Abstract
Shiga toxin–producing Escherichia coli (STEC) causes an estimated 265 000 infections in the United States annually. Of emerging non-O157:H7 STEC serotypes, O26 is the most commonly recognized. During an outbreak of STEC O26 in Oregon in 2015, we used syndromic surveillance data to supplement case finding by laboratory reporting. From 157 records retrieved by querying syndromic surveillance data, we detected 4 confirmed and 5 suspected cases. However, none of the suspected cases were confirmed by stool culture, and by the time that the data were being analyzed, the confirmed cases were already known to investigators. Syndromic surveillance data can potentially supplement case finding during outbreaks of foodborne disease. To be an effective case-finding strategy, timely completion of all steps, including collecting specimens from suspected cases, should be performed in real time.
Keywords: Shiga toxin–producing Escherichia coli, syndromic surveillance, disease outbreaks, foodborne diseases, epidemiologic methods
Escherichia coli subtypes that produce Shiga toxin can cause severe gastroenteritis1 and are responsible for an estimated 265 000 infections in the United States annually.2 The incubation period is 1 to 10 days, and illness is classically characterized by bloody diarrhea. The most common Shiga toxin–producing E coli (STEC) serotype in the United States is O157:H7; however, of the emerging non-O157:H7 STEC serotypes, O26 is the most commonly recognized.3,4
During October-December 2015, a strain of STEC O26 caused an outbreak of infections across the United States.5 The outbreak was detected in late October by public health laboratories in Oregon and Washington State when they noted an increase in STEC isolates submitted for pulsed-field gel electrophoresis (PFGE) testing by clinical laboratories. A total of 55 STEC O26 isolates with a PFGE pattern indistinguishable from that of the outbreak strain (ie, confirmed cases) were reported to local health departments in 11 states, including 13 confirmed cases in Oregon.5,6 Although patient interviews by local health departments implicated a restaurant chain as the source of infections, investigators never identified a specific vehicle of transmission.
The primary objective of this study was to evaluate if emergency department (ED) syndromic surveillance data could be used for case finding during a STEC O26 outbreak in Oregon. A secondary objective was to compare our approach with the traditional approach of reviewing clinical laboratory reports.
Methods
The Oregon Public Health Division collects excerpts daily of ED provider notes taken during ED visits in all 60 nonfederal hospitals in the state and stores these syndromic surveillance data in the Electronic Surveillance System for the Early Notification of Community-Based Epidemics (ESSENCE).7 On October 31, 2015, 2 days after the outbreak was detected, we queried ESSENCE fields for chief complaint and clinical impression to identify people who complained of “bloody diarrhea” or “bloody stool” in the Portland metropolitan area (where all known Oregon patients had dined) from October 7 (the date indicating the longest incubation period before symptom onset—October 17—in the earliest known suspected case at that time) through October 30 (the last day of complete data). We also tested the addition of “abdominal pain” to the query. Because ESSENCE contains medical record numbers, we were able to match retrieved ESSENCE records to the medical records from which they were excerpted. In accordance with ESSENCE data use agreements, we requested permission from health systems with patients identified by the ESSENCE query before searching their medical records system.
We defined a confirmed case as a STEC O26 infection with an isolate having a PFGE pattern indistinguishable from the outbreak strain PFGE pattern in an Oregonian from October 7 to October 30, 2015. We defined a suspected case as acute-onset diarrhea (ie, sudden onset of ≥3 loose stools per day for <2 weeks duration) in an Oregonian who had dined at the restaurant chain during the incubation period. (Of people with non–outbreak-related STEC cases in Oregon from 2013 to 2016, 4 of 207 [2%] reported having eaten at the implicated restaurant chain.) We reviewed patients’ medical charts and attempted to interview those whose charts listed acute-onset diarrhea to find out if they had dined at the implicated restaurant chain during their incubation periods. If they had, we collected additional food history using an outbreak-specific questionnaire and encouraged them to submit a stool sample to their health care provider or local health department. We did not interview people whose stool culture results were negative, because we considered them to be noncases; because of the delay in obtaining approval to link ESSENCE records with medical records and the time taken to review medical charts, people with a STEC-positive culture had already been interviewed by local health departments.
We performed additional ESSENCE queries to identify all people with confirmed cases who sought care in an ED (because ESSENCE data are limited to ED visits). We then compared the ESSENCE records of the subset of people with confirmed cases that were retrieved by the initial ESSENCE query (ie, “bloody diarrhea” or “bloody stool”) with the ESSENCE records of all people with confirmed cases who sought care in an ED. For confirmed cases detected by the query, we compared the date that clinical laboratories reported the infection to local health departments with (1) October 31, the date that we performed the initial query (actual timeliness), and (2) the date that the patient visited the ED and might have been detected by the case-finding strategy through syndromic surveillance data (hypothetical timeliness). This project was reviewed by the Centers for Disease Control and Prevention for human subjects protection and deemed to be nonresearch.
Results
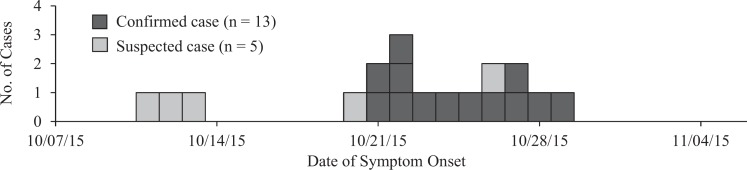
The ESSENCE query identified 157 people who reported bloody diarrhea or bloody stool in Portland metropolitan-area EDs from October 7 to October 30, 2015. Of the 157 medical charts, we reviewed 113 (72%); the others were unavailable. Of the 113 available charts, 52 (46%) contained reports of acute-onset diarrhea symptoms. Of these 52, 15 had stool cultures performed: 4 (8%) were STEC positive, and 11 (21%) were STEC negative. From the remaining 37 charts, we interviewed 28 (76%) people, of whom 5 (18%) reported dining at the restaurant chain (ie, were suspect cases; Figure); none submitted stool samples after interview. Obtaining approval to link syndromic surveillance data with medical records took 11 to 16 days. Reviewing medical charts took 3 days. We spent approximately 30 hours on this project, primarily reviewing medical charts.
Figure.
Confirmed and suspected cases from an outbreak of Shiga toxin–producing Escherichia coli O26, Oregon, 2015. A confirmed case was defined as a Shiga toxin–producing E coli O26 infection with an isolate having a pulsed-field gel electrophoresis pattern indistinguishable from the outbreak strain pattern in an Oregonian from October 7 to October 30, 2015. A suspected case was defined as acute-onset diarrhea (ie, sudden onset of ≥3 loose stools per day for <2 weeks) in an Oregonian who had dined at the restaurant chain during the incubation period.
Of 13 people with confirmed cases in Oregon, 11 sought care at a Portland-area ED. The query detected 4 of these, for a sensitivity of 36%. Adding the complaint “abdominal pain” to the ESSENCE query would have detected 5 additional confirmed cases but would have added 10 093 medical records to review. Had we followed up with patients in real time, 2 of these infections would have been detected 1 or 2 days earlier (actual timeliness) by ESSENCE than by reviewing clinical laboratory reports; all 4 could have been detected 3 or 4 days earlier (hypothetical timeliness) if the ESSENCE query had been performed the day after patients presented to the ED.
Discussion
During a STEC O26 outbreak in Oregon, we detected 4 of 13 confirmed and 5 suspected cases by using ED syndromic surveillance data. However, none of the suspected cases were confirmed by stool culture, and the confirmed cases were already known to investigators by the time that the data were being analyzed. Had stool cultures been performed on the suspected cases (and if results were positive), using syndromic surveillance data for case finding could have increased the confirmed case count in Oregon by more than one-third. This strategy has been reported elsewhere8–12; however, our study is only the second to report case finding with syndromic surveillance data during a foodborne outbreak.10
Sensitivity for detecting confirmed cases was low, but increasing sensitivity would have necessitated the review of thousands of additional charts, rendering this strategy untenable. The improvement in actual timeliness was small, but examining ED syndromic surveillance data frequently during an outbreak and promptly following up on suspected cases could improve the timeliness of case detection.
This study had several limitations. First, we did not immediately contact people who were identified through syndromic surveillance data, because we needed health system permission before linking data sources and because reviewing charts was time-consuming. Therefore, by the time that we interviewed people identified by the syndromic surveillance data, those with STEC-positive stool cultures had already been identified by clinical laboratories reporting to local health departments, and suspected cases were unlikely to have positive stool culture results. Although 3 people with suspected cases dined at the implicated restaurant >1 week before the first confirmed case, given the low prevalence of dining at the restaurant among people with non–outbreak-associated STEC cases, we believe that these cases were related to this outbreak. Additionally, we tested our strategy on a single foodborne outbreak; as such, our results may not be generalizable to other outbreaks.
Syndromic surveillance data can potentially supplement case finding during foodborne outbreaks. Situations in which they might prove particularly useful include outbreaks in which identifying all cases is imperative (eg, Ebola), for a disease with pathognomonic symptoms (eg, acute viral hepatitis), or when information on nonreportable diseases is sought. We offer 3 recommendations to increase the feasibility of this strategy. First, when using syndromic surveillance data for case finding, we recommend designing a query that strikes a balance between sensitivity and work burden. Second, because chart review is time intensive, we recommend using charts only to obtain telephone numbers and then interviewing all identified patients; a data use agreement that a priori permits linkage to medical records for outbreak investigations can facilitate this work. Third, to be an effective case-finding strategy, all steps, including collecting specimens, should be performed in real time.
Footnotes
Authors’ Note: The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the Oregon Health Authority.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Smith JL, Fratamico PM, Gunther NW., IV Shiga toxin-producing Escherichia coli. Adv Appl Microbiol. 2014;86:145–197. [DOI] [PubMed] [Google Scholar]
- 2. Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne illness acquired in the United Sates—major pathogens. Emerg Infect Dis. 2011;17(1):7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bielaszewska M, Mellmann A, Bletz S, et al. Enterohemorrhagic Escherichia coli O26: H11/H-: a new virulent clone emerges in Europe. Clin Infect Dis. 2013;56(10):1373–1381. [DOI] [PubMed] [Google Scholar]
- 4. Mingle LA, Garcia DL, Root TP, et al. Enhanced identification and characterization of non-O157 Shiga toxin-producing Escherichia coli: a six-year study. Foodborne Pathog Dis. 2012;9(11):1028–1036. [DOI] [PubMed] [Google Scholar]
- 5. Centers for Disease Control and Prevention. Multistate outbreaks of Shiga toxin-producing Escherichia coli O26 infections linked to Chipotle Mexican Grill restaurants (final update). http://www.cdc.gov/ecoli/2015/o26-11-15. Published 2016. Accessed February 16, 2017.
- 6. Oregon Public Health Division. Outbreak 2015-3740: Shiga-toxin-producing Escherichia coli O26 infections. https://public.health.oregon.gov/DiseasesConditions/CommunicableDisease/Outbreaks/Documents/outbreak_reports/2015/Outbreak-Report-2015-3740.pdf. Published 2016. Accessed February 16, 2017. [DOI] [PMC free article] [PubMed]
- 7. Lombardo JS, Burkom H, Pavlin J. ESSENCE II and the framework for evaluating syndromic surveillance systems. MMWR Suppl. 2004;53:159–165. [PubMed] [Google Scholar]
- 8. Schwarz ES, Wax PM, Kleinschmidt KC, et al. Multiple poisonings with sodium azide at a local restaurant. J Emerg Med. 2014;46(4):491–494. [DOI] [PubMed] [Google Scholar]
- 9. Healy JM, Burgess MC, Chen TH, et al. Notes from the field: outbreak of Zika virus disease—America Samoa, 2016. MMWR Morb Mortal Wkly Rep. 2016;65(41):1146–1147. [DOI] [PubMed] [Google Scholar]
- 10. Klekamp BG, Bodager D, Matthews SD. Use of surveillance systems in detection of a ciguatera fish poisoning outbreak—Orange County, Florida, 2014. MMWR Morb Mortal Wkly Rep. 2015;64(40):1142–1144. [DOI] [PubMed] [Google Scholar]
- 11. Kotzen M, Sell J, Mathes RW, et al. Using syndromic surveillance to investigate tattoo-related skin infections in New York City. PLoS One. 2015;10(6):e0130468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Atrubin D, Hamilton J, Culpepper A, Mulay PR. Utilizing Florida’s syndromic surveillance system for active case finding to support the Zika virus response. Presented at: Counsel of State and Territorial Epidemiologists Annual Conference; June 19-23, 2016; Anchorage, AK https://cste.confex.com/cste/2016/webprogram/Paper7353.html. Accessed February 22, 2017. [Google Scholar]