Abstract
Background
The lipidome is rapidly garnering interest in the field of psychiatry. Recent studies have implicated lipidomic changes across numerous psychiatric disorders. In particular there is growing evidence that the concentrations of several classes of lipids are altered in those diagnosed with MDD. However, for lipidomic abnormalities to be considered potential treatment targets for MDD (rather than secondary manifestations of the disease), a shared etiology between lipid concentrations and MDD should be demonstrated.
Methods
In a sample of 567 individuals from 37 extended pedigrees (average size 13.57 people, range = 3–80), we used mass-spectrometry lipidomic measures to evaluate the genetic overlap between twenty-three biologically distinct lipid classes and a dimensional scale of MDD.
Results
We found that the lipid class with the largest endophenotype ranking value (ERV, a standardized parametric measure of pleiotropy) were ether-phosphodatidylcholines (alkylphosphatidylcholine, PC(O) and alkenylphosphatidylcholine, PC(P) subclasses). Furthermore, we examined the cluster structure of the twenty-five species within the top-ranked lipid class, and the relationship of those clusters with MDD. This analysis revealed that species containing arachidonic acid generally exhibited the greatest degree of genetic overlap with MDD.
Conclusions
Thisstudy is the first to demonstrate a shared genetic etiology between MDD and ether-phosphatidylcholine species containing arachidonic acid, an omega-6 fatty acid that is a precursor to inflammatory mediators, such as prostaglandins. The study highlights the potential utility of the well-characterized linoleic/arachidonic acid inflammation pathway as a diagnostic marker and/or treatment target for MDD.
Keywords: Affective Disorders, Unipolar Depression, Genetics
Introduction
Major Depressive Disorder (MDD) is a common and potentially life-threatening disorder of mood (1). It affects 16.2% of individuals in the US during their lifetime (2) and as such it incurs great economic cost ($83.1 billion per annum in the US) (3). This is not to mention the personal cost where the impact of MDD on wellbeing and functioning is in line with that seen in arthritis and diabetes mellitus (4). Moreover, functional impairments remain after the remission of a depressive episode (5). Unsurprisingly, the World Health Organization (WHO) cites MDD as a leading cause of disability worldwide (6). However, despite decades of research, the etiology of the illness remains largely unknown.
Lipidomic alterations have been reported in numerous psychiatric disorders, including schizophrenia (7), autism (8, 9), and bipolar disorder (10–12). In particular, changes in the lipidome (the complete lipid profile of an organism) have been most consistently associated with MDD (13). The first indication of this association came from early trials of statins, statins are cholesterol-lowering drugs prescribed to individuals with increased lipid levels (14). During the statin trials, the lipid-lowering benefits of statin therapy (i.e. reduced cardiovascular disease risk) were offset, in some cases, by an increase in suicidality (15–20). Though, it should be noted that others have reported beneficial effects of statins on depressive symptomatology when combined with anti-depressant medications including SSRIs (21, 22). The obvious overlap between suicidality and MDD led some to propose a direct link between lipids and MDD. Indeed, subsequent studies have reported differences between depressed and healthy subjects in the concentrations of fatty acids in both animal models of depression (23–27) and also in clinical populations of humans (28–31); and also alterations in lipid classes including phospholipids (e.g., phosphatidylcholines (PCs), lysophosphatidylcholines (LPCs), lysophosphatidylethanolamine (LPEs), phosphatidylethanolamines (PEs), sphingolipids, and cholesterol esters (32–35). However, despite strong evidence linking lipid concentrations and MDD, it is currently unclear whether the lipidomic alterations observed in MDD are secondary to the manifestation of the illness or its treatment, or whether lipid concentrations are related to the genetic predisposition for depression. If the latter supposition were true, lipids could be considered a promising diagnostic and/or treatment target for MDD.
In the present study, we aimed to provide evidence for a shared etiology between lipidomic concentrations and MDD, and determine which lipid classes, and which species within those classes, might be most informative when attempting to isolate potential diagnostic and treatment targets for MDD. To achieve these aims we completed three steps: (1) we ranked sum concentrations of twenty-three lipid classes by their genetic overlap with MDD and isolated those classes with the greatest degree of overlap; (2) we took the top-ranked lipid classes and investigated the structure of the species within them using cluster analysis; (3) we evaluated the degree of genetic overlap between each species cluster and MDD in an attempt to characterize the relationships between the lipids and MDD at the species level.
Methods
Participants
Lipidomic and psychiatric data were available from a total 567 participants from 37 families (average family size = 13.57, range = 3–80) the sample was 64% female and had a mean age of 49.47 years (SD = 13.31, range = 27–97). The lipidomic data was collected as part of the San Antonio Family Study (SAFS), diagnostic data were also available in these same individuals as part of assessments conducted in overlapping individuals as part of the Genetics of Brain Structure and Function (GOBS) study. GOBS data collection occurred between 2006 and 2016. Individuals from the SAFS cohort have actively participated in research for over 18 years. Participants were randomly selected from the community with the constraints that they were of Mexican American ancestry, part of a large family, and lived in the San Antonio, TX, region. All participants provided written informed consent in compliance with the institutional review board at the University of Texas Health Science Center of San Antonio (36).
Continuous Index of MDD
All participants received the Mini-International Neuropsychiatric Interview (MINI) (37), a semi-structured interview augmented to include items on lifetime diagnostic history. Masters- and doctorate-level research staff, with established reliability for diagnosing affective disorders (κ ≥ .85), conducted the interviews. All subjects with possible psychopathology were discussed in case conferences that included licensed psychologists or psychiatrists. Lifetime consensus diagnoses were determined based on available medical records, the MINI interview, and the interviewer’s narrative. Consistent with previous work (38), all items from the Past Major Depressive Episode (A3a–g) section of the MINI were entered into a confirmatory factor analysis with a single factor, and maximum-likelihood estimates of the latent factor scores were used as the dimensional scale of MDD. In our previous study, we demonstrated that this continuous index conferred multiple advantages for gene-finding efforts over the conventional dichotomous (present-absent) diagnosis of MDD (for details, see (38). Using conventional diagnoses, 216 individuals endorsed a major depressive episode in their lifetime while 115 had experienced two or more episodes (recurrent MDD).
Lipid Extraction and Analysis Procedure
The lipid extraction procedure used in this sample has been described in detail elsewhere (see (39,40). Briefly, the San Antonio Family study is part of an ongoing longitudinal observational investigation comprising four phases of data collection during a 23-year period. The plasma samples used for lipidomic analysis in the present study were collected during the first phase, between the years 1992–1996. The order of the plasma samples was randomized prior to lipid extraction and analysis. Quality control plasma samples were included at a ratio of 1:18. Total lipid extraction from a 10 mL aliquot of plasma was performed by a single phase chloroform:methanol (2:1) extraction after the addition of 15 μL of internal standard mix containin 16 non-physiological or stable isotope lipid standards (Supplementary Table 1) (41).
Lipid analysis was performed by liquid chromatography, electrospray ionisation-tandem mass spectrometry using an Agilent 1200 liquid chromatography system combined with an Applied Biosystems API 4000 Q/TRAP mass spectrometer with a turboionspray source (350°C) and Analyst 1.5 data system (41). Liquid chromatography was performed on a Zorbax C18, 1.8μm, 50 × 2.1 mm column (Agilent Technologies) using the following gradient conditions (300μL/min) 0% solvent B to 100% solvent B over 8.0 min, 2.5 min at 100% solvent B, a return to 0% solvent B over 0.5 min then 10.5 min at 0% solvent B prior to the next injection. Diacylglycerol (DG) and triacylglycerol (TG) species (1μL injection) were analysed in a separate chromatographic run using an isocratic flow (100μL/min) of 85% solvent B over 6 min. Solvents A and B consisted of tetrahydrofuran:methanol:water in the ratio (30:20:50) and (75:20:5) respectively, both containing 10 mM ammonium formate. Columns were heated to 50°C and the auto-sampler regulated to 25°C. All other lipid species (5μL injection) were separated under gradient conditions.
Multiple reaction monitoring (MRM) experiments were used to analyses lipid species in the following classes and subclasses: dihydroceramide (dhCer), ceramide (Cer), monohexosylceramide (MHC), dihexosylceramide (DHC), trihexosylcermide (THC), GM3 ganglioside (GM3), sphingomyelin (SM), phosphatidylcholine (PC), alkylphosphatidylcholine (PC(O)), alkenylphosphatidylcholine (plasmalogen, PC(P)), lysophosphatidylcholine (LPC), lysoalkylphosphatidylcholine (lysoplatelet activating factor, LPC(O)), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylserine (PS), phosphatidylglycerol (PG), cholesterol ester (CE), free cholesterol (COH), diacylglycerol (DG) and triaclyglycerol (TG) (41–43). A total of 65 diacylglycerol and triacylglycerol species and 257 other lipid species were analyzed. The mass spectrometry conditions are shown in Supplementary Table 1. The listed abbreviations are used to refer to individual lipid species e.g. LPC 22:6, which defines a lysophosphatidylcholine with a fatty acid containing 22 carbons and six double bonds. A number of lipids contain two fatty acid chains, for these the mass spectrometry based measurements reflect the sum of the number of carbons and the sum of the number of double bonds across both fatty acids, rather than directly determining the constituent fatty acids. In accordance with this, for these species we denote the combined length and number of double bonds (e.g. PC 36:4). However, it is of note that the identity of at least the major fatty acids making up such a species in plasma may be reasonably inferred. Relative lipid amounts were calculated by relating the peak area of each species to the peak area of the corresponding stable isotope or non-physiological internal standard. Total lipid classes were calculated from the sum of the individual lipid species within each class (39).
Quantitative Genetic Analyses
All genetic analyses were performed in SOLAR (39). SOLAR implements maximum-likelihood variance decomposition to determine the contributions of genetic and environmental influences to a trait by modeling the covariance among family members as a function of expected allele sharing given the pedigree (see (40) for a detailed description of the variance components methods). The genetic analysis was done at the class levels rather than at the species level in the first instance. We did this because regulation of the lipid metabolic pathway occurs at the class level, and within each class regulation occurs at the level of the fatty acid. Thus by focusing at the class level we hoped to constrain the search space of the lipidome to a set of species and fatty acids in which we could search for associations with MDD. The genetic analyses at the class level were conducted in two steps.
First, univariate polygenic analysis was applied to the individual lipid class sum scores and the MDD index, as part of this step all traits were converted to ranks and were normalized using an inverse Gaussian transformation in addition to being residualized for relevant covariates. Age, age2, sex and their interactions were included as covariates for all traits while some additional covariates were included only for either the lipid classes or for MDD. For the lipid classes, some combination of the following metabolic covariates, collected at the time of blood sampling as part of the SAFS assessment (41), were included: BMI; antilipid (statin) medication; diabetes status; heart attack; heart surgery; smoking status; hypertension status. Inclusion of the metabolic covariates was dependent on the significance of the covariate with the lipid class in question, a liberal threshold of p < 0.10 was applied in order to increase confidence that important covariates were included. For MDD we included any alcohol and any substance use disorder.
Second, bivariate polygenic analysis was applied to each residualized lipid class sum score combined with the residualized MDD index, wherein the phenotypic covariance between the lipid score and MDD was decomposed into its genetic and environmental constituents to determine the extent to which they were influenced by shared genetic effects. Parameter estimates from the bivariate analyses were used to calculate ERVs for each MDD/lipid class pairing.
ERV Calculation
The ERV statistic has been described in detail elsewhere (40), but briefly the ERV for the ith lipid class and MDD is given by:
Where hi2 denotes the heritability of the ith lipid class, hMDD2 denotes the heritability of the MDD index, and ρg denotes the genetic correlation between the two traits. The ERV is simply an effect size bounded between zero and one, it is useful for prioritizing phenotypes in terms of their shared genetic overlap with a disease of interest. In the present study we ranked lipid classes by their genetic overlap with MDD. After ranking was performed, we tested the statistical significance of the genetic correlation between the top-ranked class and MDD. This approach involved only one null-hypothesis significance test, because we did not test (and indeed, it was never our intention to test) whether each lipid class was associated with MDD or not. Instead, we treated this as a parameter-estimation problem, with the ERV associated with each lipid class as the parameters of interest.
Cluster Analysis of Top-Ranked Lipid Class
This set of analysis was done using the lipid species encapsulated by the top-ranked lipid classes revealed by the above analysis step. This species-level analysis was done to more finely investigate the genetic overlap of the top-ranked lipid class and MDD. In order to do this we first applied bivariate polygenic models to all pairs of lipid species and then, using the genetic correlations estimated from these models, created a genetic correlation matrix of all species. Next we applied hierarchical cluster analysis, as implemented in R (42) to the genetic correlation matrix in order to establish clusters of genetically related species. In more detail, the genetic correlation matrix was converted into a matrix of dissimilarity scores by subtracting the absolute value of each correlation from 1. Agglomerative clustering was then applied to this matrix of distance scores. This method of clustering begins with n clusters where each cluster represents a single item then, at each step, two clusters are fused together in accordance with the distance values. This analysis was interpreted using a dendrogram plot where similar traits are on the same limb of the tree and distinctly different traits are placed on other limbs (43, 44). Scores for the resultant clusters were derived using principal components analysis where, for each of the clusters all lipid classes were entered into PCA and the first unrotated principal component scored was extracted for bivariate polygenic analysis with MDD.
Assignment of fatty acids to phosphatidylcholine species
Fatty acid assignments were performed on the quality control pooled plasma sample (n=6 healthy volunteers) used during the lipid analysis for this cohort. The pooled plasma samples were extracted in the same conditions replacing 10mM ammonium formate with 200μM lithium acetate in the process. Assignments were made based on the fragmentation patterns of the lithium adducts as described by Hsu et al (45) using the same chromatography system as described but with 200μM lithium acetate instead of 10mM ammonium formate. Scheduled MRMs for the possible fatty acid specific fragments for each phosphatidylcholine species were used over several injections, resulting in qualitative data of possible combinations of fatty acids for each of the species. PC(O) species of very low abundance were not able to be characterized using this approach. Throughout the present manuscript we follow the naming convention of lipid classes and species outlined by the LIPID MAPS consortium (46).
Results
Heritability of MDD
As has been previously reported the dimensional scale of MDD was deemed to be significantly heritable (h2 = 0.20, se = 0.06, p = 2.6×10−05) (38).
ERV: Ranking of Lipid Classes by Genetic Overlap with MDD
The endophenotype ranking results are presented in Table 1, which includes a list of the metabolic covariates that were included in the analysis of each class. The top ranked lipid class was PC(O) for which ERV = 0.13 (h2 = 0.39, se = 0.06, p = 1.99×10−16). The second best ranked lipid class using ERV was the PC(P). The genetic correlation between PC(O) and PC(P), which are both phosphatidylcholine lipid classes, was high and significant (ρg = −0.75, p = 1.4×10−07), consequently we elected to sum the two and treat them as a single trait. This sum score of PC(O) and PC(P) was significantly heritable (h2 = 0.39, se = 0.06, p = 4.89×10−15), and the genetic correlation between this score and MDD was significant (ρg = −0.51, p = 0.01). Therefore the lipid classes exhibiting the greatest degree of genetic overlap with MDD were the ether-phosphatidylcholine classes PC(O) and PC(P), a sum score of which shared a significant genetic correlation with MDD.
Table 1.
Ordered Endophenotype Ranking Values (ERVs), Heritability Estimates, Genetic Correlations and Included Covariates for All Lipid Classes Tested Against MDD
| Lipid Class | h2 | ρg | ERV | Metabolic Covariates Included |
|---|---|---|---|---|
| PC(O) | 0.39 | −0.46 | 0.13 | Smoking |
| PC(P) | 0.42 | −0.40 | 0.12 | Diabetes; Smoking; BMI |
| SM | 0.30 | −0.45 | 0.11 | BMI |
| COH | 0.38 | −0.39 | 0.11 | BMI |
| DHC | 0.25 | −0.43 | 0.10 | None |
| MHC | 0.49 | −0.26 | 0.09 | None |
| PC | 0.54 | −0.25 | 0.08 | BMI |
| PE(P) | 0.28 | −0.34 | 0.08 | Smoking |
| LPC(O) | 0.38 | −0.28 | 0.08 | BMI |
| THC | 0.51 | −0.21 | 0.07 | BMI |
| dhCer | 0.49 | −0.21 | 0.07 | Diabetes; BMI |
| Cer | 0.30 | 0.20 | 0.05 | Diabetes |
| PS | 0.38 | −0.18 | 0.05 | None |
| GM | 0.35 | −0.17 | 0.05 | Diabetes; BMI |
| PE(O) | 0.37 | −0.14 | 0.04 | None |
| LPC | 0.34 | −0.13 | 0.04 | Diabetes; BMI |
| CE | 0.32 | −0.12 | 0.03 | Diabetes; BMI |
| LPE | 0.34 | −0.09 | 0.03 | Diabetes; BMI |
| DG | 0.31 | −0.09 | 0.02 | Diabetes; BMI |
| TG | 0.41 | 0.05 | 0.01 | Diabetes; BMI |
| PI | 0.40 | 0.04 | 0.01 | Diabetes; BMI |
| PE | 0.34 | −0.03 | 0.01 | Diabetes; BMI |
| PG | 0.40 | 0.01 | 0.00 | Smoking |
Clustering of PC(O) and PC(P) Lipid Species
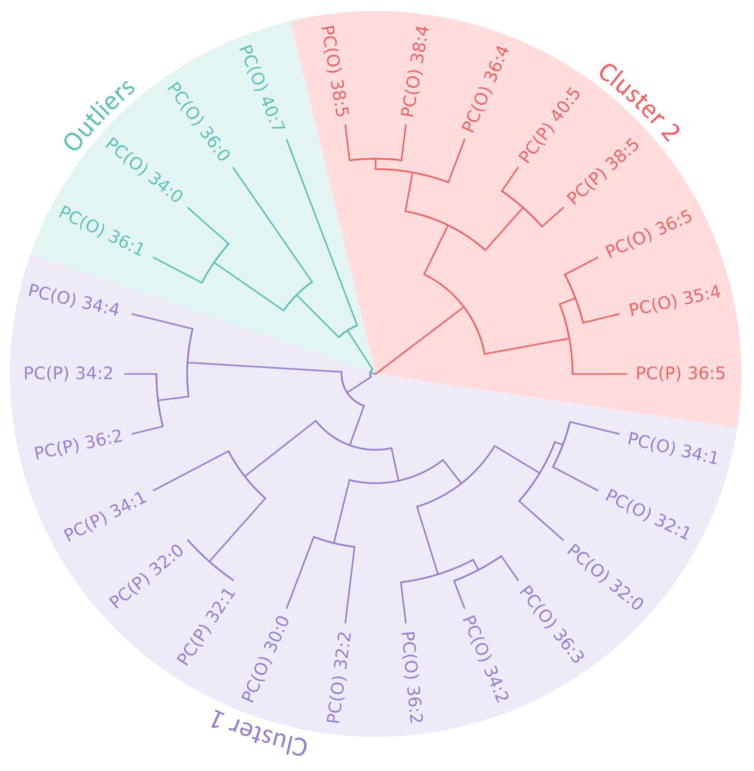
In order to identify clusters of genetically related species we applied cluster analysis to the genetic correlation matrix of all ether-phosphatidylcholine species in our top-ranked lipid classes, PC(O) and PC(P), for MDD. Figure 1 shows the results of the hierarchical cluster analysis applied to the genetic correlation matrix of all lipid species within the PC(O) and PC(P) classes. This analysis revealed three primary clusters, one of which is primarily characterized by those PC(O) and PC(P) species with a relatively lower number of carbon atoms and double bonds, shown in purple, Cluster 1. Cluster 2, shown in orange, encapsulates those species with a relatively higher number of carbon atoms and double bonds. Finally, PC(O-40:7), PC(O-36:0), PC(O) 34:0 and PC(O) 36:1 were deemed to be outliers, belonging to neither cluster, given their position on a separate branch of the dendrogram.
Figure 1.
Dendrogram of the cluster analysis of all lipid species contained in the PC(O) and PC(P) classes. Two main clusters emerged: Cluster 1 (purple), and Cluster 2 (orange), plus two outliers (green).
Genetic Overlap Between Clusters 1 and 2 and MDD
Both Clusters 1 and 2 were shown to be significantly heritable (Cluster 1: h2 = 0.3684, se = 0.06, p = 4.68×10−15; Cluster 2: h2 = 0.3965, se = 0.06, p =2.16×10−15). In order to determine which species might be driving the relationship between MDD and the PC(O) and PC(P) lipid classes we applied bivariate polygenic analysis. This analysis revealed that only Cluster 2 (ρg = −0.4852, p = 0.01) shared significant genetic overlap with MDD, while the overlap with Cluster 1 was not significant (ρg = −0.3015, p = 0.1035).
Fatty Acid Assignment of Phosphatidylcholine Species in Cluster 2
Given that Cluster 2 exhibited a significant genetic overlap with MDD we performed fatty acid assignments for all ether-phosphatidylcholine species in the cluster. In general, phosphatidylcholine species consist of 3 different classes; diacyl, alkyl and alkenyl, with ether-lipids consisting of the latter two. Our initial experiments only determined the total chain length of the phospholipid (i.e. the sum of carbons and double bonds) as is represented in Figure 1. The subsequent reanalysis of a pooled plasma sample allowed us to determine the acyl/alkyl and alkenyl chains and their relative abundance. The majority of the species observed in Cluster 2 contained either a 20:4 (eicosatetraenoic acid; ETA), a 22:5 (docosapentaenoic acid; DHA) or a 20:5 (eicosapentaenoic acid; EPA) as their sn2 side chain with as 16:0, 18:0 or 18:1 alkyl/alkenyl chain in the sn1 position (Table 2). It is well established that in humans ETA (20:4) exists mainly as the omega-6 fatty acid (or, arachidonic acid) while EPA (20:5) is an omega-3 fatty acid. DHA (22:5) however exists as both forms, with the majority existing as an omega-3 (47). This means that Cluster 2 represents alkyl- and alkenyl phosphatidylcholine species containing omega 6 and omega 3 fatty acids in the sn2 position. While arachidonic acid is represented by only three of the eight species in the cluster, in terms of lipid concentration these species represent approximately 75% of the total lipids within this cluster. Therefore, Cluster 2 is mostly characterized by those ether-phosphatidylcholine species containing arachidonic acid.
Table 2.
Fatty acid assignments of the phosphatidylcholine species in cluster 2
| Class | Species | Assignment | Fatty Acid |
|---|---|---|---|
| PC(O) | 36:4 | 20:4 | arachidonic acid |
| PC(O) | 38:4 | ||
| PC(O) | 38:5 | >70% 20:4 | |
| PC(P) | 38:5 | Mix (20:4/22:5/20:5) | arachidonic/docosapentaenoic/eicosapentaenoic acid |
| PC(P) | 40:5 | 22:6 | docosahexaenoic acid |
| PC(P) | 36:5 | 20:5 | eicosapentaenoic acid |
| PC(O) | 35:4 | likely 20:41 | arachidonic acid |
| PC(O) | 36:5 | likely 20:51 | eicosapentaenoic acid |
No product ions observed in mass spectra due to low abundance.
Discussion
The aims of the present study were to provide evidence for shared genetic overlap between lipidomic concentrations and MDD, and to determine which lipid classes, and species, in particular, might be most informative when attempting to isolate potential biomarkers for MDD. Numerous studies have highlighted an association between MDD and the lipidome (32, 34, 35), indeed it has been previously shown that reductions phosphatidylcholine (and sphingomyelin) concentrations are associated with symptoms of depression (33). However, previous research has not shown whether the lipid alterations observed in MDD are secondary to the manifestation of the illness or its treatment, or whether lipid concentrations are related to the genetic predisposition for depression. Therefore, the present study extends those findings by showing that: (1) the majority of lipid classes share at least some degree of genetic overlap with MDD; (2) the classes exhibiting the greatest degree of genetic overlap with MDD were phospholipid classes PC(O) and PC(P) which are ether-phosphatidylcholines; and (3) of those top ranked ether-phosphatidylcholine classes the species which appeared to be driving the genetic overlap with MDD were mostly those containing arachidonic acid. These findings are intriguing because they imply that rather than alterations in phospholipids being secondary to the manifestation of MDD, they might have a shared etiology with the illness, and as such these lipids, their fatty acids, and their molecular pathways, might be fruitful candidates when looking to improve diagnostic and treatment efforts in MDD. Moreover, because the pathways underling arachidonic acid synthesis and metabolism are well characterized, the present study provides an empirically testable set of hypotheses for MDD risk, namely the utility of those genes and proteins encapsulated by arachidonic acid pathways in diagnosing and treating the illness.
Lipids fulfill a plethora of biological functions (48); they can be stored as forms of energy (as fats and oils), they play a key role in membrane structure and scaffolding, and they may actively influence metabolic traffic via roles in cellular regulation, signaling, and intracellular messaging (49–51). Lipids make up 50% of the weight of the brain, in fact, the lipid concentration of the brain is second only to adipose tissue (52). Phospholipids can be broken down into two broad categories, glycerophospholipids (which include phosphatidylcholines) and sphingolipids, both of which are critical in membrane structure (49). By virtue of their amphiphilic nature these classes of lipids are able to form a semipermeable bilayer around a cell and its contents, which consists of a hydrophobic core of fatty acid tails facing each other and the phospholipid head groups pointing outwards towards the cell surfaces (49). Thus, these lipid classes are perfectly placed to modulate signal transduction, molecular recognition processes, and the transportation of ions across the cell membrane (53). Moreover, the length of the acyl lipid tails) affects bilayer width which unsurprisingly influences properties such as ion permeability in addition to the structure and function of membrane proteins (54, 55). Thus ether-phosphatidylcholines, the class of lipids most strongly associated with MDD in the present study, have an established role in cell structure and function in the brain.
The ether-phosphatidylcholine cluster that showed the greatest degree of genetic overlap with MDD is characterized by those species that contain omega 6 and omega 3 fatty acids (arachidonic acid, EPA and DHA), but the majority of fatty acids at the sn 2 position of these contains an arachidonic acid. In addition, ERV estimates for the individual species in Cluster 2 show that the species sharing the greatest genetic overlap with MDD is one containing arachidonic acid, species PC(O) 38:4 (Table S2). Arachidonic acid is an omega-6 fatty acid that is a precursor to a number of eicosanoids (e.g. prostaglandins), which are crucial for the progression and resolution of inflammatory responses (56). Inflammation has been linked to the onset of many diseases including, for example, diabetes (57), heart disease (58) and cancer (59). A number of meta-analyses have implicated the role of inflammation, and specifically pro-inflammatory cytokines (e.g. IL-6, IL-1β, TNF-α, and CRP), in the etiology of MDD (60–65). Cytokines feature upstream in the immune response to phosphatidylcholines and arachidonic acid. Specifically, eicosanoid production may be triggered when a cell is activated via the release of cytokines, this in turn triggers the release of a phospholipase (e.g. cytosolic phospholipase A2; cPLA2) at the cell membrane, which liberates arachidonic acid from the cell membrane phospholipid, rendering the fatty acid available for eicosanoid production via cyclooxygenase-2 (COX-2) (Figure S1). Thus, the association of AA containing PC(O) and PC(P) species with MDD may relate to an underlying chronic inflammation as suggested by the previous literature on cytokines and depression, and potentially highlights a downstream event underlying the relationship between inflammation, cytokines and MDD, namely the release of arachidonic acid from the cell membrane and subsequent eicosanoid synthesis.
Much attention has been paid to the relationship between dietary intake of omega-3 fatty acids and depressive symptoms (65–67), and to a lesser extent with a focus specifically on omega-6 fatty acids (68). For omega-3 fatty acids the results have been largely positive, although some controversy remains regarding the clinical subgroup for which omega-3 fatty acids are most beneficial (i.e. sub-clinical versus severe) (69–71). Moreover, two meta-analyses suggest that EPA, as opposed to an alternative fatty acid DHA, which ameliorates depressive symptoms (72, 73). As highlighted by Kiecolt-Glaser and colleagues in their recent review (65) this is consistent with the greater anti-inflammatory properties of EPA. Arachidonic acid is synthesized from dietary intake of linoleic acid (Figure S1) (74). Our findings suggest that arachidonic acid shares a genetic overlap with MDD, which might seem contradictory as dietary intake of fatty acids is an environmental factor. However, there might exist a gene-environment interaction. Or, alterations in the linoleic/arachidonic acid pathway might disrupt the downstream metabolism of arachidonic acid. Such alterations might be revealed by a focussed search of genetic variation within those pathways.
The findings of the present manuscript rely on a peripheral index of lipid levels in the form of extractions performed on plasma samples. This allows us only to speculate on the ways in which these findings might be interpreted in the brain. Phosphorous-31 magnetic resonance spectroscopic (31P MRS) imaging is a method that allows non-invasive measurement of biological compounds (e.g. phospholipids) in vivo. Alterations in peripheral lipids, including in phosphatidylcholines, have been noted in psychiatric disorders other than MDD, including in schizophrenia and bipolar disorder (75, 76). Studies employing MRS techniques have documented brain-based alterations in membrane phospholipid characteristics in schizophrenia (77), bipolar disorder (78), and in MDD (79, 80). Future work might follow up on the findings in the present manuscript using similar methods.
It is possible that lipidomic abnormalities in relation to affective disorders may be characterized differently in other ethnic populations. For example, non-Hispanic populations exhibit altered lipidomic profiles and associated risk for myocardial infarcation relative to Hispanics (81). Thus it is important that the generalizability of the findings in the present manuscript should be further tested in future research.
In summary, the findings presented here highlight ether-phosphatidylcholines, and in particular those species containing arachidonic acid, as having a sizeable genetic overlap with MDD. While it has been previously demonstrated that those with MDD exhibit altered levels of phospholipids, and also arachidonic acid, this is the first study to highlight a shared genetic etiology between the two. When taken within the context of previous research demonstrating the role of phospholipids and their fatty acids (and in particular arachidonic acid) in inflammation, and the wealth of literature linking inflammation and MDD; the present study, at the very least, highlights the potential utility of ether-phosphatidylcholines and their biochemical pathways as potentially interesting avenues of research for MDD. Going further than that, the findings of the present study generate a tentative but testable hypothesis, which is that the well-characterized linoleic/arachidonic acid inflammation pathway is a potential diagnostic marker and/or treatment target for MDD.
Supplementary Material
Lipid species internal standards and mass spectrometry conditions used for plasma lipidomic analysis.
Ordered Endophenotype Ranking Values (ERVs), Heritability Estimates, Genetic Correlations and Included Covariates for All Lipid Species Included in Cluster 2.
Arachidonic acid metabolism. Arachidonic acid is synthesized from dietary intake in linoleic acid and stored in the cell membrane. It is released from the cell membrane by PLA2 rendering the fatty acid available for eicosanoid production via COX-2.
Acknowledgments
Grant sponsor: National Institute of Mental Health; Grant numbers:MH078143, MH078111, MH083824; Grant sponsor: SOLAR NIMH; Grant number: MH059490.
Footnotes
Conflicts of Interest: none.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: Review and meta-analysis. Am J Psychiatry. 2000 Oct;157(10):1552–62. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 2.Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR, et al. The epidemiology of major depressive disorder: Results from the national comorbidity survey replication (NCS-R) JAMA. 2003 Jun 18;289(23):3095–105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- 3.Greenberg PE, Kessler RC, Birnbaum HG, Leong SA, Lowe SW, Berglund PA, et al. The economic burden of depression in the united states: How did it change between 1990 and 2000? J Clin Psychiatry. 2003 Dec;64(12):1465–75. doi: 10.4088/jcp.v64n1211. [DOI] [PubMed] [Google Scholar]
- 4.Wells KB, Stewart A, Hays RD, Burnam MA, Rogers W, Daniels M, et al. The functioning and well-being of depressed patients. results from the medical outcomes study. JAMA. 1989 Aug 18;262(7):914–9. [PubMed] [Google Scholar]
- 5.Hays RD, Wells KB, Sherbourne CD, Rogers W, Spritzer K. Functioning and well-being outcomes of patients with depression compared with chronic general medical illnesses. Arch Gen Psychiatry. 1995 Jan;52(1):11–9. doi: 10.1001/archpsyc.1995.03950130011002. [DOI] [PubMed] [Google Scholar]
- 6.Depression fact sheet number 369 [Internet] 2012 []. Available from: http://www.who.int/mediacentre/factsheets/fs369/en/index.html.
- 7.Fenton WS, Hibbeln J, Knable M. Essential fatty acids, lipid membrane abnormalities, and the diagnosis and treatment of schizophrenia. Biol Psychiatry. 2000 Jan 1;47(1):8–21. doi: 10.1016/s0006-3223(99)00092-x. [DOI] [PubMed] [Google Scholar]
- 8.Ming X, Stein TP, Brimacombe M, Johnson WG, Lambert GH, Wagner GC. Increased excretion of a lipid peroxidation biomarker in autism. Prostaglandins Leukot Essent Fatty Acids. 2005 Nov;73(5):379–84. doi: 10.1016/j.plefa.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Wiest MM, German JB, Harvey DJ, Watkins SM, Hertz-Picciotto I. Plasma fatty acid profiles in autism: A case-control study. Prostaglandins Leukot Essent Fatty Acids. 2009 Apr;80(4):221–7. doi: 10.1016/j.plefa.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Stoll AL, Severus WE, Freeman MP, Rueter S, Zboyan HA, Diamond E, et al. Omega 3 fatty acids in bipolar disorder: A preliminary double-blind, placebo-controlled trial. Arch Gen Psychiatry. 1999 May;56(5):407–12. doi: 10.1001/archpsyc.56.5.407. [DOI] [PubMed] [Google Scholar]
- 11.Versace A, Andreazza AC, Young LT, Fournier JC, Almeida JR, Stiffler RS, et al. Elevated serum measures of lipid peroxidation and abnormal prefrontal white matter in euthymic bipolar adults: Toward peripheral biomarkers of bipolar disorder. Mol Psychiatry. 2014 Feb;19(2):200–8. doi: 10.1038/mp.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ranjekar PK, Hinge A, Hegde MV, Ghate M, Kale A, Sitasawad S, et al. Decreased antioxidant enzymes and membrane essential polyunsaturated fatty acids in schizophrenic and bipolar mood disorder patients. Psychiatry Res. 2003 Dec 1;121(2):109–22. doi: 10.1016/s0165-1781(03)00220-8. [DOI] [PubMed] [Google Scholar]
- 13.Parker G, Gibson NA, Brotchie H, Heruc G, Rees AM, Hadzi-Pavlovic D. Omega-3 fatty acids and mood disorders. Am J Psychiatry. 2006 Jun;163(6):969–78. doi: 10.1176/ajp.2006.163.6.969. [DOI] [PubMed] [Google Scholar]
- 14.Taylor FC, Huffman M, Ebrahim S. Statin therapy for primary prevention of cardiovascular disease. JAMA. 2013 Dec 11;310(22):2451–2. doi: 10.1001/jama.2013.281348. [DOI] [PubMed] [Google Scholar]
- 15.Morgan RE, Palinkas LA, Barrett-Connor EL, Wingard DL. Plasma cholesterol and depressive symptoms in older men. Lancet. 1993 Jan 9;341(8837):75–9. doi: 10.1016/0140-6736(93)92556-9. [DOI] [PubMed] [Google Scholar]
- 16.Muldoon MF, Manuck SB, Matthews KA. Lowering cholesterol concentrations and mortality: A quantitative review of primary prevention trials. BMJ. 1990 Aug 11;301(6747):309–14. doi: 10.1136/bmj.301.6747.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neaton JD, Blackburn H, Jacobs D, Kuller L, Lee DJ, Sherwin R, et al. Serum cholesterol level and mortality findings for men screened in the multiple risk factor intervention trial. multiple risk factor intervention trial research group. Arch Intern Med. 1992 Jul;152(7):1490–500. [PubMed] [Google Scholar]
- 18.Maes M, Smith R, Christophe A, Vandoolaeghe E, Van Gastel A, Neels H, et al. Lower serum high-density lipoprotein cholesterol (HDL-C) in major depression and in depressed men with serious suicidal attempts: Relationship with immune-inflammatory markers. Acta Psychiatr Scand. 1997 Mar;95(3):212–21. doi: 10.1111/j.1600-0447.1997.tb09622.x. [DOI] [PubMed] [Google Scholar]
- 19.Salter M. Low serum cholesterol and suicide. Lancet. 1992 May 9;339(8802):1169. [PubMed] [Google Scholar]
- 20.Yang CC, Jick SS, Jick H. Lipid-lowering drugs and the risk of depression and suicidal behavior. Arch Intern Med. 2003 Sep 8;163(16):1926–32. doi: 10.1001/archinte.163.16.1926. [DOI] [PubMed] [Google Scholar]
- 21.Kohler O, Gasse C, Petersen L, Ingstrup KG, Nierenberg AA, Mors O, et al. The effect of concomitant treatment with SSRIs and statins: A population-based study. Am J Psychiatry. 2016 Aug 1;173(8):807–15. doi: 10.1176/appi.ajp.2016.15040463. [DOI] [PubMed] [Google Scholar]
- 22.Salagre E, Fernandes BS, Dodd S, Brownstein DJ, Berk M. Statins for the treatment of depression: A meta-analysis of randomized, double-blind, placebo-controlled trials. J Affect Disord. 2016 Aug;200:235–42. doi: 10.1016/j.jad.2016.04.047. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Zhao T, Qiu Y, Su M, Jiang T, Zhou M, et al. Metabonomics approach to understanding acute and chronic stress in rat models. J Proteome Res. 2009 May;8(5):2511–8. doi: 10.1021/pr801086k. [DOI] [PubMed] [Google Scholar]
- 24.Li ZY, Zheng XY, Gao XX, Zhou YZ, Sun HF, Zhang LZ, et al. Study of plasma metabolic profiling and biomarkers of chronic unpredictable mild stress rats based on gas chromatography/mass spectrometry. Rapid Commun Mass Spectrom. 2010 Dec 30;24(24):3539–46. doi: 10.1002/rcm.4809. [DOI] [PubMed] [Google Scholar]
- 25.Zheng S, Yu M, Lu X, Huo T, Ge L, Yang J, et al. Urinary metabonomic study on biochemical changes in chronic unpredictable mild stress model of depression. Clin Chim Acta. 2010 Feb;411(3–4):204–9. doi: 10.1016/j.cca.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Zhang F, Jia Z, Gao P, Kong H, Li X, Lu X, et al. Metabonomics study of urine and plasma in depression and excess fatigue rats by ultra fast liquid chromatography coupled with ion trap-time of flight mass spectrometry. Mol Biosyst. 2010 May;6(5):852–61. doi: 10.1039/b914751a. [DOI] [PubMed] [Google Scholar]
- 27.Liu XJ, Li ZY, Li ZF, Gao XX, Zhou YZ, Sun HF, et al. Urinary metabonomic study using a CUMS rat model of depression. Magn Reson Chem. 2012 Mar;50(3):187–92. doi: 10.1002/mrc.2865. [DOI] [PubMed] [Google Scholar]
- 28.Peet M, Murphy B, Shay J, Horrobin D. Depletion of omega-3 fatty acid levels in red blood cell membranes of depressive patients. Biol Psychiatry. 1998 Mar 1;43(5):315–9. doi: 10.1016/s0006-3223(97)00206-0. [DOI] [PubMed] [Google Scholar]
- 29.Maes M, Christophe A, Delanghe J, Altamura C, Neels H, Meltzer HY. Lowered omega3 polyunsaturated fatty acids in serum phospholipids and cholesteryl esters of depressed patients. Psychiatry Res. 1999 Mar 22;85(3):275–91. doi: 10.1016/s0165-1781(99)00014-1. [DOI] [PubMed] [Google Scholar]
- 30.Logan AC. Omega-3 fatty acids and major depression: A primer for the mental health professional. Lipids Health Dis. 2004 Nov 9;3:25. doi: 10.1186/1476-511X-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lotrich FE, Sears B, McNamara RK. Elevated ratio of arachidonic acid to long-chain omega-3 fatty acids predicts depression development following interferon-alpha treatment: Relationship with interleukin-6. Brain Behav Immun. 2013 Jul;31:48–53. doi: 10.1016/j.bbi.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Reedt Dortland AK, Giltay EJ, van Veen T, van Pelt J, Zitman FG, Penninx BW. Associations between serum lipids and major depressive disorder: Results from the netherlands study of depression and anxiety (NESDA) J Clin Psychiatry. 2010 Jun;71(6):729–36. doi: 10.4088/JCP.08m04865blu. [DOI] [PubMed] [Google Scholar]
- 33.Demirkan A, Isaacs A, Ugocsai P, Liebisch G, Struchalin M, Rudan I, et al. Plasma phosphatidylcholine and sphingomyelin concentrations are associated with depression and anxiety symptoms in a dutch family-based lipidomics study. J Psychiatr Res. 2013 Mar;47(3):357–62. doi: 10.1016/j.jpsychires.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Zheng P, Zhao X, Zhang Y, Hu C, Li J, et al. Discovery and validation of plasma biomarkers for major depressive disorder classification based on liquid chromatography-mass spectrometry. J Proteome Res. 2015 May 1;14(5):2322–30. doi: 10.1021/acs.jproteome.5b00144. [DOI] [PubMed] [Google Scholar]
- 35.Oliveira TG, Chan RB, Bravo FV, Miranda A, Silva RR, Zhou B, et al. The impact of chronic stress on the rat brain lipidome. Mol Psychiatry. 2015 Mar 10; doi: 10.1038/mp.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olvera RL, Bearden CE, Velligan DI, Almasy L, Carless MA, Curran JE, et al. Common genetic influences on depression, alcohol, and substance use disorders in mexican-american families. Am J Med Genet B Neuropsychiatr Genet. 2011 Jul;156B(5):561–8. doi: 10.1002/ajmg.b.31196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, et al. The mini-international neuropsychiatric interview (M.I.N.I.): The development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59( Suppl 20):22, 33. quiz 34–57. [PubMed] [Google Scholar]
- 38.Knowles EE, Kent JW, Jr, McKay DR, Sprooten E, Mathias SR, Curran JE, et al. Genome-wide linkage on chromosome 10q26 for a dimensional scale of major depression. J Affect Disord. 2015 Nov 17;191:123–31. doi: 10.1016/j.jad.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998 May;62(5):1198–211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glahn DC, Curran JE, Winkler AM, Carless MA, Kent JW, Jr, Charlesworth JC, et al. High dimensional endophenotype ranking in the search for major depression risk genes. Biol Psychiatry. 2012 Jan 1;71(1):6–14. doi: 10.1016/j.biopsych.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitchell BD, Almasy LA, Rainwater DL, Schneider JL, Blangero J, Stern MP, et al. Diabetes and hypertension in mexican american families: Relation to cardiovascular risk. Am J Epidemiol. 1999 Jun 1;149(11):1047–56. doi: 10.1093/oxfordjournals.aje.a009750. [DOI] [PubMed] [Google Scholar]
- 42.R Development Core Team. R: A language and environment for statistical computing. 2011. [Google Scholar]
- 43.Crawley MJ. The R book. Chichester: Wiley; 2007. [Google Scholar]
- 44.Knowles EE, Carless MA, de Almeida MA, Curran JE, McKay DR, Sprooten E, et al. Genome-wide significant localization for working and spatial memory: Identifying genes for psychosis using models of cognition. Am J Med Genet B Neuropsychiatr Genet. 2014 Jan;165(1):84–95. doi: 10.1002/ajmg.b.32211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu FF, Turk J, Thukkani AK, Messner MC, Wildsmith KR, Ford DA. Characterization of alkylacyl, alk-1-enylacyl and lyso subclasses of glycerophosphocholine by tandem quadrupole mass spectrometry with electrospray ionization. J Mass Spectrom. 2003 Jul;38(7):752–63. doi: 10.1002/jms.491. [DOI] [PubMed] [Google Scholar]
- 46.Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, et al. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009 Apr;50( Suppl):S9–14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abdelmagid SA, Clarke SE, Nielsen DE, Badawi A, El-Sohemy A, Mutch DM. Comprehensive profiling of plasma fatty acid concentrations in young healthy canadian adults. PloS one. 2015;10(2) doi: 10.1371/journal.pone.0116195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramaniam S, Fahy E, Gupta S, Sud M, Byrnes RW, Cotter D, et al. Bioinformatics and systems biology of the lipidome. Chem Rev. 2011 Oct 12;111(10):6452–90. doi: 10.1021/cr200295k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. 6. W. H. Freeman; 2012. Lipids; pp. 343–68. [Google Scholar]
- 50.Brown HA, Murphy RC. Working towards an exegesis for lipids in biology. Nat Chem Biol. 2009 Sep;5(9):602–6. doi: 10.1038/nchembio0909-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Regehr WG, Carey MR, Best AR. Activity-dependent regulation of synapses by retrograde messengers. Neuron. 2009 Jul 30;63(2):154–70. doi: 10.1016/j.neuron.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watkins PA, Hamilton JA, Leaf A, Spector AA, Moore SA, Anderson RE, et al. Brain uptake and utilization of fatty acids: Applications to peroxisomal biogenesis diseases. J Mol Neurosci. 2001 Apr-Jun;16(2–3):87, 92. doi: 10.1385/JMN:16:2-3:87. discussion 151–7. [DOI] [PubMed] [Google Scholar]
- 53.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol. 2008 Feb;9(2):112–24. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewis BA, Engelman DM. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J Mol Biol. 1983 May 15;166(2):211–7. doi: 10.1016/s0022-2836(83)80007-2. [DOI] [PubMed] [Google Scholar]
- 55.Tillman TS, Cascio M. Effects of membrane lipids on ion channel structure and function. Cell Biochem Biophys. 2003;38(2):161–90. doi: 10.1385/CBB:38:2:161. [DOI] [PubMed] [Google Scholar]
- 56.Lawrence T, Willoughby DA, Gilroy DW. Anti-inflammatory lipid mediators and insights into the resolution of inflammation. Nat Rev Immunol. 2002 Oct;2(10):787–95. doi: 10.1038/nri915. [DOI] [PubMed] [Google Scholar]
- 57.Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005 May;115(5):1111–9. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pearson TA, Mensah GA, Alexander RW, Anderson JL, Cannon RO, 3rd, Criqui M, et al. Markers of inflammation and cardiovascular disease: Application to clinical and public health practice: A statement for healthcare professionals from the centers for disease control and prevention and the american heart association. Circulation. 2003 Jan 28;107(3):499–511. doi: 10.1161/01.cir.0000052939.59093.45. [DOI] [PubMed] [Google Scholar]
- 59.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002 Dec 19–26;420(6917):860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller AH, Maletic V, Raison CL. Inflammation and its discontents: The role of cytokines in the pathophysiology of major depression. Biol Psychiatry. 2009 May 1;65(9):732–41. doi: 10.1016/j.biopsych.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raison CL, Capuron L, Miller AH. Cytokines sing the blues: Inflammation and the pathogenesis of depression. Trends Immunol. 2006 Jan;27(1):24–31. doi: 10.1016/j.it.2005.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Howren MB, Lamkin DM, Suls J. Associations of depression with C-reactive protein, IL-1, and IL-6: A meta-analysis. Psychosom Med. 2009 Feb;71(2):171–86. doi: 10.1097/PSY.0b013e3181907c1b. [DOI] [PubMed] [Google Scholar]
- 63.Dowlati Y, Herrmann N, Swardfager W, Liu H, Sham L, Reim EK, et al. A meta-analysis of cytokines in major depression. Biol Psychiatry. 2010 Mar 1;67(5):446–57. doi: 10.1016/j.biopsych.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 64.Liu Y, Ho RC, Mak A. Interleukin (IL)-6, tumour necrosis factor alpha (TNF-alpha) and soluble interleukin-2 receptors (sIL-2R) are elevated in patients with major depressive disorder: A meta-analysis and meta-regression. J Affect Disord. 2012 Aug;139(3):230–9. doi: 10.1016/j.jad.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 65.Kiecolt-Glaser JK, Derry HM, Fagundes CP. Inflammation: Depression fans the flames and feasts on the heat. Am J Psychiatry. 2015 Nov 1;172(11):1075–91. doi: 10.1176/appi.ajp.2015.15020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giles GE, Mahoney CR, Kanarek RB. Omega-3 fatty acids influence mood in healthy and depressed individuals. Nutr Rev. 2013 Nov;71(11):727–41. doi: 10.1111/nure.12066. [DOI] [PubMed] [Google Scholar]
- 67.Lin PY, Huang SY, Su KP. A meta-analytic review of polyunsaturated fatty acid compositions in patients with depression. Biol Psychiatry. 2010 Jul 15;68(2):140–7. doi: 10.1016/j.biopsych.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 68.Kiecolt-Glaser JK, Belury MA, Porter K, Beversdorf DQ, Lemeshow S, Glaser R. Depressive symptoms, omega-6:Omega-3 fatty acids, and inflammation in older adults. Psychosom Med. 2007 Apr;69(3):217–24. doi: 10.1097/PSY.0b013e3180313a45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grosso G, Pajak A, Marventano S, Castellano S, Galvano F, Bucolo C, et al. Role of omega-3 fatty acids in the treatment of depressive disorders: A comprehensive meta-analysis of randomized clinical trials. PLoS One. 2014 May 7;9(5):e96905. doi: 10.1371/journal.pone.0096905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bloch MH, Hannestad J. Omega-3 fatty acids for the treatment of depression: Systematic review and meta-analysis. Mol Psychiatry. 2012 Dec;17(12):1272–82. doi: 10.1038/mp.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Appleton KM, Rogers PJ, Ness AR. Updated systematic review and meta-analysis of the effects of n-3 long-chain polyunsaturated fatty acids on depressed mood. Am J Clin Nutr. 2010 Mar;91(3):757–70. doi: 10.3945/ajcn.2009.28313. [DOI] [PubMed] [Google Scholar]
- 72.Sublette ME, Ellis SP, Geant AL, Mann JJ. Meta-analysis of the effects of eicosapentaenoic acid (EPA) in clinical trials in depression. J Clin Psychiatry. 2011 Dec;72(12):1577–84. doi: 10.4088/JCP.10m06634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martins JG. EPA but not DHA appears to be responsible for the efficacy of omega-3 long chain polyunsaturated fatty acid supplementation in depression: Evidence from a meta-analysis of randomized controlled trials. J Am Coll Nutr. 2009 Oct;28(5):525–42. doi: 10.1080/07315724.2009.10719785. [DOI] [PubMed] [Google Scholar]
- 74.Marcel YL, Christiansen K, Holman RT. The preferred metabolic pathway from linoleic acid to arachidonic acid in vitro. Biochim Biophys Acta. 1968 Sep 2;164(1):25–34. doi: 10.1016/0005-2760(68)90067-2. [DOI] [PubMed] [Google Scholar]
- 75.Kraguljac NV, Reid M, White D, Jones R, den Hollander J, Lowman D, et al. Neurometabolites in schizophrenia and bipolar disorder - a systematic review and meta-analysis. Psychiatry Res. 2012 Aug-Sep;203(2–3):111–25. doi: 10.1016/j.pscychresns.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schneider M, Levant B, Reichel M, Gulbins E, Kornhuber J, Muller CP. Lipids in psychiatric disorders and preventive medicine. Neurosci Biobehav Rev. 2016 Jun 16; doi: 10.1016/j.neubiorev.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 77.Milev P, Miranowski S, Lim KO. Magnetic resonance spectroscopy: 31Phosphorous magnetic resonance spectroscopy (31P MRS) In: Lajtha A, Javitt DC, Kantrowitz J, editors. Handbook of Neurochemistry. 3. Springer US; 2009. pp. 425–30. [Google Scholar]
- 78.Strakowski SM, Delbello MP, Adler CM. The functional neuroanatomy of bipolar disorder: A review of neuroimaging findings. Mol Psychiatry. 2005 Jan;10(1):105–16. doi: 10.1038/sj.mp.4001585. [DOI] [PubMed] [Google Scholar]
- 79.Yildiz-Yesiloglu A, Ankerst DP. Review of 1H magnetic resonance spectroscopy findings in major depressive disorder: A meta-analysis. Psychiatry Res. 2006 Jun 30;147(1):1–25. doi: 10.1016/j.pscychresns.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 80.Maddock RJ, Buonocore MH. MR spectroscopic studies of the brain in psychiatric disorders. Curr Top Behav Neurosci. 2012;11:199–251. doi: 10.1007/7854_2011_197. [DOI] [PubMed] [Google Scholar]
- 81.Willey JZ, Rodriguez CJ, Carlino RF, Moon YP, Paik MC, Boden-Albala B, et al. Race-ethnic differences in the association between lipid profile components and risk of myocardial infarction: The northern manhattan study. Am Heart J. 2011 May;161(5):886–92. doi: 10.1016/j.ahj.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lipid species internal standards and mass spectrometry conditions used for plasma lipidomic analysis.
Ordered Endophenotype Ranking Values (ERVs), Heritability Estimates, Genetic Correlations and Included Covariates for All Lipid Species Included in Cluster 2.
Arachidonic acid metabolism. Arachidonic acid is synthesized from dietary intake in linoleic acid and stored in the cell membrane. It is released from the cell membrane by PLA2 rendering the fatty acid available for eicosanoid production via COX-2.