Abstract
Oxidative DNA-protein crosslinks have received less attention than other types of DNA damage and remain as one of the least understood types of oxidative lesion. A model system using Ribonuclease A and a 27-nucleotide DNA was used to determine the propensity of oxidative crosslinking to occur in the presence of oxidants. Crosslink formation was examined using four different oxidation systems that generate singlet oxygen, superoxide, and metal-based Fenton reactions. It is shown that oxidative crosslinking occurs in yields ranging from 14% to a maximal yield of 61% in all oxidative systems when equivalent concentrations of DNA and protein are present. Because singlet oxygen is the most efficient oxidation system in generating DNA-protein crosslinks, it was chosen for further analyses. Crosslinking occurred with single-stranded DNA binding protein and not with bovine serum albumin. Addition of salt lowers non-specific binding affinity and lowered crosslink yield by up to 59%. The yield of crosslinking increased with increased ratios of protein compared to DNA. Crosslinking was highly dependent on the number of guanines in a DNA sequence. Loss of guanine content on the 27-nucleotide DNA led to nearly complete loss in crosslinking, while primer extension studies showed crosslinks to predominantly occur at guanine base on a 100-nucleotide DNA. The chemical species generated were examined using two peptides derived from the Ribonuclease A sequence: N-acetyl-AAAKF and N-acetyl-AYKTT that were crosslinked to 2’-deoxyguanosine. The crosslink products were spiroiminodihydantoin, guanidinohydantoin, and tyrosyl-based crosslink adducts. Formation of tyrosine based adducts may be competitive with the more well-studied lysine-based crosslinks. We conclude that oxidative crosslinks may be present at high levels in cells since the propensity to oxidatively crosslink is high and so much of the genomic DNA is coated with protein.
Keywords: 1 DNA oxidation, 2 crosslinking, 3 spiroiminodihydantoin, 4 guanidinohydantoin, 5 reactive oxygen species, 6 tyrosine crosslink
Introduction
Too much reactive oxygen disrupts cellular functions via deleterious modification of DNA, protein and other biomolecules.1, 2 Reactions centered at DNA are especially important because if left unrepaired permanent mutations or cytotoxic replication stops are formed.3 Increases in the amount of reactive oxygen have been linked to aggressive forms of certain cancers and reduced activity of tumor suppressors.4, 5 Additionally, environmental pollutants like hexavalent chromium, a known carcinogen, induce oxidative stress via generation of reactive oxygen.6 Chromium has been shown to induce a type of lesion known as DNA-protein crosslinks.7, 8 These crosslinks are poorly characterized and have unknown potential to stop replication. Additionally, crosslink lesions are repaired by digestion of the protein into small peptides that could be mutagenic.9 The lack of biochemical characterization stems from the inherent complexity, unknown stability, and lack of a model system to systematically evaluate these lesions. In this work, we identified and used an appropriate model system to begin to evaluate oxidative crosslinks.
Several variants of DNA-protein crosslinks are thought to occur. Despite the lack of understanding in structure, oxidative crosslinks can be isolated from cells and are known to occur as mixed populations.10 One recognized class of crosslinks are ternary complexes.11 These forms are mutagenic with a metal ion directly attached to DNA and the protein.12 The second class of crosslinks are those induced by reactive oxygen species. This class is diverse with several structural variants proposed based on small molecule studies. One form involves bonding between 2’-deoxythymidine and tyrosine stemming from a hydroxyl radical mediated reaction.13 Most of the remaining structures are postulated to occur between the guanine and lysine or thymine and lysine.14, 15 Lysine has been found to add to both positions five and eight of guanine.15, 16 Multiple lysine side chains can serve to sequentially add to guanine,17 while other amino acids, like tyrosine, may also be incorporated to guanine.18 In addition, it has been shown that crosslinks may derive from reaction of proteins with abasic ribose hemiacetals that form upon depurination of oxidized DNA bases.19 Because of this complex mixture of possible products we wanted to determine both the propensity and structure of oxidative DNA protein crosslinks in a systematic manner.
We hypothesize that guanine base is the most likely addition site of oxidative crosslinking. Guanine is the most oxidation prone DNA base with its oxidation proceeding via several mechanisms.20, 21 The divergent mechanisms begin to coalesce upon the formation of oxygen (via either hydroxyl or peroxyl) addition products to either C5 or C8.22 The fate of these addition products give rise to many of the guanine products like mutation-prone 8-oxo-7,8-dihydro-2’-deoxyguanosine.23,24 Crosslink products likely form during these oxidative events as it has been shown that guanine oxidation leads to crosslinking 25. Oxidative DNA-protein crosslinks increase concurrently with the levels of 8-oxo-7,8-dihydro-2’-deoxyguanosine.26 In addition to competing with guanine oxidation, crosslinks can form directly from oxidation of 8-oxo-7,8-dihydro-2’-deoxyguanosine since it has an oxidation potential that is ~0.5V lower than the 2’-deoxyguanosine radical cation.27, 28 Major products of hyper-oxidation of guanine base include guanidinohydantoin and spiroiminodihydantoin rings.29 A recent crystal structure demonstrates that hydantoin lesions adopt a syn conformation with the Hoogsteen face base pairing.30 These lesions have been found to be mutagenic with insertion of purines opposite the damaged base.31, 32 Hydantoin-based oxidative crosslinks are formed when a lysine on a protein adds to DNA instead of water and we hypothesize that they may be a major type of oxidative crosslinks.
It is within this context that we began to investigate the types of DNA-protein crosslinks formed in order to elucidate their consequences and ultimately facilitate more specific cellular detection of oxidative crosslinks. In this work we utilize a model protein, Ribonuclease A, to form crosslinks. Crosslink formation was examined using four different oxidation systems: the photo-oxidant rose bengal, the photo-oxidant riboflavin, iron- and copper-based oxidation systems. Additionally, we determined that crosslinks are produced both on single-stranded and double stranded DNA with a high dependence of crosslink yield on the number of guanines in the DNA. Crosslink between Ribonuclease A and DNA were used as a substrate for primer extension. Using LTQ-FT mass spectrometry on a model reaction, it was found that both lysyl and tyrosyl products are formed.
Material and Methods
All experiments were accomplished in at least duplicates. All chemical reagents were purchased from Acros Organics unless otherwise noted. Oligonucleotides were purchased from Eurofins MWG operon. The sequence of the 27-nucleotide DNA was 5’-GGGGCCCCGTCGTTTTACAACGTCGTG-3’ and its complement was 5’-CACGACGTTGTAAAACGACGGGGCCCC-3’. In studies on the effect of guanine displacement on crosslinking, guanines are methodically removed from the 5’- end of the 27-mer and replaced with adenines. The DNA with four guanines in it has the sequence: 5’- AAAACCCCATCGTTTTACAACGTCGTG-3’, while the sixth, seventh and eighth guanines were successively replaced with adenines for the DNA sequences with three, two and one guanine/s, respectively. All DNAs used in these experiments were purified by gel electrophoresis, cutting out the band and overnight elution of the desired DNA band into water, followed by ethanol precipitation.
DNA-Ribonuclease A Oxidative Crosslinking
Crosslinking experiments were carried out by mixing the 27-nucleotide DNA, Ribonuclease A and the photo-oxidant rose bengal [27.5 μL; 25 mM sodium phosphate, 500 μM DNA, 35 μM Ribonuclease A and 100 μM photo-oxidant, pH 8] in a in a 1.5 mL Microcentrifuge tube with the lid open and irradiated with a LED with 3 mW of radiative flux lamp at 575 nm. During photolysis, the solution is held at a distance of 3 cm from the irradiation source for 20 minutes. Experiments using CuCl2 or ammonium iron(II) sulfate based oxidation systems were accomplished in a similar manner but with 3 mM sodium ascorbate, 30 mM hydrogen peroxide and 50 μM Cu(II) or Fe(II) and with no irradiation. A 12% PAGE was performed. Once complete, the gel was stained with ethidium bromide and visualized using the MultiImage II FC UV transilluminator. Yield was calculated via intergration using the manufacturer’s software. In experiments where the protein, salt, or DNA was changed the concentrations are as described. Protein staining was accomplished according to standard protocols and visualized using the 800-channel on a Licor Imager. For double stranded DNA experiments, the same conditions were employed except that an equimolar concentration of the 27-nucleotide DNA and its complement were added followed by heating at 95°C for 3 minutes and gradual cooling to room temperature to facilitate annealing.
Primer extension experiments
Crosslinking in the presence of rose bengal is carried out as before and separated using a PAGE gel. The corresponding crosslink species were directly visualized using a thin layer chromatography plate and cut out. Overnight elution in 1 mL of ddH2O followed by precipitation yielded the purified crosslinks at a concentration of 5 μM. A 100 μL volume of the purified crosslinks was added to a 5X primer extension mix (5x vent buffer, 0.5 mM dNTPs, 500 nM primer, 0.01 U/μL vent exo-DNA polymerase). A fluorescently labeled primer (5’-IrDye700-cacgacgttgtaaaacgac) is utilized in the primer extension. The samples were subsequently cycled 5 times for primer extension (95°C for 3 min, 55°C for 1 min, 72°C for 2 min). The extended samples were then denatured and a 12% denaturing PAGE gel was performed. The gel is visualized using the Odyssey Infrared Imager System and the 700-laser channel source. Synthetic oligonucleotides shorter than the 27-mer, with two nucleotides systematically removed, were treated in the same manner as the purified crosslinks.
Studies utilizing the 100-nucleotide sequence used a final concentration of 5 μM DNA that was greater than 95% pure. The 100-nucleotide sequence is 5’-ATGACCATGATTACGCCAAGCTTGCATGCCTGCAGGTCGACTCTAGAGGATC CCCGGGTACCGAGCTCGAATTCACTGGCCGTCGTTTTACAACGTCGTG and derives from the puc19 plasmid. To limit oxidation of DNA from direct photo-oxidation, all samples contained 0.5 mM BSA. DNA crosslinking and extension was performed in a similar manner as above with the exception that the DNA was not purified after irradiation but instead used directly for primer extension by adding 1 μL of the reaction mix to 24 μL of 1.05X extension mix. Sequencing is performed on the puc19 plasmid that has the same sequence using standard extension conditions with an appropriate acyclo-terminator added at 100 μM.
LC-MS product analysis
The N-terminally acetylated peptides N-Ac-AAAKF and N-Ac-AYKTT (Neo BioScience) are identical to residues 7-12 and 99-104 of the Ribonuclease A sequence. Peptides were greater than 95% pure based on HPLC. Peptide-dG reactions (50 μL) were similar as the Ribonuclease A-DNA reactions except that 5 mM peptide and 5 mM dG were used. For the N-Ac-AAAKF-dG reaction, the samples were separated by injection into a Beckman Coulter HPLC with a Cosmosil Waters 5C18-PAQ [4.6 × 150 mm] column. The gradient was 0% B for 1 m, 14% B over 35 m, 100% over 16 m with Solvent A (98% H2O, 2%ACN) and Solvent B (96% ACN, 4% H2O). Absorbance was monitored at 240 nm. Controls excluded the listed reagent. HPLC fractions were directly infused into the MS at 5 μL/min. The mass spectrometer is a Thermo Scientific LTQ-FT™, a hybrid instrument consisting of a linear ion trap and a Fourier transform ion cyclotron resonance mass spectrometer. The entire elutant was introduced into the LTQ-FT™ using the standard electrospray ionization source for the instrument with a spray voltage of 5 kV and a capillary temperature of 275 °C. Autogain control, AGC, was used and set at 500,000 with a maximum injection time of 1250 ms for FT-ICR full scans. Collision induced dissociation, MSn, was executed in the linear trap with an AGC setting of 10,000 and a maximum injection time of 500 ms. FT-ICR full scans were acquired in the positive ion mode at 100,000 resolving power at m/z 400. Mass accuracy errors were below 200 ppb for full scan and below 800 ppb for MSn. The positive ion MSn experiments were performed in the linear trap portion of the instrument using helium as a collision gas, isolation widths of 2 amu, a normalized collision energy of 35, a q value of 0.250 and an excitation time of 30 ms. The same conditions are employed for the addition of the peptide AYKTT to dG except that LC was performed on-line. Liquid chromatography separation of the sample components used a Waters Symmetry C18 5-μm, 2.1 × 150 mm column, Finnigan Surveyor MS pump and Finnigan Micro AS autosampler both from Thermo Scientific. A 200 μL/min gradient elution of water and acetonitrile took place as follows: 2 to 15% acetonitrile over 35 minutes.
Results
Characterizing oxidative DNA-Protein crosslinks using a model system
This work aims to shed light on oxidative DNA-protein crosslinks. To accomplish this objective, the employment of an appropriate protein and DNA sequence model system is essential. In cells, much of the DNA is bound by histones, which are lysine-rich. Our initial evaluation found that histones are not suitable as model proteins because their multimeric nature makes product identification challenging and their high pI causes poor band formation upon electrophoretic separation. Examination of several proteins showed Ribonuclease A as a fitting model protein. It is a monomeric protein with a small size. In addition, Ribonuclease A binds to DNA in a non-specific manner similar to most non-specific DNA-binding proteins (Figure S1). Ribonuclease A does not cleave DNA since a 2’-hydroxyl is required for nuclease activity.33, 34 It also serves as a strong template for each type of oxidative DNA-protein crosslink since it possesses ten lysine and six tyrosine residues, representing each reactive side chain of interest. Additionally, it has eight cysteine, sixteen serine, ten threonine, and four arginine residues that are known to be nucleophilic as well as three phenylalanine residues that has been shown to crosslink to 2’-deoxyribose as well as to thymine.35 An unmodified DNA sequence that can predominantly form crosslinks was designed. Multiple guanines are placed in the sequence, including four consecutive guanines that are known to lower redox potential in double stranded DNA.36, 37 In addition, a run of four thymines is located at the center of the sequence to allow for formation of thymine-based crosslinks. Based on this observation, the oligonucleotide sequence, 5’-GGGGCCCCGTCGTTTTACAACGTCGTG, 27-mer, was selected as the DNA substrate.
Oxidative crosslink formation is visualized using electrophoresis (Figure 2). Reactions and controls were electrophoresed on two gels simultaneously. One gel was visualized with ethidium bromide to detect DNA, while the other is stained with Coomassie Blue to detect protein. The oxidant used in Figure 2 is rose bengal, a singlet oxygen generator. The reaction and controls were photo-irradiated using a LED lamp emitting light at 575 nm, which corresponds to the wavelength of maximum absorption for rose bengal. Staining with ethidium bromide shows that DNA and protein irradiated in the absence of oxidant resulted to a single band at the bottom of the gel corresponding to the 27-mer (Figure 2 left). Piperidine treatment shows that irradiation is not generating any DNA sites susceptible to cleavage (Figure S7). Integration of the total peak intensity reveals that no DNA direct strand cleavage is occurring. Addition of rose bengal to DNA in the absence of protein followed by irradiation reveals slower moving bands that are not present when no oxidant is added. These bands are likely DNA-DNA crosslinks that have recently been described.38 The intensity of these crosslink bands are higher in the absence of Ribonuclease A (Figure 2, left, lane 2). Quantification of all the DNA bands shows that the 27-mer band intensity is not reduced, indicating that photo-degradation of DNA is not taking place. Irradiation of protein and DNA in the presence of rose bengal produced novel bands with higher molecular weight (Figure 2, asterisks). These bands were deduced to be covalent in nature since they could not be denatured. Loss of the high molecular weight bands were observed following trypsin digestion indicating that the resulting bands are due to protein associated with the DNA (data not shown). Initial optimization showed more DNA than protein minimizes the formation of multimeric crosslinks but lowers overall yield. Multimeric crosslinks are secondary, higher molecular weight adducts that form from more than one DNA strand crosslinked to one molecule of protein (see next section). Concentrations of 35 μM Ribonuclease A and 500 μM 27-mer promote ample crosslink formation while minimizing multimeric crosslinking.
Figure 2. Crosslink visualization with DNA and protein stain.
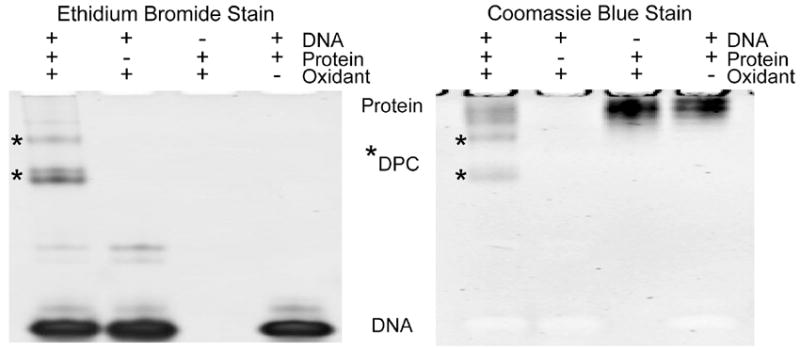
Ribonuclease A and a 27-mer DNA were incubated in the presence of the singlet oxygen generator rose bengal and irradiated. After which, the sample along with the controls were simultaneously run on two gels. One gel visualized DNA by ethidium bromide staining (left) and the second gel visualized protein by Coomassie Blue staining (right). In the lane containing all the reaction constituents, several bands are observed that contain both the protein and DNA components (bands denoted by asterisks). These bands are identified as DNA-protein crosslinks since exclusion of DNA, protein, or oxidant leads to loss of the product. Higher molecular weight DNA-only bands are also observed.
The Coomassie Blue stained gel further confirms that the new bands consist of a protein component in addition to DNA (Figure 2, right). Protein incubated without oxidant and DNA showed a single band at the top of the gel, corresponding to Ribonuclease A (data not shown). Ribonuclease A, having an isoelectric pH of 8.68, will have a net positive charge at pH 8 and is hampered from migrating into the gel. No novel bands were observed after photo-irradiation of protein and DNA in the absence of rose bengal. Rose bengal initiated photo-oxidation of Ribonuclease A without DNA results in band broadening, indicating that some protein oxidation is occurring. Rose bengal initiated photo-oxidation when both Ribonuclease A and 27-mer are present results in new bands that migrate into the gel. The product has a net negative charge from the addition of DNA, allowing the bands to move into the gel. These bands coincide precisely with the product bands observed in the ethidium bromide visualized gel, indicating that these new lesions are made up of both DNA and protein. Exclusion of DNA, protein or oxidant results in loss of product indicating that these are oxidative DNA-protein crosslinks. The yield for the reaction relative to DNA is 7.0 +/- 0.9% for total crosslink and 6.0 +/- 1.7% for the primary crosslink band relative to DNA. Taking into account the molar ratio of one to fifteen for protein-to-DNA, a yield of 6.6% is the maximum. Thus, the reaction is highly efficient for Ribonuclease A.
To determine selectivity of crosslinking for single or double stranded DNA, a crosslinking experiment was conducted to elucidate which DNA structure is more likely to participate. For crosslinks involving double-stranded DNA, equimolar concentrations of the 27-mer and its complement were annealed to promote double strand formation. To ensure that equal amounts of DNA were used in both reactions, doubly concentrated 27-mer is employed for comparison. The two forms of DNA were then incubated with Ribonuclease A and photo-oxidized in the presence of rose bengal. Controls including without DNA, without protein or minus oxidant (data not shown) were used to ensure crosslink identification. When staining, all DNA was single-stranded since a denaturing polyacrylamide gel was used. Visualization with ethidium bromide (Figure 3A) indicated that crosslinks are formed in both cases (bands denoted by asterisk). Single and double stranded DNA produced 7.0 +/-0.4% and 6.5 +/-0.3 % DNA-protein crosslinks, respectively. Again these values are at the maximum possible yield. Setting the value that forms the most product (SS) as 1.0, double stranded DNA only lowers the yield to 92% relative to single-stranded DNA. The two-tailed p-value for comparing these two analyses is 0.09 indicating that the difference is not statistically significant. This experiment reveals that there is no significant difference in the propensity of single and double stranded DNA to form crosslinks. This is mechanistically interesting since it is well known that singlet oxygen reacts less efficiently with DS DNA,39 though oxidation initiated at easy to oxidize amino acid residues like tyrosine would not depend on the DS versus SS nature of DNA.
Figure 3. Dependence of DNA-protein crosslinking on oligonucleotide structure and oxidant type.
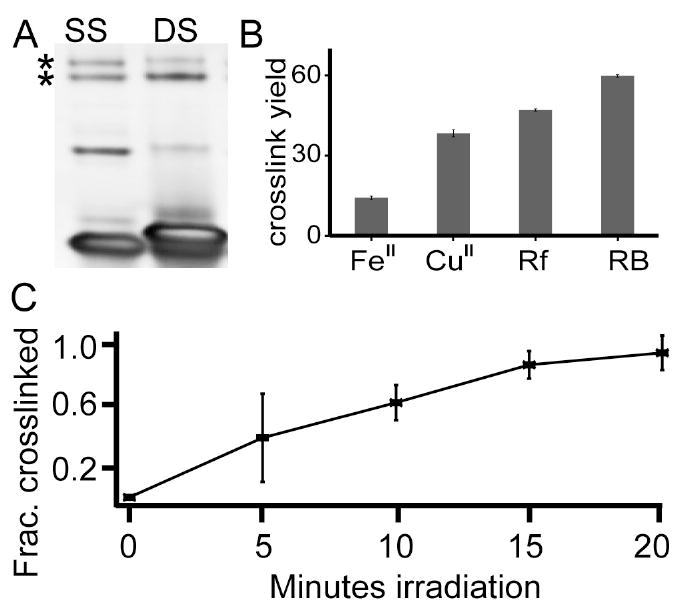
(A) The degree of oxidative crosslinking with a single stranded DNA, SS, is compared with that of double stranded, DS, DNA. The two DNA structures have a comparable tendency to form DNA-protein crosslinks. (B) Four different oxidant systems were used to generate crosslinks at equimolar concentrations of protein to DNA. Fe(II)-H2O2, Cu(II)-H2O2, riboflavin and rose bengal oxidation systems all generated high yields of crosslink despite the divergent oxidation mechanisms. Error bars are standard deviations with n=3. (C) Rose bengal induced crosslinks occur rapidly and linearly increase with irradiation time within the first twenty minutes.
Two major forms of reactive oxygen are common in the cell: superoxide and hydroxyl radical. Several oxidants were chosen to mimic these types of oxidative stress. Superoxide is the most abundant cellular reactive oxygen species and is produced by leakage of the mitochondrial electron transport chain and certain oxidases. Hydroxyl radical on the other hand is a potent cellular oxidant that reacts with all biomolecules. It is produced in the cell via Fenton reactions when hydrogen peroxide produced from superoxide dismutation, reacts with transition metals like iron and copper. The photo-oxidant riboflavin was used to produce superoxide as well as direct oxidation of substrates in a Type I fashion. Copper(II), hydrogen peroxide, and ascorbate mixtures produce species that behave as singlet oxygen, hydroxyl radical, and metal bound radicals.40 Iron(II) produces hydroxyl radical exclusively.
To determine the effect of different cellular oxidant mimics to the amount of DNA-protein crosslinks formed, equivalent concentrations of the 27-mer and Ribonuclease A were allowed to react in the presence of Fe(II)-H2O2, Cu(II)-H2O2, riboflavin or rose bengal oxidation systems (Figure 3B). For photo-oxidants rose bengal and riboflavin, 100 μM photo-oxidant was added to 500 μM concentrations of 27-mer and Ribonuclease A, followed by irradiation. The concentration of DNA and protein was equimolar since one to one binding increases yield (Figure 4C). Unlike for rose bengal, the reaction mixture containing riboflavin is irradiated using a LED lamp that emits at 375 nm, corresponding to the wavelength of maximum absorption for riboflavin. PAGE analysis of the riboflavin-mediated reaction shows the parent DNA band is smaller when no protein is present, which is indicative that strand scission is occurring. These strand scission products are observed to be less prominent when Ribonuclease A is added which is consistent with the protein being preferentially oxidized (see supporting information Figure S2 for gel image).
Figure 4. Effect of binding on DNA-protein crosslinking.
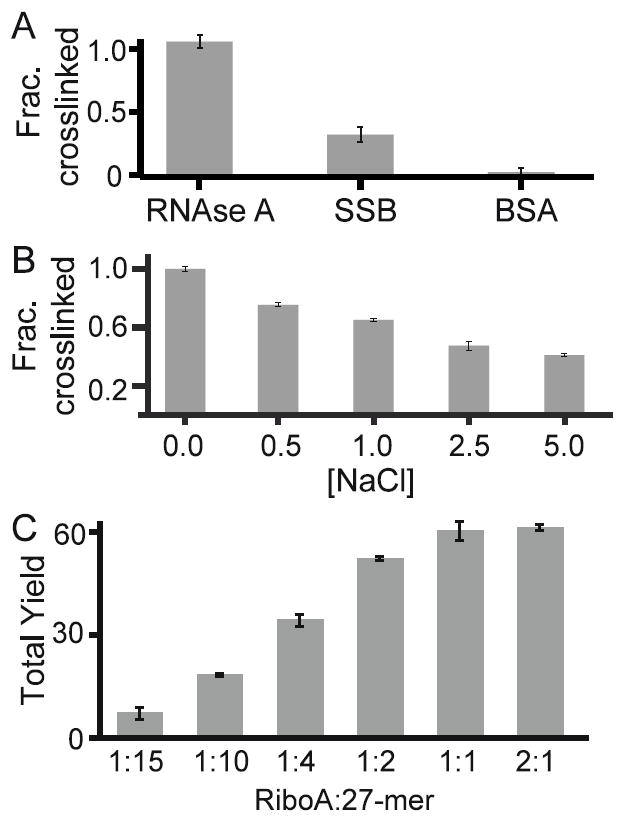
To determine the importance of binding on DNA-protein crosslink formation, two experiments were conducted. (A) The tendency of RNAse A, SSB, and BSA to form crosslinks are shown. RNase A and SSB utilize non-specific binding modes, while BSA does not bind to DNA. Crosslink yield for SSB and BSA relative to RNAse A is 31% and 3.4%, respectively. (B) Addition of NaCl is known to interrupt RNAse A:DNA binding. Increasing concentrations of NaCl result in decreasing formation of DNA-protein crosslinks. (C) Increasing ratios of Ribonuclease A to DNA lead to greater crosslink yields.
For Fe(II)-H2O2 and Cu(II)-H2O2 oxidation systems, experiments were performed as before but with 50 μM Fe(II) or Cu(II), 30 mM hydrogen peroxide and 3 mM sodium ascorbate. Samples were immediately visualized by PAGE because preliminary experiments show no noticeable increase in yield with longer incubation time. Initial optimization of reactions involving Cu(II)-H2O2 and Fe(II)-H2O2 show that lower yields of crosslinks are formed at high metal cation concentrations since DNA strand scission is extensive. In addition, significant degradation of the parent DNA is observed when allowed to incubate for several days, indicating that a large portion of the oxidative events are producing strand scission (see Figure S2 of supporting information). The Fe(II)-H2O2 oxidation system behaves in a similar manner but with no detectable DNA band degradation. All reactions described are done in at least triplicates.
Comparing the level of crosslinking among the oxidants showed that the photo-oxidants rose bengal and riboflavin produce more DNA-protein crosslinks compared to the metal-based oxidants, with rose bengal producing the most crosslinks of up to 59.7 +/-0.5% and Fe(II)-H2O2 oxidation system producing the least amount of crosslinks, generating only 14.2 +/-0.6%. Riboflavin and copper-H2O2 oxidation system produced 47.0 +/-0.4 and 38.3 +/-1.3% DNA-protein crosslinks, respectively. These findings illustrate the ubiquity of DNA-protein crosslinking under conditions of high oxidative stress as all cellular oxidant mimics utilized in these experiments were able to produce DNA-protein crosslinks.
For oxidative crosslinks to be important cellular lesions, they must form rapidly upon induction of oxidative stress. To determine if crosslinks occur rapidly Ribonuclease A and the 27-mer were exposed to rose bengal at different time periods. Crosslink formation increases linearly in the first fifteen minutes (Figure 3C) thus yield is related to the amount of oxidative events. The linear relation implies that the crosslinks are relatively stable and not being further degraded or oxidized in the first fifteen minutes. The same pattern is observed with riboflavin oxidation while the maximum yield is achieved immediately upon exposure to Cu(II) and Fe(II) oxidation systems (data not shown).
The importance of binding in DNA-protein crosslink formation was assessed (Figure 4). SSB and BSA were selected for this purpose because previous studies show that they can both form DNA-protein crosslinks.25, 41 Looking at the properties of the proteins used in these experiments, Ribonuclease A is a monomeric protein, composed of 128 amino acid residues, has a molecular weight of 13,700 Da, contains ten lysine and six tyrosine residues and is known to bind to both double-stranded and single-stranded DNA. SSB is composed of four identical subunits with a molecular weight of 18,900 Da and 178 residues in each subunit. SSB contains five lysine and four tyrosine residues in its sequence and is known to bind to single-stranded DNA. Whereas BSA is also monomeric, has a molecular weight of 66,400 Da, with 583 residues and has no known affinity for DNA. It contains fifty-eight lysine and twenty tyrosine residues in its sequence.
Experiments were performed as before. The concentration of SSB calculated is based only on one sub-unit. The product bands were subsequently quantified and normalized so that the reaction with Ribonuclease A is 1.0. The results show that the crosslink yield for SSB and BSA is 31.2 +/- 5.4% and 3.4 +/-3.3%, relative to RNAse A respectively (Figure 4A). BSA produced a significantly lower level of DNA-protein crosslinks compared to the two DNA-binding proteins suggesting that binding play an important role in crosslink formation. To further probe the significance of binding in the formation of oxidative DNA-protein crosslinks, increasing concentrations of NaCl were added to reaction mixtures of the 27-mer, Ribonuclease A, and rose bengal (Figure 4B). Setting the crosslink level when no salt is added as 1.0, the most drastic decrease in crosslink level is observed when 0.5M NaCl is added as the yield is reduced by 25 +/- 1.5%. When the concentration of NaCl was 1, 2.5, and 5 M, crosslinking dropped by 10 +/- 2.0, 18 +/- 0.7, and 5 +/-1.2 % respectively. Thus, increasing concentrations of NaCl decrease formation of DNA-protein crosslinks and demonstrates the importance of a close interaction between protein and DNA.
The yield relative to the amount of protein bound will further support binding as a key parameter in oxidative crosslinking. The ratio of Ribonuclease A/DNA was increased from as low as 1:15 up to 2:1 (Figure 4C). At low ratios most DNA is unbound and the total crosslink yield is low. As the ratio increases total crosslink yield increases such that when protein concentration equals DNA, 60.0 +/-3.3% of the DNA is crosslinked. When there is twice as much protein as DNA, 61.0 +/-0.1% of DNA participates in DNA-protein crosslinking. At Ribonuclease A/ DNA ratios above 1:10, multiple crosslink bands were observed. The crosslink band migrating the farthest into the gel is identified as the primary crosslink, while higher molecular weight crosslink bands are referred to as multimeric crosslinks. In addition to total crosslink quantification, primary crosslinks were quantified as well to determine the effects of increased protein concentration. Primary crosslink yield also increases with protein concentration. At lower protein concentrations, most of the products are primary crosslinks, with 85% of the total crosslinks due to primary crosslinks when 35 μM protein is added. Addition of more protein results to more multimeric crosslinks and less primary crosslinks. Only 47% of the total crosslinks are primary crosslinks when there is an equivalent concentration of DNA and protein in the reaction mixture. At Ribonuclease A/ DNA ratios of 1:10 and 1:4, the total crosslinks formed exceeds the amount of protein, which is the limiting component in the reaction. For instance, 1:4 Ribonuclease A/DNA ratio, with 25% protein relative to DNA results in 19% of the DNA participating in primary crosslinks and a combined level of crosslinks reaching 34%, which is more than the amount of protein present. This suggests that the higher molecular weight crosslinks contain multiple DNA molecules bound to a single protein molecule. Considering that at protein concentrations higher than 50% relative to DNA, not all proteins participate in crosslinking, as 60 +/-3.3% and 61 +/-0.1% of the DNA is crosslinked at 1:1 and 2:1 protein-DNA ratio, respectively. These three experiments highlight the importance of binding in oxidative crosslinking.
Crosslink formation predominately occurs at guanine
Experiments were accomplished to determine if oxidative crosslinks occur at either thymine or guanine. The formation of guanine-based crosslinks by capture of protein lysyl residues would be sensitive to loss of guanine base in the sequence. Guanine bases within the 27-mer were removed and replaced with adenine (Figure 5). The amount of DNA and protein was held to a ratio of 15:1 and the crosslink bands were quantified. When all the guanines were substituted with adenine the yield dropped from 7.2 +/-0.5% to 0.7 +/-0.6% (Figure 5, left). Thymines within the 27-mer were then replaced by cytosine. In this case no change in the overall yield was observed (Figure 5, left, lane 2). Guanines were systematically replaced with adenine from the 5’- to the 3’-end (Figure 5, right). The 27-mer has nine guanines. Substitution of the first five and six guanines with adenine led to a decrease in yield to 6.5 +/-0.8% and 6.2 +/-1.0%, respectively. When two and one guanine/s remained, the oxidative crosslink yield fell to 3.2 +/-0.5% and 2.1 +/-0.6%, respectively. The change in crosslink yield is highly sensitive to the number of guanines. Thus, the strong correlation between the number of guanines and the yield infers crosslinks are occurring predominantly at guanine sites. It should be noted that the terminal guanine repeat is not the most sensitive site of crosslinking. Rather, removal of the 6th guanine from the 5’ position significantly decreases the crosslink yield the most, which indicates that this particular guanine is important for crosslinking in this DNA. This experiment was also accomplished for the Cu(II)-H2O2 oxidation system. Similar results are observed (See Figure S3 of supporting information). Even though Fenton reactions are known to induce thymine-based crosslinks, we find guanine content to be essential for oxidative crosslinking in both systems.
Figure 5. Guanine content correlates with crosslink yield.
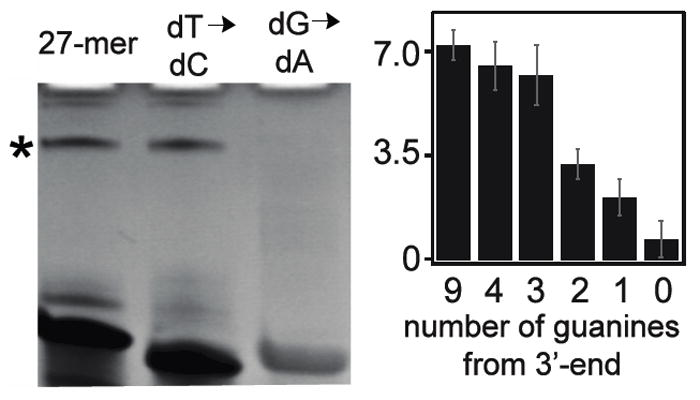
(Left) Ethidium bromide stained gel comparing the crosslinking yield of three 27-nucleotide DNA strands. The first strand is a mixed sequence. The second and third strands lack thymine and guanine, respectively. Loss of guanine leads to loss of product. (Right) Guanines from the 27-mer DNA model were sequentially replaced by adenine. Removal of guanines occurred from the 5’-end toward the 3’-end. The yield decreases as less guanines are present indicating that the crosslink product is likely between guanine and the protein.
To further elucidate the dependence of crosslinking on guanine, primer extension studies were accomplished. A primer for the 27-mer was utilized for these experiments to ensure significant replication stopping and oxidative crosslink stability during extension conditions. The 27-mer was incubated with Ribonuclease A, photo-oxidized using rose bengal, and separated by PAGE, followed by crosslink purification. Analysis of the Ribonuclease A-DNA crosslink band showed that it did not degrade (see supporting information, Figure S4). We inferred that crosslinking at sites on the primer binding site would block primer annealing and would not be observed in the assay. The only remaining guanine site on the 27-mer is the 5’-polyguanine repeat. The crosslink was then annealed to a fluorescently labeled primer and extended (Figure 6) to determine whether or not the presence of a DNA-protein crosslink will halt replication. The location of the replication stop was determined based on comparison to migration of the un-reacted 27-mer and synthetic oligonucleotides that were shorter than the 27-mer. The shorter oligonucleotides possess the intact primer binding site but have two nucleotides systematically removed. For instance, the sequence C4G2 (Figure 6A) lacks the 5’-GG and is 25 nucleotides long. These markers were also extended and used to locate the site where crosslinking induced a replication stop. After primer extension, samples were separated by PAGE and visualized. When no polymerase (Figure 6A, No ENZ) is added, a single band is observed at the bottom of the gel, indicating the un-reacted primer. Addition of polymerase to unmodified DNA gave a full-length product (Figure 6A, NO RXN). In contrast, extension of the crosslinked product (Figure 6A, DPC) led to a stop precisely one nucleotide before the poly-guanosine tract. Notice that C4 and DPC migrate equally. Extension occurred but could not go past the site of crosslinking. Additionally, crosslinks can be accurately identified using primer extension as they are stable toward elevated temperatures and basic conditions. Furthermore, this result underscores that these large lesions are strong replication stops: we observed no extension past the crosslink site.
Figure 6. Guanine is the major crosslink site.
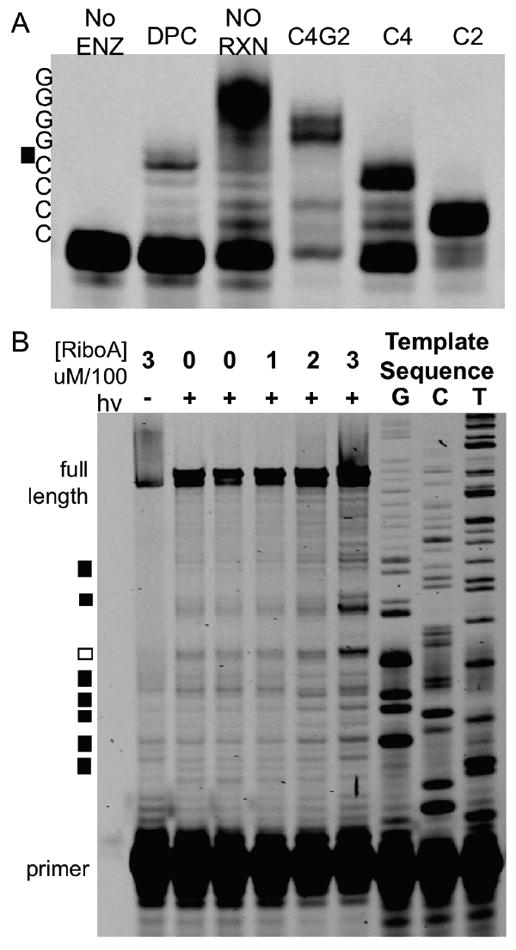
(A) The 27-nucleotide DNA was crosslinked to Ribonuclease A, the product subsequently purified, and annealed to a labeled primer. Isolation and extension of a no reaction control (lane NO RXN) led to full extension when compared to the truncated sequences on the right lanes (C4G2, C4, C2). C4G2 lacks the last two guanosine nucleotides from the 5’-end of the 27-mer, while C4 lacks the last 4 guanosine nucleotides and C2 does not have the last six nucleotides. Extension of the crosslinked product resulted in a stop before the poly-G tract (C4G2). (B) A 100-nucleotide mixed sequence DNA, rose bengal, bovine serum albumin, and Ribonuclease A at the listed concentrations (top) were incubated. Lack of irradiation or Ribonuclease A led to extension predominantly to the full-length product. As the Ribonuclease A concentration increases, new, shorter molecular weight bands are observed. Boxes to the left mark the new bands. Black boxes (7 of 8 new bands) represent crosslinks at guanine. The remaining white box occur two nucleotides before a guanine.
A synthetic 100-nucleotide DNA was used as a substrate for oxidative crosslinking (Figure 6B). This substrate is a mixed sequence and therefore will determine which nucleotides are crosslinked on a large DNA. The 100-nucleotide DNA was present at 5 μM, while the level of BSA was held constant at 500 μM to limit direct DNA photo-oxidation. To these samples, increasing concentrations of RNase A were added. BSA does not induce crosslinks while Ribonuclease A does (Figure 4A), thus, as more Ribonuclease A is present, the more crosslinks should be observed. After irradiation at 575 nm, samples were immediately extended for a single cycle and separated via PAGE. Samples were compared to thymine, guanine, and cytosine sequencing ladders. The “G” ladder corresponds to modified guanine sequences on the 100-nucleotide DNA template. The yield of full-length product is above 90% in all cases. When Ribonuclease A is incubated with the 100-nucleotide template without oxidation full length products predominate since the elevated temperatures during extension facilitate its denaturation. Incubation of DNA with BSA and subsequent oxidation led to a small about of damage owing to direct reaction of singlet oxygen with DNA. The concentration of Ribonuclease A was changed from 100 to 200 to 300 μM. Eight new shorter DNA bands are observed to increase in intensity with increasing concentrations of Ribonuclease A (Figure 6, boxes). Seven of the eight new bands are located one nucleotide from guanine (black box, compare to guanine ladder). One of the new products observed is not located at a guanine, but rather at the 5’-cytosine in the sequence 5’-GGGCCC (white box). The three-fold increase in Ribonuclease A concentration causes a four-fold increase of each polymerase stop with little variation (the increase at each site is plus or minus 30%) at the eight sites. It should be noted that each of the eight sites has a unique propensity to form crosslinks, with the single non-guanine crosslink being the second most intense. This data shows that guanine is the major site of oxidative crosslinking.
Mass spectrometry of 2’-deoxyguanosine-peptide crosslinks
Due to the difficulty in assessing the structure of an intact Ribonuclease A, a strategy where the product structure was determined using small molecule models was employed. Because several types of guanine-lysine crosslinks are possible, two smaller peptides obtained from the Ribonuclease A sequence, both containing a lysine residue were utilized in these experiments. Furthermore, tyrosine is known to add to thymine, which is why one of the peptides selected contains a tyrosine residue.14 The peptide N-Acetyl-AAAKF was used as a substrate in the oxidative crosslinking reaction in Figure 7 while N-Acetyl-AYKTT was used as a substrate in Figure 8. The peptide N-Acetyl-AAAKF is only capable of forming lysyl type crosslinks and the peptide N-Acetyl-AYKTT can form each of the three types of oxidative crosslinks: those between lysine and guanine base, those formed by addition of lysine to depurinated nucleotides, and between tyrosine and thymine base.19 The two peptides comprise residues 7-12 and 99-104 of Ribonuclease A, respectively. Because binding is an important factor in crosslink formation, the concentrations of both peptide and nucleoside were increased to 5 mM as lower concentrations resulted in much lower yields. The yield was below 5% for both peptides despite the one-to-one ratio and the elevated concentration. The peptide was incubated with the nucleoside and irradiated as before. Products were analyzed by mass spectrometry. The mass spectrometer is a hybrid instrument that consists of a linear ion trap for low-resolution spectra, and a fourier transform ion cyclotron resonance mass spectrometer for high-resolution spectra. Thus, an introduced sample has both spectra acquired. The low-resolution MS samples are sampled faster and, therefore, have a more accurate TIC. The high-resolution instrument is used to obtain elemental compositions. Mass accuracy for high-resolution mass spectrometry has errors below 500 ppb for full scan and below 1000 ppb for MSn. The MSn fragmentation is accomplished in the linear ion trap and analyzed in both modes. Therefore, MSn also has high-resolution spectra to accurately identify fragments. It is important to note that many guanine hyperoxidation products are not stable in acid. Hydantions are modified under acidic conditions, thus the addition of acid can potentially affect results.42 We optimized LC and MS conditions in water with no acid added since we have found that low pH, even during direct injection, does lead to severe degradation (data not shown).
Figure 7. LC-MS analysis of rose bengal oxidized adduct of dG and the peptide N-Ac-AAAKF.
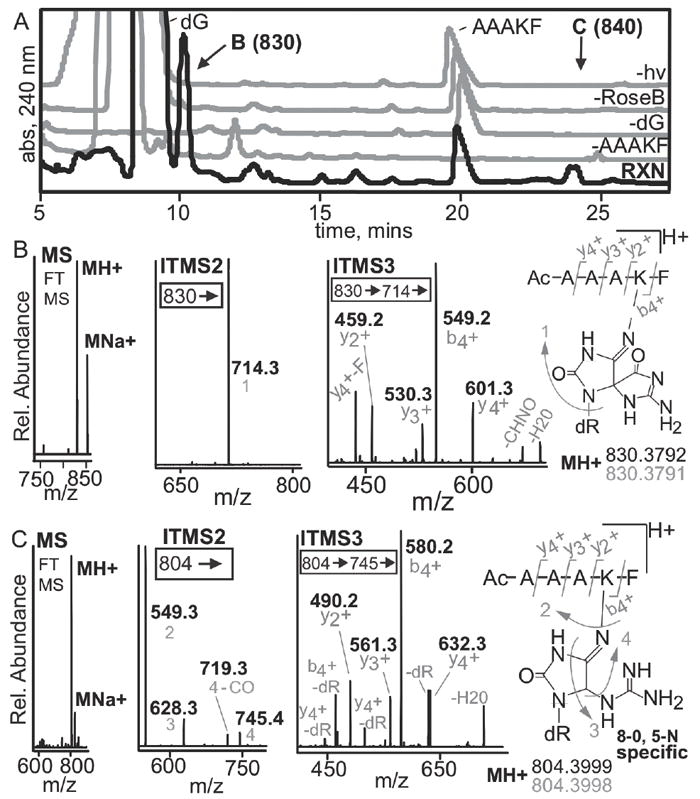
(A) A peptide containing a single crosslink active lysine and 2’-deoxyguanosine were utilized to assess if high-resolution, multi-dimensional MS could be used to establish the location of a crosslink. The HPLC-UV trace of the reaction and the controls are shown. The reaction chromatogram is in black while the controls lacking N-Ac-AAAKF, dG, rose bengal or light are in grey. The 10.5 and 24-minute peaks were unique to the reaction, denoted by B and C respectively. The products were isolated and analyzed by mass spectrometry. (B) Mass spectral analysis of the 10.5-minute peak revealed that it has an observed mass and elemental composition consistent with formation of a spiroiminodihydantoin. MS2 analyses show exclusive fragmentation at guanine. MS3 can be used to determine the site of crosslinking based on the y2+ and b4+ ions. (C) The 24-min peak shows similar results except that more DNA fragmentation is seen in MS2. The 24-min peak is a 5-lys-guanidinohydantoin crosslink based on elemental composition and MS3 fragmentation. All fragments (grey) are acquired using high resolution MS with errors below 1 ppm.
Figure 8. LC-MS identification of rose bengal oxidized adduct between dG and the peptide N-Ac-AYKTT.
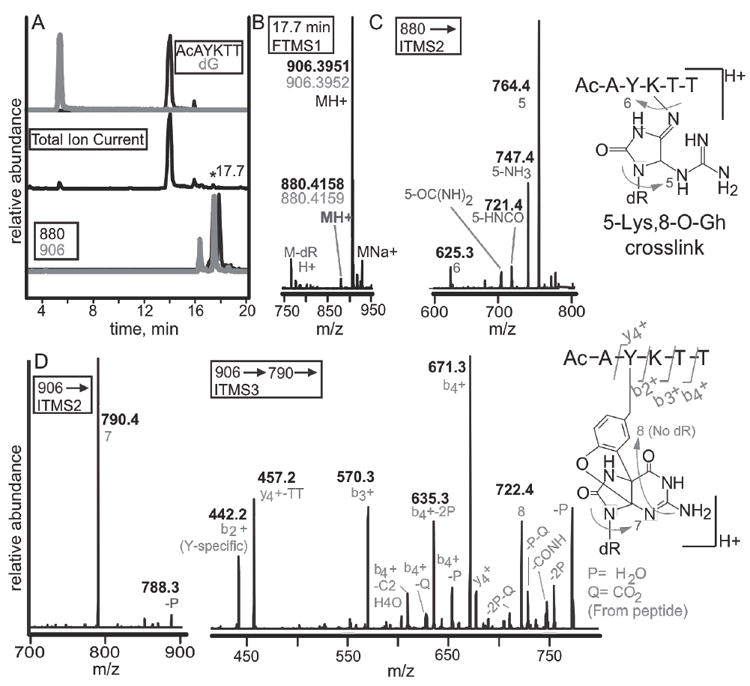
(A) A peptide containing both a tyrosine and lysine were reacted with 2’-deoxyguanosine. Selected ion chromatograms from the reaction are shown. The top two chromatograms are N-acetyl-AYKTT (black) and 2’-deoxyguanosine (grey). The total ion chromatogram (middle) shows the starting materials as the most abundant ions. Monitoring of ions with masses of 880 (black) and 906 (grey) show several peaks eluting within one minute (bottom). (B) Analysis of the 17.7-minute peak shows masses of 880.4158 and 906.3951. Experimental masses are in black while theoretical masses are in grey. (C) MS2 analysis of the 880 peak show that the lesion is guanidinohydantoin. (D) The 906 peak can either be a lysine-spiroiminodihydantoin or a tyrosine-guanine crosslink. The b2+ ion bisects the tyrosine and lysine revealing that tyrosine is modified.
The reaction with 2’-deoxyguanosine with N-Acetyl-AAAKF led to formation of hyperoxidized guanine derivatives (Figure 7). An example of a HPLC-UV trace is shown in Figure 7A. Two new prominent product bands are observed when comparing the reaction chromatogram (black) and the controls, which excluded a reaction component. Control reactions include without the peptide, 2’-deoxyguanosine, rose bengal, or irradiation (grey traces). The clearly observable products migrate at 10.5 and 24 min (denoted by B or C). The reaction trace shows that these products are not in low abundance and that LC-MS is not being used to detect a minor product. The reaction products were analyzed. The MS spectra for the 10.5 min peak is shown in Figure 7B. The 10.5 min product has an observed mass of 830.3792 that corresponds to an elemental composition C36H52N11O12 + with an error of 44 ppb. Because our conditions do not use acid, a strong sodium adduct is observed with an error of 202 ppb. The 10.5 min product was fragmented in MS2. The ion trap spectra are shown, while the high resolution MS is in the supporting information. A single major fragment was observed with a mass of 714.3 or a loss of C5H8O3. The 714.3 fragment is due to the loss of 2’-deoxyribose indicating adduct formation is between the guanine and the peptide. Further fragmentation during MS3 can be used to accurately assign the location of modification on a peptide. The seven most abundant ions during MS3 fragmentation are shown. The peptide fragments b4 +, y4 +, y2 +, y3 +, and loss H2O all have a mass that includes the hyperoxidized guanine base. The loss of H2O likely derives from the carboxyl terminus. Additionally, y4 + is observed to also lose the C-terminal phenylalanine. The ion at 549.3 could have been loss of guanine, 549.2528, or the b4 + ion, 549.3031. High resolution MS determined the ion to be by b4 +. Only a single modification of guanine is observed with a loss of CHNO. This ion stems from ring cleavage at either C8-N7 or C6-N1. Importantly, the y2 + and b4 + show that the guanine must be added to the lysyl residue since the other peptide positions are lost without the modified guanine base. The mass and fragmentations identify the lesions as a lysyl-spiroiminodihydantoin lesion but the isomeric forms cannot be determined.
We then analyzed the 24 min product in a similar manner as the 10.5 min product. The 24 min product has an observed mass of 804.3999 that corresponds to an elemental composition C35H54N11O11 + with an error of 101 ppb. Fragmentation gave a different result as compared to lysyl-spiroiminodihydantoin lesion. During MS2 we observe the major ions formed as losses from the 2’-deoxyguanosine and a single peptide modification, the loss of H2O. The largest ion observed in MS2 is loss of the 2’-deoxyguanosine (549.25 not 549.30). This ion cannot be used to determine the crosslink site. The next most abundant ion was 628.3. This ion is loss of 2’-deoxyguanosine and urea. This fragment has been previously described as 5-lysyl, 8-oxo specific.37 The fragmentation of this ion could not determine the crosslink site. We therefore, analyzed the 745.4 ion, which corresponds to loss of CH5N3 or removal of the guanidine. The resulting fragment has a double bond between C4 and N9. Thus, N9 is cationic. MS3 determined the site as the lysine residue. Because of the instability of the glycosidic bond in the charged state, loss of 2’-deoxyribose is a major ion observed. In addition, the peptide fragments are observed as both the ion and the ion without the 2’-deoxyribose. The peptide fragments are the same as the lysyl-spiroiminodihydantoin adduct. The mass and fragmentations identify the lesion as a 5-lysyl-8-oxo-guanidinohydantoin.
We then determined the crosslink product formed using the peptide N-Acetyl-AYKTT and 2’-deoxyguanosine. This peptide can be used to compare the propensity of a tyrosine-guanine and lysyl-guanine to form in a single peptide. The LC-MS of the reaction is shown in Figure 8A. Two slowly migrating products were observed with elemental compositions of C37H58N11O14 + and C38H56N11O15 + and a mass of 880.4158 and 906.3951 respectively. The largest error in all cases was 276 ppb. We looked for a crosslink formed from an abasic nucleoside and lysine that has been recently observed.43 The product would have an elemental composition of C33H53N6O13 + with a mass of 741.37, which we did not detect by selective ion monitoring. Additionally, we also did not observe any thymine-tyrosine oxidative crosslinks when a similar reaction with 2’-deoxythymidine was performed. The total ion chromatogram shows the most abundant ions to be unmodified peptide N-Ac-AYKTT and 2’-deoxyguanosine (Figure 8, compare top two traces). When 880.4 and 906.4 were monitored, we found several bands all eluting within one minute (compare traces). This data is consistent with formation of both conformational isomers and stereoisomers of hydantoin rings. The MS2 of 880.4 reveals that it is a guanidinohydantoin-peptide adduct with the major product having the peptide appended on C5 of Gh via lysine addition (data not shown). The 906.4 ion was ten times as abundant as the 880.4 ion. The 906.4 ion can be either a lysine-spiroiminodihydantoin or a tyrosine-guanine(ox) crosslink. The two most abundant ions that result when the 906 ion was fragmented, were the 790.4 ion, which corresponds to the loss of 2’-deoxyribose, which confirms that the adduct is between guanine and the peptide, and the 788.3 ion which is due to the loss of H2O (Figure 8D). In order to determine the amino acid involved and to identify the adduct structure, an MS3 of the 790.4 ion was obtained. The fifteen most abundant ions during MS3 fragmentation are shown. Two of the resulting ions are due to guanine modifications. The 722.4 peak corresponds to the loss of C2N2O, arising from the removal of C6-N1-C2. This fragment can occur for both the lysine-spiroiminodihydantoin or the tyrosine-guanine(ox) adduct. The loss of CHNO is also observed which can arise from elimination of either C8-N7 or C6-N1, also possible for both lys-sp or the tyrosine-guanine(ox) adduct. The peptide fragmentations are b4 +, b3 +, b2 +, y4 +, in addition to y4-TT, b4-CO2 and b4-C2H4O. Loss of one or two H2O molecules, by itself, along with CO2 or from the b4 + ion is also observed. The H2O molecules can come from the two threonine residues while the CO2 molecule can come from the carboxy terminal. The C2H4O lost from b4 + is likely from the remaining threonine residue. The b3 +, y4 + and the y4-TT together imply that either lysine or tyrosine is adding to guanine, since the b3 + ion is due to the loss of the two threonine residues adjacent to lysine, the y4 + ion is due to the loss of the N-acetyl-alanine next to tyrosine and the y4-TT ion is due to the loss of both. The b2 + ion on the other hand is indicative of tyrosine substitution to guanine since the observed mass, which is 442.2 is 165.1 mass units greater than the predicted mass when guanine is not included. This mass corresponds to the mass of the hyperoxidized guanine, thus the b2 + ion is N-Acetyl-AY-guanine(ox). A 497.2 peak, which is a y3 + ion for lysine substitution to guanine is found but is buried in the background. This data confirm that tyrosine modification is occurring but does not rule out lysine modification since the ions may be differentially detected by MS ionization. These data show that oxidative crosslinking is facile and product characterization shows similar trends. First, the MS2 generally cleaves at 2’-deoxyguanine leaving a peptide and a small portion of the DNA used to identify the site of crosslinking. Second, these data illustrate that both tyrosine-guanine(ox) and lysine-guanine(ox) readily form in good yields.
Conclusions
We show that oxidation-induced DNA-protein crosslinks have a high propensity to form. Earlier studies on DNA-protein crosslinks utilized oxidation prone 8-oxo-2’-deoxyguanosine-substituted oligonucleotides, the presence of which increases the propensity for oxidative crosslinking at a single site on the DNA. This study shed light on oxidative crosslinks by determining key parameters in the reaction. The first key observation is that unmodified DNA can readily lead to DNA-protein crosslink reactions in high yields of up to 61%. Despite the very different oxidation mechanisms of the four oxidants used they all have the potential to form oxidative crosslinks. These oxidants are classic mimics of cellular oxidative stress. A key and underappreciated factor in successful crosslinking is binding. Addition of salt or co-incubation with non-binding proteins severely reduces crosslink efficiency. A second key observation is that these lesions likely derive from guanine modification. We show by substituting guanine with adenine in the 27-nucleotide DNA and from primer extension studies that these products are forming at guanine sites. Structural analysis shows that guanine forms guanidinohydantoin, spiroiminodihydantoin, and an aromatic addition product from guanine hyperoxidation.
The story from the amino acid components is much different. We were able to isolate both tyrosine and lysine based crosslinks. Due to its more favorable oxidation potential, tyrosine oxidation leads to formation of tyrosine-based crosslink even in the presence of an adjacent lysine. This indicates that the formation of these tricyclic tyrosyl-adducts can compete with the more well known lysine-based crosslinks. Mechanistically, it appears that lysine crosslinks stem from DNA oxidation as we observe the formation of 5-lys that is diagnostic of DNA oxidation,16 while tyrosine-adducts stem from protein oxidation. These divergent reaction products may have little consequence when a whole protein is attached to DNA since all crosslinks are likely strong replications stops for replicative polymerases. In fact, DNA-protein crosslinking mediated by the anti-tumor trans-[PtCl2(E-iminoether)2] was shown to constitute to a fairly strong replication block for exonuclease deficient Klenow Fragment and RT HIV-1, although a small amount of translesion synthesis was observed.44 A family polymerase, DNA polymerase •, translesion synthesis may be involved in cellular tolerance of DNA-protein crosslink. 45 polymerase • can bypass exceptionally large lesions whose linkages are through the DNA-major groove, but not the minor groove. This polymerase can polymerize passed extremely large major groove lesions including DNA-protein and DNA-DNA crosslinks. It is established that crosslinks are repaired by digestion of the protein into small peptides.9 At the stage of a DNA-peptide crosslink the exact structure of a crosslink will be more important as the smaller lesions may show differential mutagenicity since the smaller lesion will be easier to bypass. The high yields of product formation illustrates that oxidative crosslinks are underappreciated as DNA lesions. Oxidative crosslinks may be present at high relative levels in cells since the propensity to oxidatively crosslink is high and so much of the genomic DNA is coated with protein.
Supplementary Material
Figure 1. Oxidative DNA-protein crosslink can form hydantion rings.
When oxidized, DNAs and proteins can crosslink due to the generation of strong electrophiles. DNA-protein crosslinks are known to occur in cells with several structures as products. Proteins are known to crosslink via tyrosyl and lysyl groups, while guanine, thymidine, and abasic sites in DNA undergo the crosslinking reaction.
Acknowledgments
We would like to thank the University of Cincinnati for startup funding, a faculty research award to E.J.M., and a summer research fellowship to M.J.S. We would also like to thank NIH-RR19900 for acquisition of the LTQ-MS.
Funding Support University of Cincinnati, startup funding
University of Cincinnati, University Research Council to EJM and MJS
NIH-RR19900 for acquisition of the LTQ-MS
non-standard abbreviations
- Gh
guanidinohydantoin
- Sp
spiroiminodihydantoin
- SSB
Single Stranded Binding Protein
- RNaseA
Ribonuclease A
- BSA
Bovine Serum Albumin
Citations
- 1.Cadet J, Douki T, Ravanat JL. Oxidatively generated damage to the guanine moiety of DNA: Mechanistic aspects and formation in cells. Acc Chem Res. 2008;41:1075–1083. doi: 10.1021/ar700245e. [DOI] [PubMed] [Google Scholar]
- 2.Valko M, Leibfritz D, Moncol J, Cronin MTD, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Berdis AJ. Mechanisms of DNA Polymerases. Chem Rev. 2009;109:2862–2879. doi: 10.1021/cr800530b. [DOI] [PubMed] [Google Scholar]
- 4.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: A mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 5.Kumar B, Koul S, Khandrika L, Meacham RB, Koul HK. Oxidative stress is inherent in prostate cancer cells and is required for aggressive phenotype. Cancer Res. 2008;68:1777–1785. doi: 10.1158/0008-5472.CAN-07-5259. [DOI] [PubMed] [Google Scholar]
- 6.Wang BJ, Sheu HM, Guo YL, Lee YH, Lai CS, Pan MH, Wang YJ. Hexavalent Chromium-induced ROS Formation, Subsequent Akt, NF-kappa B, and MAPK Activation, and TNF-alpha, and IL-1 alpha Production in Keratinocytes. Toxicol Lett. 2010;198:216–224. doi: 10.1016/j.toxlet.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 7.O’Brien TJ, Mandel HG, Sugden KD, Komarov AM, Patierno SR. Hypoxia impedes the formation of chromium DNA-adducts in a cell-free system. Biochem Pharmacol. 2005;70:1814–1822. doi: 10.1016/j.bcp.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 8.Macfie A, Hagan E, Zhitkovich A. Mechanism of DNA-Protein Cross-linking by Chromium. Chem Res Toxicol. 2010;23:341–347. doi: 10.1021/tx9003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat Res -Rev Mutat Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Medeiros MG, Rodrigues AS, Batoreu MC, Laires A, Rueff J, Zhitkovich A. Elevated levels of DNA-protein crosslinks and micronuclei in peripheral lymphocytes of tannery workers exposed to trivalent chromium. Mutagenesis. 2003;18:19–24. doi: 10.1093/mutage/18.1.19. [DOI] [PubMed] [Google Scholar]
- 11.Zhitkovich A, Voitkun V, Costa M. Formation of the amino acid-DNA complexes by hexavalent and trivalent chromium in vitro: Importance of trivalent chromium and the phosphate group. Biochemistry. 1996;35:7275–7282. doi: 10.1021/bi960147w. [DOI] [PubMed] [Google Scholar]
- 12.Zhitkovich A, Song Y, Quievryn G, Voitkun V. Non-oxidative mechanisms are responsible for the induction of mutagenesis by reduction of Cr(VI) with cysteine: Role of ternary DNA adducts in Cr(III)-dependent mutagenesis. Biochemistry. 2001;40:549–560. doi: 10.1021/bi0015459. [DOI] [PubMed] [Google Scholar]
- 13.Dizdaroglu M, Gajewski E, Reddy P, Margolis SA. Structure of a hydroxyl radical induced DNA protein cross-link involving thymine and tyrosine in nucleohistone. Biochemistry. 1989;28:3625–3628. doi: 10.1021/bi00434a071. [DOI] [PubMed] [Google Scholar]
- 14.Morimoto S, Hatta H, Fujita S, Matsuyama T, Ueno T, Nishimoto S. Hydroxyl radical-induced cross-linking of thymine and lysine: Identification of the primary structure and mechanism. Bioorg Med Chem Lett. 1998;8:865–870. doi: 10.1016/s0960-894x(98)00124-3. [DOI] [PubMed] [Google Scholar]
- 15.Xu XY, Muller JG, Ye Y, Burrows CJ. DNA-protein cross-links between guanine and lysine depend on the mechanism of oxidation for formation of C5 vs C8 guanosine adducts. J Am Chem Soc. 2008;130:703–709. doi: 10.1021/ja077102a. [DOI] [PubMed] [Google Scholar]
- 16.Gremaud JN, Martin BD, Sugden KD. Influence of Substrate Complexity on the Diastereoselective Formation of Spiroiminodihydantoin and Guanidinohydantoin from Chromate Oxidation. Chem Res Toxicol. 2010;23:379–385. doi: 10.1021/tx900362r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perrier S, Hau J, Gasparutto D, Cadet J, Favier A, Ravanat JL. Characterization of lysine-guanine cross-links upon one-electron oxidation of a guanine-containing oligonucleotide in the presence of a trilysine peptide. J Am Chem Soc. 2006;128:5703–5710. doi: 10.1021/ja057656i. [DOI] [PubMed] [Google Scholar]
- 18.Xu XY, Fleming AM, Muller JG, Burrows CJ. Formation of tricyclic 4.3.3.0 adducts between 8-oxoguanosine and tyrosine under conditions of oxidative DNA-protein cross-linking. J Am Chem Soc. 2008;130:10080–10081. doi: 10.1021/ja803896d. [DOI] [PubMed] [Google Scholar]
- 19.Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle. Proc Natl Acad Sci U S A. 2010;107:22475–22480. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 21.Margolin Y, Shafirovich V, Geacintov NE, DeMott MS, Dedon PC. DNA sequence context as a determinant of the quantity and chemistry of guanine oxidation produced by hydroxyl radicals and one-electron oxidants. J Mol Biol. 2008;283:35569–35578. doi: 10.1074/jbc.M806809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pratviel G, Meunier B. Guanine oxidation: one- and two-electron reactions. Chemistry. 2006;12:6018–6030. doi: 10.1002/chem.200600539. [DOI] [PubMed] [Google Scholar]
- 23.Delaney JC, Essigmann JM. Biological properties of single chemical-DNA adducts: A twenty year perspective. Chem Res Toxicol. 2008;21:232–252. doi: 10.1021/tx700292a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan W, Chen B, Wang L, Taghizadeh K, Demott MS, Dedon PC. Quantification of the 2-deoxyribonolactone and nucleoside 5’-aldehyde products of 2-deoxyribose oxidation in DNA and cells by isotope-dilution gas chromatography mass spectrometry: differential effects of gamma-radiation and Fe2+-EDTA. J Am Chem Soc. 2010;132:6145–6153. doi: 10.1021/ja910928n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansen ME, Muller JG, Xu XY, Burrows CJ. Oxidatively induced DNA-protein cross-linking between single-stranded binding protein and oligodeoxynucleotides containing 8-oxo-7,8-dihydro-2’-deoxyguanosine. Biochemistry. 2005;44:5660–5671. doi: 10.1021/bi047580n. [DOI] [PubMed] [Google Scholar]
- 26.Izzotti A, Cartiglia C, Taningher M, De Flora S, Balansky R. Age-related increases of 8-hydroxy-2’-deoxyguanosine and DNA-protein crosslinks in mouse organs. Mutat Res. 1999;446:215–223. doi: 10.1016/s1383-5718(99)00189-8. [DOI] [PubMed] [Google Scholar]
- 27.Stover JS, Ciobanu M, Cliffel DE, Rizzo CJ. Chemical and electrochemical oxidation of C8-arylamine adducts of 2 ’-deoxyguanosine. J Am Chem Soc. 2007;129:2074–2081. doi: 10.1021/ja066404u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall DB, Holmlin RE, Barton JK. Oxidative DNA damage through long-range electron transfer. Nature. 1996;382:731–735. doi: 10.1038/382731a0. [DOI] [PubMed] [Google Scholar]
- 29.Luo WC, Muller JG, Rachlin EM, Burrows CJ. Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-oxo-7,8-dihydroguanosine. Org Lett. 2000;2:613–616. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 30.Aller P, Ye Y, Wallace SS, Burrows CJ, Doublie S. Crystal Structure of a Replicative DNA Polymerase Bound to the Oxidized Guanine Lesion Guanidinohydantoin. Biochemistry. 2010;49:2502–2509. doi: 10.1021/bi902195p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 32.Leipold MD, Muller JG, Burrows CJ, David SS. Removal of hydantoin products of 8-oxoguanine oxidation by the Escherichia coli DNA repair enzyme, FPG. Biochemistry. 2000;39:14984–14992. doi: 10.1021/bi0017982. [DOI] [PubMed] [Google Scholar]
- 33.McPherson A, Brayer GD, Morrison RD. Crystal structure of RNase A complexed with d(pA)4. J Mol Biol. 1986;189:305–327. doi: 10.1016/0022-2836(86)90512-7. [DOI] [PubMed] [Google Scholar]
- 34.McPherson A, Brayer G, Cascio D, Williams R. The mechanism of binding of a polynucleotide chain to pancreatic ribonuclease. Science. 1986;232:765–768. doi: 10.1126/science.3961503. [DOI] [PubMed] [Google Scholar]
- 35.Sun G, Fecko CJ, Nicewonger RB, Webb WW, Begley TP. DNA-Protein Cross-linking: Model Systems for Pyrimidine-Aromatic Amino Acid Cross-linking. Org Lett. 2006;8:681–683. doi: 10.1021/ol052876m. [DOI] [PubMed] [Google Scholar]
- 36.Saito I, Takayama M, Sugiyama H, Nakatani K. Photoinduced DNA cleavage via electron-transfer- demonstration that guanine residues located 5’ to a guanine are the most electron-donating sites. J Am Chem Soc. 1995;117:6406–6407. [Google Scholar]
- 37.Solivio MJ, Joy TJ, Sallans L, Merino EJ. Copper generated reactive oxygen leads to formation of lysine-DNA adducts. J Inorg Biochem. 2010;104:1000–1005. [PubMed] [Google Scholar]
- 38.Yun BH, Geacintov NE, Shafirovich V. Generation of Guanine-Thymidine Cross-Links in DNA by Peroxynitrite/Carbon Dioxide. Chem Res Toxicol. 2011;24:1144–1152. doi: 10.1021/tx200139c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hickerson RP, Prat F, Muller JG, Foote CS, Burrows CJ. Sequence and Stacking Dependence of 8-Oxoguanine Oxidation: Comparison of One-Electron vs Singlet Oxygen Mechanisms. J Am Chem Soc. 1999;121:9423–9428. [Google Scholar]
- 40.Frelon S, Douki T, Favier A, Cadet J. Hydroxyl Radical Is Not the Main Reactive Species Involved in the Degradation of DNA Bases by Copper in the Presence of Hydrogen Peroxide. Chem Res Toxicol. 2003;16:191–197. doi: 10.1021/tx025650q. [DOI] [PubMed] [Google Scholar]
- 41.Kurbanyan K, Nguyen KL, To P, Rivas EV, Lueras AMK, Kosinski C, Stery M, Gonzales A, Mah DA, Stemp EDA. DNA-Protein Cross-Linking via Guanine Oxidation: Dependence upon Protein and Photosensitizer. Biochemistry. 2003;42:10269–10281. doi: 10.1021/bi020713p. [DOI] [PubMed] [Google Scholar]
- 42.Ye Y, Munk BH, Muller JG, Cogbill A, Burrows CJ, Schlegel HB. Mechanistic Aspects of the Formation of Guanidinohydantoin from Spiroiminodihydantoin under Acidic Conditions. Chem Res Toxicol. 2009;22:526–535. doi: 10.1021/tx800402y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guan LR, Greenberg MM. An Oxidized Abasic Lesion as an Intramolecular Source of DNA Adducts. Aust J Chem. 2011;64:438–442. doi: 10.1071/CH10420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novakova O, Kasparkova J, Malina J, Natile G, Brabec V. DNA-protein cross-linking by trans-[PtCl2(E-iminoether)2]. A concept for activation of the trans geometry in platinum antitumor complexes. Nucleic Acids Res. 2003;31:6450–6460. doi: 10.1093/nar/gkg863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamanaka K, Minko IG, Takata K, Kolbanovskiy A, Kozekov ID, Wood RD, Rizzo CJ, Lloyd RS. Novel Enzymatic Function of DNA Polymerase • in Translesion DNA Synthesis Past Major Groove DNA-Peptide and DNA-DNA Cross-links. Chem Res Toxicol. 2010;23:689–695. doi: 10.1021/tx900449u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


