Abstract
Background
Leptospirosis is a highly endemic bacterial zoonosis in French Polynesia (FP). Nevertheless, data on the epidemiology of leptospirosis in FP are scarce. We conducted molecular studies on Leptospira isolated from humans and the potential main animal reservoirs in order to identify the most likely sources for human infection.
Methodology/Principal findings
Wild rats (n = 113), farm pigs (n = 181) and domestic dogs (n = 4) were screened for Leptospira infection in Tahiti, the most populated island in FP. Positive samples were genotyped and compared to Leptospira isolated from human cases throughout FP (n = 51), using secY, 16S and LipL32 sequencing, and MLST analysis. Leptospira DNA was detected in 20.4% of rats and 26.5% of pigs. We identified two Leptospira species and three sequence types (STs) in animals and humans: Leptospira interrogans ST140 in pigs only and L. interrogans ST17 and Leptospira borgpetersenii ST149 in humans and rats. Overall, L. interrogans was the dominant species and grouped into four clades: one clade including a human case only, two clades including human cases and dogs, and one clade including human cases and rats. All except one pig sample showed a unique L. interrogans (secY) genotype distinct from those isolated from humans, rats and dogs. Moreover, LipL32 sequencing allowed the detection of an additional Leptospira genotype in pigs, clearly distinct from the previous ones.
Conclusions/Significance
Our data confirm rats as a major potential source for human leptospirosis in FP. By contrast to what was expected, farm pigs did not seem to be a major reservoir for the Leptospira genotypes identified in human patients. Thus, further investigations will be required to determine their significance in leptospirosis transmission in FP.
Author summary
Leptospirosis, a zoonosis caused by Leptospira spp. bacteria, is an important but neglected disease in the Pacific region. The bacteria can lead to human infections, either through direct contact with infected animals or through water and soil contaminated with urine. Although animals play a key role in the transmission of the disease, the relative importance of animal reservoirs for human leptospirosis infections in the Pacific have not been investigated. In French Polynesia, the animal species acting as sources for human infection are unknown. In our study, we isolated leptospires from rats, farm pigs and pet dogs from Tahiti, the most populated island of French Polynesia, and compared them to those isolated from human leptospirosis cases. The types of leptospires identified in human leptospirosis cases were also detected in rats and dogs, whereas leptospires found in pigs were genetically different. These results indicate that rats are a major potential source for human leptospirosis in Tahiti. Further investigations will be required to clarify the relative contribution of dogs and pigs to human leptospirosis in French Polynesia, to properly design or adapt interventions to reduce the burden of human leptospirosis.
Introduction
Leptospirosis is a disease caused by leptospires, bacteria belonging to the order Spirochaetales, family Leptospiraceae, genus Leptospira [1]. Leptospirosis is the most widespread zoonosis worldwide [2], but a neglected disease in most of the tropics, especially in the Pacific region [3]. Incidence of leptospirosis range from 0.1 to 1 cases per 100,000 inhabitants per year in temperate climates, 10 to 100 cases per 100,000 in the humid tropics, to over 100 cases per 100,000 in high risk groups and during outbreaks [4,5]. The clinical spectrum of human infections ranges from mild flu-like illness to severe or even fatal outcome. The genus Leptospira is divided into 22 species classified into saprophytic, intermediate and pathogenic groups [6]. Numerous animals, including rodents (considered as the main reservoir), domestic mammals (including livestock) and wildlife, are reservoirs for leptospires [7]. Leptospires are maintained in the proximal tubules of the kidneys of infected animals (chronically in animal reservoirs, or temporarily during acute infection) and excreted in urine, from which they contaminate soil, surface water, streams and rivers. Humans are infected through direct contact with urine or tissues from infected animals, or indirectly via a contaminated environment, particularly if there are abrasion or cuts in the skin. Prolonged immersion in, or swallowing of, contaminated water can also increase the risk of infection [2].
French Polynesia (FP) is part of the 22 Pacific Countries and Territories, with a population of ~270,000 inhabitants living on five archipelagos (Society, Marquesas, Tuamotu, Gambier and Austral Islands) and about 70% residing on the island of Tahiti. The annual incidence of leptospirosis in FP range from 30 to 55 per 100,000, with one to four fatal cases per year [8,9]. The incidence of leptospirosis in the Pacific region is not well-documented, mainly due to a lack of diagnostic facilities [10,11]. In addition, leptospirosis is sometimes misdiagnosed as other infections, especially arbovirus infections [12]. Data on the Leptospira serovars circulating in humans in FP are scarce. The most recent serotyping information was from cases confirmed in 2007–2010 from FP; 100 isolates were identified as serovars Icterohaemorrhagiae (serogroup Icterohaemorrhagiae) (50%), Australis (serogroup Australis) (32%), Canicola (serogroup Canicola) (7%), Ballum (serogroup Ballum) (4%), Hebdomadis (serogroup Hebdomadis) (4%) and Weilii (serovar Topaz serogroup Tarassovi) (3%) [8]. However, we cannot rule out that other serovars may be present as serotyping was available from only 20% of cases, and because only the serovars included in the panel of antigens can be detected. In addition, molecular studies have never been conducted in FP, so the genetic diversity of Leptospira is unknown.
Specific serovars are more commonly associated with particular reservoir hosts, such as Ballum with mice, Canicola with dogs, or Icterohaemorrhagiae with rats [13]. For this reason, rats, which are very common in FP, were believed to be the main reservoir of leptospires in this country. However, results of epidemiological surveys have suggested that other species might also act as important reservoirs, e.g. dogs and pigs in Society Islands, wild pigs in the upper islands, horses and goats in Marquesas [14]. Until now, animal leptospirosis studies had never been conducted in FP, and the role of animal reservoirs has therefore remained speculative.
The identification of animal carriers and reservoirs of pathogenic leptospires in FP is key to understanding the most probable routes of human exposure, and to recommend public health interventions to reduce leptospirosis disease burden. The aim of our study was (i) to estimate the prevalence of Leptospira carriage in rodents, pet dogs and domestic pigs, and (ii) to identify the Leptospira genotypes circulating in humans and animals, and thereby determine which host species may serve as important reservoirs for pathogenic leptospires in FP.
Materials and methods
Sample collection of human sera
Human sera (n = 44) were retrospectively analysed from leptospirosis cases confirmed by molecular diagnosis from January 2014 to April 2015 at the medical diagnosis laboratory of the “Institut Louis Malardé” (ILM). Cases originated throughout FP: Tahiti (n = 15), Raiatea (n = 9), Moorea (n = 8), Tahaa (n = 7), Huahine (n = 3). The island of origin was missing for two samples. Human sera from seven confirmed cases from November 2011 to January 2012 were further added to our analysis to match the collection date of pig samples. Those cases originated from Tahiti (n = 3), Raiatea (n = 1) or Moorea (n = 3).
Sample collection of animal specimens
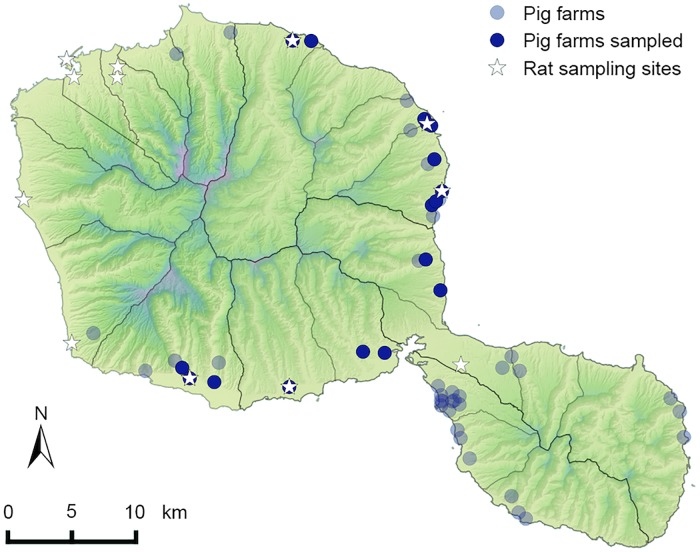
Clinically healthy pigs (Sus scrofa; n = 181) originating from 17 herds representing 16 farms across Tahiti (Fig 1) and one farm in Moorea (Society Islands) were included. Their kidneys were collected from November 2011 to March 2012 at the slaughterhouse of FP (SAEM Abattage de Tahiti) in collaboration with the Agriculture Department of FP.
Fig 1. Geographic localization of piggeries and rat sampling sites on Tahiti Island.
The pig kidneys tested for Leptospira infection originated from 17 herds representing 16 farms. Rats were trapped at 12 sampling sites, including five farms from which pig kidneys originated.
Rats (n = 113) were trapped from 12 sample sites throughout the island of Tahiti (Fig 1) from February to April 2015. Five of the sampling sites were farms from which previously collected pig kidneys originated. Trapping was conducted with wire cage live traps baited using either grilled coconut pieces or a mixture of peanut butter and canned sardine oil. At each sampling site, 40 traps were placed in the afternoon, in line when possible and at least 15 meters apart; trapped animals were collected the following morning and brought back to the laboratory for dissection. If less than ten individuals were trapped at one site, traps were left open in the same place for a second night.
Dog sera (n = 2) were collected from animals with clinical suspected leptospirosis. Kidneys from other dogs (n = 2) were collected by a veterinarian immediately after the animals died from leptospirosis.
Rat identification
Rat species were identified using morphological criteria [15] followed by a molecular confirmation by amplification of the COI gene with the primers BatL5310 / R6036R [16].
Ethics statement
The study was approved by the “Ethics Committee of French Polynesia” (n°61/CEPF). All animal procedures carried out in our study were performed in accordance with the European Union legislation for the protection of animals used for scientific purposes (Directive 2010/63/EU). Human sera were clinical samples collected for diagnostic purposes, and all patient data were anonymized prior to analysis.
Total nucleic acids extraction
All kidney samples were dissected aseptically spanning the corticomedullary junction to collect 20–25 mg of tissue. Tissue sample was lysed for 30 minutes at 56°C with 50 μl of proteinase K (600 mAU/ml, Novagen) and 50 μl of TRIS-HCL solution at 20mM, pH 8.3, 0.5% sodium dodecyl sulphate. Samples were vortexed every 10 min and then centrifuged at 9,000 rpm for 3 min to collect the supernatants.
Total nucleic acids (i.e. both DNA and RNA) were extracted from 100 μl of kidney supernatants and from 200 μl of human and dog sera using the NucliSENS easyMAG automated platform following the manufacturer’s instructions (BioMérieux, France). The elution volume per sample was 50μl.
Detection of Leptospira spp.
Previous studies have shown a better performance of RT-qPCR compared to qPCR for Leptospira detection [17–19]. In addition, RT-qPCR allows the detection of both DNA and RNA, increasing the overall sensitivity of the assay [20]. Thus, we performed qualitative RT-qPCR to detect (i) the 87-bp region of rrs (16S) gene with the primers Lepto-F, Lepto-R, Lepto-S probe for the rat samples [21], or (ii) the 241-bp region of LipL32 gene with the primers LipL32-45F, LipL32-286R and probe LipL32-189P [22] for the other samples. Preliminary sensitivity assays were conducted to select the gene to be targeted for Leptospira detection in the samples (S1 Table). Amplifications were done using the iScript One-Step RT-PCR kit for probes or the IQ Supermix (Bio-Rad Laboratories, France). PCRs were carried out on CFX96 Touch Real-Time PCR Detection System instrument (Bio-Rad Laboratories, France) using the following conditions: initial denaturation at 95°C for 3 min, followed by 45 cycles of denaturation for 3 sec at 95°C and annealing/elongation for 15 sec at 58°C. PCR detection in animal samples was performed in duplicate, or in triplicate when necessary. Using a conservative approach, samples were considered positive when the cycle threshold (Ct) value was inferior to 41 cycles for two replicates, keeping in mind that detection rates may be underestimated. All PCRs were run with negative and positive controls (L. interrogans serovar Australis and L. kirschneri serovar Grippotyphosa reference strains).
Leptospira genotyping
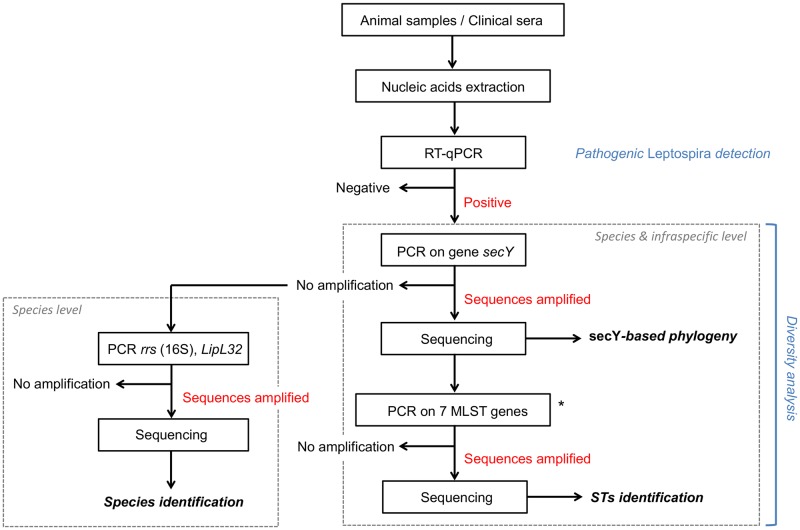
Leptospira positive samples were subjected to different genotyping methods in order to determine and to compare the Leptospira genetic diversity at the specific and infra-specific levels (see Fig 2). First, they were tested by secY sequencing, a gene which has been shown to be suitable for species identification and phylogenetic studies [23,24]. Primers secY-F / secY-R were used to amplify 549-bp fragments [25]. When secY PCR amplification failed, sequencing of alternative short regions was performed to allow Leptospira species identification: a 331-bp sequence from the rrs (16S) gene using primers LA / LB [26] and if necessary a 241-bp sequence from the LipL32 gene using primers LipL32-45F / LipL32-286R [22]. All PCRs were run with negative and positive controls as detailed above.
Fig 2. Flowchart describing the steps of the molecular analysis.
Methods used to detect and to identify Leptospira species and genotypes from samples collected in French Polynesia are detailed. Seven-loci MLST was attempted only for a selected subgroup of the animal and human samples successfully amplified on secY locus using MLST scheme #1.
Based on the observed diversity within the secY gene, multilocus sequence typing (MLST) amplifications were attempted on a subset of positive samples. Three major MLST schemes exist for Leptospira spp. typing worldwide, all supported by the online database http://pubmlst.org/leptospira/ [25,27,28]. MLST was attempted for glmU, pntA, sucA, tpiA, pfkB, mreA, and caiB genes, i.e. following typing scheme #1 [27]. This scheme was chosen to allow the comparison with published strains from the Pacific region, which used the same targets.
Similarly to the detection of Leptospira spp., we used a RT-PCR methodology to increase the sensitivity [20,29] for the Leptospira genotyping. All PCR amplifications were performed in 25 μl containing 2 μl of nucleic acids template, 5 μl of one-step RT-PCR buffer, 1 μl of dNTP mix, 1.5 μl (10 μM) of each primer, 12.75 μl of RNase-free water, 1 μl of RT-PCR enzyme mix and 0.25 μl of RNasin (Promega), using QIAGEN one-step RT-PCR kit (QIAGEN Inc., USA) and the Mastercycler Gradient instrument (Eppendorf, Germany). Primers and PCR conditions are provided as supplementary information (S2 Table).
Sanger sequencing was carried out at ILM on the ABI PRISM 310 Sequence Detection System (Applied Biosytems, USA) or PCR samples were sent to Genoscreen company (France).
Phylogenetic analysis
Consensus sequences and alignments were performed with Geneious Pro version 5.4 [30]. For MLST analyses, sequence alignments were constructed separately for all seven considered loci. A seven-loci concatenate was generated using SEQMATRIX v1.7.8 [31]. Unique allele identifiers for all seven loci were assigned and corresponding allelic profiles (or sequence types STs) were defined using the established Leptospira MLST website (http://pubmlst.org), focusing on MLST scheme #1. Phylogenetic trees were constructed based on the maximum-likelihood (ML) method with 1,000 bootstraps, using PhyML 3.1 [32]. Trees were visualized in FigTree v1.3.1 (http://tree.bio.ecd.ac.uk/). GenBank accession numbers of the sequences produced in the frame of the present study are provided as supplementary information (S3 Table).
Results
Rat identification
Three rat species were identified: Rattus norvegicus (79/113, 70%), Rattus rattus (28/113, 25%) and Rattus exulans (6/113, 5%).
Leptospira detection and prevalence in animal specimens
Kidneys from 26.5% (48/181) of pigs were positive for Leptospira (Table 1). Pigs carrying leptospires originated from 11 out of the 17 selected herds, with a prevalence varying from 10 to 89% between herds.
Table 1. Leptospira detection and identification by host species.
The prevalence of renal infection by Leptospira in farm pigs and rodents is based on RT-qPCR detection results. Infecting Leptospira species were determined by PCR, targeting either secY, rrs (16S) or LipL32 gene in positive samples. The number of successful amplification for secY is provided. Based on the observed diversity within the secY gene, MLST amplifications were attempted on a subset of positive samples; the number of successful complete MLST on seven (or six) loci is given.
| Source | Leptospira prevalence (%) | Identification success (%) | Infecting Leptospira | secY amplification | 7-loci (or 6a) MLST |
|---|---|---|---|---|---|
| Pig | 48/181 (26.5) | 30/48 (62.5) | Li (n = 30), L* (n = 10) | 30 | 4/14 (5) |
| RR | 3/28 (10.7) | 2/3 (66.7) | Li (n = 2) | 2 | 0/2 (1) |
| RN | 19/79 (24.1) | 13/19 (68.4) | Li (n = 8), Lb (n = 5) | 12 | 3/12 (6) |
| RE | 1/6 (16.7) | 1/1 (100) | Lb (n = 1) | 1 | 1/1 (1) |
| All rats | 23/113 (20.4) | 16/23 (69.6) | Li (n = 10), Lb (n = 6) | 15 | 4/15 (8) |
| Dog | - | 2/4 (50) | Li (n = 2) | 2 | 0 |
| Human | - | 39/51 (76.5) | Li (n = 37), Lb (n = 2) | 33 | 4/9 (4) |
RR: Rattus rattus;
RN: Rattus norvegicus;
RE: Rattus exulans.
Li: Leptospira interrogans;
Lb: Leptospira borgpetersenii;
L*: undetermined species closely related to Leptospira mayottensis and Leptospira alexanderi.
a Amplification and sequencing success for the 6-loci MLST scheme excluding the locus caiB.
Kidneys from 20.4% (23/113) of trapped rats were positive for Leptospira (Table 1). Positive rats were found at five sites, where three to 34 animals were trapped per location. Prevalence varied between these sites from 8.7 to 67%. No Leptospira-positive rats were found in the remaining seven sites (where one to seven rats were trapped per site). The Leptospira prevalence varied between rat species: 24.1% in R. norvegicus, 10.7% in R. rattus, and 16.7% in R. exulans (Table 1) but not significantly (Fisher’s exact test, P = 0.31). Interestingly, rat species occurred sympatrically in five sampling sites, including one piggery where specimens from the three Rattus species were trapped, and where Leptospira detection revealed that all three species were carrying Leptospira (one R. rattus, one R. exulans and six R. norvegicus; see S3 Table).
The five sampling sites where both pig and rat samples were collected showed various prevalence results: three sites showed positive prevalences for both species, one site showed zero prevalence for both species, and the last piggery revealed one infected pig (out of ten) but no infected rat (out of seven). Detailed Leptospira prevalences per site and host species are provided as supplementary information (S3 Table).
Prevalence could not be assessed for dogs or humans, as only positive samples were included in the study.
Leptospira species identification
From the 51 positive human samples, Leptospira species were successfully identified in 39 samples (76.5%): Leptospira interrogans in 37 cases and Leptospira borgpetersenii in two cases (Table 1).
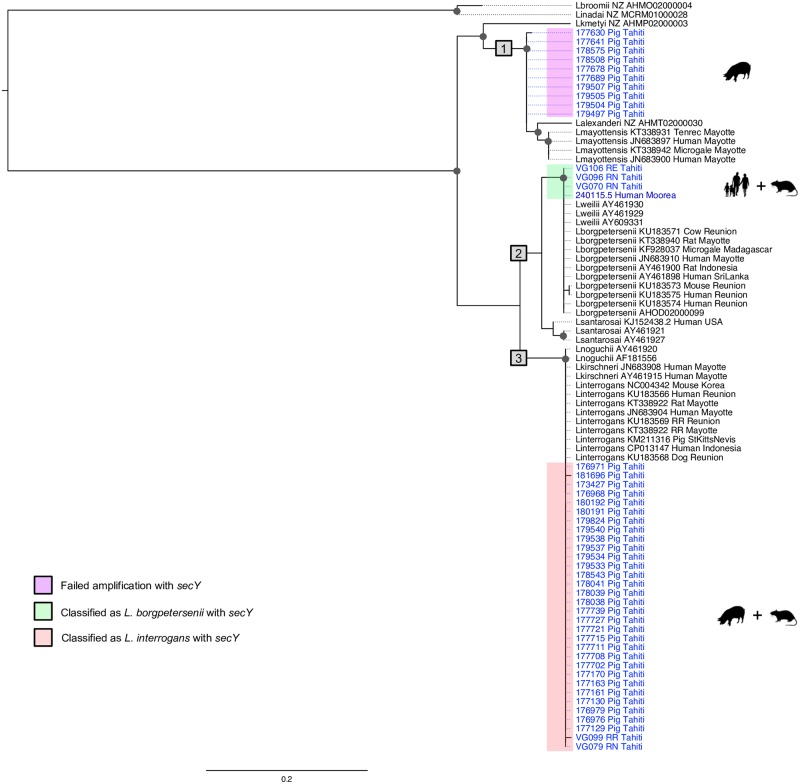
From the 48 positive pig kidneys, 30 samples (62.5%) were successfully identified as infected by L. interrogans (Table 1). For the remaining samples, PCR amplification of secY, rrs (16S) or MLST genes was not successful, but a 241-bp fragment of the LipL32 gene was sequenced from ten additional samples. The obtained LipL32 sequences (40 in total) were included in a ML tree (Fig 3) and showed a well-marked separate cluster closely related to Leptospira mayottensis and Leptospira alexanderi. PCR failure or a lack of nucleic acids template prevented Leptospira identification from the eight remaining swine positive samples.
Fig 3. Maximum-likelihood phylogenetic tree (model TN93+I; 1,000 replicates) inferred from LipL32 gene (229-bp sequence).
Clinical and animal samples from Tahiti are shown in light blue, clinical samples from other islands in French Polynesia (FP) in dark blue; referred to using identifiers accompanied by the host name. Corresponding GenBank accession numbers are reported in S5 Table. Published sequences included in the phylogeny are shown in black using the identified Leptospira species followed by GenBank accession numbers, the host species and the country of origin. The major genetic groups are highlighted with grey boxes (numbered 1 to 3). Bootstrap values higher than 70% are indicated by a dark circle. Black silhouettes represent host groups from FP (i.e. human, dog, pig or rat).
From the 23 positive rat kidneys, Leptospira species were successfully identified in 16 samples: L. interrogans in 10 cases and L. borgpetersenii in six cases. Rattus rattus was infected by L. interrogans, Rattus exulans by L. borgpetersenii and Rattus norvegicus infected with either of the two Leptospira species (Table 1).
From the dog specimens, L. interrogans was successfully identified from two animals.
Genetic diversity based on secY gene and MLST
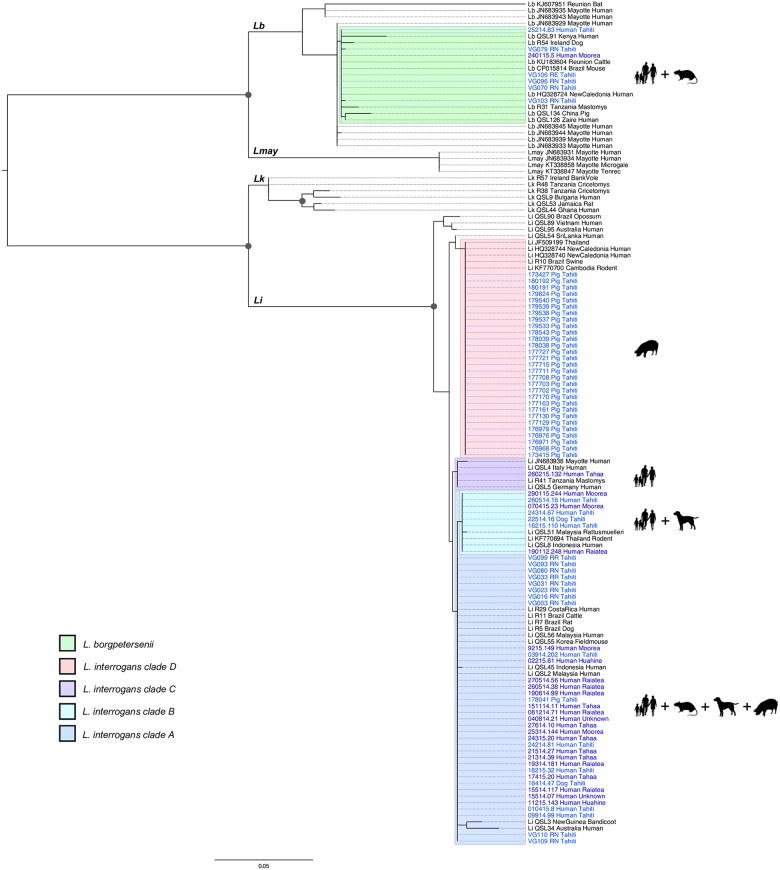
Diversity analyses were conducted on a 444-bp length fragment of the secY gene including 30 sequences from pigs, 15 from rats, two from dogs and 32 from humans (one human sample was removed from the 33 available because of its shorter sequence; Table 1). The maximum likelihood phylogenetic tree showed four L. interrogans clusters, as shown in Fig 4. Most of the clinical cases (n = 23), 10 rats, one pig and one dog clustered together into the main clade A. Human cases were from various geographical origins from Society Islands: Tahiti (n = 5), Moorea (n = 2), Raiatea (n = 6), Huahine (n = 2) and Tahaa (n = 6); two were from unknown origin in FP. A second cluster B included clinical samples from Tahiti (n = 3), Moorea (n = 2) and Raiatea (n = 1) and one dog sample. The third cluster C included one clinical sample from Tahaa. Finally, the last L. interrogans cluster D included pigs only. Almost all pigs (n = 29) clustered together in clade D, except for one pig sample that belonged to the clade A. Leptospira borgpetersenii was identified from the remaining five rat and two human samples (one from Tahiti and one from Moorea), forming a single cluster showing the same 444-bp secY sequence except for two sequences from rats which differed by one nucleotide each (Fig 4).
Fig 4. Maximum-likelihood phylogenetic tree (model TN93+G; 1,000 replicates) inferred from secY gene (444-bp sequence).
Human and animal samples from Tahiti are shown in light blue and clinical samples from other islands in French Polynesia (FP) in dark blue; they are referred to using identifiers accompanied by the host name. Corresponding GenBank accession numbers are reported on S6 Table. Published sequences included in the phylogeny are written in black using GenBank accession numbers or ID (for samples from [33]) followed by the country of origin and the host species. Leptospira species are indicated for published sequences: Li: Leptospira interrogans; Lb: Leptospira borgpetersenii; Lk: Leptospira kirschneri; Lmay: Leptospira mayottensis. Bootstrap values higher than 70% are indicated by a dark circle. Black silhouettes represent host groups from FP (i.e. human, dog, pig or rat). Legend refers to the different clusters including samples from FP, with four clades (A to D) inside the species Leptospira interrogans.
For four human samples, eight rats and five pigs, six to seven loci of the MLST scheme were successfully amplified, sequenced and concatenated for subsequent analyses (Table 2). The samples that allowed successful amplification at all MLST loci and identified as L. interrogans, were distributed in two clusters corresponding to previously described sequence types: ST17 was found in human and rats (corresponding to clade A in the secY-based phylogeny) and ST140 in pigs (corresponding to clade D). The samples that allowed successful amplification at all MLST loci and identified as L. borgpetersenii, corresponded to ST149 (one human and one rat).
Table 2. MLST results by host species.
For each of the 17 samples referred to by a unique identifier (ID), the table provides the exact or most probable sequence type (ST or allelic profile), the infecting Leptospira species and the presumptive serovar associated with the ST. GenBank accession numbers of sequences for each of the seven MLST loci are reported in the supplementary information (S4 Table).
| STs | Leptospira spp. | Presumptive serovarsa (host, country) |
ID | Host species |
|---|---|---|---|---|
| 17 | L. interrogans | Icterohaemorrhagiae (human, Japan Belgium) Copenhageni (human, Brazil; rat, Denmark Russia) |
40814.21 | Homo sapiens |
| 17415.20 | Homo sapiens | |||
| 21514.27 | Homo sapiens | |||
| VG023* | R. norvegicus | |||
| VG031* | R. norvegicus | |||
| VG033* | R. rattus | |||
| VG080 | R. norvegicus | |||
| VG093 | R. norvegicus | |||
| VG109 | R. norvegicus | |||
| 140 | L. interrogans | Pomona (human, Australia) Grippotyphosa (human, Sri Lanka) Guaratuba (possum, Brazil) |
177702* | Sus scrofa |
| 179533 | Sus scrofa | |||
| 179540 | Sus scrofa | |||
| 179824 | Sus scrofa | |||
| 180192 | Sus scrofa | |||
| 149 | L. borgpetersenii | Ballum (mouse, Denmark) Castellonis (mouse, Spain) |
240115.5 | Homo sapiens |
| VG070* | R. norvegicus | |||
| VG106 | R. exulans |
STs: sequence types.
a The presumptive serovars corresponding to STs are based on information from the PubMLST database (http://pubmlst.org/leptospira/).
*Samples with incomplete MLST for which the nearest ST match is drawn from available loci and other complete MLST.
Discussion
Our study provides the first description of circulating Leptospira species and genotypes in humans and potential animal reservoirs and/or carriers of leptospires pathogenic for humans in FP. This region presents a low diversity of pathogenic Leptospira compared to continental countries, but a similar diversity as found in other tropical islands like New Caledonia (Pacific) [34] or Reunion Island (Indian Ocean) [29] where the same species, i.e. L. interrogans and L. borgpetersenii, were identified in human and animal samples. Leptospira interrogans was the dominant species, found in 95% of human cases for which the infecting species could be genotypically determined. This is consistent with previous serological surveys showing the predominance of Icterohaemorrhagiae in patients diagnosed in FP [8].
Rodents, especially rats, are recognized as one of the most significant reservoir of leptospires worldwide [2,35]. Our results give the first direct confirmation that rats are actual reservoirs of two pathogenic Leptospira species (L. interrogans and L. borgpetersenii), the two same species identified from human leptospirosis cases in FP. The Leptospira prevalence in rats reported in our study (20.4%) is similar to rates reported in New Caledonia (20.1%) [36] or Mayotte (Indian Ocean) (15.9%) [37], but contrasting with other tropical islands like Reunion island (36.3%) [29] or Seychelles (Indian Ocean) (7.7%) (Léon Biscornet, pers. comm.). Our study also gives the first description of rat species present in Tahiti, confirming the occurrence of all three species (R. rattus, R. norvegicus and R. exulans) that were suspected to be present based on rodent surveys from other FP Islands, i.e. Huahine, Raiatea, Marquesas [16] and Tetiaroa atoll [38], where one to three of the same three Rattus spp. were previously identified. Leptospira prevalences obtained in our study varied between rat species and sampling sites, but zero prevalences corresponded to sites where the number of trapped rats was low (two to seven), possibly leading to a bias. In particular, rats were difficult to trap in urban areas (high incidence of bait taken with no captures), and R. exulans specimens (n = 6) were captured from only two sites. Results from the site where all three rat species were present and found infected do not seem to indicate a clustering associated with rat species: L. interrogans was genotyped from one R. rattus and four R. norvegicus, and L. borgpetersenii was genotyped from one R. exulans and two R. norvegicus.
The results of our investigations in slaughtered pigs originating from 16 farms on the islands of Tahiti and Moorea suggest that Leptospira infection is quite common among fattened pigs (26.5%), with infection prevalence up to 89% in one farm. Our result is similar to Leptospira prevalence reported in farmed pigs from rural Ecuador (21.1%) [39] but significantly higher than prevalences reported in New Caledonia (10.2%) [34] or Thailand (7.9%) [40]. A single infected pig was shown to carry the same genotype as found in most human samples (clade A). Of note, this pig originated from a farm that was not part of the rat sampling sites. Most pigs (n = 29) were infected by another genotype (ST140, clade D), which was not found in any human cases or other tested animal species despite the close contact of rats and pigs inside the five farms where both species were sampled. The fact that “working in a piggery” has been previously reported as a risk factor in FP [8] might be related to the density of rats found around the piggeries, rather than to the pigs themselves, as shown in American Samoa [41]. However, ST140 is not restricted to pigs in other countries as it was associated with symptomatic human leptospirosis in New Caledonia [24], Australia and Sri Lanka (http://pubmlst.org/leptospira/) (presumptive serovars Pomona and Grippotyphosa, see Table 2). The lack of matching genotypes between humans/rats and pigs (except for one pig) has to be considered with caution as the pig sampling dates back to 2011–2012, while the human cases included in our study were diagnosed in 2014–2015 (and the rats sampled in 2015). In order to assess the possible impact of this time gap, we investigated further sera from human cases collected in late 2011 and early 2012 (n = 7). Unfortunately, we successfully amplified and genotyped one sample only, isolated in January 2012 from Raiatea (Society Islands); the corresponding secY gene sequence was 100% identical to those isolated from human samples collected in 2014–2015 clustering in clade B, thus including no pig samples. The fact that clade D genotype was not identified from humans in FP could also be explained by a recent introduction of this particular genotype concomitantly with an infected pig. The last importation of breeding pigs into Tahiti occurred in 2006, with 156 animals imported from New Zealand (Hervé Bichet, pers. comm.), where pigs are known maintenance hosts for L. interrogans serovars Pomona and Tarassovi [42,43], but no MLST analysis has been performed in this country. In conclusion, there is currently insufficient evidence to definitively exclude farm pigs as a source of occupational leptospirosis in FP and further studies are required to clarify their role, especially as the genotypes found in domestic pigs in Tahiti have been responsible for human infections in the Asia-Pacific region (see [21] and Table 2).
Using Lipl32 gene sequencing, we identified a cluster closely related to Leptospira mayottensis and Leptospira alexanderi including ten pig samples. Lipl32 is a highly conserved gene [44] with more than 94% amino acid sequence identity across the main pathogenic species L. interrogans, L. borgpetersenii, L. kirschneri, L. noguchii, L. santarosai, and L. weilii [45], and Lipl32-based phylogenies generally fail to properly discriminate between these species [25,44,45]. Our Lipl32-based phylogeny shows the same clustering of the six species cited above as previously described, with one clutser including L. interrogans, L. kirschneri, and L. noguchii (clade 3, Fig 3), and another cluster including L. borgpetersenii, L. weilii and L. santarosai (clade 2, Fig 3). However, our pig samples are clearly separate from these two main clusters, and are closely related to L. alexanderi and L. mayottensis (clade 1, Fig 3). Leptospira mayottensis, first described in 2012 from human clinical cases on the tropical island of Mayotte, Indian Ocean [46,47], has been further identified in small mammals in Mayotte [37] and Madagascar [48]. This finding not only shows the potential circulation of an undescribed Leptospira species in Tahiti, but also highlights the rich biodiversity of this phylum yet to be discovered.
Results about dogs are weakened by the small sample size. The sampling was fortuitous, with four samples sent from veterinary clinics for biological confirmation of a clinical diagnosis of leptospirosis. Nevertheless, our phylogeny results show that the two dogs for which we identified the infecting Leptospira share two L. interrogans genotypes with human cases from Tahiti (clades A and B; Fig 4). Interestingly, clade B includes six human cases (all originating from Tahiti or Moorea) and one dog, but no rats. Although a more robust sampling design will be required to estimate the prevalence and the genotypic diversity of leptospires in dogs, the finding of Leptospira genotypes common to both dogs and humans raises questions about the potential role of dogs as a source for human leptospirosis infections in FP. This hypothesis is supported by previous identifications of serogroup Canicola in human cases from FP [8,49], suggesting that dogs, as the reservoir for this serogroup, have probably been the source of infection. The dogs included in our study were pets, but the population of stray dogs in Tahiti might also be important. If future investigations found that stray dogs were infected with pathogenic leptospires, it would imply that they could significantly contaminate the environment and thus be an indirect source of human infection.
Risk factors for human leptospirosis infections in FP are the same than those reported elsewhere: contact with infected animals, as well as recreational and occupational activities [9,50]. Our study highlights the need to reinforce control measures in the country to prevent human leptospirosis. We showed rodents as the probable main source of human infection, highlighting the control of rat populations as a priority. We also raised questions about the contribution of stray dogs and farm pigs to the local epidemiology of the disease. Future studies should be designed to precisely determine the extent of this contribution, so that control measures can be tailored accordingly. Finally, this study confirms that serotyping alone is insufficient to decipher the transmission patterns and reservoirs species responsible for human infections, and that direct genotyping is an important additional tool for epidemiological studies of leptospirosis.
Supporting information
Sensitivity assays included nine rat samples, three human clinical samples, three pig samples and five controls (reference strains).
(XLS)
A single multilocus sequence typing (MLST) scheme for seven pathogenic Leptospira species. PLoS Negl Trop Dis. 2013;7: e1954.
(DOC)
(RN: Rattus norvegicus; RR: Rattus rattus; RE: Rattus exulans).
(DOC)
The 50 sequences from LipL32 gene and the 79 sequences from secY gene are listed in supplementary S5 and S6 Tables respectively.
(DOC)
The sequence lengths range from 198 to 244 bp. Overlapping aligned sequences shorter than 229 bp were removed from the phylogenetic analysis (i.e. pig samples 173415, 177703, 180190 and 181453).
(XLS)
The sequence lengths range from 444 to 551 bp. Overlapping aligned sequences shorter than 444 bp were removed from the phylogenetic analysis (i.e. human sample 15414.76).
(XLS)
Acknowledgments
The authors thank the “Direction de la santé de la Polynésie Française” and Valérie Antras from the “Ministère de l’agriculture et de l’environnement / Service du développement rural”. We acknowledge the slaughterhouse of FP for providing the pig samples and the five farmers for generously allowing the trapping of rats. We also thank François Chaumette from the Port of Papeete; Pascal Correia from the Urbanism Department of Tahiti; Eliane Mama, Geoffrey Vidal, Rémy Tanguy, Edouard Suhas and Philippe Branaa from ILM. We are grateful to Colleen Lau and four anonymous reviewers whose comments have helped us improve the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files except for the sequences obtained from clinical and animal samples which are available from GenBank under the accession numbers KY356889-KY357131. Data about leptospirosis in Seychelles are from Léon Biscornet that may be contacted at: leon.biscornet@gmail.com. Data related to pork production and pig imports in Tahiti are from Hervé Bichet that may be contacted at: herve.bichet@rural.gov.pf.
Funding Statement
This work was supported by the “Ministère de l’agriculture et de l’environnement / Service du développement rural” (SDR), Tahiti – agreement n°2560/MAE/SDR, modified by the amendment n°5593/MAE/SDR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization. Human leptospirosis: guidance for diagnosis, surveillance and control. World Health Organization; 2003. [Google Scholar]
- 2.Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14: 296–326. doi: 10.1128/CMR.14.2.296-326.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlioz-Arthaud A, Kiedrzynski T, Singh N, Yvon J-F, Roualen G, Coudert C, et al. Multicentre survey of incidence and public health impact of leptospirosis in the Western Pacific. Trans R Soc Trop Med Hyg. 2007;101: 714–721. doi: 10.1016/j.trstmh.2007.02.022 [DOI] [PubMed] [Google Scholar]
- 4.Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, et al. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl Trop Dis. 2015;9: e0003898 doi: 10.1371/journal.pntd.0003898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pappas G, Papadimitriou P, Siozopoulou V, Christou L, Akritidis N. The globalization of leptospirosis: worldwide incidence trends. Int J Infect Dis. 2008;12: 351–357. doi: 10.1016/j.ijid.2007.09.011 [DOI] [PubMed] [Google Scholar]
- 6.Fouts DE, Matthias MA, Adhikarla H, Adler B, Amorim-Santos L, Berg DE, et al. What Makes a Bacterial Species Pathogenic? Comparative Genomic Analysis of the Genus Leptospira. PLoS Negl Trop Dis. 2016;10: e0004403 doi: 10.1371/journal.pntd.0004403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adler B, editor. Leptospira and Leptospirosis. Berlin, Heidelberg: Springer; 2015. [Google Scholar]
- 8.Daudens E, Mallet H-P, Buluc A, Frogier E. Situation épidémiologique de la leptospirose en Polynésie française (2006–2010). BISES. 2011;Décembre 2011: 6–8. [Google Scholar]
- 9.Mallet H-P. Le point sur la leptospirose en Polynésie française en 2013. BISES. 2014;Novembre 2014: 3–4. [Google Scholar]
- 10.Victoriano AF, Smythe LD, Gloriani-Barzaga N, Cavinta LL, Kasai T, Limpakarnjanarat K, et al. Leptospirosis in the Asia Pacific region. BMC Infect Dis. 2009;9: 147 doi: 10.1186/1471-2334-9-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musso D, Roche C, Marfel M, Bel M, Nilles EJ, Cao-Lormeau VM. Improvement of leptospirosis surveillance in remote Pacific islands using serum spotted on filter paper. Int J Infect Dis. 2014;20: 74–6. doi: 10.1016/j.ijid.2013.12.002 [DOI] [PubMed] [Google Scholar]
- 12.Nhan TX, Bonnieux E, Rovery C, De Pina JJ, Musso D. Fatal leptospirosis and chikungunya co-infection: Do not forget leptospirosis during chikungunya outbreaks. IDCases. 2016;5: 12–14. doi: 10.1016/j.idcr.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolin CA. Leptospirosis In: Fowler ME, Miller RE, editors. Zoo and wild animal medicine. Philadelphia, PA: Elsevier Science; 2003. pp. 699–702. [Google Scholar]
- 14.Gendron Y, Prieur J, Gaufroy X, Gras C. Leptospirosis in French Polynesia: 120 case reports. Médecine Trop Rev Corps Santé Colon. 1992;52: 21–27. [PubMed] [Google Scholar]
- 15.Matisoo-Smith E, Allen JS. Name that rat: Molecular and morphological identification of Pacific rodent remains. Int J Osteoarchaeol. 2001;11: 34–42. [Google Scholar]
- 16.Robins JH, Hingston M, Matisoo-Smith E, Ross HA. Identifying Rattus species using mitochondrial DNA. Mol Ecol Notes. 2007;7: 717–729. [Google Scholar]
- 17.Waggoner JJ, Balassiano I, Mohamed-Hadley A, Vital-Brazil JM, Sahoo MK, Pinsky BA. Reverse-Transcriptase PCR Detection of Leptospira: Absence of Agreement with Single-Specimen Microscopic Agglutination Testing. PLoS One. 2015;10: e0132988 doi: 10.1371/journal.pone.0132988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waggoner JJ, Pinsky BA. Molecular diagnostics for human leptospirosis. Curr Opin Infect Dis. 2016;29: 440–445. doi: 10.1097/QCO.0000000000000295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Backstedt BT, Buyuktanir O, Lindow J, W EA Jr, Reis MG, Usmani-Brown S, et al. Efficient Detection of Pathogenic Leptospires Using 16S Ribosomal RNA. PLoS One. 2015;10: e0128913 doi: 10.1371/journal.pone.0128913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Li X, Liu S, Ren H, Yang M, Ke Y, et al. Ultrasensitive Detection of Bacteria by Targeting Abundant Transcripts. Sci Rep. 2016;6: 20393 doi: 10.1038/srep20393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smythe LD, Smith IL, Smith GA, Dohnt MF, Symonds ML, Barnett LJ, et al. A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infect Dis. 2002;2: 13 doi: 10.1186/1471-2334-2-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis. 2009;64: 247–255. doi: 10.1016/j.diagmicrobio.2009.03.014 [DOI] [PubMed] [Google Scholar]
- 23.Victoria B, Ahmed A, Zuerner RL, Ahmed N, Bulach DM, Quinteiro J, et al. Conservation of the S10-spc-alpha locus within otherwise highly plastic genomes provides phylogenetic insight into the genus Leptospira. PloS One. 2008;3: e2752 doi: 10.1371/journal.pone.0002752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez J, Goarant C. Rapid Leptospira identification by direct sequencing of the diagnostic PCR products in New Caledonia. BMC Microbiol. 2010;10: 325 doi: 10.1186/1471-2180-10-325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed N, Devi SM, Valverde M de los A, Vijayachari P, Machang’u RS, Ellis WA, et al. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann Clin Microbiol Antimicrob. 2006;5: 28 doi: 10.1186/1476-0711-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mérien F, Amouriaux P, Perolat P, Baranton G, Saint Girons I. Polymerase chain reaction for detection of Leptospira spp. in clinical samples. J Clin Microbiol. 1992;30: 2219–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boonsilp S, Thaipadungpanit J, Amornchai P, Wuthiekanun V, Bailey MS, Holden MTG, et al. A single multilocus sequence typing (MLST) scheme for seven pathogenic Leptospira species. PLoS Negl Trop Dis. 2013;7: e1954 doi: 10.1371/journal.pntd.0001954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varni V, Ruybal P, Lauthier JJ, Tomasini N, Brihuega B, Koval A, et al. Reassessment of MLST schemes for Leptospira spp. typing worldwide. Infect Genet Evol. 2014;22: 216–222. doi: 10.1016/j.meegid.2013.08.002 [DOI] [PubMed] [Google Scholar]
- 29.Guernier V, Lagadec E, Cordonin C, Le Minter G, Gomard Y, Pagès F, et al. Human Leptospirosis on Reunion Island, Indian Ocean: Are Rodents the (Only) Ones to Blame? PLoS Negl Trop Dis. 2016;10: e0004733 doi: 10.1371/journal.pntd.0004733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, et al. Geneious v5.4. 2011. http://www.geneious.com/
- 31.Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27: 171–180. [DOI] [PubMed] [Google Scholar]
- 32.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52: 696–704. [DOI] [PubMed] [Google Scholar]
- 33.Nalam K, Ahmed A, Devi SM, Francalacci P, Baig M, Sechi LA, et al. Genetic affinities within a large global collection of pathogenic Leptospira: implications for strain identification and molecular epidemiology. PLoS One. 2010;5: e12637 doi: 10.1371/journal.pone.0012637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gay N, Soupé-Gilbert ME, Goarant C. Though not reservoirs, dogs might transmit Leptospira in New Caledonia. Int J Environ Res Public Health. 2014;11: 4316–4325. doi: 10.3390/ijerph110404316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adler B, de la Peña Moctezuma A. Leptospira and leptospirosis. Vet Microbiol. 2010;140: 287–296. doi: 10.1016/j.vetmic.2009.03.012 [DOI] [PubMed] [Google Scholar]
- 36.Perez J, Brescia F, Becam J, Mauron C, Goarant C. Rodent abundance dynamics and leptospirosis carriage in an area of hyper-endemicity in New Caledonia. PLoS Negl Trop Dis. 2011;5: e1361 doi: 10.1371/journal.pntd.0001361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagadec E, Gomard Y, Minter GL, Cordonin C, Cardinale E, Ramasindrazana B, et al. Identification of Tenrec ecaudatus, a Wild Mammal Introduced to Mayotte Island, as a Reservoir of the Newly Identified Human Pathogenic Leptospira mayottensis. PLoS Negl Trop Dis. 2016;10: e0004933 doi: 10.1371/journal.pntd.0004933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell JC, Faulquier L, Tonione MA. Rat invasion of Tetiaroa Atoll, French Polynesia In: Veitch CR, Clout MN and Towns DR, editors. Island invasives: eradication and management. Gland, Switzerland: IUCN; 2011. pp. 118–123. [Google Scholar]
- 39.Barragan V, Chiriboga J, Miller E, Olivas S, Birdsell D, Hepp C, et al. High Leptospira Diversity in Animals and Humans Complicates the Search for Common Reservoirs of Human Disease in Rural Ecuador. PLoS Negl Trop Dis. 2016;10: e0004990 doi: 10.1371/journal.pntd.0004990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurilung A, Chanchaithong P, Lugsomya K, Niyomtham W, Wuthiekanun V, Prapasarakul N. Molecular detection and isolation of pathogenic Leptospira from asymptomatic humans, domestic animals and water sources in Nan province, a rural area of Thailand. Res Vet Sci. 2017;115: 146–154. doi: 10.1016/j.rvsc.2017.03.017 [DOI] [PubMed] [Google Scholar]
- 41.Lau CL, Clements AC, Skelly C, Dobson AJ, Smythe LD, Weinstein P. Leptospirosis in American Samoa—estimating and mapping risk using environmental data. PLoS Negl Trop Dis. 2012;6: e1669 doi: 10.1371/journal.pntd.0001669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall RB, Manktelow BW. Fifty years of leptospirosis research in New Zealand: a perspective. N Z Vet J. 2002;50: 61–63. [DOI] [PubMed] [Google Scholar]
- 43.Thornley CN, Baker MG, Weinstein P, Maas EW. Changing epidemiology of human leptospirosis in New Zealand. Epidemiol Infect. 2002;128: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haake DA, Suchard MA, Kelley MM, Dundoo M, Alt DP, Zuerner RL. Molecular evolution and mosaicism of leptospiral outer membrane proteins involves horizontal DNA transfer. J Bacteriol. 2004;186: 2818–2828. doi: 10.1128/JB.186.9.2818-2828.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murray GL. The lipoprotein LipL32, an enigma of leptospiral biology. Vet Microbiol. 2013;162: 305–314. doi: 10.1016/j.vetmic.2012.11.005 [DOI] [PubMed] [Google Scholar]
- 46.Bourhy P, Collet L, Lernout T, Zinini F, Hartskeerl RA, van der Linden H, et al. Human Leptospira isolates circulating in Mayotte (Indian Ocean) have unique serological and molecular features. J Clin Microbiol. 2012;50: 307–311. doi: 10.1128/JCM.05931-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bourhy P, Collet L, Brisse S, Picardeau M. Leptospira mayottensis sp. nov., a pathogenic species of the genus Leptospira isolated from humans. Int J Syst Evol Microbiol. 2014;64: 4061–4067. doi: 10.1099/ijs.0.066597-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dietrich M, Wilkinson DA, Soarimalala V, Goodman SM, Dellagi K, Tortosa P. Diversification of an emerging pathogen in a biodiversity hotspot: Leptospira in endemic small mammals of Madagascar. Mol Ecol. 2014;23: 2783–2796. doi: 10.1111/mec.12777 [DOI] [PubMed] [Google Scholar]
- 49.Coudert C, Beau F, Berlioz-Arthaud A, Melix G, Devaud F, Boyeau E, et al. Human leptospirosis in French Polynesia. Epidemiological, clinical and bacteriological features. Med Trop Mars. 2007;67: 137–144. [PubMed] [Google Scholar]
- 50.Hirschauer C, Daudens E, Coudert C, Frogier E, Melix G, Fleure M, et al. Épidémiologie de la leptospirose en Polynésie française de 2006 à 2008. BEH Thématique. 2009;48-49-50: 508–511. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sensitivity assays included nine rat samples, three human clinical samples, three pig samples and five controls (reference strains).
(XLS)
A single multilocus sequence typing (MLST) scheme for seven pathogenic Leptospira species. PLoS Negl Trop Dis. 2013;7: e1954.
(DOC)
(RN: Rattus norvegicus; RR: Rattus rattus; RE: Rattus exulans).
(DOC)
The 50 sequences from LipL32 gene and the 79 sequences from secY gene are listed in supplementary S5 and S6 Tables respectively.
(DOC)
The sequence lengths range from 198 to 244 bp. Overlapping aligned sequences shorter than 229 bp were removed from the phylogenetic analysis (i.e. pig samples 173415, 177703, 180190 and 181453).
(XLS)
The sequence lengths range from 444 to 551 bp. Overlapping aligned sequences shorter than 444 bp were removed from the phylogenetic analysis (i.e. human sample 15414.76).
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files except for the sequences obtained from clinical and animal samples which are available from GenBank under the accession numbers KY356889-KY357131. Data about leptospirosis in Seychelles are from Léon Biscornet that may be contacted at: leon.biscornet@gmail.com. Data related to pork production and pig imports in Tahiti are from Hervé Bichet that may be contacted at: herve.bichet@rural.gov.pf.