Abstract
The Chinese citrus fly, Bactrocera minax (Enderlein), is a devastating citrus pest in Asia. This univoltine insect enters obligatory pupal diapause in each generation, while little is known about the course and the molecular mechanisms of diapause. In this study, the course of diapause was determined by measuring the respiratory rate throughout the pupal stage. In addition, the variation of transcriptomic and metabolomic profiles of pupae at five developmental stages (pre-, early-, middle-, late-, and post-diapause) were evaluated by next-generation sequencing technology and 1H nuclear magnetic resonance spectroscopy (NMR), respectively. A total of 4,808 genes were significantly altered in ten pairwise comparisons, representing major shifts in metabolism and signal transduction as well as endocrine system and digestive system. Gene expression profiles were validated by qRT-PCR analysis. In addition, 48 metabolites were identified and quantified by 1H NMR. Nine of which significantly contributed to the variation in the metabolomic profiles, especially proline and trehalose. Moreover, the samples collected within diapause maintenance (early-, middle-, and late-diapause) only exhibited marginal transcriptomic and metabolomic variation with each other. These findings greatly improve our understanding of B. minax diapause and lay the foundation for further pertinent studies.
Introduction
The Chinese citrus fly, Bactrocera minax (Enderlein), is a devastating oligophagous pest of citrus plants in the temperate areas of Asia, especially in China [1–3]. Larval feeding can cause serious yield losses [4–6], as such this pest has become a focus of interest in citrus-growing regions in China. Given the economic importance of B. minax, a number of prior studies have been carried out [7–13]. However, the long-lasting pupal stage in which diapause occurs has severely hindered the laboratory-rearing of this pest and restricted further scientific research.
Diapause is a life history stage that allows insects to mitigate acute environmental stresses [14,15]. Some univoltine insects enter obligatory diapause at specific stages in each generation, but require no token stimuli for diapause induction and preparation [14,16]. Similarly, univoltine tephritid fly B. minax also enters the pupal diapause to overwinter. However, the course of diapause has yet to be reported. Generally, diapause in insects features intense metabolic depression. Respiratory rate is therefore a useful marker for determining diapause initiation and termination [17]. For example, the precise time of diapause termination of Rhagoletis pomonella was identified based on the fitted trajectory of metabolic rate estimated by CO2 production. Subsequently, samples during the diapause termination were collected to investigate the molecular mechanisms underlying diapause termination and resumption of development [16]. By determining the course of diapause in B. minax through respiratory rate measurement, several other investigations into diapause can be conducted.
Next-generation sequencing has widely been used to characterize genomes and transcriptomes. This approach has facilitated studies on biological processes in organisms, such as the discovery of differentially expressed genes (DEGs) [18–20]. A B. minax transcriptome that was previously assembled and annotated can provide a foundation for further DEG analysis [21]. Similarly, metabolomic profiling has increasingly been used worldwide to investigate the qualitative and quantitative variation of metabolites in tissues and biofluids in response to biotic or abiotic factors [22]. The most widely used methods for metabolomic analysis are 1H nuclear magnetic resonance spectroscopy (NMR), gas-chromatography coupled with mass spectrometry (GC-MS), and liquid chromatography coupled with mass spectrometry (LC-MS) [23,24]. Of these methods, NMR has significant advantages, including near-universal detection of metabolites, easy quantitation, high reproducibility, minimal sample preparation, and rapidity [25,26]. So far, metabolomic analysis has been performed in many fields such as diagnostics [27], pharmacology [28], microbiology [29], and nutrition [30]. Recently, a limited number of metabolomic analyses have also been conducted on insect diapause [31–33].
In this study, the respiratory rate of B. minax was periodically measured throughout the pupal stage in order to clarify the course of diapause. To better understand the molecular mechanisms underlying diapause, high-throughput next-generation sequencing technology and 1H NMR analysis were performed to analyze respectively the transcriptomic and metabolomic profiles of B. minax at five pupal developmental stages. The results have revealed variations in the physiological pathways utilized throughout the course of diapause and provided new insights into diapause programming.
Materials and methods
Ethics statement
The owner of the orchard in Wulong County, Chongqing Municipality, China, provided permission to collect the samples for our scientific research.
Insect rearing
Fallen oranges infested with larvae were brought back to the lab from an orchard (N 29o 34.373', E 107o 54.564') in Wulong County, Chongqing Municipality, China. Third-instar larvae (15~18 mm in length) collected from the oranges were placed over sand in plastic dishes and allowed to pupate. All dishes were placed outdoors in the nylon insect rearing cage under natural temperature and light/dark cycle in the Beibei district, Chongqing Municipality, China. The sand was changed weekly and regularly watered to maintain moisture.
Measurement of respiratory rate of pupae
The respiratory rate of each group was measured 46 times from pupation to the first adult emergence, using a self-designed portable respirometry system with a high-accuracy infrared CO2 detector (S1 Fig Wosaite, Shenzhen, China). Prior to measuring the pupal respiratory rate, thirty pupae were randomly selected and divided into three groups. Each group was weighed and placed in a 50 mL sealed chamber for 2 h and the CO2 concentration in the chamber was recorded. The respiratory rate of pupae was calculated by:
| (1) |
Where R is the calculated respiratory rate in μL CO2/mg/h, C is the raised CO2 concentration in μL/L, V is the volume of the sealed chamber in L, M is the weight of the pupae in mg, and T is the duration of CO2 measurement in h. Visual inspection of respiratory rate data strongly suggested an exponential decay followed by a logistic increase and an exponential increase, based on which an eight parameter non-linear model describing this trajectory was constructed:
| (2) |
Where R is the respiratory rate, t is the time in days since investigation, α is the respiratory rate at the starting point, β is the respiratory rate at the transition between exponential decay and logistic increase, k is the rate constant, γ is the respiratory rate at the transition between logistic and exponential increase, a is the constant, c is the scaling parameter, r and b are parameters that determine the timing of two transitions, respectively. The non-linear model was fitted using GraphPad Prism 5 and Microsoft Excel 2010, and the goodness of fit was determined by χ2 test using Excel 2010. The initiation, maintenance, and termination of pupal diapause were determined in relation to the fitted model.
Insect sample collection
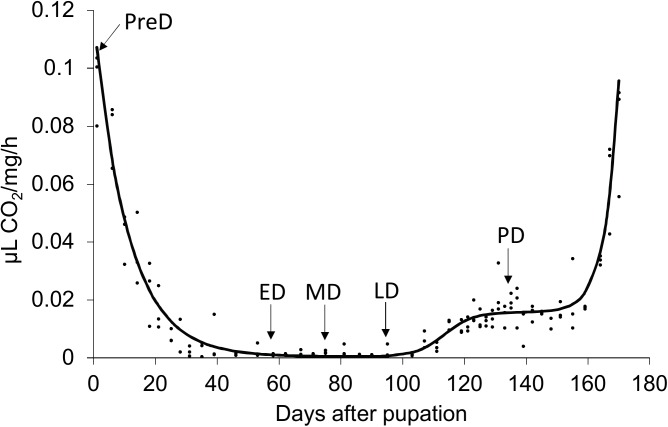
Samples were collected at five time points, pre-diapause (PreD), early-diapause (ED), middle-diapause (MD), late-diapause (LD), and post-diapause (PD) (Fig 1), as determined by respiratory rate. Prior to sample collection, all pupae were reared separately in 15 dishes, three dishes for each of the five developmental stages, under the conditions described above. At each sampling point, all of the pupae in the four dishes were collected and stored separately in liquid nitrogen for subsequent transcriptomic and metabolomic analysis.
Fig 1. Non-linear regression of respiratory rate of Bactrocera minax pupae.
Arrows indicate the time points when the samples were collected for transcriptomic and metabolomic analysis.
RNA isolation, library construction and Illumina sequencing
Total RNA from each pupa was isolated using TRIZOL Reagent (Life technologies, Carlsbad, CA, US) according to the manufacturer's instructions. The quantity and quality of total RNA was assessed with a NanoVue spectrophotometer (GE Healthcare Bio-Science, Uppsala, Sweden) and 1% agarose gel electrophoresis, respectively. The cDNA libraries were constructed using TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA) following the manufacturer’s protocol. Briefly, oligo (dT) magnetic beads were used to purify poly (A) mRNA. The resulting mRNA was mixed with fragmentation buffer and cleaved into short fragments. The first-strand cDNAs were generated with random hexamer primers. Second-strand cDNAs were synthesized using DNA polymerase I (New England BioLabs, Ipswich, MA) and RNase H (Invitrogen, Carlsbad, CA). These cDNA fragments were end-repaired, followed by single nucleotide A (adenine) addition and litigation of adaptors. After purification with AMPureXP beads, the ligated products were amplified by PCR to generate cDNA libraries, which were sequenced on an Illumina NextSeq500 (Illumina). The raw reads were filtered to remove adaptor sequences, low-quality sequences with unknown nucleotides N, reads shorter than 50 bp, and reads with more than 20% low quality bases, using the NGS QC toolkit package [34]. The clean reads were assessed for quality using FastQC (http://www.bioinformatiH2.babraham.ac.uk/projects/fastqc/) and then mapped to our previously generated transcriptome reference database [21]. Three biological replicates were generated for each developmental stage.
Discovery of differentially expressed genes and KEGG pathway analysis
Gene expression levels were determined by reads per kb of exon model per million mapped reads (RPKM) values [35], which were calculated based on the number of reads mapping to each unigene obtained previously [21]. The DEseq package was used to identify the DEGs [36]. Unigenes with P value < 0.05 and fold change value > 2 in each comparison were considered to be significantly differentially expressed genes. Benjamini-Hochberg correction was then conducted to reduce the false discovery rate (FDR). DEG cluster analysis was performed using cluster [37] and visualized by Java Tree view [38]. Principal component analysis (PCA) was carried out using pcaMethods [39] to evaluate the variation in gene expression profiles among samples and visualize the clustering of samples. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analyses were conducted using KEGG mapper to categorize the DEGs (http://www.genome.jp/kegg/tool/map_pathway2.html), and the P value < 0.01 was set as a threshold to determine the significantly regulated pathway in each comparison.
Quantitative real-time PCR (qRT-PCR) verification
qRT-PCR was performed to verify the accuracy of the DEG analysis. Total RNA from the five pupal stages described above was extracted using TRIZOL Reagent. The first-strand cDNA was synthesized using PrimeScript™ RT Master Mix (Perfect Real Time) Kit (Takara, Shiga, Japan). Twenty-one pairs of specific primers were designed to amplify the genes selected from multiple comparison (S1 Table). Ubiquitin was used as a reference gene for normalization [40]. qRT-PCR was conducted in 25 μL volumes containing 12.5 μL SYBR® Premix Ex Taq II (Takara), 2 μL primers (10 μM), 1μL cDNA, and 9.5 μL ddH2O, using a CFX96™ Real-Time PCR Detection System thermal cycler (BIO-RAD, Hercules, CA, USA). Amplification conditions were as follows: initial denaturation at 95°C for 30s; followed by 40 cycles of denaturation at 95°C for 5s, 60°C for 30s. Pearson’s r correlation coefficient was calculated to evaluate the correlation between the qRT-PCR and DEG data. Three biological and three technical replicates were performed for each gene.
1H NMR spectroscopy
About 500 mg pupae were ground in liquid nitrogen and lyophilized in a vacuum freeze dryer. More than 50 mg lyophilized sample was weighed and resuspended in 1 ml water. After vortexing for 1 min, samples were sonicated and centrifuged at 13000 rpm at 4°C for 10 min. The supernatant was filtered using 3-kDa microcentrifuge filters to remove proteins and insoluble impurities. A 450 μL filtrate was then mixed with 50 μL DSS standard solution (4.088 mM), an internal NMR chemical shift standard, for subsequent NMR analysis. One-dimensional NMR spectra of samples were acquired using a Bruker AV III 600 MHz spectrometer (Bruker Biospin Ltd., Coventry, UK) equipped with an inverse cryoprobe, operating at an NMR frequency of 600.13 MHz, and a data acquisition temperature of 298 K. A total of 64 transients were acquired in 32,768 data points using a spectral width of 8000 Hz. The free induction decay (FID) signal was multiplied by an exponential window function with 1 Hz line broadening factor before Fourier transform. Metabolites were assigned by Chenomx (Chenomx Inc., Edmonton, Canada) on the basis of chemical shifts, coupling constants, and relative intensities, against a Chenomx database that contained the unique NMR spectra of each standard compound. The absolute concentration of each compound was normalized to the sample weight. Five biological replicates were set for each selected developmental stage. PCA and partial-least squares discriminant analysis (PLS-DA) [41] were carried out to evaluate the variation in metabolite profiles among developmental stages and to visualize sample clustering. Based on PLS-DA analysis, the Variable Importance in Projection (VIP) scores were obtained to indicate the metabolites that significantly contributed to the intergroup differentiation. The concentrations of nine identified metabolites (VIP score > 1) at five developmental stages were compared using a non-parametric Kruskal-Wallis test, followed by Bonferroni correction [42]. The analyses were performed using the statistical package STATISTICA version 10 (StatSoft Inc. 2011, Tulsa, OK).
Results
Respiratory rate trajectory
The model describing the respiratory rate trajectory was fitted well according to Chi-square test (χ2 = 0.0824, d.f. = 44, P > 0.05). The exponential decay, logistic increase, and exponential increase in this model represent periods of diapause preparation, diapause termination, and preparation for adult emergence, respectively (Fig 1). According to this model, respiration was suppressed after pupation and reached its lowest level approximately 40 days later, when pupae had completely entered diapause. Approximately two months after initiation, diapause terminated and pupal development resumed with the respiratory rate increasing by 39-fold. Post-diapause development lasted for over a month until adults started to emerge.
Identification of differentially expressed genes (DEGs) among the differing developmental stages
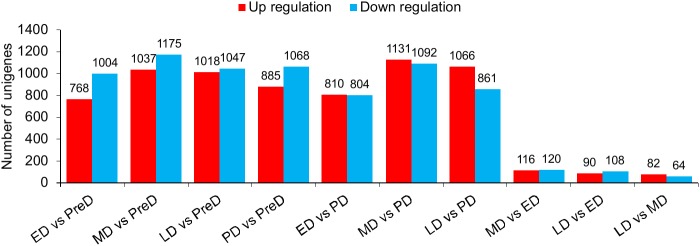
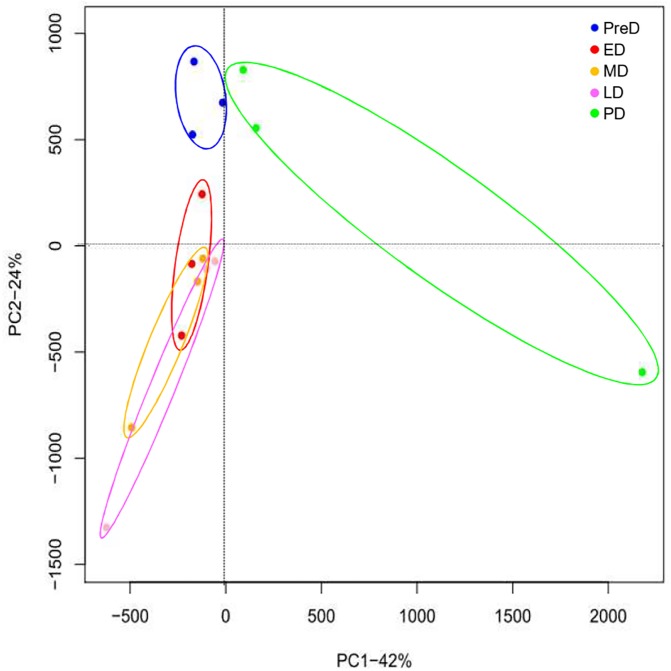
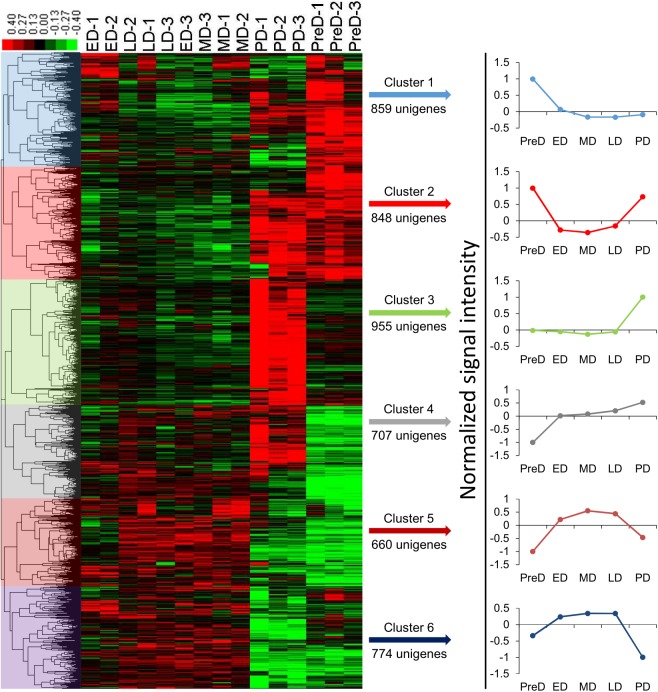
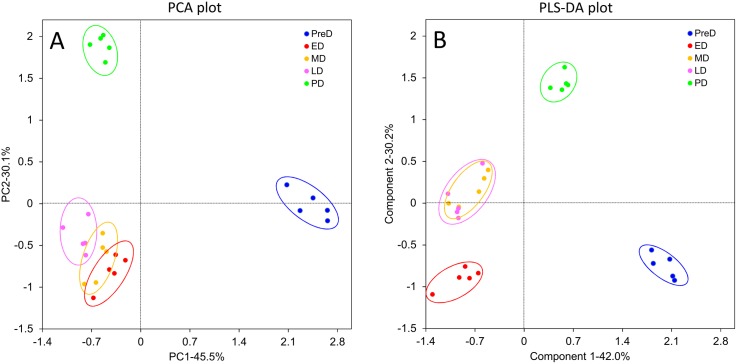
Fifteen mRNA libraries, three replicates for each developmental stage, were sequenced. At least 18 million raw reads were generated in each library (S2 Table). All raw data have been deposited in the GeneBank Sequence Read Archive (Accession number: SRP083788). After removing low quality reads, the number of clean reads ranged from 18.37 million to 51.87 million, and the proportion of useful reads exceed 97% in all libraries. Gene expression profiles at different pupal developmental stages were calculated to identify DEGs. A total of 4,808 genes were significantly differentially expressed in ten pairwise comparisons (Fig 2 and S2 Fig), whereas the number decreased to 3,290 after Benjamini-Hochberg correction to reduce FDR (S3 Fig). Interestingly, samples collected within the maintenance of diapause (ED, MD, and LD) did not show intense variation in gene expression profiles compared to each other, with only a few genes altered. The large overlap of ED, MD, and LD in the PCA plot also showed no obvious difference in gene expression profiles among these three stages (Fig 3). However, a distinct separation in the PCA plot and a number of DEGs between PreD/PD and the other time points revealed large variations in the gene expression profiles between these two stages and others (Figs 2 and 3). All DEGs were divided into 6 groups with each exhibiting a representative expression pattern. Genes in cluster 1 and 3 were highly expressed in PreD and PD, respectively, whereas genes in cluster 4 were lowly expressed in PreD. Throughout diapause maintenance, the expression of genes in cluster 2 was suppressed, whereas those in cluster 5 and 6 were activated (Fig 4).
Fig 2. Number of significantly differentially expressed genes (DEGs) in each pairwise comparison of different Bactrocera minax pupal stages.
PreD, pre-diapause. ED, early-diapause. MD, middle-diapause. LD, late-diapause. PD, post-diapause.
Fig 3. Principal component analysis (PCA) plot of gene expression profile at different Bactrocera minax pupal stages.
PreD, pre-diapause. ED, early-diapause. MD, middle-diapause. LD, late-diapause. PD, post-diapause.
Fig 4. Cluster of differentially expressed genes (DEGs) among the different Bactrocera minax pupal stages.
Each column represents a sample, and each row represents a differentially expressed gene. Green and red color gradients indicate a decrease or increase in expression, respectively. PreD, pre-diapause. ED, early-diapause. MD, middle-diapause. LD, late-diapause. PD, post-diapause.
To understand the potential functions of identified DEGs, KEGG pathway enrichment for each pairwise comparison was performed at P value < 0.01. Pathways involved in human disease were excluded after analysis. The results showed that no KEGG pathway was enriched in pairwise comparisons among the three groups from maintenance of diapause, except for “Alanine, aspartate, and glutamate metabolism” between MD and LD. Most of the significantly altered pathways in other comparisons were related to “Carbohydrate metabolism”, “Lipid metabolism”, “Amino acid metabolism”, “Biosynthesis of other secondary metabolites”, “Metabolism of cofactors and vitamins”, “Xenobiotics biodegradation and metabolism”, “Signal transduction”, “Endocrine system”, and “Digestive system” (Table 1).
Table 1. KEGG pathway analysis of differentially expressed genes among comparisons.
| Comparison | KEGG pathway | Comparison | KEGG pathway |
|---|---|---|---|
| ED vs PreD | Pentose and glucuronate interconversions* | PD vs PreD | Pentose and glucuronate interconversions* |
| Galactose metabolism* | Galactose metabolism | ||
| Ascorbate and aldarate metabolism | Ascorbate and aldarate metabolism* | ||
| Starch and sucrose metabolism* | Starch and sucrose metabolism | ||
| Glycerolipid metabolism | Steroid hormone biosynthesis | ||
| Ether lipid metabolism | Glycerolipid metabolism | ||
| Arginine and proline metabolism | Retinol metabolism | ||
| Glutathione metabolism* | Metabolism of xenobiotics by cytochrome P450* | ||
| Retinol metabolism | Drug metabolism—cytochrome P450* | ||
| Porphyrin and chlorophyll metabolism | DNA replication | ||
| Metabolism of xenobiotics by cytochrome P450* | Cell adhesion molecules (CAMs) | ||
| Drug metabolism—cytochrome P450* | PPAR signaling pathway* | ||
| Protein processing in endoplasmic reticulum | Protein digestion and absorption | ||
| Antigen processing and presentation | ED vs PD | Biosynthesis of amino acids* | |
| Estrogen signaling pathway | Ascorbate and aldarate metabolism | ||
| Protein digestion and absorption | Starch and sucrose metabolism* | ||
| Fat digestion and absorption | Glycine, serine and threonine metabolism* | ||
| MD vs PreD | Biosynthesis of amino acids* | Retinol metabolism | |
| Pentose and glucuronate interconversions* | Porphyrin and chlorophyll metabolism | ||
| Galactose metabolism | Antigen processing and presentation | ||
| Starch and sucrose metabolism* | MD vs PD | Carbon metabolism | |
| Amino sugar and nucleotide sugar metabolism | Biosynthesis of amino acids* | ||
| Nitrogen metabolism | Pentose phosphate pathway* | ||
| Fatty acid degradation | Ascorbate and aldarate metabolism | ||
| Glycerolipid metabolism | Starch and sucrose metabolism | ||
| Glycerophospholipid metabolism | Steroid hormone biosynthesis | ||
| Alanine, aspartate and glutamate metabolism* | Glycine, serine and threonine metabolism | ||
| Arginine and proline metabolism* | Glutathione metabolism | ||
| Glutathione metabolism | Retinol metabolism | ||
| Streptomycin biosynthesis | Porphyrin and chlorophyll metabolism | ||
| Aminobenzoate degradation | Metabolism of xenobiotics by cytochrome P450 | ||
| Two-component system | Drug metabolism—cytochrome P450 | ||
| Hippo signaling pathway -fly | MAPK signaling pathway | ||
| Peroxisome | Hippo signaling pathway -fly | ||
| Ovarian Steroidogenesis | Cell adhesion molecules (CAMs) | ||
| Estrogen signaling pathway | Antigen processing and presentation | ||
| Fat digestion and absorption* | LD vs PD | Purine metabolism* | |
| LD vs PreD | Pentose and glucuronate interconversions* | Glutathione metabolism | |
| Galactose metabolism* | Wnt signaling pathway | ||
| Starch and sucrose metabolism* | Hippo signaling pathway -fly | ||
| Nitrogen metabolism | Insulin secretion | ||
| Fatty acid degradation | Adrenergic signaling in cardiomyocytes | ||
| Glycerolipid metabolism* | Pancreatic secretion | ||
| Glycerophospholipid metabolism* | MD vs LD | Alanine, aspartate and glutamate metabolism | |
| Glycosaminoglycan degradation | |||
| One carbon pool by folate | |||
| Retinol metabolism | |||
| Caffeine metabolism | |||
| Drug metabolism—other enzymes | |||
| Neuroactive ligand-receptor interaction | |||
| Peroxisome* | |||
| PPAR signaling pathway* | |||
| Protein digestion and absorption | |||
| Fat digestion and absorption |
* P value < 0.001. For KEGG pathways without asterisks, P value < 0.01.
Validation of gene expression profiles
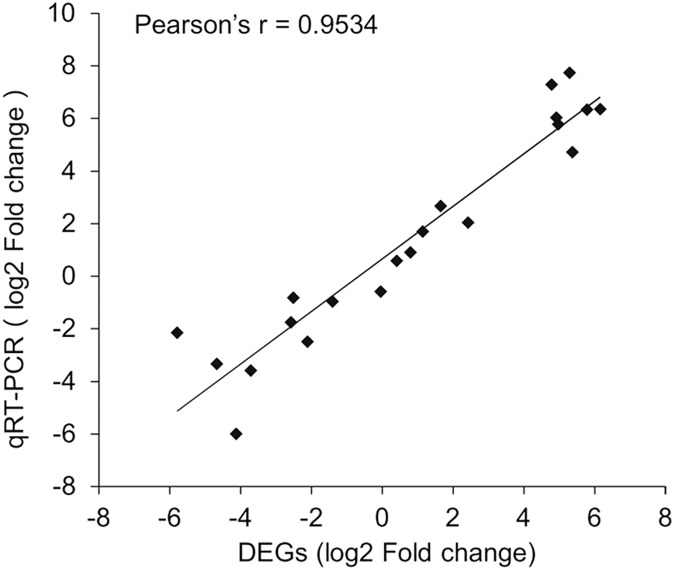
The expression levels of the 21 selected genes were measured by qRT-PCR to validate the DEG analysis. The results showed a strong correlation between the qRT-PCR and DEG data with Pearson’s correlation coefficient > 0.95 (Fig 5), indicating the reliability of using DEG data to investigate temporal-specific gene expression profiles throughout the pupal stage.
Fig 5. Correlation analysis of qRT-PCR and differentially expressed gene (DEG) data for selected genes of Bactrocera minax.
Metabolomic variation among Bactrocera minax developmental stages
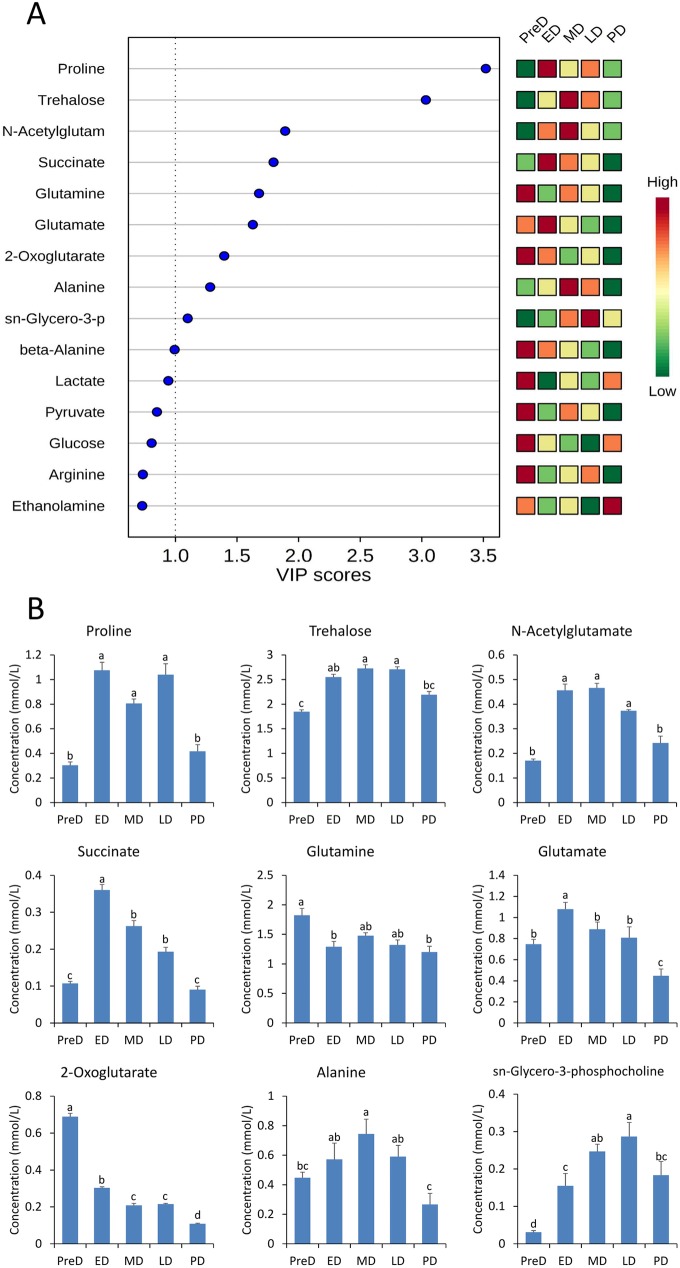
A total of 49 metabolites were identified across all samples, including amino acids and their derivatives, organic acids, nucleic acid components, sugars, and others (S4 Fig). After removing citrate, which cannot be precisely quantified due to the effect of pH value, the other 48 metabolite identities were confirmed for subsequent analysis. Both the PCA and PLS-DA plots showed close overlap between the ED, MD, and LD groups, but also showed clear separation between PreD and PD (Fig 6), indicating the metabolomic profiles changed marginally within diapause maintenance, but varied dramatically with initiation and termination of diapause. After VIP score calculation, the top 9 metabolites that contributed the most to the variation of metabolomic profiles (VIP score > 1) were identified (Fig 7A). Significant differences were found in the concentration of nine metabolites across the five developmental stages with non-parametric Krustal-Wallis test (Table 2). Of all these nine metabolites, proline, trehalose, N-acetylglutamate, succinate, glutamate, alanine, and sn-glycero-3-phosphocholine saw significant accumulations within maintenance of diapause; the glutamine concentration was higher in PreD; and the 2-oxoglutarate concentration was higher in PreD but gradually decreased along with development (Fig 7B).
Fig 6.
Principal component analysis (PCA) plot (A) and partial-least squares discriminant analysis (PLS-DA) plot (B) of metabolite profiles from different Bactrocera minax pupal stages. PreD, pre-diapause. ED, early-diapause. MD, middle-diapause. LD, late-diapause. PD, post-diapause.
Fig 7. Metabolomic variation among different Bactrocera minax pupal stages.
A. Variable importance plot showing the metabolites with the highest VIP score. B. Concentration of nine metabolites (VIP score >1) at different B. minax pupal stages. Different letters above the bars indicate significant differences determined by Bonferroni correction. PreD, pre-diapause. ED, early-diapause. MD, middle-diapause. LD, late-diapause. PD, post-diapause.
Table 2. Identification of significantly altered metabolites with non-parametric Krustal-Wallis test.
| Common name | Chemical shift (ppm) | Formula | d.f. | n | H value | P-value |
|---|---|---|---|---|---|---|
| Proline | 2.00 (m) | C5H9NO2 | 4 | 25 | 20.11 | <0.01 |
| Trehalose | 3.44(t), 3.80(m), 5.18(d) | C12H22O11 | 4 | 25 | 19.64 | <0.01 |
| N-Acetylglutamate | 2.02(s) | C7H11NO5 | 4 | 25 | 21.84 | <0.01 |
| Succinate | 2.39(s) | C4H6O4 | 4 | 25 | 22.00 | <0.01 |
| Glutamine | 2.11(m) | C5H10N2O3 | 4 | 25 | 13.23 | <0.01 |
| Glutamate | 2.04(m), 2.34(m) | C5H9NO4 | 4 | 25 | 21.00 | <0.01 |
| 2-Oxoglutarate | 2.99(t) | C5H6O5 | 4 | 25 | 21.94 | <0.01 |
| Alanine | 1.46(d) | C3H7NO2 | 4 | 25 | 18.88 | <0.01 |
| Sn-Glycero-3-phosphocholine | 4.31(m) | C8H21NO6P | 4 | 25 | 21.61 | <0.01 |
Discussion
Respiratory rate trajectory
Metabolic depression is frequently considered a universal characteristic of diapausing insects [16,17,43], hence, the respiratory rate of B. minax pupae was measured first to determine the course of diapause. The sensitivity of the CO2 detector precluded monitoring respiratory rate of individual B. minax. However, monitoring a group of pupae proved to be feasible as all adults emerged within 8 days (data not shown), indicating synchronization of pupal development. The generated curve identified important developmental landmarks throughout the pupal stage and guided our sample collection (Fig 1). To our knowledge, this is the first endeavor to describe the course of diapause in B. minax pupae, which provided the fundamental basis for subsequent transcriptomic and metabolomic analyses in this study.
Gene expression profiles throughout pupal development
Gene expression profiles varied within pupal development, but not much within the maintenance of diapause (Figs 2 and 3). When comparing PreD with others, the most significantly changed KEGG pathways were associated with metabolism, especially the “Pentose and glucuronate interconversions”, “Galactose metabolism”, “Starch and sucrose metabolism”, and “Glycerolipid metabolism” pathways, which were significantly changed in all four comparisons. These variations were consistent with the suppression in respiratory rate described above. When comparing PD with ED/MD, several metabolism-related KEGG pathways significantly changed, probably due to elevation in the metabolic rate after diapause termination. However, when comparing PD with LD, most of the enriched KEGG pathways related to signaling pathways. The “Wnt signaling pathway” and “Hippo signaling pathway—fly” is expected to be responsible for cell proliferation and differentiation in post-diapause development [44,45]. The “insulin secretion” pathway may also be involved in the insulin signaling pathway which is a conserved mechanism for controlling insect diapause [46,47].
To survive the various biotic and abiotic stresses during diapause, a series of stress-induced mechanisms in insects have to be deployed. Heat shock proteins (Hsps) are a group of well-described proteins that play vital roles in cold hardiness and normal development during diapause [48,49]. Two Hsc70s, Hsc70-1 (BmUnigene24370.co) and Hsc70-4, (BmUnigene15892.co) were found up-regulated throughout diapause in this study (S5 Fig). Hsc70s are constitutively expressed members of the Hsp70 family and function as chaperone proteins that facilitate several biological processes including signal transduction, apoptosis, protein homeostasis, and cell growth and differentiation [50]. However, their exact roles in B. minax diapause remain unknown. Diapausing insects are commonly subjected to hypoxia or anoxia which may affect the intracellular redox potential [51]. To protect cells from oxidative damages predominantly caused by reactive oxygen species (ROSs), several enzymatic mechanisms may be activated, such as glutathione S-transferase (GST), superoxide dismutase (SOD), catalase (CAT), and peroxidases (POD) [52–55]. In B. minax, a microsomal GST (mGST, BmUnigene15924.co) and a Mn-SOD (BmUnigene23806.co) were up-regulated throughout diapause (S5 Fig). Likewise, members of the GST or SOD family were also up-regulated in other insects during diapause [32,56,57]. The up-regulation of these antioxidant proteins is speculated to protect individuals from oxidative damage in hypoxia/anoxia environments. Moreover, the expression of ryanodine receptor (RyR, BmUnigene27638.co) was higher in diapause (S5 Fig). The RyR protein functions as the major component of a calcium channel that mediates the release of calcium ions from the sarco/endoplasmic reticulum [58]. Calcium is an important second messenger, and its release from intracellular stores is critical for activation of many Ca2+-dependent pathways [59]. It has been demonstrated that intracellular Ca2+ is required for basal cellular metabolism and cell survival [60], and rendered cold tolerance to organisms [61]. This may also be the case for diapausing B. minax. These proteins potentially contribute to development and stress resistance during diapause, though the exact roles they play remain unknown.
The transcriptomic profile of B. minax pupal development investigated previously [8] is partially consistent with the current study. For example, both studies demonstrated that both metabolism and hormone biosynthesis were suppressed in diapause and several HSPs exhibited a diverse expression pattern during the pupal stage. However, crucial differences between the two studies exist, most likely due to the different periods of sampling and methods of sequencing.
Metabolomic profile during pupal development
PCA and PLS-DA plots showed that the variation in metabolomic profiles throughout pupal development was consistent with that observed in the gene expression profiles (Fig 6). Based on VIP scores, the variation in the metabolomic profiles was mainly attributed to changes in nine metabolites (Fig 7 and Table 2), in particular proline and trehalose, which exhibited higher VIP scores than the others and were maintained at higher concentrations throughout diapause. Proline has been demonstrated to be an excellent cryoprotectant for insects to tolerate the formation of ice crystals in their body fluids [62]. Surprisingly, simply feeding Drosophila melanogaster larvae a proline-augmented diet can convert this chill susceptible insect to a freeze tolerant one, capable of surviving when 50% of its body water freezes [63]. Variations in the concentration of proline during pupal development were consistent with the DEG analysis which showed the “Arginine and proline metabolism” pathway was significantly altered (Table 1). Specifically, the up-regulation of two cytosol aminopeptidases (LAP3s, BmUnigene25466.co and BmUnigene21743.co) that release N-terminal proline from a peptide, and the down-regulation of three prolyl 4-hydroxylases (P4HAs, BmUnigene26736.c1, BmUnigene26352.co, and BmUnigene27483.co) that convert proline to hydroxyproline, may be conducive to proline accumulation throughout diapause (S5 Fig).
Trehalose is the principal sugar circulating in the hemolymph of most insects due to its unique chemical properties, and is an important agent for protecting cells from a variety of environmental stresses, such as dehydration, heat, freezing, and oxidation [64–66]. Trehalose accumulates in diapausing B. minax and many other winter diapausing insects, likely acting as a cryoprotectant to help organs resist temperature fluctuations [67–69]. Nevertheless, a trehalose 6-phosphate phosphatase (TPP, BmUnigene26329.co) that catalyzes the formation of trehalose was down-regulated (S5 Fig), and other TPPs were not significantly altered throughout diapause in this study. Therefore, the accumulation of trehalose may result from alterations in trehalases (Tres) that catalyze trehalose catabolism. Insect trehalase has been demonstrated to exist in two distinct forms, soluble (Tre-1) and membrane-bound (Tre-2) trehalase [70]. In the B. minax transcriptome database, only Tre-1 (BmUnigene24123.co) was identified and it did not show any significant alteration during the pupal stage (S5 Fig). Therefore, it can be inferred that another trehalase, Tre-2, may play an important role in the regulation of trehalose levels during pupal development. In addition, proline and trehalose are sources of energy in insects [64,71], accumulation of these two metabolites in diapause may facilitate energy-consuming post-diapause development.
N-acetylglutamate (NAcGlu) is an obligate activator of the urea cycle [72]. It has been reported that the accumulation of urea in diapause-destined larvae of cotton bollworm, Helicoverpa armigera, exerted a potential cryopreservative effect [32]. Thus, the elevated NAcGlu levels throughout pupal diapause may enhance urea production and contribute to cold resistance in B. minax. Moreover, the synthesis of NAcGlu by N-acetylglutamate synthase (NAGS) is stimulated by both arginine, an allosteric stimulator of NAGS, and glutamate, one of NAGS's substrates. Therefore, the high glutamate levels throughout diapause are expected to meet the requirement of NAcGlu synthesis. In this study, the elevated glutamate level may be associated with drastic down-regulation of two genes in the “Arginine, aspartate and glutamate metabolism” pathway (S5 Fig), glutamine synthetase (GlnA, BmUnigene25702.co) and glutamate dehydrogenase (GdhA, BmUnigene15155.co), which catalyze the conversion of glutamate to glutamine and 2-oxoglutarate, respectively.
Glutamine is a key metabolite playing roles in a variety of biochemical functions, such as protein and lipid synthesis, source of energy, nitrogen and carbon donation, and as a precursor to glutamate [73]. High concentrations of glutamine in PreD may be in preparation for relevant biological activities in diapause. During pupal development, the glutamine level is positively correlated to the expression level of GlnA, indicating the role of GlnA in the regulation of glutamine. Alanine is another amino acid that accumulated during diapause, which is consistent with several other studies [31,74,75]. High alanine levesl has been shown to exert synergistic and colligative effects with other solutes on cold tolerance [76].
Succinate and 2-oxoglutarate are intermediates in the tricarboxylic acid (TCA) cycle, which generates ATP and provides carbon skeletons for a range of biosynthetic processes. In diapausing B. minax, the TCA cycle may be inhibited as the concentrations of 2-oxoglutarate and two other intermediates identified in our study, pyruvate and fumarate, decreased. However, the concentrations of succinate increased in diapause, probably due to the up-regulation of succinyl-CoA synthetase (LSC1, BmUnigene28496.co) which converts succinyl-CoA to succinate and releases ATP/GTP by directly transferring a phosphoryl group to ADP/GDP (S5 Fig). This reaction is a substrate-level phosphorylation that provides a quicker but less efficient source of ATP/GTP, independent of external electron acceptors [77]. Therefore, the up-regulation of LSC1 is likely involved in maintaining the ATP/GTP levels under hypoxia and energy-limited conditions during diapause.
Conclusion
In this study, transcriptomic and metabolomic analyses were performed to discover differentially expressed genes and significantly altered metabolites that coincided with pupal development. The findings provide insights into the diapause programming of B. minax. All DEGs and significantly altered metabolites identified may collectively play roles in stress resistance and survival of B.minax during diapause. However, the functions of most of the genes and metabolites remain unknown. To elucidate the mechanisms underlying pupal diapause of B. minax and comprehensively understand this pest, further investigations will focus on the specific aspects of biological variation along with pupal development or between diapausing and non-diapausing pupae.
Supporting information
(DOC)
(DOCX)
(TIF)
Blue spot, expression of fold change > 2 and P value < 0.05. Orange spot, no difference in expression. PreD, pre-diapause. ED, early-diapause. MD, middle-diapause. LD, late-diapause. PD, post-diapause.
(TIF)
PreD, pre-diapause. ED, early-diapause. MD, middle-diapause. LD, late-diapause. PD, post-diapause.
(TIF)
1, 2-Hydroxybutyrate. 2, 2-Oxoglutarate. 3, 3-Aminoisobutyrate. 4, 3-Hydroxykynurenine. 5, Acetate. 6, Alanine. 7, Arginine. 8, Asparagine. 9, Aspartate. 10, Choline. 11, Citrate. 12, Cytidine. 13, Ethanol. 14, Ethanolamine. 15, Formate. 16, Fumarate. 17, Galactonate. 18, Glucose. 19, Glutamate. 20, Glutamine. 21, Glycine. 22, Guanosine. 23, Inosine. 24. Isoleucine. 25, Lactate. 26, Leucine. 27, Lysine. 28, Maltose. 29, Methanol. 30, N-Acetylglutamate. 31, O-Phosphocholine. 32, O-Phosphoethanolamine. 33, Pantothenate. 34, Phenylalanine. 35, Proline. 36, Pyruvate. 37, Succinate. 38, Threonate. 39, Threonine. 40, Trehalose. 41, Trigonelline. 42, Tryptophan. 43, Tyrosine. 44, UDP-N-Acetylglucosamine. 45, Uridine. 46, Valine. 47, sn-Glycero-3-phosphocholine. 48, trans-Aconitate. 49, β-Alanine. DSS, DSS Chemical Shape Indicator.
(TIF)
PreD, pre-diapause. ED, early-diapause. MD, middle-diapause. LD, late-diapause. PD, post-diapause. Hsc70, heat shock cognate protein 70. mGST, microsomal glutathione S transferase. Mn-SOD, Mn superoxide dismutase. RyR, ryanodine receptor. LAP3, cytosol aminopeptidases. P4HA, prolyl 4-hydroxylases. TPP, trehalose 6-phosphate phosphatase. Tre-1, trehalase-1. GlnA, glutamine synthetase. GdhA, glutamate dehydrogenase. LSC1, succinyl-CoA synthetase.
(TIF)
Acknowledgments
We would like to thank the technicians from Anachro Technologies Inc. for their technical support in the determination of samples. We also appreciate Dr. Charles Snart and three anonymous reviewers for their valuable comments and suggestions.
Data Availability
The clean read data are available in the GeneBank Sequence Read Archive (Accession No. SRP083788). Other data are within the paper and its Supporting Information files.
Funding Statement
This study was financially supported by the National Natural Science Foundation of China (31401742), the China Postdoctoral Science Foundation (2014M552307), and the Chongqing Research program of Basic Research and Frontier Technology (cstc2016jcyjA0203). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dorji C, Clarke AR, Drew RAI, Fletcher BS, Loday P, Mahat K, et al. Seasonal phenology of Bactrocera minax (Diptera: Tephritidae) in western Bhutan. B Entomol Res. 2006; 96: 531–538. [PubMed] [Google Scholar]
- 2.Drew RAI, Dorji C, Romig MC, Loday P. Attractiveness of various combinations of colors and shapes to females and males of Bactrocera minax (Diptera: Tephritidae) in a commercial mandarin grove in Bhutan. J Econ Entomol. 2006; 99: 1651–1656. [DOI] [PubMed] [Google Scholar]
- 3.Allwood AJ, Chinajariyawong A, Kritsaneepaiboon S, Drew RAI, Hamacek EL, Hancock DL, et al. Host plant records for fruit flies (Diptera: Tephritidae) in Southeast Asia. Raffles B Zool. 1999; supplement No. 7: 1–92. [Google Scholar]
- 4.Zhang Y, Zhang ZM. Occurrence and integrated control methods of Chinese citrus fly Bactrocera minax. Bull Agric Sci Technol. 2005; 2: 22–23. [Google Scholar]
- 5.Lv JX, Pan CK, Qi CJ. Impact of emergency on influencing mechanism of citrus circulation and its countermeasures—Taking snow disaster and citrus fruit fly events in 2008 as an example. J Huazhong Agric Univ (Soc Sci Ed). 2010; 6: 46–51. [Google Scholar]
- 6.Han P, Wang X, Niu CY, Dong YC, Zhu JQ, Desneux N. Population dynamics, phenology, and overwintering of Bactrocera dorsalis (Diptera: Tephritidae) in Hubei Province, China. J Pest Sci. 2011; 84: 289–295. [Google Scholar]
- 7.Chen EH, Dou W, Hu F, Tang S, Zhao ZM, Wang JJ. Purification and biochemical characterization of glutathione S-transferases in Bactrocera minax (Diptera: Tephritidae). Fla Entomol. 2012; 95: 593–601. [Google Scholar]
- 8.Dong YC, Desneux N, Lei CL, Niu CY. Transcriptome characterization analysis of Bactrocera minax and new insights into its pupal diapause development with gene expression analysis. Int J Biol Sci. 2014; 10: 1051–1063. doi: 10.7150/ijbs.9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong YC, Wan L, Pereira R, Desneux N, Niu CY. Feeding and mating behaviour of Chinese citrus fly Bactrocera minax (Diptera, Tephritidae) in the field. J Pest Sci. 2014; 87: 647–657. [Google Scholar]
- 10.Dong YC, Wang ZJ, Clarke AR, Pereira R, Desneux N, Niu CY. Pupal diapause development and termination is driven by low temperature chilling in Bactrocera minax. J Pest Sci. 2013; 86: 429–436. [Google Scholar]
- 11.Gao LZ, Liu YH, Wan XW, Wang J, Hong F. Screening of microsatellite markers in Bactrocera minax (Diptera: Tephritidae). Sci Agric Sin. 2013; 46: 3285–3292. [Google Scholar]
- 12.Wang J, Zhou HY, Zhao ZM, Liu YH. Effects of juvenile hormone analogue and ecdysteroid on adult eclosion of the fruit fly Bactrocera minax (Diptera: Tephritidae). J Econ Entomol. 2014; 107: 1519–1525. [DOI] [PubMed] [Google Scholar]
- 13.Zhang B, Nardi F, Hull-Sanders H, Wan XW, Liu YH. The complete nucleotide sequence of the mitochondrial genome of Bactrocera minax (Diptera: Tephritidae). PLoS ONE. 2014; 9: e100558 doi: 10.1371/journal.pone.0100558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denlinger DL. Regulation of diapause. Annu Rev Entomol. 2002; 47: 93–122. doi: 10.1146/annurev.ento.47.091201.145137 [DOI] [PubMed] [Google Scholar]
- 15.Kostal V. Eco-physiological phases of insect diapause. J Insect Physiol. 2006; 52: 113–127. doi: 10.1016/j.jinsphys.2005.09.008 [DOI] [PubMed] [Google Scholar]
- 16.Ragland GJ, Egan SP, Feder JL, Berlocher SH, Hahn DA. Developmental trajectories of gene expression reveal candidates for diapause termination: a key life-history transition in the apple maggot fly Rhagoletis pomonella. J Exp Biol. 2011; 214: 3948–3959. doi: 10.1242/jeb.061085 [DOI] [PubMed] [Google Scholar]
- 17.Hahn DA, Denlinger DL. Meeting the energetic demands of insect diapause: Nutrient storage and utilization. J Insect Physiol. 2007; 53: 760–773. doi: 10.1016/j.jinsphys.2007.03.018 [DOI] [PubMed] [Google Scholar]
- 18.Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010; 11: 31–46. doi: 10.1038/nrg2626 [DOI] [PubMed] [Google Scholar]
- 19.Kozarewa I, Ning Z, Quail MA, Sanders MJ, Berriman M, Turner DJ. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat Methods. 2009; 6: 291–295. doi: 10.1038/nmeth.1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yassour M, Kaplan T, Fraser HB, Levin JZ, Pfiffner J, Adiconis X, et al. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc Natl Acad Sci USA. 2009; 106: 3264–3269. doi: 10.1073/pnas.0812841106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Xiong KC, Liu YH. De novo transcriptome analysis of Chinese citrus fly, Bactrocera minax (Diptera: Tephritidae), by high-throughput Illumina sequencing. PLoS ONE. 2016; 11: e0157656 doi: 10.1371/journal.pone.0157656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenz EM, Wilson ID. Analytical strategies in metabonomics. J Proteome Res. 2007; 6: 443–458. doi: 10.1021/pr0605217 [DOI] [PubMed] [Google Scholar]
- 23.Farag MA, Porzel A, Wessjohann LA. Comparative metabolite profiling and fingerprinting of medicinal licorice roots using a multiplex approach of GC-MS, LC-MS and 1D NMR techniques. Phytochemistry. 2012; 76: 60–72. doi: 10.1016/j.phytochem.2011.12.010 [DOI] [PubMed] [Google Scholar]
- 24.Madji Hounoum B, Blasco H, Nadal-Desbarats L, Dieme B, Montigny F, Andres CR, et al. Analytical methodology for metabolomics study of adherent mammalian cells using NMR, GC-MS and LC-HRMS. Anal Bioanal Chem. 2015; 407: 8861–8872. doi: 10.1007/s00216-015-9047-x [DOI] [PubMed] [Google Scholar]
- 25.Tredwell GD, Edwards-Jones B, Leak DJ, Bundy JG. The development of metabolomic sampling procedures for Pichia pastoris, and baseline metabolome data. PLoS ONE. 2011; 6: e16286 doi: 10.1371/journal.pone.0016286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S, Nagana Gowda GA, Ye T, Raftery D. Advances in NMR-based biofluid analysis and metabolite profiling. Analyst. 2010; 135: 1490–1498. doi: 10.1039/c000091d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slupsky CM, Steed H, Wells TH, Dabbs K, Schepansky A, Capstick V, et al. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin Cancer Res. 2010; 16: 5835–5841. doi: 10.1158/1078-0432.CCR-10-1434 [DOI] [PubMed] [Google Scholar]
- 28.Jung J, Park M, Park HJ, Shim SB, Cho YH, Kim J, et al. (1)H NMR-based metabolic profiling of naproxen-induced toxicity in rats. Toxicol Lett. 2011; 200: 1–7. doi: 10.1016/j.toxlet.2010.09.020 [DOI] [PubMed] [Google Scholar]
- 29.Booth SC, Workentine ML, Wen J, Shaykhutdinov R, Vogel HJ, Ceri H, et al. Differences in metabolism between the biofilm and planktonic response to metal stress. J Proteome Res. 2011; 10: 3190–3199. doi: 10.1021/pr2002353 [DOI] [PubMed] [Google Scholar]
- 30.O'Sullivan A, Gibney MJ, Brennan L. Dietary intake patterns are reflected in metabolomic profiles: potential role in dietary assessment studies. Am J Clin Nutr. 2011; 93: 314–321. doi: 10.3945/ajcn.110.000950 [DOI] [PubMed] [Google Scholar]
- 31.Michaud MR, Denlinger DL. Shifts in the carbohydrate, polyol, and amino acid pools during rapid cold-hardening and diapause-associated cold-hardening in flesh flies (Sarcophaga crassipalpis): a metabolomic comparison. J Comp Physiol B. 2007; 177: 753–763. doi: 10.1007/s00360-007-0172-5 [DOI] [PubMed] [Google Scholar]
- 32.Zhang Q, Lu YX, Xu WH. Proteomic and metabolomic profiles of larval hemolymph associated with diapause in the cotton bollworm, Helicoverpa armigera. BMC Genomics. 2013; 14: 751 doi: 10.1186/1471-2164-14-751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu YX, Zhang Q, Xu WH. Global metabolomic analyses of the hemolymph and brain during the initiation, maintenance, and termination of pupal diapause in the cotton bollworm, Helicoverpa armigera. PLoS ONE. 2014; 9: e99948 doi: 10.1371/journal.pone.0099948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE. 2012; 7: e30619 doi: 10.1371/journal.pone.0030619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008; 5: 621–628. doi: 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 36.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010; 11: R106 doi: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998; 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saldanha AJ. Java Treeview-extensible visualization of microarray data. Bioinformatics. 2004; 20: 3246–3248. doi: 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- 39.Stacklies W, Redestig H, Scholz M, Walther D, Selbig J. pcaMethods-a bioconductor package providing PCA methods for incomplete data. Bioinformatics. 2007; 23: 1164–1167. doi: 10.1093/bioinformatics/btm069 [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Zhao J, Liu YH. Evaluation of endogenous reference genes in Bactrocera minax (Diptera: Tephritidae). Acta Entomol Sin. 2014; 57: 1375–1380. [Google Scholar]
- 41.Mevik BH, Wehrens R. The pls package: Principal component and partial least squares regression in R. J Stat Softw. 2007; 18: 1–23. [Google Scholar]
- 42.Broadhurst DI, Kell DB. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics. 2006; 2: 171–196. [Google Scholar]
- 43.Ragland GJ, Fuller J, Feder JL, Hahn DA. Biphasic metabolic rate trajectory of pupal diapause termination and post-diapause development in a tephritid fly. J Insect Physiol. 2009; 55: 344–350. doi: 10.1016/j.jinsphys.2008.12.013 [DOI] [PubMed] [Google Scholar]
- 44.Nusse R. Wnt signaling and stem cell control. Cell Res. 2008; 18: 523–527. doi: 10.1038/cr.2008.47 [DOI] [PubMed] [Google Scholar]
- 45.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005; 122: 421–434. doi: 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- 46.Sim C, Denlinger DL. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens. Proc Natl Acad Sci USA. 2008; 105: 6777–6781. doi: 10.1073/pnas.0802067105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sim C, Denlinger DL. Insulin signaling and the regulation of insect diapause. Front Physiol. 2013; 4: 189 doi: 10.3389/fphys.2013.00189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rinehart JP, Li A, Yocum GD, Robich RM, Hayward SAL, Denlinger DL. Up-regulation of heat shock proteins is essential for cold survival during insect diapause. Proc Natl Acad Sci USA. 2007; 104: 11130–11137. doi: 10.1073/pnas.0703538104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu Rev Physiol. 1999; 61: 243–282. doi: 10.1146/annurev.physiol.61.1.243 [DOI] [PubMed] [Google Scholar]
- 50.Mayer MP, Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci. 2005; 62: 670–684. doi: 10.1007/s00018-004-4464-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacRae TH. Gene expression, metabolic regulation and stress tolerance during diapause. Cell Mol Life Sci. 2010; 67: 2405–2424. doi: 10.1007/s00018-010-0311-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kankofer M. Antioxidative defence mechanisms against reactive oxygen species in bovine retained and not-retained placenta: activity of glutathione peroxidase, glutathione transferase, catalase and superoxide dismutase. Placenta. 2001; 22: 466–472. doi: 10.1053/plac.2001.0650 [DOI] [PubMed] [Google Scholar]
- 53.Enayati AA, Ranson H, Hemingway J. Insect glutathione transferases and insecticide resistance. Insect Mol Biol. 2005; 14: 3–8. doi: 10.1111/j.1365-2583.2004.00529.x [DOI] [PubMed] [Google Scholar]
- 54.Fridovich I. Superoxide anion radical (O2-.), superoxide dismutases, and related matters. J Biol Chem. 1997; 272: 18515–18517. [DOI] [PubMed] [Google Scholar]
- 55.Chelikani P, Fita I, Loewen PC. Diversity of structures and properties among catalases. Cell Mol Life Sci. 2004; 61: 192–208. doi: 10.1007/s00018-003-3206-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bi Z, Yang X, Yu W, Shu J, Zhang Y. Diapause-associated protein3 functions as Cu/Zn superoxide dismutase in the Chinese oak silkworm (Antheraea pernyi). PLoS ONE. 2014; 9: e90435 doi: 10.1371/journal.pone.0090435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tu X, Wang J, Hao K, Whitman DW, Fan Y, Cao G, et al. Transcriptomic and proteomic analysis of pre-diapause and non-diapause eggs of migratory locust, Locusta migratoria L. (Orthoptera: Acridoidea). Sci Rep. 2015; 5: 11402 doi: 10.1038/srep11402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santulli G, Marks AR. Essential roles of intracellular calcium release channels in muscle, brain, metabolism, and aging. Curr Mol Pharmacol. 2015; 8: 206–222. [DOI] [PubMed] [Google Scholar]
- 59.Berridge MJ, Bootman MD, Lipp P. Calcium-a life and death signal. Nature. 1998; 395: 645–648. doi: 10.1038/27094 [DOI] [PubMed] [Google Scholar]
- 60.Bround MJ, Wambolt R, Luciani DS, Kulpa JE, Rodrigues B, Brownsey RW, et al. Cardiomyocyte ATP production, metabolic flexibility, and survival require calcium flux through cardiac ryanodine receptors in vivo. J Biol Chem. 2013; 288: 18975–18986. doi: 10.1074/jbc.M112.427062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teets NM, Yi SX, Lee RE Jr., Denlinger DL. Calcium signaling mediates cold sensing in insect tissues. Proc Natl Acad Sci USA. 2013; 110: 9154–9159. doi: 10.1073/pnas.1306705110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kostal V, Zahradnickova H, Simek P. Hyperprolinemic larvae of the drosophilid fly, Chymomyza costata, survive cryopreservation in liquid nitrogen. Proc Natl Acad Sci USA. 2011; 108: 13041–13046. doi: 10.1073/pnas.1107060108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kostal V, Simek P, Zahradnickova H, Cimlova J, Stetina T. Conversion of the chill susceptible fruit fly larva (Drosophila melanogaster) to a freeze tolerant organism. Proc Natl Acad Sci USA. 2012; 109: 3270–3274. doi: 10.1073/pnas.1119986109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson SN. Trehalose—The insect 'blood' sugar. Adv Insect Physiol. 2003; 31: 205–285. [Google Scholar]
- 65.Benaroudj N, Lee DH, Goldberg AL. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J Biol Chem. 2001; 276: 24261–24267. doi: 10.1074/jbc.M101487200 [DOI] [PubMed] [Google Scholar]
- 66.Jain NK, Roy I. Effect of trehalose on protein structure. Protein Sci. 2009; 18: 24–36. doi: 10.1002/pro.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Q, Hao YJ, Li Y, Zhang YJ, Ren S, Si FL, et al. Gene cloning, characterization and expression and enzymatic activities related to trehalose metabolism during diapause of the onion maggot Delia antiqua (Diptera: Anthomyiidae). Gene. 2015; 565: 106–115. doi: 10.1016/j.gene.2015.03.070 [DOI] [PubMed] [Google Scholar]
- 68.Kostal V, Simek P. Dynamics of cold-hardiness, supercooling and cryoprotectants in diapausing and nondiapausing pupae of the cabbage root fly, Delia-Radicum L. J Insect Physiol. 1995; 41: 627–634. [Google Scholar]
- 69.Rozsypal J, Kostal V, Zahradnickova H, Simek P. Overwintering strategy and mechanisms of cold tolerance in the codling moth (Cydia pomonella). PLoS ONE. 2013; 8: e61745 doi: 10.1371/journal.pone.0061745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang J, He WB, Su YL, Bing XL, Liu SS. Molecular characterization of soluble and membrane bound trehalases of the whitefly, Bemisia tabaci. Arch Insect Biochem. 2014; 85: 216–233. [DOI] [PubMed] [Google Scholar]
- 71.Arrese EL, Soulages JL. Insect fat body: Energy, metabolism, and regulation. Annu Rev Entomol. 2010; 55: 207–225. doi: 10.1146/annurev-ento-112408-085356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caldovic L, Tuchman M. N-acetylglutamate and its changing role through evolution. Biochem J. 2003; 372: 279–290. doi: 10.1042/BJ20030002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Newsholme P, Lima MMR, Porcopio J, Pithon-Curi TC, Doi SQ, Bazotte RB, et al. Glutamine and glutamate as vital metabolites. Braz J Med Biol Res. 2003; 36: 153–163. [DOI] [PubMed] [Google Scholar]
- 74.Rivers DB, Lee RE, Denlinger DL. Cold hardiness of the fly pupal parasitoid Nasonia vitripennis is enhanced by its host Sarcophaga crassipalpis. J Insect Physiol. 2000; 46: 99–106. [DOI] [PubMed] [Google Scholar]
- 75.Li YP, Goto M, Ito S, Sato Y, Sasaki K, Goto N. Physiology of diapause and cold hardiness in the overwintering pupae of the fall webworm Hyphantria cunea (Lepidoptera: Arctiidae) in Japan. J Insect Physiol. 2001; 47: 1181–1187. [DOI] [PubMed] [Google Scholar]
- 76.Churchill TA, Storey KB. Metabolic consequences of rapid cycles of temperature-change for freeze-avoiding vs. freeze-tolerant insects. J Insect Physiol. 1989; 35: 579–585. [Google Scholar]
- 77.Lambeth DO, Tews KN, Adkins S, Frohlich D, Milavetz BI. Expression of two succinyl-CoA synthetases with different nucleotide specificities in mammalian tissues. J Biol Chem. 2004; 279: 36621–36624. doi: 10.1074/jbc.M406884200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOCX)
(TIF)
Blue spot, expression of fold change > 2 and P value < 0.05. Orange spot, no difference in expression. PreD, pre-diapause. ED, early-diapause. MD, middle-diapause. LD, late-diapause. PD, post-diapause.
(TIF)
PreD, pre-diapause. ED, early-diapause. MD, middle-diapause. LD, late-diapause. PD, post-diapause.
(TIF)
1, 2-Hydroxybutyrate. 2, 2-Oxoglutarate. 3, 3-Aminoisobutyrate. 4, 3-Hydroxykynurenine. 5, Acetate. 6, Alanine. 7, Arginine. 8, Asparagine. 9, Aspartate. 10, Choline. 11, Citrate. 12, Cytidine. 13, Ethanol. 14, Ethanolamine. 15, Formate. 16, Fumarate. 17, Galactonate. 18, Glucose. 19, Glutamate. 20, Glutamine. 21, Glycine. 22, Guanosine. 23, Inosine. 24. Isoleucine. 25, Lactate. 26, Leucine. 27, Lysine. 28, Maltose. 29, Methanol. 30, N-Acetylglutamate. 31, O-Phosphocholine. 32, O-Phosphoethanolamine. 33, Pantothenate. 34, Phenylalanine. 35, Proline. 36, Pyruvate. 37, Succinate. 38, Threonate. 39, Threonine. 40, Trehalose. 41, Trigonelline. 42, Tryptophan. 43, Tyrosine. 44, UDP-N-Acetylglucosamine. 45, Uridine. 46, Valine. 47, sn-Glycero-3-phosphocholine. 48, trans-Aconitate. 49, β-Alanine. DSS, DSS Chemical Shape Indicator.
(TIF)
PreD, pre-diapause. ED, early-diapause. MD, middle-diapause. LD, late-diapause. PD, post-diapause. Hsc70, heat shock cognate protein 70. mGST, microsomal glutathione S transferase. Mn-SOD, Mn superoxide dismutase. RyR, ryanodine receptor. LAP3, cytosol aminopeptidases. P4HA, prolyl 4-hydroxylases. TPP, trehalose 6-phosphate phosphatase. Tre-1, trehalase-1. GlnA, glutamine synthetase. GdhA, glutamate dehydrogenase. LSC1, succinyl-CoA synthetase.
(TIF)
Data Availability Statement
The clean read data are available in the GeneBank Sequence Read Archive (Accession No. SRP083788). Other data are within the paper and its Supporting Information files.