Abstract
Despite the high response rates of individuals with myelodysplastic syndrome (MDS) with deletion of chromosome 5q (del(5q)) to treatment with lenalidomide (LEN) and the recent identification of cereblon (CRBN) as the molecular target of LEN, the cellular mechanism by which LEN eliminates MDS clones remains elusive. Here we performed an RNA interference screen to delineate gene regulatory networks that mediate LEN responsiveness in an MDS cell line, MDSL. We identified GPR68, which encodes a G-protein-coupled receptor that has been implicated in calcium metabolism, as the top candidate gene for modulating sensitivity to LEN. LEN induced GPR68 expression via IKAROS family zinc finger 1 (IKZF1), resulting in increased cytosolic calcium levels and activation of a calcium-dependent calpain, CAPN1, which were requisite steps for induction of apoptosis in MDS cells and in acute myeloid leukemia (AML) cells. In contrast, deletion of GPR68 or inhibition of calcium and calpain activation suppressed LEN-induced cytotoxicity. Moreover, expression of calpastatin (CAST), an endogenous CAPN1 inhibitor that is encoded by a gene (CAST) deleted in del(5q) MDS, correlated with LEN responsiveness in patients with del(5q) MDS. Depletion of CAST restored responsiveness of LEN-resistant non-del(5q) MDS cells and AML cells, providing an explanation for the superior responses of patients with del(5q) MDS to LEN treatment. Our study describes a cellular mechanism by which LEN, acting through CRBN and IKZF1, has cytotoxic effects in MDS and AML that depend on a calcium- and calpain-dependent pathway.
LEN, a synthetic glutamic acid derivative, is approved for the treatment of hematological malignancies, including MDS and multiple myeloma1. Myelodysplastic syndromes are hematopoietic stem cell (HSC) disorders that are characterized by ineffective hematopoiesis, peripheral cytopenias, and a propensity for progression to AML. LEN is approved for the treatment of lower-risk del(5q) MDS, and it provides durable hematologic and cytogenetic responses in the majority of patients2. LEN is also effective in a subset of individuals with non-del(5q) MDS, but only for 26% of the patients3. LEN has diverse biological effects—including the suppression of inflammatory cytokines, the inhibition of MDM2 proto-oncoprotein (MDM2) activity via serine/threonine protein phosphatase 2A (PP2A) and cell division cycle 25C (CDC25C), and the stimulation of T lymphocytes4,5. More recently, LEN has been shown to bind directly to cereblon (CRBN), a substrate receptor of the CRL4CRBN E3 ligase complex6, which results in the ubiquitination and degradation of select protein targets. LEN treatment promotes CRBN-dependent degradation of IKZF1 and IKZF3, as well as degradation of casein kinase I isoform alpha (CSNK1A1; also known as CK1-α), which are key factors in hematologic diseases7–9. Despite the identification of these direct CRL4CRBN targets, the mechanistic basis for the therapeutic effects of LEN in MDS is not fully understood. Insight into LEN’s mechanism of action may improve our understanding of clinical response and inform rational combinatorial therapeutic approaches.
RESULTS
A genome-wide RNA interference screen identifies determinants of LEN sensitivity in MDS
Although very few cellular models of MDS exist, we have previously extensively characterized a LEN-sensitive cell line, MDSL, which was derived from a patient with lower-risk MDS10,11. MDSL cells show incomplete differentiation and propagate in vitro as a heterogeneous mixture of functionally distinct immature CD34+ hematopoietic stem and progenitor cells and mature CD34− myeloid cells (Supplementary Fig. 1a). Treatment with LEN suppressed colony formation of CD34+ MDSL cells in methylcellulose (Supplementary Fig. 1b), induced apoptosis (annexin V+ cells) of MDSL cells specifically within the CD34− subpopulation (Supplementary Fig. 1c), and impaired total cell growth in liquid culture12 (Supplementary Fig. 1d). Thus, LEN treatment has pleiotropic effects on MDSL cell function and viability by targeting both the CD34+ and CD34− subpopulations. As expected, the effects of LEN were mediated by CRBN, as knockdown of CRBN expression in MDSL cells completely abrogated LEN-mediated colony inhibition (Supplementary Fig. 1e,f). Overall, MDSL cells represent a tractable in vitro model to study the pleiotropic effects of LEN on MDS cell viability, proliferation, and clonogenicity.
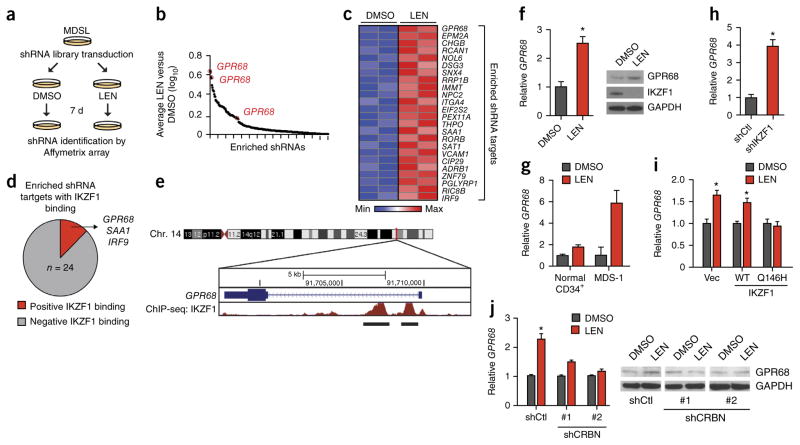
To identify genes and/or regulatory networks that are critical for a LEN-mediated response, we performed a genome-wide RNA interference (RNAi)-based loss-of-function genetic screen using LEN-treated MDSL cells (Fig. 1a). The library of 200,000 lentivi-ral-packaged shRNAs, which targets 47,400 human transcripts listed in the National Center for Biotechnology Information (NCBI) RefSeq database (3–5 shRNAs per transcript, including mRNAs and expressed sequence tags (ESTs)), was constructed in a feline immunodeficiency virus (FIV)-based vector (pSIF-H1-copGFP) that co-expresses green fluorescent protein (GFP) (referred to as the SBI library). We confirmed that lentiviral transduction of MDSL cells with an empty vector (pSIF-H1-copGFP) did not modify the response of the cells to LEN treatment (Supplementary Fig. 2a,b). The sequences of the shRNA templates corresponded to probes on the Affymetrix U133 Plus 2.0 GeneChip Array, allowing for post-screening identification of shRNAs by microarray analysis. MDSL cells were transduced with the lentiviral shRNA library and sorted for GFP-expressing cells. These cells were then treated with 10 μM LEN (or DMSO) for 7 d (Fig. 1a and Supplementary Fig. 2b). The design of the screen permitted for enrichment of shRNAs that impair LEN-induced cytotoxicity. A total of 470 shRNAs were differentially enriched after LEN treatment (Supplementary Table 1), of which 256 were positively enriched (Fig. 1b). By using a scoring criterion of ≥1.5-fold enrichment in two replicate screens (false discovery rate (FDR)-adjusted P < 0.05) by at least two independent shRNAs, we compiled a narrowed list of 24 enriched shRNAs that are presumed to target genes involved in mediating sensitivity to LEN (Fig. 1c). To assess possible interactions between these candidate LEN-sensitivity genes, pathway analysis was performed using NetWalker analysis13. One of the molecular networks identified was the tumor protein 53 (TP53; encoding p53) pathway (data not shown), which has previously been implicated in LEN resistance and relapse in patients with MDS14. Although it did not fulfill our selection criteria, an shRNA targeting CRBN was also enriched in cells that were treated with LEN, which is consistent with previous reports and which established the validity of the RNAi screen (Supplementary Fig. 3a)9. Moreover, on the basis of knockdown experiments in MDSL cells with shRNAs from a different library (the RNAi Consortium; TRC), four of eight ‘hits’ were confirmed to reduce sensitivity to LEN (Supplementary Figs. 1f and 3b).
Figure 1.
A genome-wide RNAi screen identifies determinants of LEN sensitivity in MDS. (a) Schematic outline of the LEN-resistance screen performed in MDSL cells, using whole-transcriptome human lentiviral-packaged (GFP+) shRNAs. (b) Log10 ratios for the abundance of individual shRNAs in MDSL cells treated with LEN versus those treated with DMSO, as calculated from the average of two biological replicates. Each dot represents an enriched individual shRNA. Only shRNA targets with a LEN: DMSO ratio >1.0 are shown. (c) shRNAs enriched ≥1.5-fold are shown in a heat map for two independent biological replicates of the RNAi-screen. Colors are represented as fold change of shRNA abundance in the LEN-treated group as compared to that in the DMSO-treated group. The most-enriched and significant shRNA clone for each gene is listed on the right. Significance of the listed shRNA clone for each gene was determined by FDR-adjusted P < 0.05. (d) Summary of IKZF1 binding to the promoters of the 24 genes identified in the RNAi-enrichment analysis, as assessed by ChiP-Seq from published ENCODE consortium data17. (e) Schematic of IKZF1-binding sites within the promoter of GPR68, as assessed by ChiP-seq analysis. The numbers within the chromosome schematic indicate band identifications. The numbers above GPR68 indicate the base pair positions. (f) GPR68 mRNA expression (left), and GPR68 and IKZF1 protein expression (right), in MDSL cells that were treated with DMSO or 10 μM LEN (n = 3). GAPDH was used as a loading control. (g) GPR68 mRNA expression in primary CD34+ (n = 4 per group) and primary MDS BM HSPCs (MDS-1) (n = 3 per group; error bars were derived from technical replicates) that were treated with DMSO or 10 μM LEN. (h) Relative GPR68 mRNA levels in MDSL cells expressing either an shRNA targeting IKZF1 or a control shRNA (n = 2 per group from independent experiments; error bars were derived from biological and technical replicates). (i) Relative GPR68 mRNA levels in MDSL cells that were transduced with a construct encoding either WT IKZF1 or the degradation-resistant IKZF1Q146H mutant (Q146H) and subsequently treated with DMSO or 10 μM LEN for >2 d (n = 2 per group; error bars were derived from biological and technical replicates). (j) Relative GPR68 mRNA levels (n = 3 per group from independent experiments) (left) and representative western blot analysis (of n = 2) of GPR68 protein expression (right) in MDSL cells after knockdown of CRBN with either of two independent shRNAs (shCRBN #1 and shCRBN #2) and treatment with DMSO or 10 μM LEN for 4 d. shCtl indicates cells that expressed a control shRNA. Throughout, error bars represent mean ± s.e.m. *P < 0.05 by Student’s t-test.
IKZF1 regulates GPR68 expression in MDS
Following LEN treatment, IKZF1 is degraded by the CRL4CRBN E3 ligase complex8,9,15. Because IKZF1 is a direct substrate of CRBN and can function as a transcriptional repressor or activator16, the list of hits was narrowed by examining the promoters of the genes that were targeted by the enriched shRNAs for occupancy with IKZF1 (Supplementary Fig. 4). Examination of promoter regions—by use of the Encyclopedia of DNA Elements (ENCODE) functional genomics data from a normal lymphoblastoid cell line17—revealed three genes (interferon regulatory factor 9 (IRF9), serum amyloid A1 (SAA1), and G-protein-coupled receptor 68 (GPR68) that contained binding peaks for IKZF1, as measured by chromatin immunoprecipitation sequencing (ChiP-seq) (Fig. 1d and Supplementary Fig. 4). Of the LEN-sensitivity genes identified in the screen, GPR68 emerged as the leading candidate because: (i) shRNAs targeting GPR68, which were represented by three independent RNAi sequences, showed the highest level of enrichment (Fig. 1c); (ii) the GPR68 promoter shows robust IKZF1 promoter occupancy by ChiP-seq (Fig. 1e); and (iii) GPR68 mRNA and GPR68 protein expression increased in MDSL cells, primary MDS hematopoietic stem–progenitor cells (HSPCs), and normal CD34+ cells, following LEN treatment, which coincided with IKZF1 protein degradation (Fig. 1f,g). To confirm IKZF1-mediated regulation of GPR68, IKZF1 was either knocked down in MDSL cells using shRNAs or was overexpressed. Consistent with its role as a transcriptional repressor, reduced IKZF1 expression de-repressed GPR68 mRNA expression by ~4-fold (Fig. 1h), whereas overexpression of a degradation-resistant IKZF1 mutant (IKZF1Q146H) but not of wild-type (WT) IKZF1 maintained GPR68 repression after LEN treatment (Fig. 1i). Moreover, the LEN-mediated increase in GPR68 expression is CRBN dependent, as knockdown of CRBN prevented LEN-induced GPR68 expression (Fig. 1j and Supplementary Fig. 1e). Taken together, these results demonstrate that GPR68 is a LEN- and CRBN-responsive gene that is regulated by IKZF1 in MDS cells.
GPR68 is a determinant of LEN sensitivity in MDS
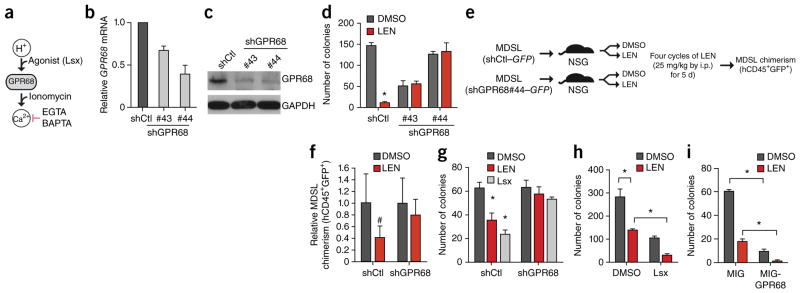
GPR68, also known as OGR1, belongs to a family of proton-sensing G-protein-coupled receptors18. After extracellular acidosis, GPR68 is activated and couples to Gq/11, resulting in signaling to the phospholipase-C–calcium (Ca2+) pathway (Fig. 2a)18,19. We validated the requirement of GPR68 for LEN sensitivity by using two additional independent shRNAs. Consistent with the results of the RNAi screen, knockdown of GPR68 reversed the inhibitory effects of LEN on MDSL colony formation in methylcellulose (Fig. 2b–d; P < 0.05) and on cell viability (Supplementary Fig. 5a; P < 0.05). Knockdown of GPR68 did not affect CRBN expression or CRBN substrate degradation (Supplementary Fig. 5b,c).
Figure 2.
GPR68 is a critical determinant of LEN sensitivity in MDS. (a) Overview of GPR68 activation by decreased pH (increased H+) or its agonist (Lsx). GPR68 induces calcium (Ca2+), which can be augmented by ionomycin (iono) or be inhibited by the calcium chelators EGTA and BAPTA. (b,c) Validation of GPR68 knockdown in MDSL cells expressing either of two independent GPR68-specific shRNAs (#43 and #44) by either qRT–PCR (n = 3 per group) (b) or immunoblotting (c). shCtl indicates MDSL cells that expressed a control shRNA. Western blot is representative of two experiments, and GAPDH was used as a loading control. (d) Colony formation by MDSL cells that were transduced with constructs expressing the indicated shRNAs, in the presence of DMSO or 10 μM LEN (n = 3 per group from independent experiments). (e) Overview of in vivo LEN treatment of NSG mice that were xenografted with MDSL cells transduced with constructs encoding either shGPR68 (#44) and GFP or shCtl and GFP. (f) Relative BM engraftment of MDSL cells (human CD45+ and GFP+ cells), as determined after 4 weeks of LEN treatment by normalizing to engraftment in vehicle-treated (DMSO) mice (shCtl: DMSO-treated, n = 7; LEN-treated, n = 7; shGPR68: DMSO-treated, n = 7; LEN-treated, n = 5). (g) Colony formation by MDSL cells expressing either shCtl or shGPR68 after treatment with DMSO, 10 μM LEN, or 10 μM Lsx (n = 3 per group from independent experiments). (h) Colony formation by MDSL cells that were treated with DMSO, 10 μM LEN, 10 μM Lsx, or a combination of 10 μM LEN and 10 μM Lsx (n = 3 per group from independent experiments). (i) Colony formation in MDSL cells that were transduced with either MIG-GPR68 or an empty vector (MIG) and that were subsequently treated with DMSO or 10 μM LEN (n = 3 per group from independent experiments). Throughout, error bars represent mean ± s.e.m. *P < 0.05 and #P < 0.1 by Student’s t-test.
To determine the requirement of GPR68 for LEN sensitivity in vivo, MDSL cells (1 × 106) expressing either a control shRNA (shCtl) or a GPR68-specific shRNA (shGPR68) were engrafted intravenously (i.v.) into immunocompromised NOD–SCID–IL2R-γ(NSG) mice (Fig. 2e). Ten days after engraftment, LEN (25 mg/per kg body weight (mg/kg)) or vehicle (DMSO) was administered by intraperitoneal (i.p.) injection for 4 weeks, as previously described20, after which the level of MDSL cell engraftment in the bone marrow (BM) (double-positive human CD45+GFP+ cells) was determined. In mice that were transplanted with control MDSL cells (expressing shCtl), BM engraftment was 60% less after LEN treatment as compared to that with DMSO treatment (0.41 ± 0.19; P = 0.1)(Fig. 2f). In contrast, mice that were transplanted with shGPR68-expressing MDSL cells were less sensitive to LEN treatment, as BM engraftment was comparable in mice treated with LEN or DMSO (0.80 ± 0.27, P = 0.4)(Fig. 2f). These data indicate that GPR68 expression is necessary for LEN sensitivity of MDS cells in vitro and in vivo.
GPR68 can be activated with a G protein receptor agonist (N-cyclopropyl-5-(thiophen-2-yl)-isoxaole-3-carboxamide (Lsx)), independently of extracellular pH levels (Fig. 2a)21. Treatment of MDSL cells with 10 μM Lsx inhibited colony formation and induced apoptosis to a similar extent as did LEN treatment (Fig. 2g and Supplementary Fig. 6a). The effect of Lsx on MDSL requires GPR68, as knockdown of GPR68 reversed the inhibitory effects of Lsx (Fig. 2g). In addition, LEN and Lsx cooperatively inhibited MDS cell function. Co-treatment of MDSL cells or primary MDS BM cells with LEN (10 μM) and Lsx (10 μM) resulted in further reductions in colony formation (Fig. 2h) and cell viability (Supplementary Fig. 6a,b), as compared to those with individual treatments. To establish whether GPR68 overexpression is sufficient to affect MDS cell function and LEN sensitivity, MDSL cells were transduced with a retroviral vector (MSCV-IRES-GFP; hereafter referred to as MIG) expressing GPR68 (MIG-GPR68) and then treated with LEN (10 μM) in methylcellulose (Supplementary Fig. 6c). GPR68 overexpression significantly suppressed MDSL colony formation, which was further suppressed in the presence of LEN (Fig. 2i; P < 0.05), suggesting that activation or overexpression of GPR68 can enhance LEN sensitivity.
LEN sensitivity is regulated by GPR68-mediated intracellular calcium flux
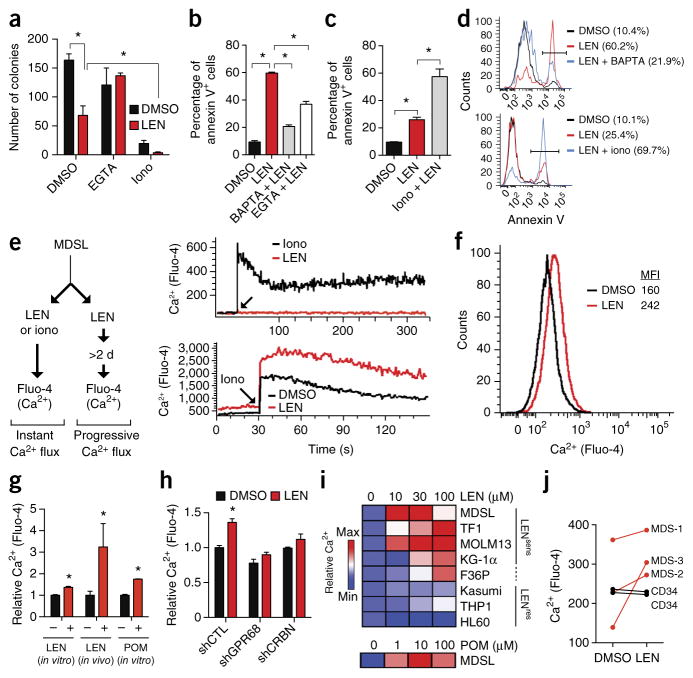
A cellular consequence of GPR68 activation is an increase in cytoplasmic calcium levels18,22. We therefore investigated whether intracellular calcium levels affected LEN sensitivity by using calcium chelators and inducers (Figs. 2a and 3a–d). Treatment with EGTA or BAPTA, which block extracellular and intracellular calcium, respectively, restored colony formation (Fig. 3a; P < 0.05) and cell survival (Fig. 3b,d; P < 0.05) of LEN-treated MDSL cells. Ionomycin treatment, which elevates cytosolic calcium levels, further reduced MDSL colony formation and survival when administered together with LEN (Fig. 3a,c,d). These findings reveal the importance of cytosolic calcium to LEN function. Next we examined whether LEN directly affects cytosolic calcium concentrations. Using a fluorescence dye that detects free cytosolic calcium (Fluo-4), we examined two mechanisms of calcium induction in MDSL cells—acutely modulated calcium flux and progressive calcium accumulation (Fig. 3e, left). Whereas treatment with ionomycin rapidly induced calcium influx within seconds, cytosolic calcium levels did not increase immediately after addition of LEN (Fig. 3e, top right). Treatment of MDSL cells with LEN for ≥2 d, however, resulted in elevated cytosolic calcium levels (Fig. 3e (bottom right) and further increased ionomycin-induced calcium levels (Fig. 3e, bottom right). To evaluate whether LEN affects calcium levels in vivo, we generated MDSL xenografts in NSG mice followed by injection of DMSO or LEN, as in Figure 2e. Consistent with the in vitro data, in vivo administration of LEN increased intracellular calcium levels in MDSL cells (Fig. 3g). In addition, pomalidomide (POM) treatment also elevated calcium levels in MDSL cells in vitro, indicating that the observed effect of LEN on calcium extends to other immunomodulating drugs (Fig. 3g). Knockdown of GPR68 or CRBN prevented increases in calcium levels in LEN-treated MDSL cells (Fig. 3h), indicating that LEN treatment gradually increases cytosolic calcium concentrations through CRBN and GPR68 signaling.
Figure 3.
LEN sensitivity is determined by GPR68-mediated intracellular calcium flux. (a) Colony formation by MDSL cells that were treated with DMSO or 10 μM LEN alone, or with either 1 mM EGTA or 2 μM ionomycin (iono) in the presence or absence of 10 μM LEN (n = 3 per group from independent experiments). (b) Annexin V staining of MDSL cells that were treated with 5 μM BAPTA or 200 μM EGTA, with or without 10 μM LEN (n = 2 per group from independent experiments; error bars were derived from biological and technical replicates). (c) Annexin V staining of MDSL cells that were treated with 400 nM ionomycin, with or without 10 μM LEN (n = 4 per group from independent experiments). (d) Representative histograms from b (top) and c (bottom). Parentheses indicate the percentage of annexin V+ cells under the bar. (e) Left, schematic of acute or progressive intracellular calcium (Ca2+) flux analysis in MDSL cells, using Fluo-4. Right, representative histograms for kinetics of intracellular calcium levels in MSDL cells after the immediate addition of 10 μM LEN or 5 μM ionomycin (top), or after treatment with DMSO or 10 μM LEN for 2 d and then treatment with 5 μM ionomycin (bottom) (n = 3). (f) Representative histogram for intracellular calcium levels (using Fluo-4) in MDSL cells that were treated with DMSO or 10 μM LEN for 2 d (n = 3). MFI, mean fluorescent intensity. (g) In vitro intracellular calcium levels in MDSL cells that were treated with DMSO, 10 μM LEN, or 1 μM POM for 4 d, or in vivo calcium levels in MDSL cells that were engrafted in NSG mice, which were subsequently injected with DMSO or LEN (25 mg/kg) for 4 d (n = 4 mice per group). (h) Intracellular calcium levels in MDSL cells that were treated with DMSO or 10 μM LEN after knockdown of GPR68 or CRBN expression (n = 3). (i) Relative intracellular calcium levels in MDS and AML cell lines that were treated with the indicated concentrations of LEN or POM for 7 d. LENsens, LEN sensitive; LENres, LEN resistant. (j) Intracellular calcium levels in BM cells from patients with MDS (n = 3) and in normal CD34+ cells from healthy control individuals (n = 2), following treatment with DMSO or 10 μM LEN for 2–4 d. Throughout, error bars represent mean ± s.e.m. *P < 0.05 by Student’s t-test.
To examine the potential correlation between LEN-mediated calcium flux and cell function, a panel of MDS and AML cell lines was evaluated for changes in cytosolic calcium, progenitor function in methylcellulose, cell viability, and CRBN substrate degradation. MDSL, TF1, and MOLM13 cells were the most sensitive to LEN treatment, as indicated by reduced colony formation and viability (Supplementary Fig. 7a,b). In contrast, HL60, Kasumi, and THP1 cells were resistant to LEN treatment, whereas KG-1α and F36P cells showed an intermediate response. In the cell lines tested, we did not observe a correlation between LEN sensitivity and basal cytosolic calcium or GPR68 mRNA levels, or between LEN sensitivity and LEN-induced GPR68 expression (Supplementary Fig. 7c–f). However, the LEN-induced increases in calcium levels were observed in the LEN-sensitive but not the LEN-resistant cell lines (Fig. 3i). LEN sensitivity of the cell lines was not simply CRBN dependent, as LEN treatment resulted in CRBN substrate degradation in nearly all of the cell lines (Supplementary Fig. 7g). POM treatment was also effective in suppressing viability and function of MDS and AML cells (Supplementary Fig. 8a), and its inhibitory effects also correlated with effects on calcium flux in the cell lines tested (Fig. 3i and Supplementary Fig. 8b). Primary del(5q) MDS cells are known to be more sensitive to LEN as compared to normal BM cells2. Although basal calcium levels were variable among primary BM samples from three individuals with del(5q) MDS and CD34+ cell samples from two normal, healthy donors, only the MDS samples showed an increase in cytosolic calcium levels after 2 d of LEN treatment in vitro (Fig. 3j). Together, these data indicate that LEN-mediated cytotoxicity in MDS and AML cell lines and in primary cells from individuals with MDS requires intracellular calcium flux.
Calcium-dependent calpain activity is required for LEN sensitivity
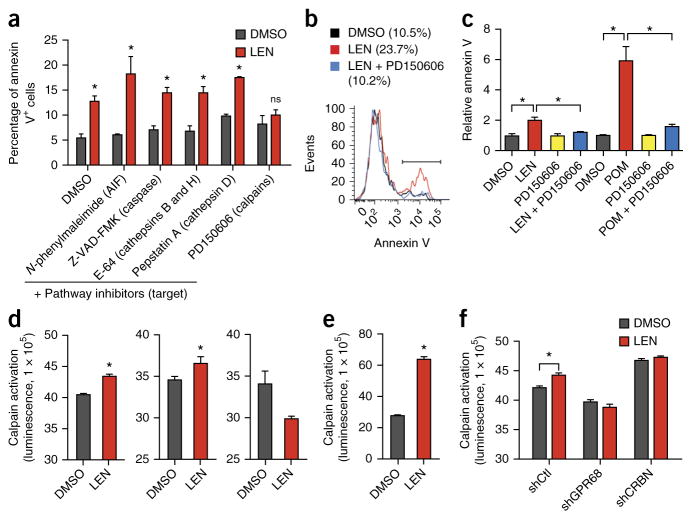
Calcium is an important second messenger that has been implicated in cell fate decisions23. Therefore, we next attempted to identify the calcium-dependent molecular mechanism that mediates the cytotoxic effects of LEN. We screened a series of pharmacological inhibitors for their efficacy in reversing LEN-mediated cytotoxicity of MDSL cells and selected five commercially available small molecules that represented inhibitors of major cell death pathways—including inhibitors of apoptosis-inducing factor (AIF), cathepsins, caspases, and calpains. Among the inhibitors, treatment with only the calpain inhibitor (PD160505) prevented LEN- and POM-induced apoptosis of MDSL cells (Fig. 4a–c), implicating calpains in immunomodulatory drug (IMiD)-mediated cell death. Calpains are a family of calcium-dependent cysteine proteases, of which calpain 1 (CAPN1) and CAPN2 are the most abundant24. Because elevated calcium levels can stimulate calpain protease activity25, we next determined whether LEN regulates calpain activation by using a luminescence assay for calpain substrate cleavage by lysates of LEN-treated cells. LEN treatment of MDSL cells for 4 d resulted in cleavage of the calpain substrate (P < 0.05) to a similar extent as that observed with ionomycin treatment (positive control) (Fig. 4d). In contrast, treatment of the LEN-resistant cell line HL60 with LEN did not result in cleavage of the calpain substrate (Fig. 4d), suggesting that LEN sensitivity correlates with calpain activation. Moreover, primary MDS BM cells also showed increased calpain substrate cleavage following treatment of the cells with LEN (Fig. 4e; P < 0.05). LEN-induced cleavage of calpain substrates was inhibited in MDSL cells after knockdown of GPR68 or CRBN (Fig. 4f). Collectively, these observations add credence to a model in which sensitivity of MDS and AML cells to LEN correlates with calcium-induced calpain activation.
Figure 4.
Calcium-dependent calpain activity is required for LEN sensitivity. (a) Annexin V staining of MDSL cells that were treated with DMSO or 10 μM LEN for 2 d, followed by concurrent treatment with compounds inhibiting AIF, caspases, cathepsins, or calpains for 2 additional days (n = 2 per group from independent experiments; error bars were derived from biological and technical replicates). (b) Representative histograms of annexin V staining of MDSL cells that were treated with DMSO, 10 μM LEN, or the combination of 10 μM LEN and the calpain inhibitor PD150606 (n = 3). Parentheses indicate the percentage of annexin V+ cells under the bar. (c) Relative levels of annexin V staining of MDSL cells that were treated with DMSO, 10 μM LEN, or 1 μM POM for 2 d, followed by concurrent treatment with PD150606 (n = 4 per group from independent experiments). (d) Calpain activity (as measured by luminescence) in lysates of MDSL (left and middle) or HL60 (right) cells that were treated with DMSO or 10 μM LEN for 4 d (left and right), or with DMSO or 5 μM ionomycin for 30 min (middle) (n = 2 per group from independent experiments; error bars were derived from biological and technical replicates). (e) Calpain activity (luminescence) in lysates of primary BM MNCs from individuals with MDS, which were treated with DMSO or 10 μM LEN for 2 d (n = 2 per group from independent experiments; error bars were derived from biological and technical replicates). (f) Calpain activity in lysates of MDSL cells that expressed shRNAs targeting GPR68 or CRBN and were treated with DMSO or 10 μM LEN for 4 d (n = 2 per group from independent experiments; error bars were derived from biological and technical replicates). Throughout, error bars represent mean ± s.e.m. *P < 0.05 by Student’s t-test.
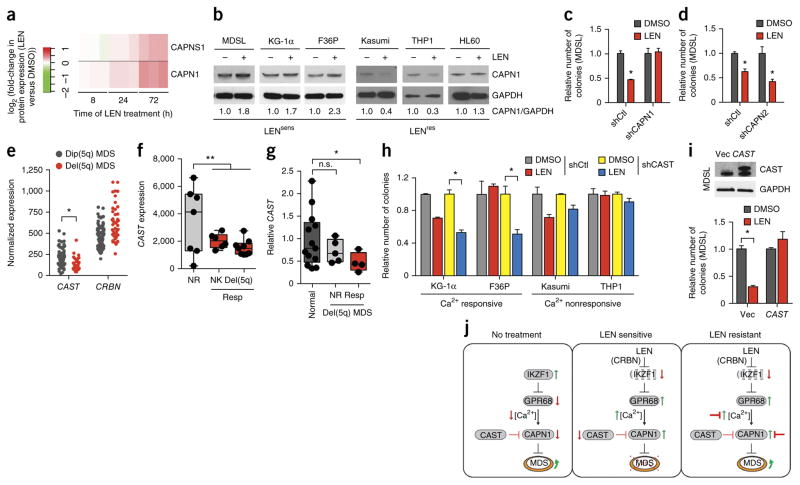
Reduced CAST and increased CAPN1 confer sensitivity of MDS to LEN
To further explore the mechanism of LEN sensitivity, we performed global proteomic analyses of LEN-treated MDSL cells. A pathway analysis of differentially expressed proteins after LEN treatment revealed enrichment of mu-calpain (calpain 1) and m-calpain (calpain 2) pathways, but without reaching statistical significance (data not shown). Assessment of individual proteins from the calpain pathway signatures showed that the levels of both CAPN1 and its small subunit 1 (CAPNS1) increased ~2-fold in a time-dependent manner in LEN-treated MDSL cells (Fig. 5a). To ascertain whether the increase in CAPN1 occurs in additional LEN-sensitive cell lines, we examined CAPN1 protein expression after LEN treatment of KG-1α and F36P cells. Consistent with the mass spectrometry findings for MDSL cells, LEN induced a ~2-fold increase in CAPN1 protein levels after a 96-h treatment in KG-1α and F36P cells (Fig. 5b). In contrast, treatment of the LEN-resistant HL60, THP1 and Kasumi1 cell lines with LEN did not result in increased CAPN1 levels (Fig. 5b). Increased CAPN1 protein expression was also observed after POM treatment of sensitive but not resistant AML cells (Supplementary Fig. 8c). Knockdown of IKZF1 or GPR68, or overexpression of GPR68, in untreated MDSL cells did not affect CAPN1 protein expression, indicating that the regulation of CAPN1 protein expression is probably independent of IKZF1 and GPR68 (Supplementary Fig. 9).
Figure 5.
Loss of CAST expression and increased CAPN1 expression confer sensitivity of MDS cells to LEN. (a) Expression of CAPNS1 and CAPN1 protein in LEN-sensitive MDSL cells after treatment with LEN for the indicated periods of time. The heat map summarizes the fold change (log2) in CAPNS1 and CAPN1 protein expression after LEN treatment, relative to that for DMSO treatment, in triplicate samples (vertical rectangles). (b) Representative immunoblot analysis (of n = 2) for CAPN1 expression in MDS and AML cell lines that were treated with DMSO or 10 μM LEN for 4 d. (c,d) Colony formation by MDSL cells that express either shCAPN1 (c) or shCAPN2 (d), in the presence of DMSO or 10 μM LEN (n = 3 per group from independent experiments). (e) Expression of CAST and CRBN mRNA in CD34 BM cells from patients with MDS who have either del(5q) (n = 29) or a normal karyotype (dip(5q); n = 93), as obtained from publicly available data27. (f) Expression of CAST mRNA in BM cells from patients with MDS patients who have either del(5q) or a normal karyotype (NK) and that either responded (Resp) (n = 15) or did not respond (NR; n = 7) to LEN treatment, as obtained from publicly available data34. The box-and-whisker plots show the median, the 25th–75th percentiles, and range of values. (g) Relative baseline expression of CAST, as determined by qRT–PCR, in BM cells from healthy control individuals (n = 13) and from either LEN-responsive (Resp; n = 4) or nonresponsive (NR; n = 5) patients with MDS. Data is shown relative to that for normal controls. (h) Colony formation by MDS and AML cell lines expressing shCAST or shCtl that were treated with DMSO or 10 μM LEN (n = 3 per group). Cell lines were grouped on the basis of their Ca2+ level increases after LEN treatment (Fig. 3i) as either Ca2+ responsive or Ca2+ nonresponsive. (i) Colony formation (bottom) by MDSL cells that were transduced with an empty lentiviral expression vector (Vec) or one that expresses CAST, as determined by immunoblot analysis (top), and that were treated with DMSO or 10 μM LEN (n = 3 per group from independent experiments). (j) Overview of the proposed mechanism by which LEN, acting through CRBN, induces IKZF1 degradation and GRP68 upregulation, leading to increased intracellular calcium levels and calpain activation. Prior to treatment (no treatment), MDS cells express IKZF1, which suppresses GPR68 expression and intracellular Ca2+ levels. In LEN-sensitive cells, LEN treatment results in IKZF1 degradation (via CRBN), which leads to de-repression of GPR68 expression, increased calcium levels, and activation of CAPN1. In LEN-resistant cells, various connections in the CRBN–IKZF1–GPR68–Ca2+–CAPN1 network could be disrupted and/or inhibited, permitting MDS cell expansion. Throughout, error bars represent mean ± s.e.m. *P < 0.05 by Student’s t-test.
To determine whether calpains are necessary for mediating LEN-induced cytotoxicity, CAPN1 and CAPN2 expression were knocked down in MDSL cells (Supplementary Fig. 10a,b). Although basal colony formation was impaired for CAPN1- or CAPN2-depleted cells, treatment with LEN did not inhibit colony formation for CAPN1-depleted MDSL cells (Fig. 5c). Resistance to LEN in CAPN1-depleted MDSL cells could not be explained by changes in CRBN expression, as CRBN protein levels were unaffected after knockdown of CAPN1 (Supplementary Fig. 10c). In contrast, knockdown of CAPN2 did not affect the ability of LEN to inhibit MDSL colony formation (Fig. 5d). These data implicate CAPN1 as a mediator of LEN-induced cytotoxicity. Notably, for one LEN-sensitive cell line (TF1 cells), increased calpain activity or CAPN1 expression was not observed following LEN treatment, suggesting that alternative mechanisms of LEN action exist (Supplementary Fig. 11).
Given that our findings implicate calcium-dependent calpain activation in LEN-induced cell death, we speculated that impaired regulation of calcium signaling or calpain activation would affect LEN sensitivity in patients with del(5q) MDS. To explore this connection, we searched for genes that encode known calcium and/or calpain regulators and that are located on chromosome 5q. Notably, the gene encoding calpastatin (CAST), a physiological inhibitor of CAPN1, is located at 5q15 (ref. 26). On the basis of publically available gene expression data27, we found CAST mRNA expression to be significantly reduced in del(5q) MDS CD34+ cells as compared to CD34+ MDS cells with a normal 5q copy number (dip(5q) MDS cells) (P < 0.05; Fig. 5e). Moreover, as compared to non-responders, patients with MDS who responded to LEN treatment expressed CAST in BM mononuclear cells (MNCs) at lower levels, independently of chromosome 5q deletion status (n ≥ 6 per group; P = 0.01; two-tailed t-test; Fig. 5f). To confirm these observations, we examined CAST expression in an independent cohort of del(5q) MDS samples from subjects who either responded to or were refractory to LEN treatment. Individuals with MDS who responded to LEN treatment expressed CAST in BM MNCs at lower levels than nonresponders or control BM cells (Fig. 5g; P = 0.01). Collectively, these findings suggest that haploid expression of CAST may be necessary to achieve sufficient calpain activation in LEN-treated cells.
To test the hypothesis that reduced CAST expression sensitizes cells to LEN, we knocked down CAST in AML cell lines that are either responsive (KG-1α and F36P) or unresponsive (Kasumi and THP1) to calcium induction by LEN. KG-1α and F36P cells were selected as they show increased intracellular calcium and calpain expression after LEN treatment but remain less sensitive to the cytotoxic effects of LEN (Supplementary Table 2). In contrast, Kasumi and THP1 cells did not show increased intracellular calcium levels or increased calpain expression after LEN treatment, and therefore they should not be sensitized to LEN following CAST knockdown. After knockdown of CAST, the cells were plated in methylcellulose with 10 μM LEN. As observed for CAPN1-depleted cells, knockdown of CAST affected basal colony formation. Nevertheless, after the knockdown of CAST, KG-1α and F36P cells acquired increased sensitivity to LEN, as relative colony formation was reduced by >50% (P < 0.05; Fig. 5h). In contrast, knockdown of CAST in Kasumi and THP1 cells did not cooperate with LEN to suppress colony formation (Fig. 5h). Finally, re-expression of CAST in LEN-sensitive MDSL cells conferred resistance to LEN, as indicated by restored colony formation (Fig. 5i). In conclusion, these findings support a model in which haploid expression of CAST sensitizes ‘primed’ cells to LEN-induced calpain activation in del(5q) MDS (Fig. 5j).
DISCUSSION
By taking a nonbiased genome-wide screening approach, our study identified a cellular mechanism of LEN action in MDS and AML that relies on a calcium–calpain-dependent pathway. LEN treatment increased cytosolic calcium levels in MDS and AML cells to a greater extent than in normal HSPCs. Elevated cytosolic calcium levels was not observed in LEN-resistant MDS or AML cell lines, suggesting that calcium signaling is associated with LEN-induced cell death. Elevated calcium levels resulted in activation of calpains, a family of calcium-dependent proteases responsible for initiating apoptosis, providing a mechanistic explanation for LEN-induced cytotoxicity. A high-throughput proteomic mass spectrometry approach identified increased CAPN1 protein abundance after LEN treatment as the strongest effect in the calpain protein signature. Collectively, these observations reveal that LEN concurrently induces cytosolic calcium and CAPN1 protein expression, maximizing the sensitivity of MDS cells to LEN-induced apoptosis. Furthermore, we propose that haploid expression of the gene encoding the physiological inhibitor of CAPN1, CAST (5q15), permits full activation of CAPN1 in LEN-treated del(5q) MDS cells (Fig. 5j). Our in vitro studies also revealed that a LEN-sensitive cell line (TF1 cells) showed an increase in cytosolic calcium levels but not in calpain activation, and that a cell line that showed only a minor cytotoxic effect of LEN treatment (F36P cells) had increased CAPN1 expression. Nevertheless, induced CAPN1 expression and cytosolic calcium flux were closely tied to LEN responsiveness. Thus, we postulate that baseline CAST and/or induced CAPN1 expression, in addition to calcium flux, may predict LEN responsiveness in individuals with MDS or AML.
LEN induces ubiquitination and degradation of IKZF1, IKZF3, and CK1-α by co-opting CRBN E3 ligase activity. IKZF1 and IKZF3 are transcription factors that are highly expressed in myeloid and lymphoid malignancies, and their degradation by LEN has been linked to T cell stimulation and cytotoxicity in multiple myeloma cells28. Here we show that LEN treatment leads to the death of MDS and AML cells, by linking degradation of IKZF1 to de-repression of GPR68 and subsequent calcium–calpain activation. In addition, CK1-α is encoded by a gene located in the deleted region for del(5q) MDS, and its haploinsufficient expression sensitizes cells to LEN treatment, providing a mechanistic explanation for the therapeutic window in patients with del(5q) MDS8. Degradation of CK1-α results in the stabilization of p53 and apoptosis of hematopoietic cells29, and it is well established that TP53 mutations confer resistance to LEN treatment in individuals with MDS30. Relevant to our findings, we found that p53 and calcium homeostasis are interdependent events, such that mutations in TP53 may also affect calcium signaling in MDS cells31,32. Collectively, these observations suggest that degradation of IKZF1 and subsequent de-repression of GPR68, as well as degradation of CK1-α, are cooperative mechanisms mediating the effects of LEN in MDS and AML cells. Given the complexity of IKZF1-, IKZF3-, and CK1-α-regulated networks and their potentially distinct effects on myeloid biology, a comprehensive assessment of the effects of LEN-mediated degradation of these proteins, both individually and in combination, in del(5q) and normal-karyotype MDS is warranted.
A characteristic of LEN therapy is the selective suppression of MDS-propagating cells in parallel with the activation of T cells5,12. The finding that LEN treatment leads to an elevation of intracellular calcium levels may provide a hint to explain this paradox. Our findings suggest that MDS and AML cells do not tolerate such an increase in intracellular calcium and undergo calpain-mediated apoptosis (Fig. 5j). In stark contrast, high intracellular calcium levels in T cells leads to their activation via calcium-dependent signaling through the transcription factor NFAT33. This discrepancy in calcium tolerance between HSPCs and T cells may explain the differential response to LEN therapy. In summary, our findings describe a new cellular mechanism of LEN action through an IKZF1 - and calcium–calpain-dependent pathway in del(5q) and non-del(5q) MDS and AML, which may improve our understanding of clinical responses and inform rational combinatorial therapeutic approaches.
ONLINE METHODS
shRNA library screen and data analysis
The loss-of-function screen was performed with GeneNet Lentiviral shRNA Libraries (System Biosciences; S12XXB-1) and analyzed by GeneNet Library Data Analysis Software (System Biosciences), according to the manufacturer’s instructions. Briefly, vesicular stomatitis virus (VSV)-G (envelope glycoprotein)-pseudotyped lentiviral particles that target ~47,000 transcripts in the human genome according to the NCBI RefSeq database (GRCh38.p6) were transduced into 2 × 106 MDSL cells with a multiplicity of infection (MOI) of 0.5–1.0. Within this range, each cell that has successfully been transduced is expected to contain only one copy of a given shRNA construct. 3 d after transduction, GFP+ transduced MDSL cells were purified for drug selection by using DMSO or 10 μM LEN treatment for 7 d. 5 × 106 viable MDSL cells were then isolated for RNA extraction. After two rounds of PCR, the amplified cDNA, corresponding to the unique shRNA hairpins, were labeled with biotin. After removing the sense strands, the biotin-labeled antisense strands were used as hybridization targets for Human Genome U133 Plus 2.0 GeneChip arrays (Affymetrix; #900470). GeneNet Library Data Analysis Software (System Biosciences) was used to generate expression values for individual shRNAs by comparing the background-corrected and normalized signal values for each shRNA in LEN-treated and control samples. The list of enriched shRNAs was calculated based on the absolute mean difference of expression values of shRNAs in LEN- versus control-treated replicate samples. Only shRNAs with >1.5-fold enrichment were further considered. P values were calculated based on the biological replicates of individual shRNA clones using the Student’s t-test. To further narrow the list of hits for adjusted P values, a false discovery rate (FDR) correction was applied.
Primary samples and CD34+ cells
MDS MNCs from bone marrow aspirates were obtained at diagnosis as part of a multicenter phase 2 trial based in Italy. Diagnoses were performed at the University of Milan and informed consent was obtained. Human CD34+ umbilical cord blood (UCB) and normal BM MNC control samples were obtained from the Translational Research Development Support Laboratory of Cincinnati Children’s Hospital under an approved Institutional Review Board protocol. Samples from both male and female subjects were analyzed. For RT–qPCR analysis, RNA was extracted from del(5q) MDS and age-matched healthy control BM MNCs. For measurement of intracellular calcium levels and calpain activity, MDS MNCs were cultured in StemSpan Serum-Free Expansion Medium (SFEM; Stemcell Technologies) supplemented with recombinant human stem cell factor (rhSCF; 02830, StemCell Technologies), human Flt3 ligand (rhFL; 02840, StemCell Technologies), human thrombopoietin (rhTPO; 02522, PeproTech), human interleukin (IL)-6 (rhIL-6; 02606, StemCell Technologies), and human recombinant IL-3 (rhIL-3; 02603, StemCell Technologies) at 10 ng/ml each. Human CD34+ UCB cells were maintained in StemSpan SFEM supplemented with 10 ng/ml of rhSCF, rhFL, rhTPO, rhIL-3, and rhIL-6.
Cell lines
HL60, THP1, Kasumi-1, KG-1α, and TF1 cells were purchased from the American Type Culture Collection. MOLM13 cells were purchased from AddexBio. F36P cells were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). The MDSL cell line was previously described10. Cell lines were cultured in RPMI 1640 medium with 10% FBS and 1% penicillin–streptomycin. KG-1α cells were cultured in 20% FBS. MDSL and TF1 cell lines were supplemented with 10 ng/ml rhIL-3. F36P cells were cultured with 10 ng/ml human recombinant granulocyte–macrophage colony-stimulating factor (rhGM-CSF; 300-03, PeproTech). All cell lines were confirmed by fingerprinting analysis and were found to be negative for mycoplasma.
Colony-forming assay
Based on a time-course analysis of LEN-treated cells in methylcellulose, the optimum time to assess colony formation was determined to be between days 7 and 14, before colony formation reached saturation. MDS or AML cells were plated in methylcellulose (4434; Stemcell Technologies) in the presence of DMSO or 10 μM LEN, and they were evaluated for colony formation after ~10 d. MDSL cells were plated in methylcellulose in the presence of DMSO, 10 μM LEN, 10 μM Lsx, 1 mM EGTA, or 2 μM ionomycin, and they were evaluated for colony formation after ~10 d. For knockdown of human CRBN, GPR68, CAPN1, and CAPN2, MDSL cells were transduced with lentivirus and sorted for GFP+CD34+ cells. For knockdown of human CAST, KG-1α and F36P cells were transduced with lentivirus and sorted for GFP+CD34+ cells. Kasumi and THP1 cells were transduced with lentivirus encoding shCAST. Transduced cells were plated in methylcellulose in the presence of DMSO, 10 μM LEN, or 10 μM Lsx, and were evaluated for colony formation after 10 d. For knockdown of human SAA1, IRF9, ribosomal RNA processing 1B (RRP1B), regulator of calcineurin 1 (RCAN1), calcium homeostasis endoplasmic reticulum protein (CHERP), epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (EPM2A), or RAR related orphan receptor B (RORB), MDSL cells were transduced with lentivirus (pLKO.1-Puro) and plated in methylcellulose that contained 2 μg/ml puromycin (61-385-RA; Fisher) and either DMSO or 10 μM LEN.
Cell death assay
Annexin V analysis was carried out as previously described35. Cultured cells were stained with Annexin V–APC (eBioscience; 88-8007-74) and 7AAD (eBioscience; 00-6993-50) according to the manufacturer’s instructions. For MDSL, TF1, KG-1α, and F36P cells, the CD34+ compartment was gated for apoptosis analysis.
Cell proliferation and viability assay
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assays were performed according to the manufacturer’s instructions (Promega, G1112).
Detection of intracellular calcium
Detection of intracellular calcium was performed according to the manufacturer’s instructions (Invitrogen, F10471). Briefly, 5 × 105 cells were resuspended in 200 μl of 1× Hank’s Balanced Salt Solution (2% FBS). 200 μl of Fluo-4 DirectTM calcium assay buffer was added to a cell suspension with 5 mM probenecid. The mixture was incubated at 37 °C for 30 min. Measurement of Fluo-4 signals was performed by using a Canto II flow cytometer with excitation at A494nm and emission at A516nm.
qRT–PCR
Total RNA was extracted and purified by using Quick-RNA MiniPrep (R1055; Zymo research), and reverse transcription was carried out by using High-Capacity RNA-to-cDNA Kit (4387406; Invitrogen). Quantitative PCR was performed with Taqman Master Mix (4324018; Life Technologies) for human GAPDH (cat. # 4351370; assay # Hs02758991_g1), human CRBN (cat. # 4453320; assay # Hs00372271_m1), human GPR68 (cat. # 4453320; assay # Hs00268858_s1), human CAPN1 (cat. # 4453320; assay # Hs00559804_m1), human CAPN2 (cat. # 4448892, assay # Hs00965097_m1), and human CAST (cat. # 4453320; assay # Hs001156280_m1).
Flow cytometry
MDSL, TF1, KG-1α and F36P cells were washed, and 1 × 106 cells were resuspended in 0.2 ml PBS with 2% FBS, 1 mM EDTA, and 0.04% NaN3. Cells were then incubated for 30 min with eFluor-450-conjugated human CD34 antibody (48-0349-41; eBiosciences) for apoptosis analysis.
Immunoblotting
Total protein lysates were obtained from cells by lysing the samples in cold RIPA buffer in the presence of phenylmethylsulfonyl fluoride (PMSF), sodium orthovanadate, and protease and phosphatase inhibitors (all from ThermoFisher Scientific). Protein concentration was evaluated by a BCA assay (23225; Pierce). Western blot analysis was performed with antibodies to the following proteins: GPR68 (sc-79622; Santa Cruz), CRBN (SAB2106014; Sigma), CAPN1 (2556; Cell Signaling Technology), CAPN2 (2539; Cell Signaling Technology), CAST (4146; Cell Signaling Technology), IKZF1 (sc-398265; Santa Cruz), CK1-α (sc-6477; Santa Cruz), and GAPDH (5174; Cell Signaling Technology). Anti-IKZF1 (1:500), anti-GPR68 (1:800), and all other primary antibodies (1:1,000) were diluted in 1× TBST buffer (Cell Signaling) containing 5% BSA or 5% milk. All secondary antibodies are conjugated with horseradish peroxidase (HRP). Goat anti-rabbit (111-035-003; Jackson ImmunoResearch) and goat anti-mouse (115-035-003; Jackson ImmunoResearch) secondary antibodies were diluted at 1:10,000 in 1× TBST buffer containing 5% milk. Donkey anti-goat (sc-2020; Santa Cruz) antibody was diluted at 1:3,000. ECL Western Blotting Substrate (32106; Pierce) and Supersignal West Femto (34095; Pierce Chemical) were used for detection of HRP on immunoblots with X-ray film or with a ChemiDoc Touch Imaging System (Bio-Rad).
Calpain activity
Calpain activity was measured according to the manufacturer’s instructions (G8501; Promega). Briefly, cells were washed in cold calcium-free 1× PBS and lysed in 10 mM HEPES (pH 7.2), 10 mM dithiothreitol (DTT), 1 mM EDTA, and 0.1% BSA by using sonication. Lysates were mixed with Calpain-Glo Buffer, Suc-LLVY-Glo Substrate, and Luciferin Detection Reagent, and the luminescence was read by using a microplate luminometer (Evision; Perkin Elmer) at 30-s intervals for 5 min.
Reagents and related assays
MDS and AML cell lines were cultured in the presence of DMSO, LEN (S1029; Selleckchem), or POM (P0018; Sigma) at the indicated concentrations for the apoptosis studies and the intracellular calcium analysis. MDSL cells were treated with DMSO or 10 μM LEN for 2 d, followed by cotreatment with 20 μM Lsx (Neuronal Differentiation Inducer III; 480743; EMD Millipore) for 1 d, 400 nM ionomycin (Calcium Ionophore, C5722, Sigma) for 1 d, 200 μM EGTA (045172; Fisher) for 5 d, or 5 μM BAPTA-AM (A1076; Sigma) for 5 d for the apoptosis assay. MDSL and TF-1 cells were treated with DMSO or 10 μM LEN for 2 d, followed by cotreatment with 5 μM N-phenylmaleimide (P27100; Sigma), 20 μM Z-VAD-FMK (a pan-caspase inhibitor; FMK001; R&D Systems), 1 μM E-64 (E3132; Sigma), 1 μM pepstatin (P5318; Sigma), and 100 μM PD150606 (D5946; Sigma) for 3 d for the apoptosis assay. MDSL cells were treated with 1 μM POM for 2 d, followed by cotreatment with 300 μM PD150606 for 3 d for the apoptosis assay. MDS and AML cells were treated with DMSO or 10 μM LEN for 4 d, or with 5 μM ionomycin for 30 min, for the measurement of calpain activity.
Lentivirus and retrovirus production
The lentiviral shRNA expression vector (pLKO.1; OpenBiosystems) was obtained from the Lentiviral core at CCHMC and was used to express shCtl (scrambled control) and shRNA specific for human GPR68 (TRCN0000011650, TRCN0000011651, and TRCN0000357255), human IKZF1 (TRCN0000107872), human CAPN1 (TRCN0000003558), human CAPN2 (TRCN0000003543), human CAST (TRCN0000073638), human CRBN (TRCN0000141985 and TRCN0000141562), human EPM2A (TRCN0000002591), human IRF9 (TRCN0000022069), human RCAN1 (TRCN0000019848), human RORB (TRCN0000022169), human RRP1B (TRCN0000130433), human SAA1 (TRCN0000083048), and human CHERP (TRCN0000053623). The lentiviral expression vector for CAST expression (pReceiver-Lv228-hCAST; EX-T9667-Lv228) and the control vector (pReceiver-Lv228; EX-NEG-Lv228) were obtained from GeneCopoeia. The lentiviral expression vector for IKZF1 expression (EGI-IKZF1) and the control vector (EGI) were previously described8. The IKZF1 degradation mutant was created by mutating Glu146 to His (IKZF1Q146H) with the QuikChange Multi Site-Directed Mutagenesis Kit (200515; Agilent Technologies). The retroviral vector for GPR68 expression (MSCV-GPR68-IRES-GFP; MIG-GPR68) was previously described36. Lentiviruses and retroviruses were generated as previously described35.
MDSL xenografts
Busulfan (30 mg/kg; B2635; Sigma) was given intraperitoneally to 8-week-old NOD–SCID–IL2R-γ(NSG) mice 1 d before xenotransplantation. Parental or transduced (shGPR68-pLKO.1-GFP or control pLKO.1 with scramble sequence) MDSL cells (1 × 106) were intravenously injected into NSG mice as previously described20. 10 d after engraftment with transduced MDSL cells, DMSO or 25 mg/kg LEN were intraperitoneally injected for four cycles (1 cycle = 5 consecutive days + 2 d off). BM cells were collected through intrafemoral aspiration at 4 weeks after transplantation, to measure MDSL cell engraftment based on coexpression of human CD45 and GFP. For in vivo calcium measurements, 6 weeks after engraftment of parental MDSL cells into recipient NSG mice, DMSO or 25 mg/kg LEN was injected (via alternating intravenous and intraperitoneal administration) for 4 d, and total BM cells were harvested to measure intracellular calcium levels in MDSL cells (gated on human CD45+ cells) as described above. All mouse experiments were subject to institutional approval by the CCHMC Institutional Animal Care and Use Committee. Male and female mice were equally assigned to the experimental groups. The sample size was determined from published studies, which was based on providing 60–80% power for a target effect size of 1.2–1.5. No randomization was performed, and no animals were excluded from the final analysis.
Mass spectrometry
MDSL cell samples for multiplexed quantitative mass spectrometry analysis were processed and analyzed at the Thermo Fisher Scientific Center for Multiplexed Proteomics at Harvard Medical School. Sample processing steps included cell lysis, tandem protein digestion by using LysC and trypsin, peptide labeling with either Tandem Mass Tag 6-plex or 10-plex reagents, and peptide fractionation. Multiplexed quantitative mass spectrometry data were collected on an Orbitrap Fusion mass spectrometer operating in a MS3 mode by using synchronous precursor selection for the MS2 to MS3 fragmentation. MS/MS data were searched against a Uniprot human database with both the forward and reverse sequences by using the SEQUEST algorithm. Additional data-processing steps included controlling for peptide- and protein-level FDRs, assembling proteins from peptides, and quantifying protein from peptides.
Statistical analysis
Results are depicted as the mean ± s.e.m. Statistical analyses were performed by using Student’s t-test. GraphPad Prism (v5, GraphPad) was used for statistical analysis.
Acknowledgments
This work was supported by Cincinnati Children’s Hospital Research Foundation (D.T.S.). We thank J. Bailey and V. Summey for assistance with transplantations. The normal bone marrow samples were received through the Normal Donor Repository in the Translational Core Laboratory at Cincinnati Children’s Research Foundation, which is supported through the NIDDK-funded Center of Excellence in Molecular Hematology (grant no. P30DK090971; to Y. Zheng (CCHMC)). We thank B. Ebert (Harvard Medical School; Dana-Farber Cancer Institute) for providing the EGI and EGI-IKZF1 vectors, and for his expertise and discussions. In addition, we thank Y. Xu (Indiana University School of Medicine) for providing the MIG-GPR68 vector and K. Tohyama (Kawasaki Medical School) for the MDSL cells.
Footnotes
AUTHOR CONTRIBUTIONS
J.F. and D.T.S. designed and interpreted data, and wrote the paper; D.T.S. conceived the study, obtained funding, and coordinated collaborations; J.F., X.L., L.B., B.B., and C.F. performed experiments and analyzed data; C.R., A.C., M.C., and E.N.O. provided and characterized patient samples; K.M. provided important reagents and conceptual input to the design of the study; H.L.G. provided conceptual input to the design of the study; and K.K. performed the bioinformatics analysis of the shRNA screen.
Accession codes. Gene Expression Omnibus: the shRNA screen sequencing data are available under accession code GSE82235.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Pan B, Lentzsch S. The application and biology of immunomodulatory drugs (IMiDs) in cancer. Pharmacol Ther. 2012;136:56–68. doi: 10.1016/j.pharmthera.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 2.List A, et al. Myelodysplastic Syndrome-003 Study Investigators. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N Engl J Med. 2006;355:1456–1465. doi: 10.1056/NEJMoa061292. [DOI] [PubMed] [Google Scholar]
- 3.Giagounidis AA. Lenalidomide for del(5q) and non-del(5q) myelodysplastic syndromes. Semin Hematol. 2012;49:312–322. doi: 10.1053/j.seminhematol.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Wei S, et al. Lenalidomide promotes p53 degradation by inhibiting MDM2 auto-ubiquitination in myelodysplastic syndrome with chromosome 5q deletion. Oncogene. 2013;32:1110–1120. doi: 10.1038/onc.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDaniel JM, Pinilla-Ibarz J, Epling-Burnette PK. Molecular action of lenalidomide in lymphocytes and hematologic malignancies. Adv Hematol. 2012;2012:513702. doi: 10.1155/2012/513702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito T, et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 7.Krönke J, et al. Lenalidomide induces ubiquitination and degradation of CK1-α in del(5q) MDS. Nature. 2015;523:183–188. doi: 10.1038/nature14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krönke J, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu G, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuoka A, et al. Lenalidomide induces cell death in an MDS-derived cell line with deletion of chromosome 5q by inhibition of cytokinesis. Leukemia. 2010;24:748–755. doi: 10.1038/leu.2009.296. [DOI] [PubMed] [Google Scholar]
- 11.Tohyama K, et al. A novel factor-dependent human myelodysplastic cell line, MDS92, contains hemopoietic cells of several lineages. Br J Haematol. 1995;91:795–799. doi: 10.1111/j.1365-2141.1995.tb05391.x. [DOI] [PubMed] [Google Scholar]
- 12.Pellagatti A, et al. Lenalidomide inhibits the malignant clone and upregulates the SPARC gene mapping to the commonly deleted region in 5q– syndrome patients. Proc Natl Acad Sci USA. 2007;104:11406–11411. doi: 10.1073/pnas.0610477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komurov K, Dursun S, Erdin S, Ram PT. NetWalker: a contextual network analysis tool for functional genomics. BMC Genomics. 2012;13:282. doi: 10.1186/1471-2164-13-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saft L, et al. p53 protein expression independently predicts outcome in patients with lower-risk myelodysplastic syndromes with del(5q) Haematologica. 2014;99:1041–1049. doi: 10.3324/haematol.2013.098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gandhi AK, et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4CRBN. Br J Haematol. 2014;164:811–821. doi: 10.1111/bjh.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rebollo A, Schmitt C. Ikaros, Aiolos, and Helios: transcription regulators and lymphoid malignancies. Immunol Cell Biol. 2003;81:171–175. doi: 10.1046/j.1440-1711.2003.01159.x. [DOI] [PubMed] [Google Scholar]
- 17.Rosenbloom KR, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig MG, et al. Proton-sensing G-protein-coupled receptors. Nature. 2003;425:93–98. doi: 10.1038/nature01905. [DOI] [PubMed] [Google Scholar]
- 19.Huang WC, Swietach P, Vaughan-Jones RD, Ansorge O, Glitsch MD. Extracellular acidification elicits spatially and temporally distinct Ca2+ signals. Curr Biol. 2008;18:781–785. doi: 10.1016/j.cub.2008.04.049. [DOI] [PubMed] [Google Scholar]
- 20.Rhyasen GW, et al. An MDS xenograft model utilizing a patient-derived cell line. Leukemia. 2014;28:1142–1145. doi: 10.1038/leu.2013.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell JL, et al. Regulated expression of pH-sensing G-protein-coupled receptor 68 identified through chemical biology defines a new drug target for ischemic heart disease. ACS Chem Biol. 2012;7:1077–1083. doi: 10.1021/cb300001m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frick KK, Krieger NS, Nehrke K, Bushinsky DA. Metabolic acidosis increases intracellular calcium in bone cells through activation of the proton receptor OGR1. J Bone Miner Res. 2009;24:305–313. doi: 10.1359/jbmr.081015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 24.Croall DE, Ersfeld K. The calpains: modular designs and functional diversity. Genome Biol. 2007;8:218. doi: 10.1186/gb-2007-8-6-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorimachi H, Hata S, Ono Y. Impact of genetic insights into calpain biology. J Biochem. 2011;150:23–37. doi: 10.1093/jb/mvr070. [DOI] [PubMed] [Google Scholar]
- 26.Jerez A, et al. Topography, clinical, and genomic correlates of 5q myeloid malignancies revisited. J Clin Oncol. 2012;30:1343–1349. doi: 10.1200/JCO.2011.36.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pellagatti A, et al. Gene expression profiles of CD34+ cells in myelodysplastic syndromes: involvement of interferon-stimulated genes and correlation to FAB subtype and karyotype. Blood. 2006;108:337–345. doi: 10.1182/blood-2005-12-4769. [DOI] [PubMed] [Google Scholar]
- 28.Zhu YX, et al. Identification of cereblon-binding proteins, and relationship with response and survival after IMiDs in multiple myeloma. Blood. 2014;124:536–545. doi: 10.1182/blood-2014-02-557819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Järås M, et al. Csnk1a1 inhibition has p53-dependent therapeutic efficacy in acute myeloid leukemia. J Exp Med. 2014;211:605–612. doi: 10.1084/jem.20131033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jädersten M, et al. TP53 mutations in low-risk myelodysplastic syndromes with del(5q) predict disease progression. J Clin Oncol. 2011;29:1971–1979. doi: 10.1200/JCO.2010.31.8576. [DOI] [PubMed] [Google Scholar]
- 31.Giorgi C, Bonora M, Pinton P. Inside the tumor: p53 modulates calcium homeostasis. Cell Cycle. 2015;14:933–934. doi: 10.1080/15384101.2015.1010973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giorgi C, et al. p53 at the endoplasmic reticulum regulates apoptosis in a Ca2+-dependent manner. Proc Natl Acad Sci USA. 2015;112:1779–1784. doi: 10.1073/pnas.1410723112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macian F. NFAT proteins: key regulators of T cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 34.Ebert BL, et al. An erythroid differentiation signature predicts response to lenalidomide in myelodysplastic syndrome. PLoS Med. 2008;5:e35. doi: 10.1371/journal.pmed.0050035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang J, et al. Cytotoxic effects of bortezomib in myelodysplastic syndrome–acute myeloid leukemia depend on autophagy-mediated lysosomal degradation of TRAF6 and repression of PSMA1. Blood. 2012;120:858–867. doi: 10.1182/blood-2012-02-407999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh LS, et al. Ovarian cancer G-protein-coupled receptor 1, a new metastasis suppressor gene in prostate cancer. J Natl Cancer Inst. 2007;99:1313–1327. doi: 10.1093/jnci/djm107. [DOI] [PubMed] [Google Scholar]