mol cm−2)−1 s−1). We assume the rate for CD36 coupling to R2 and CD47 coupling to R2 to be 3.1×1011 (mol cm−2)−1 s−1, which is two orders of magnitude smaller than the R2-NRP coupling rate. Thus, we impose that TSP1 receptors interact with the same kinetics as VEGF receptors and co-receptors; however, interactions between TSP1 and VEGF receptors occur at a lower rate. Receptors are internalized and inserted at the cell membrane such that the total number of receptors (ligated plus unbound) is constant. The internalization rate is 2.8×10−4 s−1, and the rate at which receptors are recycled back to the surface is 2.8×10−4 mol cm−3 tissue per s. TSP1 can also bind to glycosaminoglycan (GAG) chains located in the extracellular matrix or endothelial and tumor cell basement membranes with a Kd of 200 nmol/l.43
Protease activity
MMP9 is secreted in its inactive form, proMMP9, and is activated by MMP3 upon binding and cleavage.44 The model includes inhibition of MMP9 activity by TSP1; however, the exact mechanism is not known. Using a minimal model consisting of MMP3, MMP9, proMMP9, and TSP1, we implemented alternative mechanisms of TSP1-mediated inhibition and fit the parameter values to experimental data.45 The data quantify the concentration of active MMP2 in the presence or absence of TSP1, and we assume TSP1 inhibits MMP9 in the same way.
We investigated allowing TSP1 to bind directly to MMP3, proMMP9, or active MMP9, similar to the work of Vempati et al.46 in studying MMP9 inhibition by tissue inhibitors of metalloproteinases. We found that allowing TSP1 to bind directly to MMP3 provides a simple mechanism of MMP2 inhibition that fits experimental data. We set the on and off rates for MMP3 and proMMP9 and the rate of MMP2 activation by MMP3 to be 104 (mol/l)−1 s−1, 10−3 s−1, and 10−3 s−1 respectively.46 Using these values, the model captures the relative amount of active MMP2 without TSP1 inhibition measured experimentally (Supplementary Figure S1A). We then fit the model to experimental data of the relative amount of active MMP2 in the presence of different concentrations of TSP1 and estimated the rate at which TSP1 unbinds from MMP3 (assuming an on rate of 105 (mol/l)−1 s−1, based on ref. 47). The model predictions, including confidence intervals, are shown in Supplementary Figure S1B. On the basis of this fitting, the rate of unbinding of TSP1 and MMP3 is estimated to be 2.1×10−3 s−1 (±3.3×10−4); this value is used in the full model. Using the estimated parameter, the model predicts the data not used in the fitting (Supplementary Figure S1C). The equations for the minimal model are provided in Supplementary File S2.
Both MMP3 and active MMP9 are able to cleave VEGF165 to form VEGF114, and their proteolysis rates are set to 631 (mol/l)−1 s−1, based on a previous model of VEGF proteolysis by MMPs.48 MMP9 can also bind to LRP1 with a Kd value of 53 nmol/l,49 enabling removal via internalization, another means of regulating its activity.50
TSP1 is subject to proteolysis by multiple enzymes, generating inactive TSP1. As a simple mechanism, we introduce a reaction in which TSP1 is converted to a cleaved form (TSP1cleaved). The rate of cleavage is set at 3.9×10−3 s−1, assuming the same proteolysis rate as VEGF and multiplying by the total concentration of species who act to proteolyze TSP1 (plasmin, elastase, and thrombin), ~4.5 nmol/l.51,52 This is similar to a previously estimated rate.53
Implementation of the model
Generation of the reaction network
We used BioNetGen,54 a rule-based approach to formulate the reaction network. BioNetGen enables automated generation of the reaction network using a set of defined reaction rules. This approach is particularly useful in models that involve dynamic assembly of multi-protein complexes, which occurs our model, for example, in the case of complexes involving TSP1, VEGF, VEGFR2, β1, and CD36. The baseline model includes 18 molecule types (Figure 2) that can participate in 92 reaction rules to form a total of 130 molecular species based on 488 reactions. Altogether, the model is comprised of 130 non-linear ordinary differential equations (ODEs) that predict the species’ concentrations over time. The equations were implemented in MATLAB (The MathWorks, Natick, MA, USA). The model files are provided in Supplementary Files S3 and S4.
Numerical implementation
The ODEs were solved using the ode15 s solver in MATLAB. We performed a Monte Carlo (MC) analysis, in which we ran 5,000 simulations for all cases (i.e., baseline model, each change to the TSP1 unbinding rates, and each TSP1 mimetic). In the MC analysis, we utilize the eFAST analysis to determine which parameters to vary. In the simulations, the values of 12 parameters were sampled from a uniform distribution. The selected parameters were: receptor coupling rates for CD36-β1, CD36-VEGFR2, and CD47-VEGFR2; MMP cleavage rate; and the density of TSP1 receptors on tumor and endothelial cells (four densities per cell type). These are parameters that either had high individual sensitivity indices based on the eFAST analysis, had unknown values due to a lack of quantitative data from the literature, or both. The coupling rates were allowed to vary 100-fold above and below the baseline values, while the MMP cleavage rates and receptor densities varied 10-fold above and below the baseline values.
Parameter estimation
Estimation of the kinetic parameters related to MMP activation was achieved using the ‘lsqnonlin’ function in MATLAB, as previously described.55
Sensitivity analysis
The extended Fourier amplitude sensitivity test (eFAST), a global variance-based sensitivity analysis, was used to understand how different parameters (‘model inputs’) affect model predictions (‘model outputs’). In this method, the inputs are varied together within certain ranges at different frequencies and the model outputs are calculated. The Fourier transform of a model output can then be calculated to identify which inputs have the most influence based on the amplitude of each inputs’ frequency, with greater amplitudes indicating more sensitive parameters. By varying all of the inputs at the same time, this method allows for the calculation of two different sensitivity indices: the first-order FAST indices, Si, and the total FAST indices, STi. The first-order indices measure the local sensitivity of individual inputs, while the total indices measure the global sensitivity by accounting for second and higher-order interactions between multiple inputs. A greater total index compared to first-order index indicates that an input is more important in combination with other model parameters than alone.56
We implemented the eFAST method using MATLAB code developed by Kirschner and colleagues.56 We analyzed the effects of three groups of parameters (receptor numbers, kinetic parameters, and secretion/degradation rates) on seven different model outputs (the concentrations of TSP1, VEGF, proMMP9, MMP9, and the TSP1-VEGF, and MMP3-TSP1 complexes, and the ‘angiogenic ratio’, defined as the ratio of total receptor-bound VEGF to receptor-bound TSP1). In each case, the parameter values were allowed to vary 100-fold above and below the baseline values (a total range of four orders of magnitude) to account for potentially large uncertainty in the model parameters for which there are little data.
Acknowledgments
This work was supported by the National Institutes of Health (F31 CA200242 to JAR) and a USC Provost’s Undergraduate Research Fellowship (to CDS).
Footnotes
Supplementary Information accompanies the paper on the npj Systems Biology and Applications website (http://www.nature.com/npjsba)
The authors declare no conflict of interest.
References
- Fan, F., Schimming, A., Jaeger, D. & Podar, K. Targeting the tumor microenvironment: focus on angiogenesis. J. Oncol. 2012, 281261 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjaans, M. et al. VEGF pathway targeting agents, vessel normalization and tumor drug uptake: from bench to bedside. Oncotarget 5, 21247–21258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, R. K. Normalizing tumor microenvironment to treat cancer: Bench to bedside to biomarkers. J. Clin. Oncol. 31, 2205–2218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerbel, R. S. Reappraising antiangiogenic therapy for breast cancer. Breast (Edinburgh, Scotland) 20, S56–S60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando, N. T. et al. Tumor escape from endogenous, extracellular matrix-associated angiogenesis Inhibitors by up-regulation of multiple proangiogenic factors. Clin. Cancer Res. 14, 1529–1539 (2008). [DOI] [PubMed] [Google Scholar]
- Tortora, G., Ciardiello, F. & Gasparini, G. Combined targeting of EGFR-dependent and VEGF-dependent pathways: rationale, preclinical studies and clinical applications. Nat. Rev. Clin. Oncol. 5, 521–530 (2008). [DOI] [PubMed] [Google Scholar]
- Zhao, Y. & Adjei, A. A. Targeting angiogenesis in cancer therapy: Moving beyond vascular endothelial growth factor. Oncologist 20, 660–673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlatky, L., Hahnfeldt, P. & Folkman, J. Clinical application of antiangiogenic therapy: Microvessel density, what it does and doesn't tell us. J. Natl Cancer Inst. 94, 883–893 (2002). [DOI] [PubMed] [Google Scholar]
- Brostjan, C. et al. Neoadjuvant treatment of colorectal cancer with bevacizumab: the perioperative angiogenic balance is sensitive to systemic thrombospondin-1 levels. Clin. Cancer Res. 14, 2065–2074 (2008). [DOI] [PubMed] [Google Scholar]
- Xie, L. et al. Counterbalancing angiogenic regulatory factors control the rate of cancer progression and survival in a stage-specific manner. Proc. Natl Acad. Sci. USA 108, 9939–9944 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, J. C. & Lawler, J. The thrombospondins. Int. J. Biochem. Cell Biol. 36, 961–968 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good, D. J. et al. A tumor suppressor-dependent inhibitor of angiogenesis is immunologically and functionally indistinguishable from a fragment of thrombospondin. Proc. Natl Acad. Sci. USA 87, 6624–6628 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler, P. R. & Lawler, J. Molecular basis for the regulation of angiogenesis by thrombospondin-1 and -2. Cold Spring Harb. Perspect. Med. 2, a006627 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Jilani, S. M., Nikolova, G. V., Carpizo, D. & Iruela-Arispe, M. L. Processing of VEGF-A by matrix metalloproteinases regulates bioavailability and vascular patterning in tumors. J. Cell Biol. 169, 681–691 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg, J. S. et al. CD47 is necessary for inhibition of nitric oxide-stimulated vascular cell responses by thrombospondin-1. J. Biol. Chem. 281, 26069–26080 (2006). [DOI] [PubMed] [Google Scholar]
- Fukumura, D., Kashiwagi, S. & Jain, R. K. The role of nitric oxide in tumour progression. Nat. Rev. Cancer 6, 521–534 (2006). [DOI] [PubMed] [Google Scholar]
- Kaur, S. & Roberts, D. D. CD47 applies the brakes to angiogenesis via vascular endothelial growth factor receptor-2. Cell Cycle 10, 10–12 (2011). [DOI] [PubMed] [Google Scholar]
- Nicosia, R. F. & Tuszynski, G. P. Matrix-bound thrombospondin promotes angiogenesis in vitro. J. Cell Biol. 124, 183–193 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resovi, A., Pinessi, D., Chiorino, G. & Taraboletti, G. Current understanding of the thrombospondin-1 interactome. Matrix Biol. 37, 83–91 (2014). [DOI] [PubMed] [Google Scholar]
- Finley, S. D., Chu, L.-H. & Popel, A. S. Computational systems biology approaches to anti-angiogenic cancer therapeutics. Drug Discov. Today 20, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan, S. & Woo, S. Systems pharmacology approaches for optimization of antiangiogenic therapies: challenges and opportunities. Front. Pharmacol. 6, 33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley, S. D. & Popel, A. S. Effect of tumor microenvironment on tumor VEGF during anti-VEGF treatment: systems biology predictions. J. Natl Cancer Inst. 105, 802–811 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley, S. D., Angelikopoulos, P., Koumoutsakos, P. & Popel, A. S. Pharmacokinetics of anti-VEGF agent aflibercept in cancer predicted by data-driven, molecular-detailed model. CPT Pharmacometrics Syst. Pharmacol. 4, 641–649 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol, A. et al. The protease MT1-MMP drives a combinatorial proteolytic program in activated endothelial cells. FASEB J. 26, 4481–4494 (2012). [DOI] [PubMed] [Google Scholar]
- Etique, N. et al. LRP-1: a checkpoint for the extracellular matrix proteolysis. Biomed. Res. Int. 2013, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolkman, J. A. & Stemmer, W. P. C. Directed evolution of proteins by exon shuffling. Nat. Biotechnol. 19, 423–428 (2001). [DOI] [PubMed] [Google Scholar]
- Jeanne, A., Schneider, C., Martiny, L. & Dedieu, S. Original insights on thrombospondin-1-related antireceptor strategies in cancer. Front. Pharmacol. 6, 252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, A. H. & Kyriakides, T. R. Matricellular proteins and biomaterials. Matrix Biol. 37, 183–191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohan Ehrenfest, D. M., de Peppo, G. M., Doglioli, P. & Sammartino, G. Slow release of growth factors and thrombospondin-1 in Choukroun's platelet-rich fibrin (PRF): a gold standard to achieve for all surgical platelet concentrates technologies. Growth Factors 27, 63–69 (2009). [DOI] [PubMed] [Google Scholar]
- Sharan, S. & Woo, S. Quantitative insight in utilizing circulating angiogenic factors as biomarkers for antiangiogenic therapy: systems pharmacology approach. CPT: Pharmacometrics Syst Pharmacol 3, 1–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanafelt, T. D. et al. Pretreatment angiogenic cytokines predict response to chemoimmunotherapy in patients with chronic lymphocytic leukaemia. Br. J. Haematol. 146, 660–664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusnati, M. & Presta, M. Angiogenic growth factors interactome and drug discovery: The contribution of surface plasmon resonance. Cytokine Growth Factor Rev. 26, 293–310 (2015). [DOI] [PubMed] [Google Scholar]
- Baker, E. A., Leaper, D. J., Hayter, J. P. & Dickenson, A. J. The matrix metalloproteinase system in oral squamous cell carcinoma. Br. J. Oral Maxillofac. Surg. 44, 482–486 (2006). [DOI] [PubMed] [Google Scholar]
- Byrne, G. J. et al. Angiogenic characteristics of circulating and tumoural thrombospondin-1 in breast cancer. Int. J. Oncol. 31, 1127–1132 (2007). [DOI] [PubMed] [Google Scholar]
- Poon, R. T. et al. Clinical significance of thrombospondin 1 expression in hepatocellular carcinoma. Clin. Cancer Res. 10, 4150–4157 (2004). [DOI] [PubMed] [Google Scholar]
- Manni, A. et al. Effects of α-difluoromethylornithine on thrombospondin-1 production by human breast cancer cells. J. Oncol. 31, 1187–1191 (2007). [PubMed] [Google Scholar]
- Kumar, S., Kapoor, A., Desai, S., Inamdar, M. M. & Sen, S. Proteolytic and non-proteolytic regulation of collective cell invasion: tuning by ECM density and organization. Sci. Rep. 6, 19905 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen, M. et al. Tissue-based map of the human proteome. Science 347 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- Isenberg, J. S. et al. Differential interactions of thrombospondin-1, -2, and -4 with CD47 and effects on cGMP signaling and ischemic injury responses. J. Biol. Chem. 9, 1116–1125 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailenko, I. et al. Cellular internalization and degradation of thrombospondin-1 is mediated by the amino-terminal heparin binding domain (HBD): high affinity interaction of dimeric hbd with the low density lipoprotein receptor-related protein. J. Biol. Chem. 272, 6784–6791 (1997). [DOI] [PubMed] [Google Scholar]
- Pearce, S. F. A., Wu, J. & Silverstein, R. L. Recombinant GST/CD36 fusion proteins define a thrombospondin binding domain: evidence for a single calcium-dependent binding site on CD36. J. Biol. Chem. 270, 2981–2986 (1995). [DOI] [PubMed] [Google Scholar]
- Kaur, S. et al. Thrombospondin-1 inhibits VEGF receptor-2 signaling by disrupting its association with CD47. J. Biol. Chem. 285, 38923–38932 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon, M. E., Stipp, C. S. & Lander, A. D. Interactions of neural glycosaminoglycans and proteoglycans with protein ligands: assessment of selectivity, heterogeneity and the participation of core proteins in binding. Glycobiology 9, 143–155 (1999). [DOI] [PubMed] [Google Scholar]
- Ramos-DeSimone, N. et al. Activation of matrix metalloproteinase-9 (MMP-9) via a converging plasmin/stromelysin-1 cascade enhances tumor cell invasion. J. Biol. Chem. 274, 13066–13076 (1999). [DOI] [PubMed] [Google Scholar]
- Bein, K. & Simons, M. Thrombospondin type 1 repeats interact with matrix metalloproteinase 2: regulation of metalloproteinase activity. J. Biol. Chem. 275, 32167–32173 (2000). [DOI] [PubMed] [Google Scholar]
- Vempati, P., Karagiannis, E. D. & Popel, A. S. A biochemical model of matrix metalloproteinase 9 activation and inhibition. J. Biol. Chem. 282, 17585–37596 (2007). [DOI] [PubMed] [Google Scholar]
- Northrup, S. H. & Erickson, H. P. Kinetics of protein-protein association explained by Brownian dynamics computer simulation. Proc. Natl Acad. Sci. USA 89, 3338–3342 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vempati, P., Mac Gabhann, F. & Popel, A. S. Quantifying the proteolytic release of extracellular matrix-sequestered VEGF with a computational model. PLoS ONE 5, e11860 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Steen, P. E. et al. The hemopexin and O-glycosylated domains tune gelatinase B/MMP-9 bioavailability via inhibition and binding to cargo receptors. J. Biol. Chem. 281, 18626–18637 (2006). [DOI] [PubMed] [Google Scholar]
- Herz, J. & Strickland, D. K. LRP: a multifunctional scavenger and signaling receptor. J. Clin. Invest. 108, 779–784 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, J. C. & Lawler, J. The thrombospondins. Cold Spring Harb. Perspect. Biol. 3, 1–29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iruela-Arispe, M. L. Regulation of thrombospondin1 by extracellular proteases. Curr. Drug Targets 9, 863–868 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman, L. et al. Plasmin triggers a switch-like decrease in thrombospondin-dependent activation of TGF-β1. Biophys. J. 103, 1060–1068 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faeder, J. R., Blinov, M. L. & Hlavacek, W. S. Rule-based modeling of biochemical systems with BioNetGen. Methods Mol. Biol. 500, 113–167 (2009). [DOI] [PubMed] [Google Scholar]
- Finley, S. D., Dhar, M. & Popel, A. S. Compartment model predicts VEGF secretion and investigates the effects of VEGF Trap in tumor-bearing mice. Front Oncol. 3, 196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino, S., Hogue, I. B., Ray, C. J. & Kirschner, D. E. A methodology for performing global uncertainty and sensitivity analysis in systems biology. J. Theor. Biol. 254, 178–196 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.