Abstract
Protein glycosylation can have an enormous variety of biological consequences, reflecting the molecular diversity encoded in glycan structures. This same structural diversity has imposed major challenges on the development of methods to study the intact glycoproteome. We recently introduced a method termed isotope-targeted glycoproteomics (IsoTaG), which utilizes isotope recoding to characterize azidosugar-labeled glycopeptides bearing fully intact glycans. Here, we describe the broad application of the method to analyze glycoproteomes from a collection of tissue-diverse cell lines. The effort was enabled by a new high-fidelity pattern-searching and glycopeptide validation algorithm termed IsoStamp v2.0, as well as by novel stable isotope probes. Application of the IsoTaG platform to 15 cell lines metabolically labeled with Ac4GalNAz or Ac4ManNAz revealed 1375 N- and 2159 O-glycopeptides, variously modified with 74 discrete glycan structures. Glycopeptide-bound glycans observed by IsoTaG were found to be comparable to released N-glycans identified by permethylation analysis. IsoTaG is therefore positioned to enhance structural understanding of the glycoproteome.
Keywords: glycoprotein, glycoconjugate, glycoproteomics, chemical biology, LC–MS/MS, metabolic labeling, chemical enrichment, glycomics, IsoStamp
Graphical Abstract

INTRODUCTION
Glycosylation is a protein post-translational modification (PTM) that generates extreme proteomic diversity and contributes to a plethora of cellular functions.1,2 Protein glycosylation most commonly occurs at Asn (N-linked) or Ser/Thr (O-linked) residues, with the pendant glycan capable of displaying immense structural variability. These glycans can modulate protein structure, stability and recognition.3 Dysregulation of the glycoproteome is often correlated with disease states including cancer,4,5 inflammation,1,6 neurological disorders, 7,8 and diabetes.9,10 The ability to define the intact glycoproteome—that is, the assignment of specific glycan structures to protein sites in a whole proteome—would accelerate structure–function studies and serve to identify potential disease biomarkers and therapeutic targets.
While there has been much interest in profiling the intact glycoproteome, the complexity of glycoproteoforms (and more broadly, all proteoforms11) remains challenging to completely define. Mass spectrometry (MS) is commonly employed for characterization of complex proteomic samples. However, in order to profile the whole cell glycoproteome by MS, enrichment of specific glycoprotein subtypes and reduction in glycan heterogeneity are typically required. Enrichment can be achieved by metabolic labeling,12–14 chemical manipulation, 15–17 enzymatic labeling,18 lectin affinity chromatography, 19,20 or other methods.21,22 Reduction in glycan complexity is achieved for N-glycoproteins by enzymatic digestion with PNGase F or Endo H,23 and for O-glycoproteins by genetic engineering in cell lines.24,25 Glycan truncation simplifies detection by MS and computational analysis via database searching, but at the expense of lost information encoded within the glycan. Several glycoproteomics protocols have shown promise for intact glycoproteomics;26–31 however, these methods require separate analyses of glycans and their peptide scaffolds, and at present are restricted to N-glycans.
To address the unique challenges in global characterization of the intact glycoproteome, we developed a mass-independent chemical glycoproteomics platform, termed isotope targeted glycoproteomics (IsoTaG, Figure 1).32–34 IsoTaG is performed by first, isotopic recoding and enrichment of metabolically labeled glycoproteins and, second, directed tandem MS (MS2 or MSn) analysis and intact glycopeptide assignment. Isotopic recoding is accomplished by metabolic labeling of cell or tissue samples with azide- or alkyne-functionalized sugars, followed by chemical conjugation with a biotin probe bearing a unique isotopic signature. Examples of sugar labels are peracetylated N-azidoacetylmannosamine (Ac4ManNAz), which is converted to the corresponding azidosialic acid (SiaNAz), and peracetylated N-azidoacetylgalactosamine (Ac4GalNAz), which is metabolized to label glycans possessing N-acetylglucosamine (GlcNAc) or N-acetylgalactosamine (GalNAc).
Figure 1.
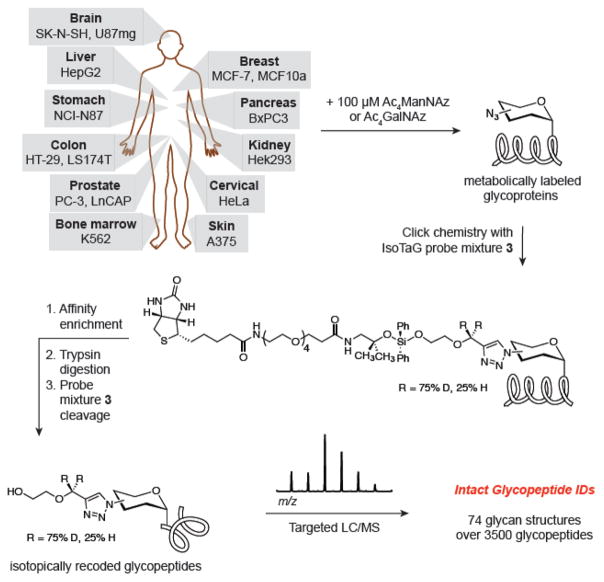
Schematic of IsoTaG method. Cell lines derived from various human tissues were treated with Ac4ManNAz or Ac4GalNAz to generate metabolically labeled glycoproteins. The glycoproteins were tagged with IsoTaG probe mixture 3 and affinity enriched. Tryptic digestion and cleavage of probe mixture 3 recovered isotopically recoded glycopeptides for analysis. Glycopeptides were analyzed by targeted LC–MS using IsoStamp v2.0 to direct selection of isotopically recoded species. A survey of 15 human cell lines was performed to demonstrate the broad applicability of IsoTaG and revealed 74 glycan structures over 3500 intact glycopeptides.
Through the probe, a unique isotopic signature is embedded exclusively into glycopeptides. The isotopic signature serves as a computationally recognizable full-scan MS reporter. We automated its detection with a pattern searching algorithm, termed isotopic signature transfer and mass pattern prediction (IsoStamp).35 In an initial report, we demonstrated that the use of dibrominated probes led to the reproducible identification of over 500 intact glycopeptides across 250 glycoproteins from three cell lines.32
Here, we report an extensive investigation into the scope of mass-independent MS to enable chemical glycoproteomics and demonstrate its application to 15 cell lines. These efforts have produced IsoStamp v2.0, a high-fidelity pattern-searching algorithm for both detection and validation of isotopically recoded species, provided on a web-based platform for public access. A total of eight stable isotope probe mixtures were prepared to experimentally optimize pattern recognition. The probe mixture that resulted in the best pattern recognition (probe mixture 3) was applied to 15 metabolically labeled cell lines, analysis of which yielded a database of 74 glycan structures and over 3500 glycopeptides. The glycan structures observed by IsoTaG from one cell line (PC-3) were reflective of the types and frequencies of structures observed by a conventional analysis of released N-glycans. IsoTaG therefore enables the analysis of intact metabolically labeled glycopeptides across a range of cell lines and will accelerate structure and function studies of the dynamic glycoproteome.
EXPERIMENTAL PROCEDURES
IsoStamp v2.0
Isostamp v2.0 is available at www.isostamp.org as an interactive web interface.
Cell Culture Procedures
All cell lines were obtained from the American Type Culture Collection (ATCC) and maintained at 37 °C and 5% CO2 in a water-saturated incubator. Cell lines were metabolically labeled between passages 4–24. Cell culture experiments were performed as described.32,33 Briefly, cells were seeded at 2 × 105 cells/mL in complete media and the presence of 100 μM Ac4GalNAz, Ac4ManNAz, or DMSO vehicle control containing Ac4GalNAc (3.0 μL). Cells were incubated for 48 h at 37 °C. The media was aspirated and the cells washed with PBS (1 × 10 mL). Washed dishes were resuspended in media containing 100 μM glycan metabolite without FBS additive (15 mL), and the cells were incubated for an additional 48 h at 37 °C. Media was harvested by centrifugal concentration (10 kDa molecular weight cutoff spin filter), washed with 1% triton ×100/PBS (3 × 15 mL), and the remaining residue collected as the “conditioned media”. Cells were harvested, washed, and resuspended in lysis buffer (10 mM HEPES, pH 7.9, 15 mM MgCl2, 10 mM KCl, 0.5% triton ×100, 1× protease inhibitors, 1 μM thiamet G, 2 mL), swelled for 5 min on ice, and broken by Dounce homogenization. The homogenized lysate was transferred to a centrifuge tube, and insoluble material was pelleted by centrifugation (3700g, 10 min, 4 °C). The supernatant was collected as the “soluble fraction” and the pellet kept as the “insoluble fraction.” The pellet was separately solubilized in 1% Rapigest/PBS (400 μL). Protein concentrations from the three fractions were measured by bicinchonic acid assay (Pierce) and normalized to 4.5 mg/mL.
Chemical Glycoproteomics Enrichment
Chemical enrichment were performed analogously to those previously reported32,33 with silane probe mixtures 1–8. Briefly, click chemistry reagents (40.0 μL, 200 μM probe mixture, 300 μM CuSO4, 600 μM BTTP,36 2.50 mM sodium ascorbate, mixed immediately before addition to lysates) were added and the reaction was incubated for 3.0 h at 24 °C. Tagged proteins were methanol precipitated at −80 °C, pelleted by centrifugation and the supernatant was discarded. Protein pellets were resuspended in 400 μL 1% RapiGest/PBS and solubilized by probe sonication (Misonix, 1.5 min, 4 °C). Streptavidin–agarose resin [200 μL, washed with PBS (3 × 1 mL)] was added, and the resulting mixture was incubated for 12 h at 24 °C with rotation. The beads were pelleted by centrifugation and the supernatant containing uncaptured proteins was separated. The beads were washed with 1% RapiGest/PBS (1 mL), 6 M urea/H2O (2 × 1 mL), and PBS (5 × 1 mL), and the beads were pelleted by centrifugation (3000g, 3 min) between washes.
Washed beads were resuspended in 5 mM DTT/PBS (200 μL) and incubated for 30 min at 24 °C with rotation. Ten mM iodoacetamide/PBS (4.0 μL, 500 mM stock solution) was added to the reduced proteins, and allowed to react for 30 min at 24 °C with rotation, in the dark. Beads were pelleted by centrifugation (3000g, 3 min) and resuspended in 0.5 M urea/PBS (200 μL). Trypsin (1.5 μg) was added to the resuspended beads, and digestion proceeded for 12 h at 37 °C. Beads were pelleted by centrifugation (3000g, 3 min), and the supernatant digest was collected. The beads were washed with PBS (1 × 200 μL) and H2O (2 × 200 μL). Washes were combined with the supernatant digest to form the trypsin digest. Probe mixture 3 was cleaved with two treatments of 2% formic acid/H2O (200 μL) for 30 min at 24 °C with rotation and the eluent was collected. The beads were washed with 50% acetonitrile–water +1% formic acid (2 × 400 μL), and the washes were combined with the eluent to form the cleavage fraction. The trypsin digest and cleavage fraction were concentrated using a vacuum centrifuge (i.e., a speedvac, 40 °C) to 50–100 μL. Samples were desalted with a ZipTip P10 and stored at −20 °C until analysis.
α-Biotin Immunoblotting
Aliquots collected during the enrichment procedure (10.0 μL) were reduced and separated by standard SDS-PAGE (Bio-Rad, Criterion system), electroblotted onto nitrocellulose, blocked in 5% bovine serum albumin (Sigma) in Tris-buffered saline with Tween (10 mM Tris pH 8.0, 150 mM NaCl, 0.1% Tween-20), and analyzed by standard enhanced chemiluminescence immunoblotting methods (Pierce). The staining agent was high-sensitivity streptavidin–HRP (Pierce, 1:100 000).
Mass Spectrometry Procedures
Alpha-Crystallin Glycoproteomics
Tryptic digested alpha-crystallin samples were analyzed by a LTQ-Orbitrap Elite mass spectrometer equipped with electron transfer dissociation (ThermoFisher Scientific). Samples were loaded onto a C18 trap column coupled to an UltiMate Rapid Separation LC (Dionex) system at 5 μL/min for 10 min. Samples were then loaded onto a 25 cm length C18 analytical column (Picofrit 75 μm ID, New Objective) packed in-house with Magic C18AQ resin (Michrom Bioresources). Tryptic peptides were eluted using a multistep gradient at a flow rate of 0.6 μL/min from 0.1% formic acid in water to 85–0.1% formic acid in acetonitrile over 120 min.
Peptides were fragmented using higher energy collisional dissociation (HCD), electron transfer dissociation (ETD), and collision-induced dissociation (CID). The electrospray ionization voltage was set to 2.25 kV and the capillary temperature was set to 200 °C. MS1 scans were performed over m/z 400–1800 and the top three most intense ions (2+ or higher charge states) were subjected to three subsequent fragmentation methods including HCD with 27 eV, default charge state +4, for 0.1 s, ETD for 200 ms with supplemental activation of 35 eV, and CID at 35 eV for 10 ms.
Targeted Glycoproteomics on Metabolically Labeled Tissue Culture Samples
Trypsin-digested proteins were analyzed using a Thermo Dionex UltiMate3000 RSLCnano liquid chromatograph that was connected in-line with a LTQ-Orbitrap-XL mass spectrometer equipped with a nanoelectrospray ionization (nanoESI) source (Thermo Fisher Scientific, Waltham, MA). The LC was equipped with a C18 precolumn (Acclaim PepMap 100, 20 mm length × 0.075 mm inner diameter, 3 μm particles, 100 Å pores, Thermo), a C18 analytical column (Acclaim PepMap 300, 150 mm length × 0.075 mm inner diameter, 5 μm particles, 300 Å pores, Thermo) and a 1 μL sample loop. Acetonitrile (Fisher Optima grade, 99.9%), formic acid (1 mL ampules, 99+%, Thermo Pierce), and water purified to a resistivity of 18.2 MΩ·cm (at 25 °C) using a Milli-Q Gradient ultrapure water purification system (Millipore, Billerica, MA) were used to prepare mobile phase solvents. Solvent A was 99.9% water/0.1% formic acid and solvent B was 99.9% acetonitrile/0.1% formic acid (v/v). Samples contained in polypropylene autosampler vials with septa caps (Agilent, Santa Clara, CA) were loaded into the autosampler compartment prior to analysis. The autosampler compartment was maintained at 4 °C. The elution program consisted of isocratic flow at 2% B for 4 min, a linear gradient to 50% B over 98 min, isocratic flow at 95% B for 6 min, and isocratic flow at 2% B for 12 min, at a flow rate of 300 nL/min. The column exit was connected to the nanoESI emitter in the ion source of the mass spectrometer using polyimide-coated, fused-silica tubing (20 μm inner diameter × 280 μm outer diameter, Thermo).
Full-scan mass spectra were acquired in the positive ion mode over the range m/z = 400 to 1800 using the Orbitrap mass analyzer, in profile format, with a mass resolution setting of 60 000 (at m/z = 400, measured at full width at half-maximum peak height, fwhm). The lock mass feature was enabled to provide real-time internal mass calibration using known background ions.37 Inclusion lists were generated using the IsoStamp algorithm (www.isostamp.org). In the data-dependent mode, the three most intense ions exceeding an intensity threshold of 50 000 counts were selected from each full-scan mass spectrum for tandem mass spectrometry (MS/MS, i.e., MS2) analysis using CID. MS2 spectra were acquired using the linear ion trap or the Orbitrap analyzer (in the latter case, with a resolution setting of 7500 at m/z = 400, fwhm), in centroid format, with the following parameters: isolation width 4 m/z units, normalized collision energy 28%, default charge state 3+, activation Q 0.25, and activation time 30 ms. The three most intense fragment ions in each MS2 spectrum exceeding an intensity threshold of 1000 counts were selected for MS3 analysis using CID. MS3 spectra were acquired using the linear ion trap, in centroid format, with the same parameters as those used for MS2. When MS2 spectra were acquired using the Orbitrap analyzer, real-time charge state screening was enabled to exclude unassigned charge states from MS/MS analysis. To avoid the occurrence of redundant MS/MS measurements, real-time dynamic exclusion was enabled to preclude reselection of previously analyzed precursor ions, with the following parameters: repeat count 1, repeat duration 30 s, exclusion list size 500, exclusion duration 90 s, and exclusion mass width ±1.5 m/z units. Global precursor mass lists (i.e., inclusion lists) were enabled to specify the m/z values and retention times of glycopeptide precursor ions detected in full-scan mass spectra by the IsoStamp isotope pattern-searching algorithm. Data acquisition was controlled using Xcalibur software (version 2.0.7, Thermo).
Targeted Data Analysis Procedures
The raw data was processed using Proteome Discoverer software (version 1.4, Thermo Fisher Scientific) and searched against the human-specific SwissProt-reviewed protein database downloaded on July 18, 2014. Indexed databases for tryptic digests were created allowing for up to three missed cleavages, one fixed modification (carboxyamidomethylcysteine, + 57.021 Da), and variable modifications (methionine oxidation, + 15.995 Da; and others as described below). Precursor ion mass tolerances for spectra acquired using the Orbitrap and linear ion trap (LTQ) were set to 10 ppm and 1.5 Da, respectively. The fragment ion mass tolerance was set to 0.8 Da. Raw data was processed through IsoStamp v2.0 to generate an mzXML file filtered for potential glycopeptide spectra. The filtered mzXML file was processed using the SEQUEST HT search engine to initially identify species from exact mass measurements using a modified HexNAc, termed “Si2HexNAz” (C13H18D2N4O7, + 346.1458 Da), with variable attachment to serine, threonine, or asparagine residues. Tandem MS data were screened for glycopeptide signifiers including isotopically recoded precursor ion in full-scan mass (i.e., MS1) spectra, and neutral or charged glycan losses in MS2 spectra. Selected MS2/MS3 spectra were documented and saved separately. Saved spectra were manually annotated for glycan structure and peptide mass. Saved spectra were then searched iteratively using the Byonic search algorithm v2.0 as a node in Proteome Discoverer 1.4. Initial searches allowed for singly tagged N- and O-glycan variable modifications (see input file below) using precursor masses designated from the MS1 (e.g., MS1 precursor for both MS2 and MS3 spectra) or MS(n − 1) (e.g., MS1 precursor for MS2 spectra, and MS2 precursor for MS3 spectra). Computational assignments of all spectra were validated by manual inspection for glycan and peptide fragments. High probability assignments were inspected for validity, and unassigned spectra were archived for further analysis. For MS(n − 1) assignments, the assignment was validated for exact mass from the MS1 (Δmass = <5 ppm). Unassigned spectra from the initial searches were sorted by glycan type based on fragment ions observed in the MS2 spectra (e.g., Si2HexNAz, elaborated O-glycan, or elaborated N-glycan) and searched with variable modification using a focused glycan database. Finally, spectra that remained low confidence assignments were then manually inspected for similarities to assigned spectra (i.e., characteristic peptide fragment ions) or searched against the UniprotKB database (downloaded September 30, 2014) with variable modification on the specific glycan. Only spectra for which the glycan structure was fully characterized were included in the final analysis.
All annotated spectra are made available at www.isostamp.org/pdfs.
Released N-Linked Profiling
Direct MS analysis of the released N-glycans was performed on whole PC-3 cells treated with DMSO (as control), Ac4ManNAz or Ac4GalNAz, based on the method of Aoki et al.38 Briefly, the cell pellets (7.5 × 106 cells each) were homogenized in chloroform/methanol/water (4:8:3, v/v/v) by dounce homogenization. Delipidated protein-rich precipitate was collected by cold centrifugation. The protein-rich pellet was further washed with cold acetone and dried. The released N-glycans were prepared and purified from the protein pellet as described elsewhere.38 Released glycans were permethylated based on the method of Anumula and Taylor39 and profiled directly by mass spectrometry. MALDI-TOF mass spectra of the permethylated N-glycans were acquired in the reflector positive ion mode using α-dihyroxybenzoic acid (DHBA, 20 mg/mL solution in 50% methanol–water) as a matrix. The spectra were acquired using a TOF/TOF 5800 System (AB SCIEX).
RESULTS
Development of IsoStamp v2.0 for High Fidelity Pattern Matching and Validation
Broad implementation of mass-independent MS is predicated on automated recognition of isotopically recoded species with low false-positive and false-negative rates. To automate pattern recognition of isotopically recoded species, we developed the computer program IsoStamp, which compares observed and predicted isotopic envelopes to identify chemically tagged species in full-scan mass spectra.35 However, our first rendition of this program, IsoStamp v1.0, was not able to identify many tagged glycopeptides from samples as complex as whole cell glycoproteomes. The false negative rate of IsoStamp v1.0 proved to be too high for intact glycoproteome analysis.
To improve algorithm fidelity, we developed IsoStamp v2.0 and provided it as open-source software on a fully integrated web interface for public access (see Methods). IsoStamp v2.0 features improvements to peak picking, model generation, and the scoring algorithm. On program initiation, IsoStamp v2.0 enters a peak picking phase in which centroided, full-scan mass spectra in mzXML format are combined into bundles from which isotopically related peaks are extracted (Figure 2A). The algorithm next enters a model generation phase (Figure 2B). The default peptide model (“base pattern”) is derived from a modified averagine distribution40 convolved with and without the pattern of a user-defined chemical tag. The averagine distribution was modified to integrate a special handling for sulfur, wherein the predictive model is generated iteratively for 0–3 sulfur atoms.
Figure 2.
Workflow for model generation and pattern matching by IsoStamp v2.0. (A) Full scan MS data are bundled across scans and peaks are selected for analysis by charge state. (B) A predicted distribution is generated by convolution of a base pattern derived from a modified averagine model and an overlaid tag pattern. (C) Observed and predicted distributions are compared by subtraction to obtain linear and MSQR errors. Examples of errors used in the error function are shown in red overlaid on the observed pattern match shown in gray, where a denotes the abundance of the peak contributing to the error measurement. Error types: m = misaligned, pr = precursor, N = noise, d = m/z deviation, M = missing.
Predicted isotopic distributions are generated by the algorithm, compared to measured isotopic distributions, and scored for match quality. The differences between predicted and observed distributions are fit by linear error and mean square quantization error (MSQR). Matches that pass a minimum error threshold for both (<0.85) are then scored using eq 1:
| (1) |
where Reference is an adjustable base value and Error is defined by eq 2:
| (2) |
where k values are weighted coefficients, and a values are abundances measured from the bundled MS1 data. Variable definition and visual representation of these error types are illustrated in Figure 2C. IsoStamp v2.0 accounts for several types of error during pattern matching, including: extra peaks (e.g., due to overlapping isotopic distributions, or electronic or chemical noise artifacts), fewer (or “missing”) peaks, or poor match quality (e.g., due to misalignment of peaks). Weights for each error type were optimized iteratively to minimize false positive and false negative rates against model system and complex glycopeptide data (see below). Matches are ranked by score and detailed match outputs are generated alongside for validation of algorithm performance. Examples of good and poor match outputs are presented in the Supporting Information. Finally, scored matches are summarized as output files containing precursor ion inclusion lists and chart summaries of matches.
The algorithm performance was evaluated with bovine serum albumin (BSA) chemically modified on cysteine residues with halogenated arenes, namely, monobromoarene (tag 1) dichloroarene (tag 2), and dibromoarene (tag 3) (Figure 3A and B). Tagged BSA was spiked into Jurkat whole cell lysates at amounts in the range of 3.00–0.03 pmol.35 The first generation IsoStamp algorithm had false-negative rates of 28–67% based on manual validation of observed tagged peptides in 30 mol tagged BSA. By comparison, IsoStamp v2.0 displayed false-negative rates of 13–21%. IsoStamp v2.0 produced an overall increase in true positive identification rates across tags 1–3 at 3.00 pmol amounts (Figure 3C). This fidelity was maintained at amounts of BSA as low as 0.03 pmol across tags (Figure 3C). As a measure of false-positive rate, we found that scores for 3.0 pmol BSA displaying tag 3 in Jurkat whole cell lysate increased by up to 90 points as compared to Jurkat whole cell lysate alone (Figure 3D). In aggregate, 100% of tag 3-carrying true positive species scored higher than the average score from Jurkat whole cell lysate alone. Elevated score distributions for BSA peptides carrying tags 1 and 2 in Jurkat whole cell lysate were likewise observed with IsoStamp v2.0 (Figure 3D).
Figure 3.
Fidelity of pattern recognition by IsoStamp. (A) Model system design. Bovine serum albumin (BSA) was tagged with halogen tags, digested, and spiked into Jurkat lysates. (B) Structures of halogen tags include tag 1 (monobromide), tag 2 (dichloride), and tag 3 (dibromide). (C) Comparison of false negative rates between IsoStamp v1 and IsoStamp v2 across tags 1–3. Bovine serum albumin tagged with a halogenated arene (tags 1–3) was added to Jurkat whole cell lysate and analyzed by MS. Data were obtained from ref 35. (D) Score distribution of pattern matches produced by IsoStamp v2 searching for halogen tagged peptides in complex mixtures. Jurkat lysates with halogen tagged BSA peptides generated better scored pattern matches than Jurkat lysate alone. Higher score indicates better match.
The substantial improvement obtained with IsoStamp v2.0 prompted further adaptation as a tool for pattern-dependent filtration of tandem MS data. The MS2 filtration feature computationally filters for MS2 (and MSn) spectra that carry glycopeptide signatures. Specifically, MS2 spectra that derive from a patterned species in the MS1 and/or exhibit characteristic glycan fragment ions (i.e., glycan oxonium ions and ions due to charged or neutral losses)41 are scored according to eq 3:
| (3) |
where k values are weighted coefficients, a values are abundances of observed glycosidic fragment ions in the MS2 spectra, “ox” represents glycan oxonium ions (e.g., Hex and HexNAc), and “loss” represents fragment ions derived from charged or neutral losses (e.g., M–Hex and M–HexNAc). Interestingly, although detection of charged or neutral loss fragment ions was useful for identifying MS2 spectra derived from glycopeptides, it was often insufficient, by itself, for definitive glycopeptide identification without the accompanying precursor ion isotopic pattern from the full-scan mass spectrum. MS2 spectra with ScoreMS2 greater than a user-defined threshold are filtered to produce a new mzXML file, which can then be used to facilitate spectral assignment of glycopeptides by database searching. Implementation of the MS2 filtration algorithm removed approximately 70% of spectra derived from untagged peptides. Automated filtration of tandem MS data was critical during analysis of the glycoproteome from 15 cell lines.
The ability of isotope recoding to increase confidence in spectral assignments has been previously noted.35,42,43 Such confidence is particularly critical in glycoproteomics, due to cleavage of labile glycosidic bonds during collision-induced dissociation (CID) and the lower fragmentation efficiencies of ETD.41 We quantified the validation advantage of isotope recoding by spectral counting. Across three Ac4ManNAz-labeled samples analyzed on an Orbitrap Elite, a total of 150–490 spectra derived from isotopically recoded precursors. Glycopeptide assignments by database searching at a 1% false discovery rate (FDR) were found to have 25–58 validated assignments (up to 88% false negative rate) and 1–3 false positives (4% FDR actual).44
Development of Stable Isotope-Encoded IsoTaG Probe Mixtures
Although the dibromide tag was highly recognizable with IsoStamp v2.0, we sought to explore alternative probes with smaller, less sterically congested structures. A primary motivation was the absence of core fucosylation in the N-glycan structures from our initial studies.32 The majority of the N-glycan structures carried GlcNAz incorporation at the reducing end of the glycan structure. Thus, the absence of core fucosylation was potentially caused by lack of access of the probe to the sterically congested GlcNAz. To probe this hypothesis, we developed a series of IsoTaG probes encoded by zero [M], two [M + 2], or four [M + 4] deuterium atoms. The IsoTaG probe with zero, and that with two deuterium atoms [M, M + 2] were mixed in different proportions; 1:1, 1:2, 1:3 and 1:4 (abbreviated as probe mixtures 1, 2, 3 and 4, respectively, Figure 4A). Alongside, the probes with zero and four deuterium atoms [M, M + 4] were mixed and prepared in the same manner (mixtures 5, 6, 7 and 8, for the mixture ratios 1:1, 1:2, 1:3 and 1:4, respectively).
Figure 4.
Evaluation of stable isotope probes and pattern diversity. (A) Stable isotope probe mixtures 1–8 were prepared by NMR titration and validated by MS. (B) Manual validation of the top 50 scored pattern matches by IsoStamp v2.0 produced by the stable isotope mixtures 1–8. BSA was chemically modified on its lysine residues with azidoacetate, conjugated with the stable isotope mixtures by CuAAC, and analyzed on an Orbitrap XL.
To test pattern recognition, BSA was chemically modified on its lysine residues with azidoacetate and then conjugated with the isotope recoding probe mixtures 1–8 via copper-catalyzed azide–alkyne cycloaddition (CuAAC). The biotinylated BSA samples were digested with trypsin, treated with mild acid for linker cleavage, and analyzed by MS. The top 50 matches produced by IsoStamp v2.0 were manually validated. The observed false-positive rates indicated that the highest recognition was achieved by isotope mixture 3 with a ratio of [M, M + 2] of 1:3 (Figure 4B). Furthermore, isotope mixture 3 produced higher overall scores than other probe mixtures. Wider isotopic distributions (e.g., [M, M + 4]) produced higher false-positive rates.
The fragmentation of a glycopeptide recoded with mixture 3 was compared to that using the previously employed dibromide probe.32 For this experiment, we used as a substrate an O-GlcNAcylated peptide derived from alpha-crystallin which was enzymatically elaborated with GalNAz.18 Little difference in HCD, CID, or ETD fragmentation was observed between the deuterated and brominated glycopeptides (Figures S1–S3). Intriguingly, doubly charged positive glycopeptide precursor ions were readily fragmented by ETD and, in some cases, fragmented to a greater degree than the unmodified glycopeptide. A degree of internal tag fragmentation was observed with ETD, although this was not seen with all precursor ions.45 A maximum of 0.1 min chromatographic retention time shift between differentially deuterated isotopes was observed by nanoflow liquid chromatography. This was negligible for isotopic pattern matching due to implementation of spectral bundling (Figure S4). These results demonstrate that deuterated or brominated probes are readily compatible with a variety of fragmentation methods, including ETD.
Intact Glycopeptides Found in a Survey of 15 Cell Lines
A survey of intact glycoproteomes from 15 cell lines was pursued using isotope recoding mixture 3 (Figure 1). Cell lines were metabolically labeled with 100 μM Ac4GalNAz or Ac4ManNAz, and cell lysates and conditioned media were collected. The intact glycoproteome was tagged and affinity enriched with mixture 3. Inclusion list-directed MS2 and MS3 measurements, using CID, were performed on an LTQ-Orbitrap-XL mass spectrometer. Each MS data file was manually annotated for the number of spectra derived from isotopically recoded species. A quantitative measure of these efficiencies by spectral counting is presented in Figure S5. Metabolic labeling efficiency was observed to be metabolite and cell line dependent. The cell lines PC-3, MCF10a, and LnCAP produced the greatest number of spectra and thus glycopeptide assignments from Ac4GalNAz labeling. Spectral counting revealed that the cell lines HepG2, SK-N-SH, and BxPC3 were labeled by Ac4ManNAz at the highest rates. In contrast, Hek293 revealed limited spectral counts from either metabolite. In general, the spectral counts were predicted by Western blot data obtained prior to MS analysis, and appears to reflect general trends in the ability to metabolically label different cell lines.46
Glycopeptide structure assignment was completed by a stepwise process. Fragmentation of glycopeptides by CID typically yield glycan fragmentation in the MS2 followed by peptide fragmentation in the MS3. Following selection of CID tandem mass spectra derived from precursor ions exhibiting the characteristic isotopic pattern, glycan structures were manually assigned, and the underlying peptide mass was extracted from the MS2 data. Glycan structures from CID spectra were manually annotated for glycan neutral and charged losses (see www.isostamp.org/pdfs for annotated spectra) to provide the constitutional composition of glycan structures. With the constitutional composition in hand, linkages between glycans and the glycan structure assignments were based on homology with previously defined mammalian glycans. Peptide assignments were performed by database searching (SwissProt human proteome, 1% FDR) and validated by correlation of the exact mass of the intact glycopeptide to the observed MS1 precursor ion (within a stated mass tolerance of <10 ppm). Obtaining confident peptide assignments requires sufficient peptide backbone fragmentation during tandem MS analysis, which was limited by the number of pendant glycans. Peptide assignments were possible with up to approximately ten pendant glycans with the CID MS2/MS3 method employed. This limitation, along with the onus on manual glycan annotation, can be overcome using alternate fragmentation methods (e.g., HCD) implemented on an Orbitrap Elite or Fusion Tribrid instrument.34 Finally, glycopeptide spectra were cross-correlated by peptide mass to the database of assigned glycopeptides, and homologous spectra were assigned by inference. Only spectra with full compositional assignment of the glycan structures were included in the final data set. In aggregate, 3535 spectra were assigned defined glycan structures, of which 1825 spectra were assigned to a peptide sequence. All spectra were annotated and are provided as Supporting Information.
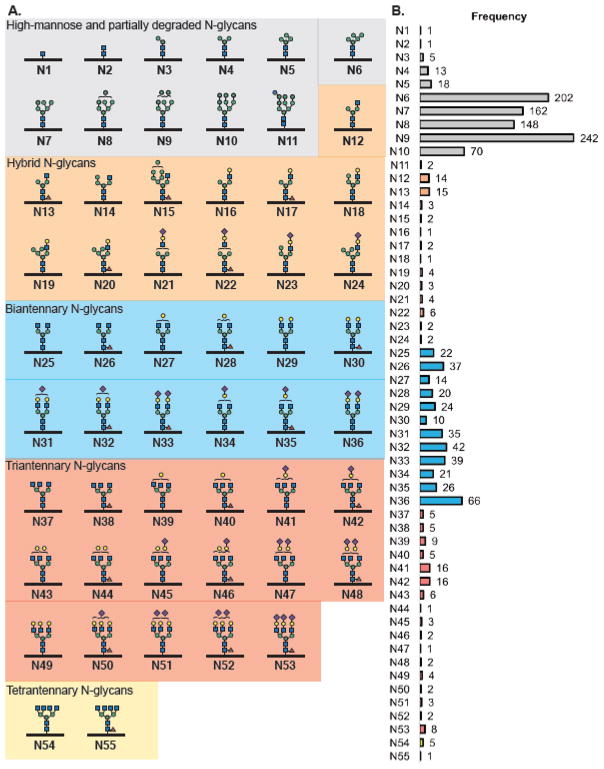
The use of isotope recoding mixture 3 with the 15 cell lines revealed a total of 74 glycan structures spanning 55 N- and 18 O-glycans. Glycan structures were coded as N1–N55 and O1–O18 to summarize constitutionally similar structures aside from the frequency of azidosugar incorporation and multiple tagging events to a glycan structure. N-Glycans ranged from high-mannose and partially degraded N-glycans to complex triantennary and tetrantennary glycan structures (Figure 5). Notably, core fucosylation was observed on a range of N-glycan structures, whereas few such structures were previously found using a dibrominated probe.32 Relative quantification of glycans by spectral counting revealed a high frequency of high-mannose N-glycans (N6–N10) followed by biantannary N-glycans (N25–N36, Figure 5B).
Figure 5.
N-Glycan structures observed in a 15-cell line survey. (A) N-Glycans ranged from high-mannose and partially degraded, hybrid, biantennary, triantennary, and tetrantennary glycans. (B) Frequency of observed metabolically labeled N-glycan structures. Bars are color coded to class of glycan structure.
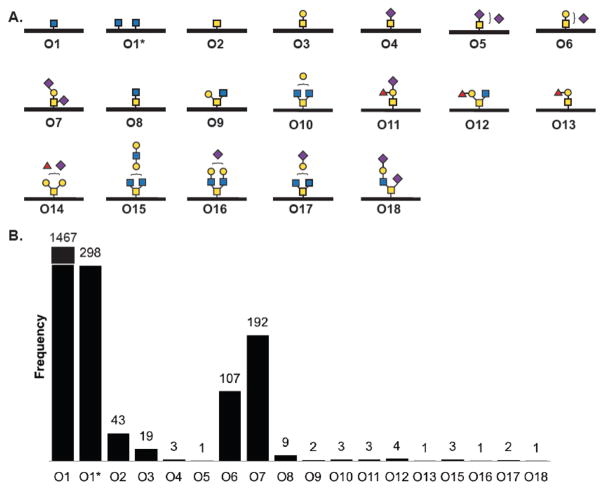
The observed O-glycans encompassed 18 structures, with O-GlcNAc (O1, O1*) comprising the preponderance of O-glycan assignments (>80%, Figure 6). The assignment of a single HexNAc residue as either O-GlcNAc (O1) or mucin-type O-GalNAc (O2) was based on known subcellular localization of the associated glycoprotein, or by the cellular fraction that was analyzed in instances where the peptide substrate remained unassigned. In these instances, cellular fractions derived from the conditioned media were assigned as mucin-type (O2), and cellular fractions derived from cytoplasmic and nuclear fractions were assigned as O-GlcNAc (O1).47 Mucin-type glycans included the Tn antigen (O2), cores 1–4 (O3, O9, O8, O10, respectively), and higher-order glycan structures (e.g., O15, O16). A diversity of sialylated glycans was observed, with a particular abundance of O-glycans O6 and O7 (Figure 6B). Furthermore, fucosylated O-glycans (O11–14) were observed when tagged and enriched using probe mixture 3.
Figure 6.
O-Glycan structures observed in a 15-cell line survey. (A) A total of 18 O-glycan structures were observed, including O-GlcNAc (O1, O1*), mucin type (O2–O18), and sialylated glycan structures (e.g., O5–O7). (B) Frequency of observed metabolically labeled O-glycan structures.
Metabolic Label Incorporation Revealed Across 15 Cell Lines
The survey of 15 cell lines herein revealed a distribution of protein localization analogous to that reported previously (Figure 7A).32 Approximately 60% of the glycoproteins were intracellularly localized, largely due to contributions from Ac4GalNAz metabolic label incorporation (Figure 7A). From Ac4GalNAz-labeled samples, intracellular O-GlcNAz constituted approximately two-thirds of glycopeptides, N-GlcNAz constituted an additional 30%, and approximately 5% of glycopeptides represented O-GalNAz incorporation (Figure 7B). Sialylated glycopeptides produced by Ac4ManNAz labeling were found on O-glycan attachment sites (78%) more frequently than N-glycan attachment sites (22%, Figure 7C). The majority of glycopeptides were only observed in a single cell line (62%, Figure 7D).
Figure 7.
Glycoproteins and glycopeptides identified across 15 cell lines. (A) Distribution of glycoprotein subcellular localization. (B) Distribution of glycans labeled by Ac4GalNAz. (C) Distribution of glycan attachment sites labeled by Ac4ManNAz. (D) Frequency of unique peptide substrate occurrence across cell lines. The majority of peptide substrates were observed in only one cell line.
By spectral counting, the ten most frequently observed glycopeptides across cell lines were obtained from Ac4GalNAz metabolic labeling (Table 1). Frequently observed glycoproteins carried glycans that included O-GlcNAc (Entry 2–5, 10), N-GlcNAc (Entry 1, 7–9) and O-GalNAz (Entry 6). Four distinct glycopeptides from host cell factor 1 (P51610), a highly O-GlcNAcylated and abundant glycoprotein, fell within the top ten glycopeptides. The extensive list of glycoproteins presented here illustrates the capacity of Ac4GalNAz to act as a reporter for metabolic pathway activity in the cell.
Table 1.
Ten Most Frequently Observed Glycopeptides, and Their Associated Glycan Structures, Across a 15-Cell Line Survey
| entry | glycopeptide (glycosite bolded) | frequency | glycan structures | protein (accession) |
|---|---|---|---|---|
| 1 | LSALDNLLNHSSMFLK | 66 | N6, N7, N8, N9, N10 | Hypoxia up-regulated protein 1 (Q9Y4L1) |
| 2 | TAAAQVGTSVSSATNTSTRPIITVHK | 61 | O1, O1* | Host cell factor 1 (P51610) |
| 3 | KTPTTVVPLISTIAGDSSR | 50 | O1, O1* | Vascular endothelial zinc finger 1 (Q14119) |
| 4 | SGTVTVAQQAQVVTTVVGGVTK | 44 | O1, O1* | Host cell factor 1 (P51610) |
| 5 | TIPMSAIITQAGATGVTSSPGIK | 43 | O1, O1* | Host cell factor 1 (P51610) |
| 6 | GAPNKEETPATESPDTGLYYHR | 41 | O2, O4, O6, O7, O17 | Nucleobindin-1 (Q02818) |
| 7 | TILVDNNTWNNTHISR | 35 | N5, N6, N7, N8, N9, N10 | Dolichyl-diphosphooligosaccharide—protein glycosyltransferase subunit STT3A (P46977) |
| 8 | TVLTPATNHMGNVTFTIPANR | 34 | N6, N7, N8, N9 | Complement C3 (P01024) |
| 9 | TTLVDNNTWNNSHIALVGK | 33 | N6, N7, N8, N9, N10, N11 | Dolichyl-diphosphooligosaccharide—protein glycosyltransferase subunit STT3B (Q8TCJ2) |
| 10 | VMSVVQTKPVQTSAVTGQASTGPVTQIIQTK | 33 | O1, O1* | Host cell factor 1 (P51610) |
Metabolic Labeling Minimally Perturbs Some Cellular Biosynthetic Pathways
Metabolic labeling marks glycoproteins during their biosynthesis, and thereby enables studies of glycosylation dynamics. However, the effect of metabolic labeling on the steady-state profile of cellular glycans has not been characterized. To assess the effects of metabolic labeling on glycan profile, we compared released N-glycans from untreated and azidosugar-treated PC-3 cells. Briefly, lysates from PC-3 cells labeled with Ac4GalNAz, Ac4ManNAz, or DMSO were homogenized and delipidated. Proteins were enriched by cold-acetone precipitation and digested by trypsin, and N-glycans were released enzymatically from the crude mixture by PNGase F. Released N-glycans were permethylated with methyl iodide and analyzed by MS.
The free N-glycans of unlabeled and azidosugar-treated samples revealed a largely unperturbed collection of native glycan structures (Figure 8A). Released glycans observed by Ac4GalNAz labeling and nonsialylated glycans from Ac4ManNAz labeled samples were present in similar distribution to the unlabeled DMSO control. These results indicate that, in the case of Ac4GalNAz, the azidosugar is incorporated at low rates with little perturbation to the underlying glycosylation pathways of native glycans.
Figure 8.
Comparison of N-glycan structures by released N-glycan analysis and glycans observed by IsoTaG from a single cell line. (A) Released permethylated N-glycan analysis by MALDI. Glycan structures from PC-3 cells also observed by IsoTaG are marked with “*”. (B) Intact glycan analysis by IsoTaG of PC-3 cells. Glycan structures from PC-3 cells that are also found by free glycan analysis are shown in black. Structure codes correspond to the glycan structures in Figure 5.
Sialylated glycans in the DMSO control sample were complex biantennary and triantennary glycans. These sialylated glycans were not observed in Ac4ManNAz-treated samples. The relative abundances of N-glycans on intact glycopeptides characterized using IsoTaG were consistent with the relative abundances of glycans of control sample observed by free N-glycan analysis (Figure 8B). Optimization of methods to analyze the azide-incorporated glycan structures directly are currently underway.
DISCUSSION
We describe the development of IsoTaG for intact glycoproteomics analysis of metabolically labeled samples. IsoTaG uses efficient enrichment and accurate isotopic pattern matching to facilitate detection and assignment of metabolically labeled glycopeptides by MS.
IsoStamp v2.0 is a high fidelity pattern matching algorithm that directs tandem MS analysis to isotopically recoded precursor ions, and subsequently filters MS2 spectra for glycopeptides. Significant improvement to false positive and false negative rates were achieved by optimization of data handling and model generation. Input full-scan MS data are bundled to average ion abundances over multiple scans prior to pattern matching. Isotopic model generation was improved to account for the impact of sulfur isotopes on the averagine model. Sulfur isotopes contribute the greatest isotopic perturbation to the averagine model among the elements that comprise natural glycopeptides. The base pattern is generated iteratively for 0–3 sulfur atoms to improve the predictive accuracy of the model. With these improvements, false negative rates fell to as low as 13% in model systems. Without implementation of the IsoTaG pattern recognition, up to 88% of HCD spectra derived from isotopically patterned species were not identified as glycopeptides by database searching alone (using Sequest or Byonic algorithms). While some of these spectra may have eluded assignment due to other factors (e.g., low signal-to-noise ratios), a majority of the glycopeptides would be undiscovered without the isotopic pattern recognition and targeted approach. Isotope recoding provides an orthogonal handle for detection and characterization of glycopeptides in a chemical proteomics platform. In addition to our investigations, there has been growing interest in the use of isotope recoding for validation of chemically modified substrates.42,43,48–50
Investigation of probe design and pattern recognition revealed that brominated and deuterated probes fragment similarly by CID, HCD, and ETD (Figures S1–S3). However, use of the dibromide tag may sterically inhibit tagging of sugars embedded at the reducing end of a glycan in metabolically labeled glycoproteins. The broader array of glycan structures tagged by stable isotope probe mixture 3 supports the possibility of steric occlusion caused by dibrominated tags in earlier reports.32 Pattern recognition by IsoStamp v2.0 showed the highest fidelity for an isotopic ratio of 1:3 over [M, M + 2], and lower fidelity for patterns over [M, M + 4] spacing. The higher false positive rates for [M, M + 4] potentially derive from the wider isotopic envelope under evaluation.
Fifteen cell lines were metabolically labeled with Ac4GalNAz or Ac4ManNAz and surveyed by IsoTaG. A total of 55 N-glycans and 18 O-glycans were observed with isotope probe mixture 3. In general, Ac4GalNAz labeling incorporated only one azidosugar per glycopeptide with the exception of a subset of O-GlcNAcylated glycopeptides. Higher replacement rates of sialic acid by Ac4ManNAz was observed, which produced glycopeptides with up to four azidosugar substitutions. Multiple tagging events are readily accounted for by searching for the expected isotope pattern due to multiple tags within the IsoStamp v2.0 software.
Ac4ManNAz labeling yielded sialylated glycans, of which the majority were O-glycosylated. The higher rates of sialylated O-glycans suggest the existence of a wider array of O-glycoproteins that have yet to be discovered. Ac4GalNAz labeling yielded O-GlcNAz as the primary glycan structure observed (>60%). The preponderance of O-GlcNAz glycopeptides may be related to the ease of identification of O-GlcNAc as a single HexNAc, potentially different permissiveness of O-glycosyltransferase (OGT) as compared to the ppGalNAcTs, and the rate of epimerization of UDP-GlcNAc and UDPGalNAc by the GALE pathway. Notably, a total of 21 fucosylated N-glycan structures were observed, as compared to a single fucosylated N-glycan previously observed using brominated probes on similar cell lines (PC-3).32 Manual annotation of the observed glycan structures revealed core-fucosylation, due to localization of fucose to the reducing-end of the glycan structure (see Experimental Procedures). In conjunction, the majority of GlcNAz incorporation was observed to occur at the reducing end of N-glycan structures, supporting our hypothesis that the isotope probe mixture 3 would be able to tag a broader spectrum of N-glycan structures.
Released N-linked glycan analysis of whole PC-3 cells, which were treated with DMSO (control), Ac4GalNAz or Ac4ManNAz, was carried out without introducing IsoTaG labeling, for comparison with glycopeptide profiling with IsoTaG. The results showed native glycans carried homologous glycan structures to those identified by the IsoTaG glycopeptide analysis. Furthermore, glycan abundances observed by the conventional free N-glycan analysis were similar irrespective of the metabolic labeling, with the exception of sialylated glycans from Ac4ManNAz samples. Unobserved sialylated glycan structures in the conventional N-linked profiling is indicative of high levels of replacement of sialic acid by SiaNAz or possible suppression of sialyl transferases. Direct observation of azide-labeled glycans by the conventional procedure may be precluded by chemical destruction and derivatization of the azide during permethylation. High replacement rates of sialic acid by SiaNAz are in line with previous reports by direct sialic acid quantification.14
In conclusion, we report the evolution of IsoTaG aided by IsoStamp v2.0 and the probe mixture 3 for high fidelity pattern matching and characterization of intact glycopeptides that represent, at least for N-glycans in PC-3 cells, the distribution of glycan structures observed by PNGase F-released glycan analysis. We applied this improved system to a broader set of cell lines and quantified the relative abundances of glycan structures observed, as well as labeling efficiency, using spectral counting. Application of isotopic recoding uniformly improved the selection and confidence of our results across sample types and metabolic labels. These results set the foundation for broader applications of IsoTaG and mass-independent MS. With a generally applicable method to discern glycosylation in a site-specific manner, further correlation of structure to function for particular glycosites may now be uncovered and studied. In addition to the acceleration of structure–function studies of the glycoproteome, the methodology employed in IsoTaG may potentially be applied toward studies of other chemically tagged post-translational modifications.
Supplementary Material
Acknowledgments
We would like to thank Chris Doty-Humphrey for computational support. Financial support from the US National Institutes of Health (CA200423, C.R.B.; U01 CA207702, C.R.B. and S.J.P.), (P41GM10349010, 1S10OD018530, P.A.), (1S10OD020062-01, A.T.I.), Jane Coffin Childs Memorial Fund (C.M.W.), Burroughs Wellcome Fund Career Awards at the Scientific Interface (C.M.W.), and Stanford Undergraduate Advising and Research Student Grant (A.F.) are gratefully acknowledged.
Footnotes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome. 6b01053.
Synthetic methods, examples, and Figures S1–S6 (PDF)
Tables S1 and S2 (XLSX)
Author Contributions
C.R.B. and C.M.W. conceived and directed the project. W.B. and D.Z. developed IsoStamp web platform and open source support. C.M.W. and A.F. performed cell culture and glycopeptide enrichment. C.M.W., A.T.I., and S.J.P. collected data. C.M.W. and A.F. analyzed data. M.I. and P.A. performed free glycan analysis. C.M.W. composed the manuscript. All authors edited and approved the manuscript.
References
- 1.Dube DH, Bertozzi CR. Glycans in Cancer and Inflammation - Potential for Therapeutics and Diagnostics. Nat Rev Drug Discovery. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 2.Adamczyk B, Tharmalingam T, Rudd PM. Glycans as cancer biomarkers. Biochim Biophys Acta, Gen Subj. 2012;1820:1347–1352. doi: 10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Parodi AJ. Protein glucosylation and its role in protein folding. Annu Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- 4.Radhakrishnan P, Dabelsteen S, Madsen FB, Francavilla C, Kopp KL, Steentoft C, Vakhrushev SY, Olsen JV, Hansen L, Bennett EP, Woetmann A, Yin G, Chen L, Song H, Bak M, Hlady RA, Peters SL, Opavsky R, Thode C, Qvortrup K, Schjoldager KT, Clausen H, Hollingsworth MA, Wandall HH. Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proc Natl Acad Sci U S A. 2014;111:E4066–4075. doi: 10.1073/pnas.1406619111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 6.Lasky L. Selectins: interpreters of cell-specific carbohydrate information during inflammation. Science. 1992;258:964–969. doi: 10.1126/science.1439808. [DOI] [PubMed] [Google Scholar]
- 7.Yuzwa SA, Shan X, Macauley MS, Clark T, Skorobogatko Y, Vosseller K, Vocadlo DJ. Increasing O-Glcnac slows neurodegeneration and stabilizes tau against aggregation. Nat Chem Biol. 2012;8:393–399. doi: 10.1038/nchembio.797. [DOI] [PubMed] [Google Scholar]
- 8.Marotta NP, Lin YH, Lewis YE, Ambroso MR, Zaro BW, Roth MT, Arnold DB, Langen R, Pratt MR. O-GlcNAc modification blocks the aggregation and toxicity of the protein α-synuclein associated with Parkinson’s disease. Nat Chem. 2015;7:913–920. doi: 10.1038/nchem.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma J, Hart GW. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev Proteomics. 2013;10:365–380. doi: 10.1586/14789450.2013.820536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagerlöf O, Slocomb JE, Hong I, Aponte Y, Blackshaw S, Hart GW, Huganir RL. The nutrient sensor OGT in PVN neurons regulates feeding. Science. 2016;351:1293–1296. doi: 10.1126/science.aad5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durbin KR, Fornelli L, Fellers RT, Doubleday PF, Narita M, Kelleher NL. Quantitation and Identification of Thousands of Human Proteoforms below 30 kDa. J Proteome Res. 2016;15:976–982. doi: 10.1021/acs.jproteome.5b00997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 13.Hang HC, Yu C, Kato DL, Bertozzi CR. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc Natl Acad Sci U S A. 2003;100:14846–14851. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang PV, Chen X, Smyrniotis C, Xenakis A, Hu T, Bertozzi CR, Wu P. Metabolic Labeling of Sialic Acids in Living Animals with Alkynyl Sugars. Angew Chem, Int Ed. 2009;48:4030–4033. doi: 10.1002/anie.200806319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 16.Nilsson J, Ruetschi U, Halim A, Hesse C, Carlsohn E, Brinkmalm G, Larson G. Enrichment of glycopeptides for glycan structure and attachment site identification. Nat Methods. 2009;6:809–811. doi: 10.1038/nmeth.1392. [DOI] [PubMed] [Google Scholar]
- 17.Song X, Ju H, Lasanajak Y, Kudelka MR, Smith DF, Cummings RD. Oxidative release of natural glycans for functional glycomics. Nat Methods. 2016;13:528–534. doi: 10.1038/nmeth.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, Hsieh-Wilson LC. Direct In-Gel Fluorescence Detection and Cellular Imaging of O-GlcNAc-Modified Proteins. J Am Chem Soc. 2008;130:11576–11577. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zielinska DF, Gnad F, Wiśniewski JR, Mann M. Precision Mapping of an In Vivo N-Glycoproteome Reveals Rigid Topological and Sequence Constraints. Cell. 2010;141:897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Trinidad JC, Schoepfer R, Burlingame AL, Medzihradszky KF. N- and O-Glycosylation in the Murine Synaptosome. Mol Cell Proteomics. 2013;12:3474–3488. doi: 10.1074/mcp.M113.030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandler KB, Costello CE. Glycomics and glycoproteomics of membrane proteins and cell-surface receptors. Electrophoresis. 2016;37:1407–1419. doi: 10.1002/elps.201500552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thaysen-Andersen M, Packer NH, Schulz BL. Maturing Glycoproteomics Technologies Provide Unique Structural Insights into the N-glycoproteome and Its Regulation in Health and Disease. Mol Cell Proteomics. 2016;15:1773–1790. doi: 10.1074/mcp.O115.057638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagglund P, Bunkenborg J, Elortza F, Jensen ON, Roepstorff P. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J Proteome Res. 2004;3:556–566. doi: 10.1021/pr034112b. [DOI] [PubMed] [Google Scholar]
- 24.Steentoft C, Vakhrushev SY, Vester-Christensen MB, Schjoldager KT, Kong Y, Bennett EP, Mandel U, Wandall H, Levery SB, Clausen H. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat Methods. 2011;8:977–982. doi: 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- 25.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, Bennett EP, Mandel U, Brunak S, Wandall HH, Levery SB, Clausen H. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarentino AL, Gomez CM, Plummer TH. Deglycosylation of asparagine-linked glycans by peptide:N-glycosidase F. Biochemistry. 1985;24:4665–4671. doi: 10.1021/bi00338a028. [DOI] [PubMed] [Google Scholar]
- 27.Parker BL, Thaysen-Andersen M, Solis N, Scott NE, Larsen MR, Graham ME, Packer NH, Cordwell SJ. Site-Specific Glycan-Peptide Analysis for Determination of N-Glycoproteome Heterogeneity. J Proteome Res. 2013;12:5791–5800. doi: 10.1021/pr400783j. [DOI] [PubMed] [Google Scholar]
- 28.Yin X, Bern M, Xing Q, Ho J, Viner R, Mayr M. Glycoproteomic analysis of the secretome of human endothelial cells. Mol Cell Proteomics. 2013;12:956–978. doi: 10.1074/mcp.M112.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu SW, Pu TH, Viner R, Khoo KH. Novel LC-MS(2) product dependent parallel data acquisition function and data analysis workflow for sequencing and identification of intact glycopeptides. Anal Chem. 2014;86:5478–5486. doi: 10.1021/ac500945m. [DOI] [PubMed] [Google Scholar]
- 30.Shah P, Wang X, Yang W, Toghi Eshghi S, Sun S, Hoti N, Chen L, Yang S, Pasay J, Rubin A, Zhang H. Integrated Proteomic and Glycoproteomic Analyses of Prostate Cancer Cells Reveal Glycoprotein Alteration in Protein Abundance and Glycosylation. Mol Cell Proteomics. 2015;14:2753–2763. doi: 10.1074/mcp.M115.047928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toghi Eshghi S, Shah P, Yang W, Li X, Zhang H. GPQuest: A Spectral Library Matching Algorithm for Site-Specific Assignment of Tandem Mass Spectra to Intact N-glycopeptides. Anal Chem. 2015;87:5181–5188. doi: 10.1021/acs.analchem.5b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woo CM, Iavarone AT, Spiciarich DR, Palaniappan KK, Bertozzi CR. Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat Methods. 2015;12:561–567. doi: 10.1038/nmeth.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo CM, Bertozzi CR. Curr Protocols Chem Biol. John Wiley & Sons, Inc; 2016. [Google Scholar]
- 34.Woo CM, Felix A, Zhang L, Elias JE, Bertozzi CR. Isotope-targeted glycoproteomics (IsoTaG) analysis of sialylated Nand O-glycopeptides on an Orbitrap Fusion Tribrid using azido and alkynyl sugars. Anal Bioanal Chem. 2017;409:579–588. doi: 10.1007/s00216-016-9934-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palaniappan KK, Pitcher AA, Smart BP, Spiciarich DR, Iavarone AT, Bertozzi CR. Isotopic Signature Transfer and Mass Pattern Prediction (IsoStamp): An Enabling Technique for Chemically-Directed Proteomics. ACS Chem Biol. 2011;6:829–836. doi: 10.1021/cb100338x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Hong S, Tran A, Jiang H, Triano R, Liu Y, Chen X, Wu P. Sulfated Ligands for the Copper(I)-Catalyzed Azide–Alkyne Cycloaddition. Chem - Asian J. 2011;6:2796–2802. doi: 10.1002/asia.201100385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keller BO, Sui J, Young AB, Whittal RM. Interferences and contaminants encountered in modern mass spectrometry. Anal Chim Acta. 2008;627:71–81. doi: 10.1016/j.aca.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 38.Aoki K, Perlman M, Lim JM, Cantu R, Wells L, Tiemeyer M. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J Biol Chem. 2007;282:9127–9142. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- 39.Anumula KR, Taylor PB. A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal Biochem. 1992;203:101–108. doi: 10.1016/0003-2697(92)90048-c. [DOI] [PubMed] [Google Scholar]
- 40.Senko MW, Beu SC, McLafferty FW. Determination of monoisotopic masses and ion populations for large biomolecules from resolved isotopic distributions. J Am Soc Mass Spectrom. 1995;6:229–233. doi: 10.1016/1044-0305(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 41.Leymarie N, Zaia J. Effective use of mass spectrometry for glycan and glycopeptide structural analysis. Anal Chem. 2012;84:3040–3048. doi: 10.1021/ac3000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarkar M, Pascal BD, Steckler C, Aquino C, Micalizio GC, Kodadek T, Chalmers MJ. Decoding split and pool combinatorial libraries with electron-transfer dissociation tandem mass spectrometry. J Am Soc Mass Spectrom. 2013;24:1026–1036. doi: 10.1007/s13361-013-0633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomohiro T, Morimoto S, Shima T, Chiba J, Hatanaka Y. An Isotope-Coded Fluorogenic Cross-Linker for High-Performance Target Identification Based on Photoaffinity Labeling. Angew Chem, Int Ed. 2014;53:13502–13505. doi: 10.1002/anie.201408580. [DOI] [PubMed] [Google Scholar]
- 44.Spectra (HCD, CID, and/or EthcD) were collected on an Orbitrap Elite or Lumos and assigned by Byonic at 1% FDR.
- 45.Woo CM, Lund PJ, Davis M, Pitteri SJ, Bertozzi CR. Mapping and quantification of over 2,000 O-GlcNAcylated peptides in activated human T cells with isotope-targeted glycoproteomics (IsoTaG) doi: 10.1074/mcp.RA117.000261. unpublished. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaro BW, Batt AR, Chuh KN, Navarro MX, Pratt MR. The small molecule 2-Azido-2-deoxy-glucose is a metabolic chemical reporter of O-GlcNAc modifications in mammalian cells, revealing an unexpected promiscuity of O-GlcNAc transferase. ACS Chem Biol. 2016 doi: 10.1021/acschembio.6b00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hart GW. Dynamic O-linked glycosylation of nuclear and cytoskeletal proteins. Annu Rev Biochem. 1997;66:315–335. doi: 10.1146/annurev.biochem.66.1.315. [DOI] [PubMed] [Google Scholar]
- 48.Goodlett DR, Bruce JE, Anderson GA, Rist B, Pasa-Tolic L, Fiehn O, Smith RD, Aebersold R. Protein identification with a single accurate mass of a cysteine-containing peptide and constrained database searching. Anal Chem. 2000;72:1112–1118. doi: 10.1021/ac9913210. [DOI] [PubMed] [Google Scholar]
- 49.Hernandez H, Niehauser S, Boltz SA, Gawandi V, Phillips RS, Amster IJ. Mass defect labeling of cysteine for improving peptide assignment in shotgun proteomic analyses. Anal Chem. 2006;78:3417–3423. doi: 10.1021/ac0600407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brodie NI, Makepeace KAT, Petrotchenko EV, Borchers CH. Isotopically-coded short-range hetero-bifunctional photo-reactive crosslinkers for studying protein structure. J Proteomics. 2015;118:12–20. doi: 10.1016/j.jprot.2014.08.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.