Abstract
Sparsely tested group of platinum nanoparticles (PtNPs) may have a comparable effect as complex platinum compounds. The aim of this study was to observe the effect of PtNPs in in vitro amplification of DNA fragment of phage λ, on the bacterial cultures (Staphylococcus aureus), human foreskin fibroblasts and erythrocytes. In vitro synthesized PtNPs were characterized by dynamic light scattering (PtNPs size range 4.8–11.7 nm), zeta potential measurements (-15 mV at pH 7.4), X-ray fluorescence, UV/vis spectrophotometry and atomic absorption spectrometry. The PtNPs inhibited the DNA replication and affected the secondary structure of DNA at higher concentrations, which was confirmed by polymerase chain reaction, DNA sequencing and DNA denaturation experiments. Further, cisplatin (CisPt), as traditional chemotherapy agent, was used in all parallel experiments. Moreover, the encapsulation of PtNPs in liposomes (LipoPtNPs) caused an approximately 2.4x higher of DNA damage in comparison with CisPt, LipoCisPt and PtNPs. The encapsulation of PtNPs in liposomes also increased their antibacterial, cytostatic and cytotoxic effect, which was determined by the method of growth curves on S. aureus and HFF cells. In addition, both the bare and encapsulated PtNPs caused lower oxidative stress (determined by GSH/GSSG ratio) in the human erythrocytes compared to the bare and encapsulated CisPt. CisPt was used in all parallel experiments as traditional chemotherapy agent.
Introduction
The first anticancer drug cisplatin (cis-diamminedichloroplatinum(II)) was discovered in 1965 by Rosenberg during his studies on the effects of an electric current on bacterial growth [1]. The binding of cisplatin to DNA and the interactions with non-DNA targets with subsequent triggering of cell death through apoptosis, necrosis or both belong to the most important parameters of cisplatin cytotoxicity [2]. Since then, numerous platinum complexes of second generation (carboplatin, oxaliplatin, nedaplatin) and third generation (lobaplatin, heptaplatin) have been developed [3] and evaluated as anticancer agents [4, 5]. The undesired side effects can be eliminated by specific molecular interactions of a drug with cancer cells and by selective targeting, which can be done by their encapsulation in the molecules suitable for selective transportation. The most common transporters belong to functionalized polymers, nanoparticles with conjugated platinum drugs, carbon nanotubes and micelles. Among them, liposomes, which consist of a phospholipid bilayer surrounding an aqueous core, are a very important class of compounds [6]. Liposomes have interesting biological activities including effective drug loading capacity, biocompatibility and improved pharmacokinetics. The liposomal formulations of cisplatin and oxaliplatin (Lipoplatin™ and Lipoxal™) are now widely used to reduce drug toxicity and also improve drug targeting [7, 8]. Experimentally, many nanoparticles consisting of the polymers or copolymers and the Pt-based drugs have been tested. Among these, polyethylene glycol-functionalized poly-isobutylene-maleic acid copolymer can form complexes with cisplatin. The nanoparticles are internalized into the endolysosomal compartment of cancer cells releasing cisplatin in a pH-dependent manner. These nanoparticles have been developed for better antitumor efficacy, which was confirmed in a 4T1 breast cancer model in vivo with limited nephrotoxicity [9]. Furthermore, the nanoparticles treatment resulted in reduced systemic and nephrotoxicity, which was confirmed by the decreased biodistribution of platinum in kidney [10, 11]. The nanoparticles of oxaliplatin and carboplatin with polyisobutylene maleic acid copolymer with glucosamine and albumin-bound paclitaxel, respectively, were also tested [12, 13].
Plenty of studies discuss the effects of halogenated Pt salts and complexes and are reviewed elsewhere [14–16], whereas the effects of PtNPs are not mostly elucidated. It was found that PtNPs have anti-oxidant and anti-inflammatory effects. In this case, the platinum nanoparticles are able to scavenge superoxide anion radical (O2 -) and hydroxyl radical (OH) [11, 17, 18] and this ability was also shown for the PtNPs within apoferritin [19, 20]. The ability of the PtNPs to scavenge the reactive oxygen species was studied in mouse ischemic brain. The treatment with PtNPs significantly improved motor function and greatly reduced the infarct volume, especially in the cerebral cortex [21, 22]. The effects of the PtNPs of sizes of 20, 100 and more than 100 nm on HT29 have been confirmed and have been found to cause DNA strand breaks in a concentration, time and size-dependent manner, but appear not to translocate into the nucleus or interact with mitochondria [23].
The interaction of Pt-attached iron oxide nanoparticles with DNA of was studied previously using gel electrophoresis and transmission electron microscopy. Two types of interactions were suggested in this case: the first interaction involved the direct linkage of DNA molecules onto the surface of the nanoparticles, while the second interaction was suggested to be the detachment of small PtNPs from the iron oxide support by the strong DNA interaction [24, 25]. On the other hand, the PtNPs did not show cytotoxicity in several different cultured cells (TIG-1, MI-38, MRC-5, HeLa, and HepG2) [17]. The contradiction of these studies indicates that the biological effects of PtNPs remains unclear and seems to be dependent on the particle size and most probably surface properties.
The interaction of different nanoparticles (NPs) with bacteria and fungi was studied by Chwalibog et al. and Sawosz et al. [26, 27]. It was found that silver, gold, and PtNPs, with negative zeta potential, had cell damaging properties on Staphylococcus aureus (Gram-positive bacteria) and Candida albicans (fungi). The analysis of morphological effects of the interaction of PtNPs with Listeria monocytogenes (Gram-positive) by transmission electron microscopy (TEM) revealed that the PtNPs entered into the cells of Listeria monocytogenes. The authors even stated that the nanoparticles were located close to DNA and probably bound to it. In the case of Salmonella enteritidis (Gram-negative), the PtNPs probably bound to DNA. This was studied by confocal microscopy with Nomarski contrast and fluorescent DAPI labelling. After washing and centrifugation, the PtNPs-DNA complexes were characterized. The results indicated that the bacteria could be used as a transporter to deliver PtNPs to the specific points in the body [27, 28].
In this work, we present our results on the preparation of liposome with encapsulated platinum derivatives both PtNPs and cisplatin (CisPt). The aim of this study was to investigate the effect of these PtNPs on in vitro amplification of DNA fragment of phage λ, and on the bacterial cultures (Staphylococcus aureus), human foreskin fibroblasts (HFF) and erythrocytes.
Materials and methods
Chemicals and material
Cholesterol, 1,2-dioleoyl-sn-glycero-3-phospho-rac-(1-glycerol) sodium salt, chloroform, cisplatin, PtCl4, polyvinylpyrrolidone (PVP, 40 k), HCl (37%, w/w), NaBH4 and water were purchased from Sigma-Aldrich (St. Louis, MO, USA) in ACS purity. Hydrogenated phosphatidylcholine from soybean was a gift from Lipoid GMBH (Ludwigshafen, Germany). The deionised water was prepared using Aqual 25 (Aqual, Brno, Czech Republic) and was further purified using apparatus MilliQ Direct QUV equipped with the UV lamp (Merck Millipore, Merck KgaA, Darmstadt, Germany). The pH of the solutions was measured using a pH meter WTW inoLab (WTW GmbH, Weilheim, Germany).
Preparation of PtNPs
The PtNPs were prepared according to Oh et al. [29] with a slight modification. The PtNPs were also covered by polyvinylpyrrolidone (PVP), in order to prevent their aggregation [30]. Briefly, PtCl4 (0.034 g) was dissolved in acidic water (5 mL) with 16 μL of 37% HCl. The solution of PtCl4 (5 mL) was added, with stirring, to another solution of 0.135 g PVP in water (45 mL). The mixture was stirred for 1 h at 25°C. After the addition of NaBH4 (50 mg) the final colour of the solution became black. The mixture was stirred overnight. Prepared particles were characterized as described in S1 Text.
Preparation of liposome film
The liposomes were prepared according to the published method [31] with necessary modifications [32]. Briefly, cholesterol (100 mg), 1,2-dioleoyl-sn-glycero-3-phospho-rac-(1-glycerol) sodium salt (100 mg) and phosphatidylcholine (100 mg) were dissolved in chloroform (4.5 mL). A lipid film was obtained by rotary evaporation of chloroform. The residual chloroform was removed by nitrogen.
Preparation of liposome filled with PtNPs (LipoPtNPs)
The solutions containing 0, 0.015, 0.03131, 0.0625, 0.125, 0.25, 0.5 or 1 mL of platinum nanoparticles (PtNPs, 400 μg/mL of Pt) were diluted with water to 1 mL and added to liposome (20 mg). The samples were homogenized for 15 min in an ultrasonic bath Sonorex Digital 10P (Bandelin, Berlin, Germany). The homogenized mixtures were then heated and shaken for 15 min at 60°C using a Thermomixer Comfort (Eppendorf, Germany). The samples were then washed several times with water employing Amicon 3k centrifugation filters (Merck Millipore, Merck KgaA, Darmstadt, Germany) with a final sample volume of 1 mL [32].
Preparation of liposome filled with cisplatin (LipoCisPt)
The solutions containing 0, 0.015, 0.03131, 0.0625, 0.125, 0.25, 0.5 or 1 mL of cisplatin (CisPt, 0.4 mg Pt per mL) were diluted with water to 1 mL and added to liposome (20 mg). The samples were prepared according to the same procedure described in paragraph above.
Polymerase chain reaction (PCR)
Taq PCR kit was purchased from New England Biolabs (Ipswitch, MA, USA). The forward and reverse primers for the amplification of λ xis gene fragment were synthesized by Sigma-Aldrich and their sequences were 5'-CCTGCTCTGCCGCTTCACGC-3' and 5'-TCCGGATAAAAACGTCGATGACATTTGC-3', respectively. The volume of the reaction mixture was 25 μL, which was composed of 2.5 μL of 10× standard Taq reaction buffer, 0.5 μL of 1 mM deoxynucleotide solution, 0.5 μL of each of the primers (10 μM), 0.125 μL of Taq DNA polymerase; selected volume of water or drugs diluted with water (sterile, ACS purity, Sigma-Aldrich) and 0.5 μL of bacteriophage λ DNA.
The PCR was performed using Mastercycler ep realplex4 S (Eppendorf, AG, Hamburg, Germany) with the following cycling conditions: initial denaturation for 120 s at 95°C; 30 cycles of PCR (15 s at 95°C, 15 s at 64°C and 45 s at 72°C) with a final elongation for 5 min at 72°C. The obtained DNA fragments (498 bp) were purified by MinElute PCR Purification Kit (Qiagen, Germany). The DNA concentration was determined spectrophotometrically.
Agarose gel electrophoresis
The amplified product was analysed using agarose gel electrophoresis and the working conditions were as follows: 2% agarose gel (High melt/Medium fragment, Mercury, San Diego, CA, USA) with 1× TAE buffer, 60 V and 160 min (Bio-Rad, Hercules, CA, USA). The 100 bp DNA ladder (New England Biolabs, Ispwich, MA, USA) was used as a molecule size marker. The bands were visualized using UV transilluminator at 312 nm (Vilber-Lourmat, Marne-la-Vallée Cedex, France).
The fluorescence intensity of the ethidium bromide was detected by In-Vivo Xtreme (Carestream Health Inc., Rochester, NY, USA). The parameters were as follows: Excitation filter: 520 nm; Emission filter: 600 nm; Exposure time: 6 s; Binning: 2 × 2, f-Stop: 1.1; and Field of View: 19.5 × 19.5 cm. The images were processed by Carestream Molecular Software. The fluorescence was quantified using software and the background was subtracted. The intensity of the fluorescence of ethidium bromide was directly proportional to the concentration of the DNA.
DNA sequencing
The purified DNA fragments (50 μg/mL) were mixed with different concentrations of PtNPs (25, 50, 100, 200 and 400 μg/mLPt) in a ratio of 1:1 (v/v). The solutions of DNA with PtNPs were incubated for 1 h at 25°C (Thermomixer 5355, Eppendorf, Germany). To remove the excess of PtNPs, a dialysis was performed for 12 h at 6°C using 0.025 μm membrane filter (Millipore, Ireland) with ACS water.
CisPt interactions with DNA
The solution of the DNA fragments (50 μg/mL) was mixed with different concentrations of cisplatin (25, 50, 100, 200 and 400 μg/mLPt) in a rate of 1:1 (v/v) in the environment of 10 mM NaClO4. The solutions of DNA with the drugs were incubated for 24 h at 37°C. To remove the excess of the platinum-based drug a dialysis was performed for 24 h at 6°C using a 0.025 μm membrane filter (Millipore, Ireland).
Sequencing of platinated DNA
The conditions of sequencing reaction were as described elsewhere [33].
Comet assay
The comet assay was performed according to Singh et al. [34] with small modifications [24, 35]. Conventional slides were immersed in 0.5% normal-melting agarose (v/v, CLP, San Diego, USA) and dried at 50°C. The cells and slides were handled under dimmed light conditions during the whole process. Briefly, 75 μL of 0.5% low-melting agarose was mixed with approximately 10000 cells suspended in 10 μL of PBS and layered onto the slides and immediately covered with coverslips. After agarose solidification (5 min at 4°C), the coverslips were removed and the slides were immersed overnight in dark at 4°C in freshly prepared lysing solution (2.5 M NaCl, 100 mM Na2EDTA, 10 mM Tris, pH 10) containing 1% Triton X-100 and 10% DMSO (v/v). After the lysis, the slides were placed on a horizontal gel electrophoresis unit filled with electrophoretic buffer (300 mM NaOH, 1mM Na2EDTA, pH > 13) and left there for 30 min in dark at 4°C for DNA unwinding. Then, the electrophoresis was performed for 30 min at 1.25 V/cm (300 mA). After the electrophoresis, the slides were washed with neutralizing buffer (0.4 M Tris, pH 7.5) and the slides were stained with 2 μg/mL of ethidium bromide for 5 min in dark at 25°C. After the staining, the slides were washed with distilled water, covered with coverslips and imaged by a microscope using the inverted system fluorescence microscope Olympus IX 71S8F-3 (Tokyo, Japan). The excitation filter 520–550 nm and the emission filter of 580 nm were employed (Magnification: 100×) in this experiment. The images were captured by Camera Olympus DP73 and processed by Stream Basic 1.7 Software with the resolution of 4800 × 3600 pixels. From each slide, 10 fields were taken and the nuclei were classified in five categories ranging from 0 (no visible tail) to 4 (most DNA in tail). To quantify the effect of the drugs on DNA damage, an index of damage (ID) was calculated as a sum of the numbers of the nuclei in each category multiplied with 0.1 for category 0 and by 1, 2, 3 and 4 for other categories divided by total number of the nuclei.
Cultivation of Staphylococcus aureus
Staphylococcus aureus (NCTC 8511) was obtained from the Czech Collection of Microorganisms, Faculty of Science, Masaryk University, Brno, Czech Republic. The strains of the bacterium were stored as a spore suspension in 20% (v/v) glycerol at −20°C according to the conditions mentioned in Chudobova et al. [36, 37]. Prior to use in this study, the strains were thawed and the glycerol was removed by washing with distilled water. The cultivation medium contained: meat peptone (5 g/L), NaCl (5 g/L), bovine extract (1.5 g/L), yeast extract (1.5 gL) (HIMEDIA, Mumbai, India), and sterilized MilliQ water with 18 MΩ. Before sterilization, the pH of the cultivation medium was adjusted to 7.4 and the media were sterilized at 121°C for 30 min in a sterilizer (Tuttnauer 2450EL, Israel). The prepared cultivation media was inoculated with the bacterial culture into 25 mL Erlenmeyer flasks and subsequently the bacterial cultures were cultivated for 24 h on a shaker at 600 rpm and 37°C. The cultivated bacterial cultures were diluted by the cultivation medium to the optical density (OD600) = 0.1 AU and used in the following experiments [38].
Growth curves
The antimicrobial effect of the tested compounds was analysed by measuring the absorbance using an apparatus Multiskan EX (Thermo Fisher Scientific, Germany) according to Chudobova et al. [39]. Briefly, the concentrations of the platinum compounds (PtNPs and/or cisplatin) were 0, 0.5, 1.0, 1.9, 3.8, 7.5, 15, 30 and 60 μg/mL of Pt. The obtained results were recalculated to relative absorbance value, thus the control growth curve without the addition of the platinum derivatives refers to 100%. The values less than 100% indicated relative cell death.
Cultivation of human foreskin fibroblast
Human Foreskin Fibroblast (HFF) cell line was obtained from ATCC Cell Lines (Wesel, Germany). HFF cell line was grown as monolayers in DMEM (high glucose) medium with 10% foetal bovine serum, supplemented with penicillin and streptomycin (1 U per mL) at 37°C in 5% CO2 atmosphere. Approximately 2 × 105 cells at logarithmic growth phase were treated with free and/or encapsulated CisPt or PtNPs in concentrations 0, 12.5, 25, 50, 100 and 200 μg/mL of Pt for 2h, and harvested using a cell-scraper and resuspended in 200 μL of PBS. After the treatment, the cells were harvested using a cell-scraper and resuspended in 200 μL of PBS. All the experiments were performed in two replicates. HFF were also used for Comet assay.
Cell growth and proliferation assay of HFF using impedance measurement with xCELLigence system
The xCELLigence system was used according to the instructions of the supplier (Roche Applied Science and ACEA Biosciences). The xCELLigence system consists of four main components: a Real time cell analysis (RTCA) analyser, a RTCA DP station, a RTCA computer with integrated software and a disposable E-plate 16. Firstly, the optimal seeding concentration for the proliferation and RTCA assay of HFF was determined. After seeding the total number of the cells in 200 μL medium in each well of the E plate 16, the attachment, proliferation and spreading of the cells were monitored every 15 min. All the experiments were carried out for 100 h. The obtained results were recalculated to the relative (%) value of the growth. The method of the calculation was same as described for Staphylococcus aureus.
Preparation of erythrocytes for haematological analysis and determination of glutathione
Human blood was obtained from the Department of Physiology, Faculty of Medicine, Masaryk University, Czech Republic. We analysed the samples of healthy volunteers (n = 3), whereas the written consent of blood donors was granted. The research has been approved by the Independent ethics committee (No. 104MS451) at University Hospital, Brno, Czech Republic. The blood samples of three healthy volunteers were collected in the tubes with heparin. The blood samples were centrifuged for 10 min at 2000 rpm. After removing of plasma, the erythrocytes together with platelets were diluted in phosphate buffer pH 7 in a ratio of 1:1. The number of erythrocytes and platelets was determined using haematology analyser BC– 5800 (Mindray, Shenzhen, China). Cisplatin or cisplatin encapsulated in liposomes (LipoCisPt) were added to the erythrocytes mixture with platelets and various concentrations of the Pt derivatives (PtNPs, PtNPs encapsulated in liposomes (LipoPtNPs)) in a ratio of 1:1 to reach the desired concentrations (12.5, 25, 50, 100, 200 μg/mL of Pt and 0.6, 1.3, 2.5, 5 and 10 mg/mL of liposomes) in the resulting mixture. The erythrocytes treated with the platinum compounds were mixed with trifluoroacetic acid (TFA) in a ratio of 2:1 (100 μL of erythrocytes + 50 μL of 15% TFA (v/v)). Subsequently, the mixture was homogenized using ultrasonic needle for 2 min and vortexed for 10 min. After the centrifugation of the mixture for 20 min at 25 000 rpm, 20 μL of the supernatant was used for chromatographic analysis.
Estimation of selected haematological parameters of erythrocytes mixture with platelets exposed to platinum derivatives
The haematological analyses of the erythrocytes mixture with platelets exposed to platinum derivatives encapsulated in liposomes or alone were performed using Haematology Analyser BC-5800 (Mindray, Shenzen, China) coupled with flow cytometry. The number of erythrocytes and platelets was determined by impedance method. The flow cytometry together with fluorescence-activated cell sorting was applied to determine the number of leukocytes. The histograms of the erythrocytes mixture with platelets and Pt derivatives encapsulated in liposomes or alone were recorded. The liposomes were visualised in the Baso channel as a red rhombus.
Determination of reduced and oxidized glutathione
Reduced (GSH) and oxidized (GSSG) glutathione were assayed using a high performance liquid chromatography with electrochemical detection (HPLC-ED) according to Kleckerova et al. [40] and Skladanka et al. [41]. Two solvent delivery pumps (Model 582 ESA Inc., Chelmsford, MA, USA) operating in a range of 0.001–9.999 mL.min-1, a column Zorbax eclipse AAA C18 (150 × 4.6; 3.5 μm particle size; Agilent Technologies, Santa Clara, CA, USA) and a CoulArray electrochemical detector (Model 5600A, ESA, USA) with one flow cell were used in this experiment. The cell consists of four working carbon porous electrodes, each one with auxiliary and dry Pd/H2 reference electrodes. Both the detector and the reaction coil/column were thermostated. Twenty microliters sample was injected using autosampler (Model 542 HPLC, ESA, USA). The samples were stored in the carousel at 8°C. The column was thermostated at 32°C. The mobile phase was consisted of 80 mM TFA (A) and methanol (B). The compounds of interest were separated by the following linear gradient: 0 → 1 min (3% B, v/v), 1 → 2 min (10% B, v/v), 2 → 5 min (30% B, v/v), 5 → 6 min (98% B, v/v). The flow rate of the mobile phase was 1 mL.min-1 and working electrode potential was set to 900 mV
Descriptive statistics
Data were processed using MICROSOFT EXCEL® (USA) and STATISTICA.CZ Version 8.0 (Czech Republic). Results are expressed as mean ± standard deviation (S.D.) unless noted otherwise (EXCEL®). Statistical significances of the differences were determined using STATISTICA.CZ. Differences with p < 0.05 (labelled with “*” in figures) were considered significant and were determined by using of one way ANOVA test (particularly Scheffe test), which was applied for means comparison.
Results
Characterization of PtNPs
The synthesized PtNPs as well as the liposomal formulations were characterized by particle-size and Zeta potential measurement, transmission microscopy imaging, X-ray fluorescence and UV/Vis spectrophotometry and atomic absorption spectrometry. The size of the PtNPs determined by dynamic light scattering was mainly 6.5 nm in diameter with the Zeta potential of -15 kV (Figures A and B in S1 Fig). The X-ray spectrum of the PtNPs was recorded and the two characteristic peaks of Pt Kα (391 keV) and Pt Kβ (457 keV) were observed (Figures A in S2 Fig). The presence of Pt was also confirmed by atomic absorption spectrometry. Subsequently, the stock solution of PtNPs with Pt concentration of 400 μg/mL was prepared and used for further experiments. For these experiments, the stock solution was diluted creating solutions of PtNPs with Pt concentrations ranging from 0 to 200 μg/mL of Pt in PtNPs. The colour of the PtNPs was found to be brown. (Figures B and C in S1 Fig).
PtNPs and CisPt interactions with DNA fragment from phage λ and Taq DNA polymerase
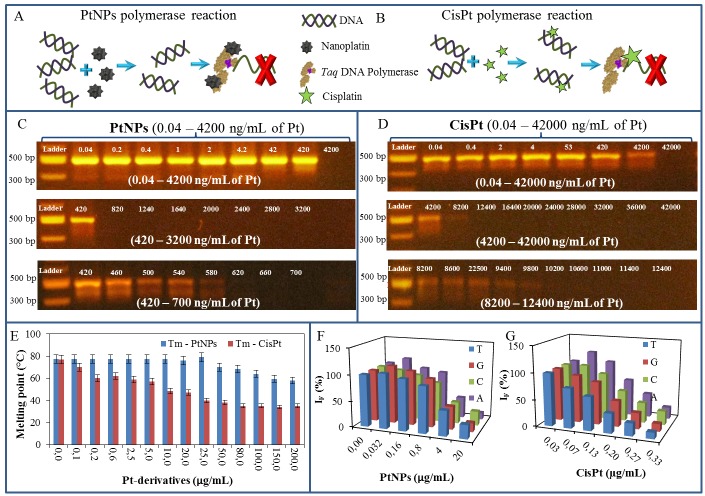
The impact of PtNPs and CisPt on DNA was studied using PCR, DNA melting point (Tm) analysis and Sanger sequencing. It was observed that the function of Taq DNA polymerase was blocked by the PtNPs more effectively than by CisPt. The suggested scheme of the possible mechanism is shown in Figs 1A and 2B.
Fig 1.
(A) The suggested effects of PtNPs on polymerase chain reaction (PCR) is based on binding of PtNPs to the Taq DNA polymerase, which leads to ceasing of PCR, (B) whereas CisPt primarily intercalates in DNA structure and stops PCR by this way. The gel electrophoregrams of PCR product mixture with particular concentration of (C) PtNPs (0.04–4 200 ng/mL of Pt) and (D) CisPt (0.04–42 000 ng/mL of Pt). (E) DNA denaturation temperature affected by the 0–200 μg/mL of Pt derivatives. Fluorescence of labelled nucleotides of DNA fragment after sequencing, which was influenced by (F) 0–20 μg/mL of PtNPs and (G) 0–0.33 μg/mL of CisPt. For all measurement n = 3.
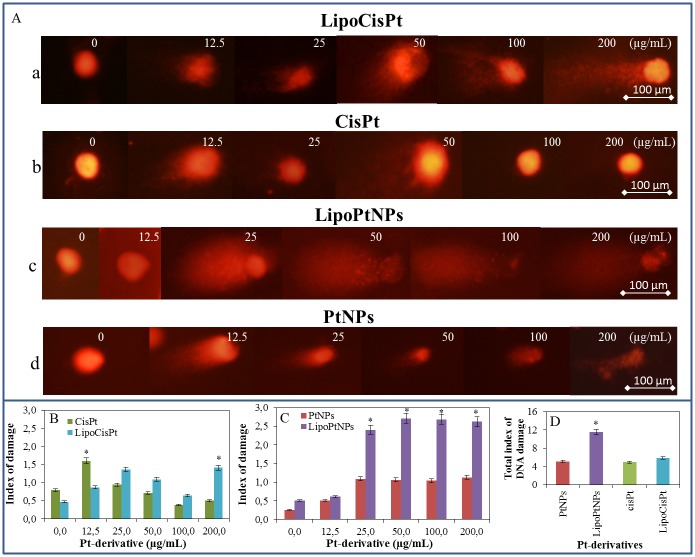
Fig 2.
(A) The alkaline comet assay of nuclear DNA from human fibroblasts (HFF) treated with (a) cisplatin (0–200 μg/mL of Pt) encapsulated in liposome LipoCisPt, (b) same concentration of CisPt only, (c) PtNPs encapsulated in liposome LipoPtNPs and (d) free PtNPs. (B) The index of damage of CisPt and LipoCisPt. (C) The index of damage of PtNPs and LipoPtNPs. (D) The nuclear DNA of human fibroblast and summarized their total damage. For all measurement n = 3.
In the first part of the experiment, it was observed that the PCR was stopped after the addition of at least 0.58 μg/mL PtNPs (Fig 1C). CisPt was studied in the same way and it was found that 9.8 μg/mL CisPt was able to stop the PCR, which is nearly 17-times higher concentration compared to that of the PtNPs (Fig 1D).
In the second part of the experiment, the changes in denaturation temperatures (Tm) were studied. To observe these changes, 5 μg/mL DNA fragments from λ bacteriophage was mixed with the PtNPs or CisPt (0–200 μg/mL). 0.1 μg/mL CisPt decreased the Tm by 7°C compared to the untreated control sample (77°C). To reach the same effect on the Tm, 500 times more PtNPs (50 μg/mL) was needed (Fig 1E). Based on this observation, it was concluded that PtNPs interact more likely with polymerase than with the DNA.
In the third part of the experiment, the DNA fragment of λ bacteriophage (5 μg/mL) was added to the CisPt or PtNPs solutions. These mixtures were subsequently dialysed to remove the particles, which were not interacted or bound to DNA. The modified DNA was used for Sanger sequencing reaction and the DNA was further analysed by the capillary gel electrophoresis with laser-induced fluorescence detection. The fluorescence signals of the probed fragments were compared to the control DNA sample (without the addition of CisPt or PtNPs). Fig 1F shows that the addition of 0.032 μg/mL PtNPs induced the increase in the fluorescence signals of the fragments terminated with the dideoxynucleotides thymine (T), guanine (G) or adenine (A) compared to the control sample. In the case of DNA fragments terminated with the cytosine (C), the decreasing fluorescence was observed even when 0.032 μg/mL PtNPs were added.
After CisPt application, the fluorescence signals of DNA fragments terminated with T and G decreased with the increasing concentration of CisPt. At the DNA fragments terminated with C and A, the fluorescence signal firstly increased (at 0.07 μg/mL CisPt), followed by the decrease in the fluorescence signal with the increasing concentration of CisPt as showed in Fig 1G. The signal decrease by 50% was reached after the application of PtNPs and CisPt in concentrations of 9.6 μg/mL and 0.13 μg/mL, respectively.
From the results of the second and third part of the experiment it is clear that the concentration of the PtNPs should be 500 times (in the case of Tm change) and 74 times (in the case of sequencing) higher than that of CisPt to achieve the same effect in DNA interaction. Conversely, as confirmed by the first experiment, where the PCR mixture contained together with DNA and Taq DNA polymerase, the PCR was stopped by the addition of 580 ng/mL PtNPs, which is 16.9 times lower concentration than that of CisPt. The results of these analyses have unequivocally demonstrated that the PtNPs have a higher affinity to Taq DNA polymerase than to DNA.
Comet assay
Based on previous experiments, we followed with the cell experiments, where we also tested liposomal forms of both drugs to mimic targeted delivery. Therefore, the extent of DNA damage in the individual somatic cells was studied. To assess the effect of both the PtNPs and CisPt on nuclear DNA of eukaryotic cells the alkaline comet assay was used. The HFF cells in exponential growth phase were exposed to bare and liposome-encapsulated Pt-derivatives in concentrations of 0, 12.5, 25, 50, 100 and 200 μg/mL. Then, the cells were harvested and immobilized into agarose, and lyzed. Later, the nuclei were electrophoresed and stained with ethidium bromide (Fig 2A). The microphotographs show the evident difference between the effect of CisPt (Fig 2Aa and 2Ab) and PtNPs (Fig 2Ac and 2Ad) on the degradation of HFF nuclei. The index of the cell nuclei damage was increased by the encapsulation of CisPt or PtNPs within liposome (Fig 2B and 2C). The liposome-encapsulated PtNPs clearly showed the most intensive degradation effects among all the studied drugs and formulations. As shown in Fig 2C, the platinum nanoparticles (25–200 μg/mL) encapsulated into liposome (LipoPtNPs) exhibited more than two-times higher damage index than that of bare PtNPs. The comparison of the total index of the DNA damage, i.e. mean of indexes of damage presented in Fig 2B and 2C, showed that the PtNPs encapsulated within liposome damaged mostly the cell nuclei (Fig 2D).
The assessment of PtNPs and CisPt cytotoxicity and the effect of their encapsulation in liposomes
In these experiments we focused on the assessment of the cytotoxicity of the platinum derivatives and also on studying of the effect of their encapsulation in liposomes. Both S. aureus and HFF were analysed after the application of the PtNPs and CisPt encapsulated in liposomes or alone by the method of growth curves using Multiskan EX (S. aureus) and xCELLigence (HFF) instruments.
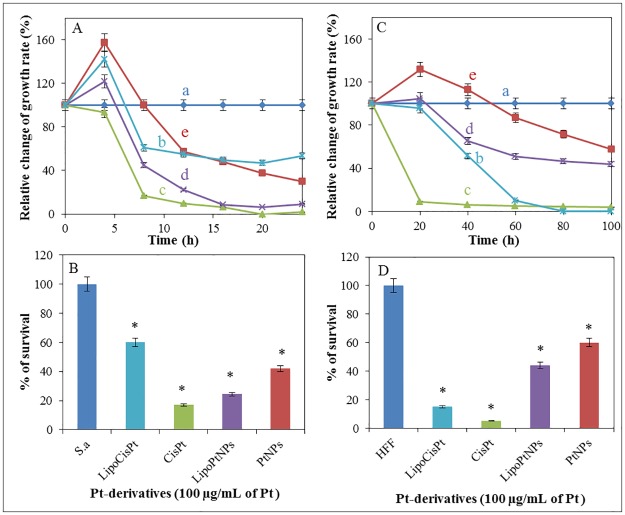
Firstly, the growth curves of the bacterial culture were measured after the addition of the Pt-derivatives encapsulated in liposome or alone in concentrations of 100 μg/mL of Pt. The obtained results were recalculated to relative absorbance value, thus, the control growth curve without the addition of the platinum derivatives was considered as 100% (Fig 3Aa). The values lower than 100% indicated relative cell death. The highest cytotoxic effect (100% dead cells) was observed in case of CisPt (Fig 3Ac), whereas a similar effect was achieved after the addition of LipoPtNPs, (Fig 3Ad). After the application of LipoPtNPs, the relative growth rate was found to be decreased for 53% (i.e. 53% dead cells) as compared to that of CisPt. The cytotoxicity of the PtNPs was found to be the lowest among all the drugs during the first 8 h of the treatment (Fig 3Ae), but after 24 h their cytostatic effect was found to be the third most intensive and its value was 20% higher and 30% lower than that of LipoCisPt and CisPt respectively. The results are summarized in Fig 3B, where the cytotoxicity of the Pt-derivatives encapsulated in liposomes or alone are shown.
Fig 3. The effect of platinum derivatives on both bacterial and human cells after application of platinum derivatives alone or encapsulated in liposomes.
Relative change of growth rate of (A) S. aureus bacterial culture and (C) HFF after treatment with Pt-derivatives (100 μg/mL of Pt) during 24 h (bacterial culture) and 100 h (HFF) long experiments. (a) Cell culture without any treatment, (b) LipoCisPt, (c) CisPt, (d) LipoPtNPs and (e) PtNPs. Percentage survival of (B) S. aureus bacterial culture and (D) human foreskin fibroblasts (HFF) after 24 and 100 h of treatment with Pt-derivatives (100 μg/mL of Pt) alone and encapsulated in liposomes. For all measurement n = 3.
The growth of HFF was monitored by xCELLigence method in the presence of the platinum derivatives (Fig 3C). The growth of the cells was expressed as percentage of control, thus, the growth curve of the control cells cultivated without the addition of any drug corresponded to 100%. For all tested platinum derivatives, the decrease of the cell growth during the cultivation was observed with exception of PtNPs (Fig 3Ce), where the growth of HFF was found to be stimulated during the first 50 h, but after 50 h the growth inhibition occurred. The highest cytostatic effect (100% decrease) was recorded for CisPt (Fig 3Cc) and LipoCisPt (Fig 3Cb). The PtNPs exhibited markedly lower cytotoxic effect compared to CisPt. After liposome encapsulation, the LipoPtNPs the rate if cell growth inhibition was higher, compared to bare PtNPs. The inhibition activity of LipoPtNPs after 100 h was comparable to the bare PtNPs (Fig 3Cd). The summary of the obtained results is shown in Fig 3D, where the cytotoxicity of the Pt-derivatives encapsulated in liposomes or alone is shown. Based on these results, CisPt had the highest cytostatic and cytotoxic effect for both S. aureus and HFF. The encapsulation of cisplatin in liposomes decreased both cytostatic and cytotoxic effects of cisplatin for the bacterial cells as well as for HFF. The PtNPs were found to be more effective in the bacterial cells than in HFF. The encapsulation in liposomes increased both the cytostatic and cytotoxic effects of the PtNPs, where one may suggest one of the following four mechanisms of liposome-cell interaction by which liposomes deliver their content in cells can occur: i) adsorption followed by extracellular release of liposome content; ii) endocytosis clathrin-(in) dependent; iii) lipid exchange by transfer of lipophilic compounds from the liposomal bilayer to the cell membrane and iv) fusion with the intracellular membrane.
The influence of Pt derivatives encapsulated in liposomes and alone on erythrocytes mixture with platelets (flow cytometry and GSH/GSSG ratio)
The commonly used cytostatics such as CisPt as well as their new alternatives such as PtNPs are often administered intravenously and therefore they may interact with erythrocytes and platelets.
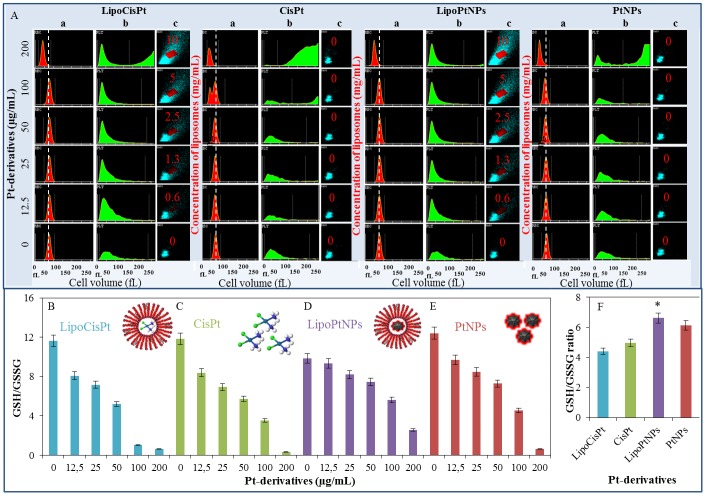
The influence of the platinum derivatives encapsulated in liposomes or alone on human erythrocytes mixed with platelets was studied by flow cytometry using haematology analyser and the oxidative stress was determined by GSH/GSSG ratio. The mixtures of the erythrocytes and platelets were exposed to LipoCisPt, CisPt, LipoPtNPs and PtNPs in concentrations of 0, 12.5, 25, 50, 100 and 200 μg/mL of Pt for 16 h. The concentrations of the liposomes in LipoPtNPs and LipoCisPt were as follows: 0, 0.6, 1.3, 2.5, 5, and 10 mg/mL. The histograms of the mixture and the Pt derivatives encapsulated in liposomes or alone were recorded by a haematology analyser. The liposomes were visualised in the Baso channel as a red rhombus (Fig 4A). No significant interaction was found for any of the Pt derivatives and the erythrocyte-platelet mixture.
Fig 4. Selected haematological parameters of erythrocytes mixture with platelets and oxidative stress (GSH/GSSG) in the mixture after application of Pt derivatives encapsulated in liposomes or alone in concentrations 0, 12.5, 25, 50, 100 and 200 μg/mL of Pt and 0, 0.6, 1.3, 2.5, 5 and 10 mg/mL of liposomes.
(A) Histograms of (a) erythrocytes and (b) platelets, and (c) visualisations of liposomes in the Baso channel after application of LipoCisPt, CisPt, LipoPtNPs and PtNPs. GSH/GSSG ratio of erythrocytes mixture with platelets in the same concentrations as in A—(B) LipoCisPt, (C) CisPt, (D) LipoPtNPs, and (E) PtNPs. (F) Mean of GSH/GSSG ratio. For all measurement n = 3, significant difference is indicated by *p<0.05.
The PtNPs encapsulated in liposomes were prepared and subsequently diluted decreasing the concentration of both components concurrently. The effect of this treatment is shown in Fig 4Ac.
The GSH and GSSG concentrations were determined by HPLC-ED (Fig 4B–4E). After treatment by all the Pt derivatives, a decrease in the GSH/GSSG ratio was observed that indicates the increasing amount of the oxidative stress in the erythrocytes. From Fig 4F, it can be clearly seen that LipoCisPt and CisPt caused higher oxidative stress than LipoPtNPs and PtNPs. By the statistical analysis using Lord´s T test, it was proven that compounds containing CisPt statistically differ from compounds containing PtNPs. However, the statistically significant difference was detected also between CisPt and PtNPs as well as between LipoCispt and LipoPtNPs. At the confidence level of 0.05.
Discussion
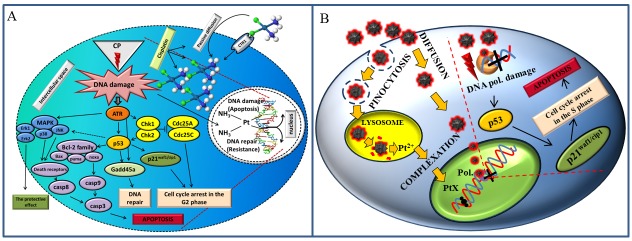
In the present experiment, the effects of PtNPs, or CisPt alone and encapsulated in liposomes (LipoPtNPs or LipoCisPt) on bacterial cultures (Staphylococcus aureus), HFF, erythrocytes and DNA (PCR product of Xis gene of λ phage) were studied. The platinum complexes are the active substance used as effective cytostatics [14]. Hundreds of Pt(II) and Pt(IV) complexes have been synthesized since the discovery the antibacterial effects of platinum in 1965 [42–44]. The effect of the platinum complexes (cisplatin, carboplatin and oxaliplatin) is most probably based on their covalent binding with DNA bases [45, 46] forming intra- and interstrand crosslinks, DNA—protein crosslinks, and monoadducts with DNA [47]. DNA secondary structures can block transcription and replication with subsequent apoptosis [48]. Pt drugs enter cells mainly via passive diffusion, although some evidence of its active transport by Ctr1 system involved in the maintaining of copper homeostasis was reported (Fig 5A) [49–52]. Another possibility is the intake of the nanoparticles via endocytosis [53]. Cisplatin-induced DNA damage activates ATR kinase, which triggers the effector molecules. One of the ATR kinase targets is a tumour suppressor protein p53, which is phosphorylated by the kinase on serine 15. This phosphorylation causes the decrease of p53 affinity to its negative regulator Mdm-2, which leads to an increased p53 concentration. The protein p53 then initiates the transcription of p21 gene, which inhibits the cyclin-dependent kinases Cdk2 and Cdk4 and consequent arrests the cell cycle. p53 also induces the expression of pro-apoptotic members of Bcl-2 family such as Bax, Puma and Noxa in response of the activation of mitochondrial apoptotic pathway.
Fig 5.
(A) Effect of CisPt on the somatic cell. (B) Proposed effect of PtNPs on the somatic cell. Figure prepared according to [54].
On the mitochondrial surface, these pro-apoptotic proteins meet the anti-apoptotic Bcl-2 family members. The main determinant of cisplatin-induced DNA damage is the ratio of pro-apoptotic and anti-apoptotic proteins. The majority of the pro-apoptotic signals lead to the release of cytochrome c from mitochondria followed by the activation of cysteine caspases selectively degrading the target proteins. Initiator caspases (Casp-8, 9 and 10) activate the effector caspases (Casp-3, 6 and 7). Casp-8 and Casp-9 are activated by outer pathway through the death receptors and inner mitochondrial pathway respectively. Both Casp-8 and 9 then activate Casp-3, which is responsible for intrinsic apoptotic process [53, 55, 56].
Among thousands of published papers aimed at the mentioned platinum-based cytostatics, only few can be found with the physiological action of the platinum nanoparticles [57, 58]. Some papers inform about the antioxidant properties of the PtNPs. Kajita et al. reported that PtNPs eliminated the anion radicals and hydrogen peroxide by a mechanism similar to superoxide-dismutase and catalase action [18]. Kim et al. [59] also confirmed the antioxidant function of PtNP,s which were shown to reduce the oxidative stress in murine osteoclasts. Onizawa et al. [60], studied the antioxidant activity of PtNPs in murine lungs after exposition to cigarette smoke. In a separate experiment, synthetic PtNPs at ppb levels were shown to extend the nematode lifespan and scavenge ROS which was induced by paraquat treatment in the nematode [61]. The anti-inflammatory effects of platinum nanoparticles on the lipopolysaccharide-induced inflammatory response in RAW 264.7 macrophages were determined by Rehman et al. [62].
Oh et al. reported the synthesis of the PtNPs of a diameter of 4.8 nm [29], which were characterized by X-ray and UV/Vis spectrophotometry [63]. In this work, the PtNPs were shown (using different techniques like PCR, DNA sequencing and denaturation) to inhibit the Taq DNA polymerase and affect the secondary structure of DNA in higher concentrations. Different papers on the interaction of heavy metals (Cu, Pb, and Cd) with DNA polymerase and thus inhibition of PCR were previously published [64, 65]. Our findings are consistent with earlier reports emphasizing intracellular release of Pt2+ ions from PtNPs, which could block cell division by binding with DNA [66]. The intracellular interactions of the PtNPs with bacterial DNA (Salmonella Enteritidis) were also previously reported [27] and it was shown using DNA-binding fluorochrome that one hour treatment of Salmonella enteritidis with PtNPs removed part of the DNA from the bacterial cell [27]. Shukla et al. showed that the nanoparticles tend to aggregate in the liposomes [67]. The aggregation of the PtNPs in liposomes may be responsible for the observed PtNPs cytotoxicity. This phenomenon can be explained by the oxidation of Pt by hydrogen peroxide according to the equation: Pt + H2O2 + 2H+ → Pt2+ + 2H2O [67]. The different mechanisms of CisPt and PtNPs action were found by single cell gel electrophoresis (comet assay) in this study. This method allows us to detect DNA damage on the single cell level [68]. Different modes of HFF nuclei degradation were observed after the treatment with CisPt or PtNPs, where the PtNPs were found to cause a higher degree of DNA degradation compared to CisPt. This finding may be caused by the fact that the alkaline comet assay is more sensitive to dsDNA breakage than to ssDNA breakage. In case of CisPt, ssDNA breakage [69], and covalent binding to DNA and interstrand crosslink [45] were observed as a result of a direct interaction with the DNA. The dsDNA breakages were caused rather by the PtNPs-induced Pt2+ ions formed during the incubation of the PtNPs with DNA [23].
The interactions of the Pt-derivatives with DNA might cause the arrest of cell cycle or have mutagenic effect [70]. The results suggest that p53 was activated in PtNPs treated cells due to the genotoxic stress with subsequent activation of p21 leading to a proliferating cell nuclear antigen-mediated growth arrest and apoptosis (Fig 5B) [54]. In this study, an attention was also paid to the antibacterial effects of the nanomaterials [71]. The PtNPs seemed to be a significant antibacterial agent [26, 72]. According to Chwalibog et al. PtNPs cause the degradation of cytoplasmic membrane and bacterial cell wall [26]. Our results indicated that the encapsulation of the nanoparticles in liposomes may lead to a more effective delivery of the nanoparticles as well as the disruption of the cellular membranes and thus increase their cytotoxicity and antibacterial action [73, 74].
In this work, the inhibition of bacterial and HFF growth by PtNPs and CisPt alone and encapsulated in liposome was compared by the method of growth curves and it was found that the inhibition effect of LipoPtNPs on S. aureus cells was higher compared to bare PtNPs. The inhibition of both keratinocytes and bacterial growth by PtNPs was also reported previously by Konieczny et al. [53], where the authors found an increased DNA damage and caspase 9 activity in human keratinocytes and a size-dependent inhibition effect of PtNPs on S. aureus and E. coli using a colony-reduction assay. No differences between PtNPs and CisPt action on human erythrocytes was found by haematological analyser. Both the bare and encapsulated PtNPs were found to cause significantly lower oxidative stress in the erythrocytes compared to the bare and encapsulated CisPt (as measured by GSH/GSSG ratio).
Conclusions
In this paper, we showed that PtNPs primarily inhibit the activity of Taq DNA polymerase and damage to the DNA structure. We tend to believe that this effect together with the transition of PtNPs to Pt2+ causes mutagenicity and increases DNA damage compared to CisPt. The cytotoxic effect of the PtNPs may be increased by their encapsulation in liposome. The results suggest that the activation of p53 in PtNPs treated cells was caused by the genotoxic stress with subsequent activation of p21 leading to a proliferating cell nuclear antigen-mediated growth arrest in S phase and following apoptosis [54]. It is conceivable that the PtNPs with the effective antitumor activity may provide an alternative treatment for cancer.
Supporting information
The following experimental details are shown there Particle size and zeta-potential analysis (PtNPs and Liposomes), Particle size assessment, Zeta potential assessment, Transmission electron microscopy (PtNPs and Liposomes), X-ray fluorescence analysis (XRF), Atomic absorption spectrometry and UV/vis spectrophotometry.
(DOCX)
A) Dynamic light scattering analysed of PtNPs in PBS, pH 7.4, with corresponding ζ potential inserted. B) TEM micrograph (length of scale bar is 50 nm) of and PtNPs. C) Dynamic light scattering analysed of Liposomes in PBS, pH 7.4, with corresponding ζ potential inserted. D) TEM micrograph (length of scale bar is 200 nm) of Liposomes.
(DOCX)
A) X-ray fluorescence (XRF) spectrum of 200 μg/mL the platinum nanoparticles (PtNPs). B) The absorption spectrum of polyvinylpyrrolidone (PVP); in inset: chemical formula of PVP basic unit—pyrrolidone. C) The absorption spectra of PtNPs (a = 200, b = 100, c = 50, d = 25, e = 12.5, f = 6.3, g = 3.2 and h = 0 μg/mL of Pt). D) PtNPs visualized in ambient light measured in the same concentrations as in S1 Fig.
(DOCX)
Acknowledgments
The authors wish to express their thanks to Martina Stankova, Lucie Dostalova and Radek Chmela for technical assistance.
Data Availability
All data are included in the paper.
Funding Statement
This research was carried out under the project CEITEC 2020 (LQ1601) with financial support from the Ministry of Education, Youth and Sports of the Czech Republic under the National Sustainability Programme II. The financial support from AZV project no. 15-28334A is also highly acknowledged. Moreover, authors acknowledge the CF CEITEC - Cryo-electron Microscopy and Tomography supported by the CIISB research infrastructure (LM2015043 funded by MEYS CR) for their support with obtaining scientific data presented in this paper.
References
- 1.Rosenberg B, Van Camp L, Krigas T. Inhibition of cell division in Escherichia coli by electrolysis products from a platinum electrode. Nature. 1965;205:698–9. [DOI] [PubMed] [Google Scholar]
- 2.Cepeda V, Fuertes MA, Castilla J, Alonso C, Quevedo C, Perez JM. Biochemical Mechanisms of Cisplatin Cytotoxicity. Anti-Cancer Agents Med Chem. 2007;7(1):3–18. doi: 10.2174/187152007779314044 [DOI] [PubMed] [Google Scholar]
- 3.Ali I, Wani WA, Saleem K, Haque A. Platinum Compounds: A Hope for Future Cancer Chemotherapy. Anti-Cancer Agents Med Chem. 2013;13(2):296–306. [DOI] [PubMed] [Google Scholar]
- 4.Pekarik V, Gumulec J, Masarik M, Kizek R, Adam V. Prostate cancer, miRNAs, metallothioneins and resistance to cytostatic drugs. Curr Med Chem. 2013;20(4):534–44. [DOI] [PubMed] [Google Scholar]
- 5.Hrabeta J, Adam V, Eckschlager T, Frei E, Stiborova M, Kizek R. Metal containing cytostatics and their interaction with cellular thiol compounds causing chemoresistance. Anti-Cancer Agents Med Chem. 2016;16(6):686–98. [DOI] [PubMed] [Google Scholar]
- 6.Edwards KA, Bolduc OR, Baeumner AJ. Miniaturized bioanalytical systems: enhanced performance through liposomes. Curr Opin Chem Biol. 2012;16(3–4):444–52. doi: 10.1016/j.cbpa.2012.05.182 [DOI] [PubMed] [Google Scholar]
- 7.Boulikas T. Low toxicity and anticancer activity of a novel liposomal cisplatin (Lipoplatin) in mouse xenografts. Oncol Rep. 2004;12(1):3–12. [PubMed] [Google Scholar]
- 8.Stathopoulos GP, Boulikas T, Kourvetaris A, Stathopoulos J. Liposomal oxaliplatin in the treatment of advanced cancer: A phase I study. Anticancer Res. 2006;26(2B):1489–93. [PubMed] [Google Scholar]
- 9.Paraskar A, Soni S, Basu S, Amarasiriwardena CJ, Lupoli N, Srivats S, et al. Rationally engineered polymeric cisplatin nanoparticles for improved antitumor efficacy. Nanotechnology. 2011;22(26):1–13. doi: 10.1088/0957-4484/22/26/265101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paraskar AS, Soni S, Chin KT, Chaudhuri P, Muto KW, Berkowitz J, et al. Harnessing structure-activity relationship to engineer a cisplatin nanoparticle for enhanced antitumor efficacy. Proc Natl Acad Sci U S A. 2010;107(28):12435–40. doi: 10.1073/pnas.1007026107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park EJ, Kim H, Kim Y, Park K. Intratracheal Instillation of Platinum Nanoparticles May Induce Inflammatory Responses in Mice. Arch Pharm Res. 2010;33(5):727–35. doi: 10.1007/s12272-010-0512-y [DOI] [PubMed] [Google Scholar]
- 12.Paraskar A, Soni S, Roy B, Papa AL, Sengupta S. Rationally designed oxaliplatin-nanoparticle for enhanced antitumor efficacy. Nanotechnology. 2012;23(7):1–17. doi: 10.1088/0957-4484/23/7/075103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conlin AK, Seidman AD, Bach A, Lake D, Dickler M, D'Andrea G, et al. Phase II Trial of Weekly Nanoparticle Albumin-Bound Paclitaxel With Carboplatin and Trastuzumab as First-line Therapy for Women With HER2-Overexpressing Metastatic Breast Cancer. Clin Breast Cancer. 2010;10(4):281–7. doi: 10.3816/CBC.2010.n.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4(4):307–20. doi: 10.1038/nrd1691 [DOI] [PubMed] [Google Scholar]
- 15.Ho YP, Au-Yeung SCF, To KKW. Platinum-based anticancer agents: Innovative design strategies and biological perspectives. Med Res Rev. 2003;23(5):633–55. doi: 10.1002/med.10038 [DOI] [PubMed] [Google Scholar]
- 16.Jung YW, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107(5):1387–407. doi: 10.1021/cr068207j [DOI] [PubMed] [Google Scholar]
- 17.Hamasaki T, Kashiwagi T, Imada T, Nakamichi N, Aramaki S, Toh K, et al. Kinetic analysis of superoxide anion radical-scavenging and hydroxyl radical-scavenging activities of platinum nanoparticles. Langmuir. 2008;24(14):7354–64. doi: 10.1021/la704046f [DOI] [PubMed] [Google Scholar]
- 18.Kajita M, Hikosaka K, Iitsuka M, Kanayama A, Toshima N, Miyamoto Y. Platinum nanoparticle is a useful scavenger of superoxide anion and hydrogen peroxide. Free Radic Res. 2007;41(6):615–26. doi: 10.1080/10715760601169679 [DOI] [PubMed] [Google Scholar]
- 19.Zhang LB, Laug L, Munchgesang W, Pippel E, Gosele U, Brandsch M, et al. Reducing Stress on Cells with Apoferritin-Encapsulated Platinum Nanoparticles. Nano Letters. 2010;10(1):219–23. doi: 10.1021/nl903313r [DOI] [PubMed] [Google Scholar]
- 20.Fan J, Yin JJ, Ning B, Wu XC, Hu Y, Ferrari M, et al. Direct evidence for catalase and peroxidase activities of ferritin-platinum nanoparticles. Biomaterials. 2011;32(6):1611–8. doi: 10.1016/j.biomaterials.2010.11.004 [DOI] [PubMed] [Google Scholar]
- 21.Takamiya M, Miyamoto Y, Yamashita T, Deguchi K, Ohta Y, Abe K. Strong Neuroprotection with a Novel Platinum Nanoparticle against Ischemic Stroke- and Tissue Plasminogen Activator-related Brain Damages in Mice. Neuroscience. 2012;221:47–55. doi: 10.1016/j.neuroscience.2012.06.060 [DOI] [PubMed] [Google Scholar]
- 22.Takamiya M, Miyamoto Y, Yamashita T, Deguchi K, Ohta Y, Ikeda Y, et al. Neurological and Pathological Improvements of Cerebral Infarction in Mice With Platinum Nanoparticles. J Neurosci Res. 2011;89(7):1125–33. doi: 10.1002/jnr.22622 [DOI] [PubMed] [Google Scholar]
- 23.Gehrke H, Pelka J, Hartinger CG, Blank H, Bleimund F, Schneider R, et al. Platinum nanoparticles and their cellular uptake and DNA platination at non-cytotoxic concentrations. Arch Toxicol. 2011;85(7):799–812. doi: 10.1007/s00204-010-0636-3 [DOI] [PubMed] [Google Scholar]
- 24.Palchoudhury S, Xu YL, Rushdi A, Bao YP. DNA Interaction of Pt-Attached Iron Oxide Nanoparticles. IEEE Trans Magn. 2013;49(1):373–6. doi: 10.1109/tmag.2012.2223660 [Google Scholar]
- 25.Fenske AE, Glaesener S, Bokemeyer C, Thomale J, Dahm-Daphi J, Honecker F, et al. Cisplatin resistance induced in germ cell tumour cells is due to reduced susceptibility towards cell death but not to altered DNA damage induction or repair. Cancer Lett. 2012;324(2):171–8. doi: 10.1016/j.canlet.2012.05.009 [DOI] [PubMed] [Google Scholar]
- 26.Chwalibog A, Sawosz E, Hotowy A, Szeliga J, Mitura S, Mitura K, et al. Visualization of interaction between inorganic nanoparticles and bacteria or fungi. Int J Nanomed. 2010;5:1085–94. doi: 10.2147/ijn.s13532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawosz E, Chwalibog A, Szeliga J, Sawosz F, Grodzik M, Rupiewicz M, et al. Visualization of gold and platinum nanoparticles interacting with Salmonella Enteritidis and Listeria monocytogenes. Int J Nanomed. 2010;5:631–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelka J, Gehrke H, Esselen M, Turk M, Crone M, Brase S, et al. Cellular Uptake of Platinum Nanoparticles in Human Colon Carcinoma Cells and Their Impact on Cellular Redox Systems and DNA Integrity. Chem Res Toxicol. 2009;22(4):649–59. doi: 10.1021/tx800354g [DOI] [PubMed] [Google Scholar]
- 29.Oh JG, Kim H. Synthesis of core-shell nanoparticles with a Pt nanoparticle core and a silica shell. Curr Appl Phys. 2013;13(1):130–6. doi: 10.1016/j.cap.2012.06.025 [Google Scholar]
- 30.Barabas R, Cziko M, Dekany I, Bizo L, Bogya ES. Comparative study of particle size analysis of hydroxyapatite-based nanomaterials. Chem Pap. 2013;67(11):1414–23. doi: 10.2478/s11696-013-0409-6 [Google Scholar]
- 31.Kunjachan S, Blauz A, Mockel D, Theek B, Kiessling F, Etrych T, et al. Overcoming cellular multidrug resistance using classical nanomedicine formulations. Eur J Pharm Sci. 2012;45(4):421–8. doi: 10.1016/j.ejps.2011.08.028 [DOI] [PubMed] [Google Scholar]
- 32.Nejdl L, Rodrigo MAM, Kudr J, Ruttkay-Nedecky B, Konecna M, Kopel P, et al. Liposomal nanotransporter for targeted binding based on nucleic acid anchor system. Electrophoresis. 2014;35(2–3):393–404. doi: 10.1002/elps.201300197 [DOI] [PubMed] [Google Scholar]
- 33.Smerkova K, Dostalova S, Skutkova H, Ryvolova M, Adam V, Provaznik I, et al. Isolation of Xis Gen Fragment of lambda Phage from Agarose Gel Using Magnetic Particles for Subsequent Enzymatic DNA Sequencing. Chromatographia. 2013;76(7–8):329–34. doi: 10.1007/s10337-012-2326-1 [Google Scholar]
- 34.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low-levels of DNA damage in individual cells Exp Cell Res. 1988;175(1):184–91. doi: 10.1016/0014-4827(88)90265-0 [DOI] [PubMed] [Google Scholar]
- 35.Mouron SA, Golijow CD, Dulout FN. DNA damage by cadmium and arsenic salts assessed by the single cell gel electrophoresis assay. Mutat Res Genet Toxicol Environ Mutagen. 2001;498(1–2):47–55. doi: 10.1016/s1383-5718(01)00266-2 [DOI] [PubMed] [Google Scholar]
- 36.Chudobova D, Dostalova S, Blazkova I, Michalek P, Ruttkay-Nedecky B, Sklenar M, et al. Effect of Ampicillin, Streptomycin, Penicillin and Tetracycline on Metal Resistant and Non-Resistant Staphylococcus aureus. Int J Environ Res Public Health. 2014;11(3):3233–55. doi: 10.3390/ijerph110303233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chudobova D, Dobes J, Nejdl L, Maskova D, Rodrigo MAM, Nedecky BR, et al. Oxidative Stress in Staphylococcus aureus Treated with Silver(I) Ions Revealed by Spectrometric and Voltammetric Assays. Int J Electrochem Sci. 2013;8(4):4422–40. [Google Scholar]
- 38.Chudobova D, Nejdl L, Gumulec J, Krystofova O, Rodrigo MAM, Kynicky J, et al. Complexes of Silver(I) Ions and Silver Phosphate Nanoparticles with Hyaluronic Acid and/or Chitosan as Promising Antimicrobial Agents for Vascular Grafts. Int J Mol Sci. 2013;14(7):13592–614. doi: 10.3390/ijms140713592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chudobova D, Dostalova S, Ruttkay-Nedecky B, Guran R, Rodrigo MAM, Tmejova K, et al. The effect of metal ions on Staphylococcus aureus revealed by biochemical and mass spectrometric analyses. Microbiol Res. 2015;170:147–56. doi: 10.1016/j.micres.2014.08.003 [DOI] [PubMed] [Google Scholar]
- 40.Kleckerova A, Sobrova P, Krystofova O, Sochor J, Zitka O, Babula P, et al. Cadmium(II) and Zinc(II) Ions Effects on Maize Plants revealed by Spectroscopy and Electrochemistry. Int J Electrochem Sci. 2011;6(12):6011–31. [Google Scholar]
- 41.Skladanka J, Adam V, Zitka O, Krystofova O, Beklova M, Kizek R, et al. Investigation into the Effect of Molds in Grasses on Their Content of Low Molecular Mass Thiols. Int J Environ Res Public Health. 2012;9(11):3789–805. doi: 10.3390/ijerph9113789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bednarski PJ, Korpis K, Westendorf AF, Perfahl S, Grunert R. Effects of light-activated diazido-Pt-IV complexes on cancer cells in vitro. Philos Trans R Soc A-Math Phys Eng Sci. 2013;371(1995):1–6. doi: 10.1098/rsta.2012.0118 [DOI] [PubMed] [Google Scholar]
- 43.Melnik M, Mikus P. Structural characterization of heterometallic platinum complexes with non-transition metals. Part II: heterotrimeric complexes. Main Group Met Chem. 2013;36(1–2):1–10. doi: 10.1515/mgmc-2012-0062 [Google Scholar]
- 44.Sgarbossa P, Scarso A, Strukul G, Michelin RA. Platinum(II) Complexes with Coordinated Electron-Withdrawing Fluoroalkyl and Fluoroaryl Ligands: Synthesis, Reactivity, and Catalytic Activity. Organometallics. 2012;31(4):1257–70. doi: 10.1021/om200953z [Google Scholar]
- 45.Chang CL, Lando DY, Fridman AS, Hu CK. Thermal stability of DNA with interstrand crosslinks. Biopolymers. 2012;97(10):807–17. doi: 10.1002/bip.22077 [DOI] [PubMed] [Google Scholar]
- 46.Brabec V, Kasparkova J. Modifications of DNA by platinum complexes—Relation to resistance of tumors to platinum antitumor drugs. Drug Resist Update. 2005;8(3):131–46. doi: 10.1016/j.drup.2005.04.006 [DOI] [PubMed] [Google Scholar]
- 47.Boulikas T, Vougiouka M. Cisplatin and platinum drugs at the molecular level (review). Oncol Rep. 2003;10(6):1663–82. [PubMed] [Google Scholar]
- 48.Theile D, Detering JC, Herold-Mende C, Dyckhoff G, Haefeli WE, Weiss J, et al. Cellular Pharmacokinetic/Pharmacodynamic Relationship of Platinum Cytostatics in Head and Neck Squamous Cell Carcinoma Evaluated by Liquid Chromatography Coupled to Tandem Mass Spectrometry. J Pharmacol Exp Ther. 2012;341(1):51–8. doi: 10.1124/jpet.111.189621 [DOI] [PubMed] [Google Scholar]
- 49.Mitchell C, Kabolizadeh P, Ryan J, Roberts JD, Yacoub A, Curiel DT, et al. Low-dose BBR3610 toxicity in colon cancer cells is p53-independent and enhanced by inhibition of epidermal growth factor receptor (ERBB1)-Phosphatidyl inositol 3 kinase signaling. Mol Pharmacol. 2007;72(3):704–14. doi: 10.1124/mol.107.038406 [DOI] [PubMed] [Google Scholar]
- 50.Summa N, Maigut J, Puchta R, van Eldik R. Possible biotransformation reactions of polynuclear Pt(II) complexes. Inorg Chem. 2007;46(6):2094–104. doi: 10.1021/ic061990d [DOI] [PubMed] [Google Scholar]
- 51.Oehlsen ME, Hegmans A, Qu Y, Farrell N. Effects of geometric isomerism in dinuclear antitumor platinum complexes on their interactions with N-acetyl-L-methionine. J Biol Inorg Chem. 2005;10(5):433–42. doi: 10.1007/s00775-005-0009-1 [DOI] [PubMed] [Google Scholar]
- 52.Williams JW, Qu Y, Bulluss GH, Alvorado E, Farrell NP. Dinuclear platinum complexes with biological relevance based on the 1,2-diaminocyclohexane carrier ligand. Inorg Chem. 2007;46(15):5820–2. doi: 10.1021/ic700410y [DOI] [PubMed] [Google Scholar]
- 53.Konieczny P, Goralczyk AG, Szmyd R, Skalniak L, Koziel J, Filon FL, et al. Effects triggered by platinum nanoparticles on primary keratinocytes. Int J Nanomed. 2013;8:3963–75. doi: 10.2147/ijn.s49612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asharani PV, Xinyi N, Hande MP, Valiyaveettil S. DNA damage and p53-mediated growth arrest in human cells treated with platinum nanoparticles. Nanomedicine. 2010;5(1):51–64. doi: 10.2217/nnm.09.85 [DOI] [PubMed] [Google Scholar]
- 55.Vavrova M, Vokurkova. Two main routes induction of apoptosis in mammalian cells. Radiation oncology. 2003;(cxc):1–9. [Google Scholar]
- 56.Jia J, Zhu F, Ma XH, Cao ZWW, Li YXX, Chen YZ. Mechanisms of drug combinations: interaction and network perspectives. Nat Rev Drug Discov. 2009;8(2):111–28. doi: 10.1038/nrd2683 [DOI] [PubMed] [Google Scholar]
- 57.Wahab R, Yang YB, Umar A, Singh S, Hwang IH, Shin HS, et al. Platinum Quantum Dots and Their Cytotoxic Effect Towards Myoblast Cancer Cells (C2C12). J Biomed Nanotechnol. 2012;8(3):424–31. doi: 10.1166/jbn.2012.1448 [DOI] [PubMed] [Google Scholar]
- 58.Manikandan M, Hasan N, Wu HF. Platinum nanoparticles for the photothermal treatment of Neuro 2A cancer cells. Biomaterials. 2013;34(23):5833–42. doi: 10.1016/j.biomaterials.2013.03.077 [DOI] [PubMed] [Google Scholar]
- 59.Kim WK, Kim JC, Park HJ, Sul OJ, Lee MH, Kim JS, et al. Platinum nanoparticles reduce ovariectomy-induced bone loss by decreasing osteoclastogenesis. Exp Mol Med. 2012;44(7):432–9. doi: 10.3858/emm.2012.44.7.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Onizawa S, Aoshiba K, Kajita M, Miyamoto Y, Nagai A. Platinum nanoparticle antioxidants inhibit pulmonary inflammation in mice exposed to cigarette smoke. Pulmonary Pharmacology & Therapeutics. 2009;22(4):340–9. http://dx.doi.org/10.1016/j.pupt.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 61.Yan HX, Kinjo T, Tian HZ, Hamasaki T, Teruya K, Kabayama S, et al. Mechanism of the Lifespan Extension of Caenorhabditis elegans by Electrolyzed Reduced Water-Participation of Pt Nanoparticles. Biosci Biotechnol Biochem. 2011;75(7):1295–9. doi: 10.1271/bbb.110072 [DOI] [PubMed] [Google Scholar]
- 62.Rehman MU, Yoshihisa Y, Miyamoto Y, Shimizu T. The anti-inflammatory effects of platinum nanoparticles on the lipopolysaccharide-induced inflammatory response in RAW 264.7 macrophages. Inflamm Res. 2012;61(11):1177–85. doi: 10.1007/s00011-012-0512-0 [DOI] [PubMed] [Google Scholar]
- 63.Zolotov YA, Petrukhin OM, Malofeeva GI, Marcheva EV, Shiryaeva OA, Shestakov VA, et al. Determination of platinum metals by X-ray-fluorescence, atomic emission and atomic-absorption spectrometry after pre-concentration with a polymeric thioether. Anal Chim Acta. 1983;148(APR):135–57. doi: 10.1016/s0003-2670(00)85160-0 [Google Scholar]
- 64.Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase beta. Biochemistry. 1996;35(39):12762–77. doi: 10.1021/bi9529566 [DOI] [PubMed] [Google Scholar]
- 65.Popenoe EA, Schmaeler MA. Interaction of human DNA polymerase β with ions of copper, lead, and cadmium. Arch Biochem Biophys. 1979;196(1):109–20. http://dx.doi.org/10.1016/0003-9861(79)90557-5. [DOI] [PubMed] [Google Scholar]
- 66.Gao JH, Liang GL, Zhang B, Kuang Y, Zhang XX, Xu B. FePt@CoS2 yolk-shell nanocrystals as a potent agent to kill HeLa cells. J Am Chem Soc. 2007;129(5):1428–33. doi: 10.1021/ja067785e [DOI] [PubMed] [Google Scholar]
- 67.Shukla R, Bansal V, Chaudhary M, Basu A, Bhonde RR, Sastry M. Biocompatibility of gold nanoparticles and their endocytotic fate inside the cellular compartment: A microscopic overview. Langmuir. 2005;21(23):10644–54. doi: 10.1021/la0513712 [DOI] [PubMed] [Google Scholar]
- 68.Fikrova P, Stetina R, Hrnciarik M, Rehacek V, Jost P, Hronek M, et al. Detection of DNA crosslinks in peripheral lymphocytes isolated from patients treated with platinum derivates using modified comet assay. Neoplasma. 2013;60(4):413–8. doi: 10.4149/neo_2013_053 [DOI] [PubMed] [Google Scholar]
- 69.Brabec V, Kasparkova J. Molecular aspects of resistance to antitumor platinum drugs. Drug Resist Update. 2002;5(3–4):147–61. doi: 10.1016/s1368-7646(02)00047-x [DOI] [PubMed] [Google Scholar]
- 70.Kosmider B, Wyszynska K, Janik-Spiechowicz E, Osiecka R, Zyner E, Ochocki J, et al. Evaluation of the genotoxicity of cis-bis(3-aminoflavone)dichloroplatinum(II) in comparison with cis-DDP. Mutat Res Genet Toxicol Environ Mutagen. 2004;558(1–2):93–110. doi: 10.1016/j.mrgentox.2003.11.006 [DOI] [PubMed] [Google Scholar]
- 71.Li Y, Chen DH, Yan J, Chen Y, Mittelstaedt RA, Zhang YB, et al. Genotoxicity of silver nanoparticles evaluated using the Ames test and in vitro micronucleus assay. Mutat Res Genet Toxicol Environ Mutagen. 2012;745(1–2):4–10. doi: 10.1016/j.mrgentox.2011.11.010 [DOI] [PubMed] [Google Scholar]
- 72.Gopal J, Hasan N, Manikandan M, Wu HF. Bacterial toxicity/compatibility of platinum nanospheres, nanocuboids and nanoflowers. Scientific Reports. 2013;3 doi: 10.1038/srep01260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paukner S, Kohl G, Jalava K, Lubitz W. Sealed bacterial ghosts—Novel targeting vehicles for advanced drug delivery of water-soluble substances. J Drug Target. 2003;11(3):151–61. [DOI] [PubMed] [Google Scholar]
- 74.Mayr UB, Walcher P, Azimpour C, Riedmann E, Haller C, Lubitz W. Bacterial ghosts as antigen delivery vehicles. Adv Drug Deliv Rev. 2005;57(9):1381–91. doi: 10.1016/j.addr.2005.01.027 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following experimental details are shown there Particle size and zeta-potential analysis (PtNPs and Liposomes), Particle size assessment, Zeta potential assessment, Transmission electron microscopy (PtNPs and Liposomes), X-ray fluorescence analysis (XRF), Atomic absorption spectrometry and UV/vis spectrophotometry.
(DOCX)
A) Dynamic light scattering analysed of PtNPs in PBS, pH 7.4, with corresponding ζ potential inserted. B) TEM micrograph (length of scale bar is 50 nm) of and PtNPs. C) Dynamic light scattering analysed of Liposomes in PBS, pH 7.4, with corresponding ζ potential inserted. D) TEM micrograph (length of scale bar is 200 nm) of Liposomes.
(DOCX)
A) X-ray fluorescence (XRF) spectrum of 200 μg/mL the platinum nanoparticles (PtNPs). B) The absorption spectrum of polyvinylpyrrolidone (PVP); in inset: chemical formula of PVP basic unit—pyrrolidone. C) The absorption spectra of PtNPs (a = 200, b = 100, c = 50, d = 25, e = 12.5, f = 6.3, g = 3.2 and h = 0 μg/mL of Pt). D) PtNPs visualized in ambient light measured in the same concentrations as in S1 Fig.
(DOCX)
Data Availability Statement
All data are included in the paper.