Abstract
This paper addresses how to evaluate the efficacy of the growing inventory of thiol-reactive inhibitors of mammalian protein disulfide isomerase (PDI) enzymes under realistic concentrations of potentially competing thiol-containing peptides and proteins. For this purpose, we introduce a variant of the widely-used reductase assay by using a commercially-available cysteine derivative (BODIPY FL L-Cystine; BD-SS) that yields a 55-fold increase in fluorescence (excitation/emission; 490/513 nm) on scission of the disulfide bond. This plate reader-compatible method detects human PDI down to 5–10 nM, can utilize a range of thiol substrates (including 5 μM dithiothreitol, 10 μM reduced RNase thiols, and 5 mM glutathione; GSH), and can operate from pH 6–9.5 in a variety of buffers. PDI assays often employ low micromolar levels of substrates leading to ambiguities when thiol-directed inhibitors are evaluated. The present work utilizes 5 mM GSH for both pre-incubation and assay phases to more realistically reflect the high concentration of thiols that an inhibitor would encounter intracellularly. Extracellular PDI faces a much lower concentration of potentially competing thiols; to assess reductase activity under these conditions, the pre-reduced PDI is treated with inhibitor and then fluorescence increase upon reduction of BD-SS is followed in the absence of additional competing thiols. Both assay modes were tested with four mechanistically diverse PDI inhibitors. Two reversible reagents, 3,4-methylenedioxy-β-nitrostyrene (MNS) and the arsenical APAO, were found to be strong inhibitors of PDI in the absence of competing thiols, but were ineffective in the presence of 5 mM GSH. A further examination of the nitrostyrene showed that MNS not only forms facile Michael adducts with GSH, but also with the thiols of unfolded proteins (Kd values of 7 and <0.1 μM, respectively) suggesting the existence of multiple potential intracellular targets for this membrane-permeant reagent. The inhibition of PDI by the irreversible alkylating agent, the chloroacetamide 16F16, was found to be only modestly attenuated by 5 mM GSH. Finally, the thiol-independent flavonoid inhibitor quercetin-3-O-rutinoside was found to show equal efficacy in reoxidation and turnover assay types. This work provides a framework to evaluate inhibitors that may target the CxxC motifs of PDI and addresses some of the complexities in the interpretation of the behavior of thiol-directed reagents in vivo.
Keywords: Assay, Binding, BODIPY FL-L Cystine, Disulfide, Dithiothreitol, Glutathione, Inhibitor, Protein disulfide isomerase, Reductase, Ribonuclease A, Thiols
Graphical Abstract
INTRODUCTION
In 1963, Anfinsen and Straub independently described an oxidative catalyst present in vertebrate liver and pancreas homogenates, that was later identified as protein disulfide isomerase [1,2]. About 20 protein disulfide isomerases have been described in humans [3,4]; the best understood, PDI, is found at nearly millimolar concentrations in the lumen of the ER and at much lower levels on the mitochondrial outer membrane, within the nuclear matrix and cytosol, and at the cell surface [5–8]. PDI contains four thioredoxin-like domains (a, b, b' and a') with the outer a and a' modules containing CxxC motifs that catalyze both disulfide oxidoreductase and redox-neutral disulfide shuffling modes [9–12]. Additionally, PDI has been shown to function as a chaperone during the folding of reduced lysozyme, glyceraldehyde-3-phosphate dehydrogenase, green fluorescent protein, and procollagen [13–16]. Extracellular PDI has been implicated in a range of key cellular events including integrin activation, viral fusion, adhesion, invasion and thrombus formation [17–20]. Although the isomerase is now a target for therapeutic intervention [21,22], the similarities in the chemical reactivities of many of the PDI family members and their multiple cellular locales presents significant challenges in the design of specific reagents that can target a single isomerase at a unique cellular location.
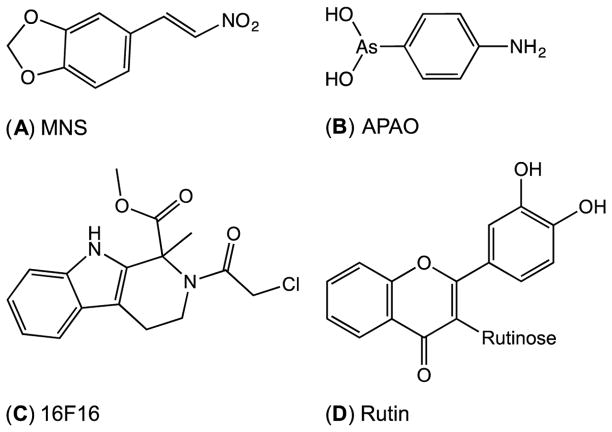
A large number of small molecule inhibitors of PDI have now been described, including 3,4-methylenedioxy-β-nitrostyrene (MNS) (Fig. 1) [19], 4-amino-phenylarsine oxide (APAO) [23], a chloroacetamide analog, 16F16 [24], and a flavonoid, quercetin-3-O-rutinoside (rutin) [25]. Compounds A–C in Fig. 1 are expected to be overtly reactive towards thiolate nucleophiles. While the enone functionality of rutin (Fig. 1D) is a potential weak Michael acceptor, its mode of inhibition appears to be unrelated to the CxxC motifs of PDI [26]. The observation that a sizable proportion of PDI inhibitors are thiol-reactive raises important issues concerning their mode of action and specificity in vivo, and how PDI inhibition should be best assessed in vitro.
Fig. 1.
A sampling of inhibitors of PDI. (A) 3,4-methylenedioxy-β-nitrostyrene (MNS). (B) 4-amino-phenylarsine oxide (APAO). (C) 16F16. (D) Quercetin-3-O-rutinoside (rutin).
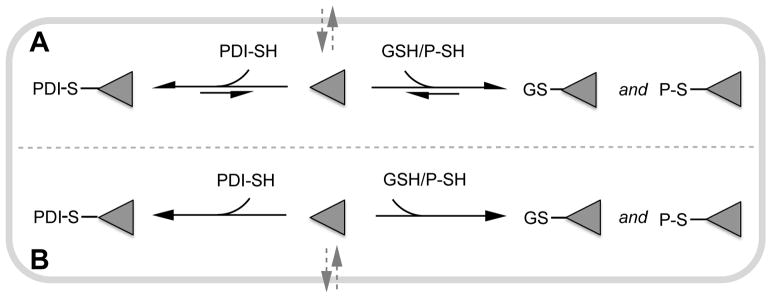
In terms of intracellular targeting of PDI, the high concentration of sulfhydryl groups in mammalian cells, notably the multimillimolar levels of GSH and exposed protein thiols [27,28], could markedly attenuate the effectiveness of membrane-permeant thiol-reactive small molecules. When these reactions involve reversible thiol adducts, the effectiveness of an inhibitor will reflect Kd values combined with the prevailing concentrations of PDI and competing thiols in the cell (Fig. 2A). For irreversible chemistry (Fig. 2B) the determinant for selectivity of labeling would be a strong kinetic preference for reaction with PDI over the competing intracellular thiols. In marked contrast to the intracellular case, cell surface PDI [29] is likely to encounter a much lower concentration of competing thiols (e.g. ~10 μM glutathione and cysteine [28,30,31]). Here, the susceptibility of PDI to inhibition by thiol directed reagents is expected to be strongly influenced by the presence of additional exofacial reductive pathways and thiol-containing proteins [32,33].
Fig. 2.
Intracellular competition between glutathione (GSH), reduced PDI (PDI-SH) and other protein thiols (P-SH) for a membrane-permeant inhibitor, I. Panels A and B represent reversible and irreversible thiol-directed inhibition, respectively. PDI is likely to be largely reduced intracellularly.
Extrapolating the in vivo effectiveness of thiol-reactive inhibitors of PDI using in vitro assays presents significant challenges. Most current steady-state assays of PDI involve thiol-containing substrates or products, with or without the inclusion of redox buffers generated from -SH/ -SS- mixtures. We suggest that an appraisal of the likely in vivo effectiveness of thiol-reactive inhibitors should include in vitro assessment of activity under realistic concentrations of potentially competing thiols. For example, PDI reduction assays are often conducted at low thiol concentrations (e.g. 5 μM DTT), making them potentially susceptible to artifacts arising from substrate depletion following the addition of thiol-reactive inhibitors at micromolar concentrations.
In view of the increasing inventory of thiol-reactive inhibitors of PDI emerging from high-throughput screening and rational design strategies, we wanted to develop general approaches to assess whether the isomerase is a prospective target of such inhibitors under conditions that may prevail intracellularly. At the outset we chose the compound 3,4-methylenedioxy-β-nitrostyrene (MNS; Fig. 1A), as it was recently suggested to suppress the growth of human triple negative breast cancer cells (MDA-MD-231) by inhibiting PDI [19]. MNS is one of many nitrostyrene derivatives that have been considered for therapeutic use [34–37], and has also been reported to suppress platelet aggregation and activation of the NLRP3 inflammasome activation [38,39], to inhibit protein tyrosine kinase [39] and monoamine oxidase B [40], to attenuate tumor growth [41], and to slow mobility and colony formation in osteosarcoma cells [42].
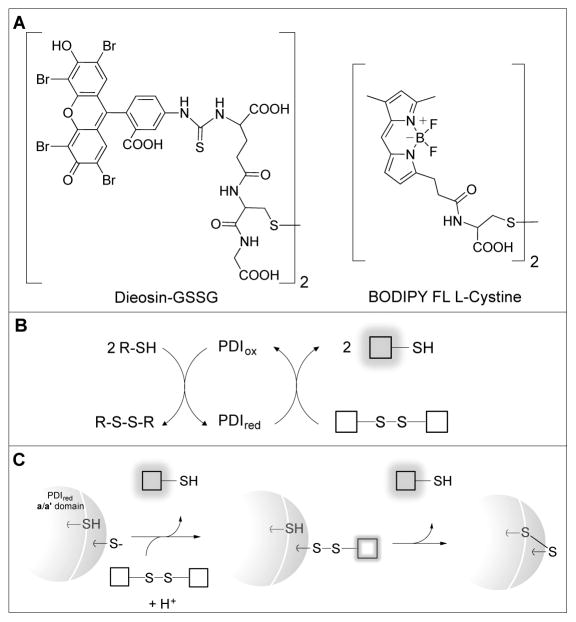
In exploring the interaction between human PDI and MNS, we initially employed a very sensitive reductase assay developed by Raturi and Mutus that utilizes a self-quenched dieosin derivative they synthesized by conjugating oxidized glutathione with eosin-isothiocyanate (Fig. 3A) [43]. Subsequently, we found that BODIPY FL-L-cystine (BD-SS) represents a very convenient commercially-available substitute for the dieosin derivative. Herein, we first validate the use of BD-SS in both turnover and reoxidation assays for the general assessment of PDI enzymatic activity. We then use these assays, and additional spectroscopic experiments, to explore the complexities and potential pitfalls in the evaluation of PDI inhibitors that are themselves thiol-reactive.
Fig. 3.
Assay reagents and depictions of multiple turnover and reoxidation assays. (A) Dieosin-GSSG (Di-E-GSSG) and BODIPY FL L-cystine (BD-SS) can be used as “turn-on” fluorophores to measure the reductase activity of PDI. (B) Multiple turnover assays are sustained by the inclusion of an exogenous reductant (depicted by R-SH). (C) The reoxidation of reduced PDI by a fluorogenic disulfide in the absence of exogenous reductant.
MATERIALS AND METHODS
Materials
BODIPY FL L-cystine was from Thermo Fisher Scientific or Setareh Biotech. 4-amino-phenylarsine oxide was synthesized as described previously [44]. Glutathione, 3,4-methylenedioxy-β-nitrostyrene, N-ethylmaleimide, 16F16, aldolase (rabbit muscle) and pyruvate kinase (rabbit muscle) were obtained from Sigma Aldrich. Quercetin-3-O-rutinoside was from Acros Organics (Fisher Scientific). Tris(hydroxypropyl)phosphine was from Calbiochem (EMD Millipore). Dithiothreitol and tris-(carboxyethyl) phosphine hydrochloride were purchased from Gold Biotechnology. Bovine pancreatic ribonuclease A was from Fisher Scientific.
General Methods
Agilent 8452A or 8453 instruments were used to record UV-Vis spectra. Fluorescence spectra were acquired using an Aminco Bowman 2 luminescence spectrometer. Stock solutions of BD-SS were prepared in phosphate buffer and stored in small aliquots at −20 °C. Reductant solutions were made fresh daily from concentrated solutions stored at −20 °C. Thiol and phosphine concentrations were confirmed using 5,5’-dithiobis(2-nitrobenzoic acid). Unless otherwise stated, the buffer used in this work was 50 mM potassium phosphate containing 1 mM EDTA, pH 7.5. BD-SH fluorescence was negligibly affected in a range of buffers spanning pH 6–9.5 (citrate, Tris, HEPES, PBS). Stock solutions of 16F16 were prepared in DMSO with a final solvent concentration of 1.5% in inhibitor and control wells. Stock solutions of MNS were prepared in ethanol and diluted ~100-fold in water before use. Rutin stocks were prepared directly in water.
BD-SS and MNS extinction coefficient determinations
Solutions containing BD-SS were incubated in phosphate buffer for 3 h with 0–170 μM THP. The mixtures were then diluted 10-fold and their absorbances at 505 nm were plotted as a function of THP concentration. Three independent experiments yielded an extinction coefficient of 101,700 ± 1,800 M−1cm−1 for BD-SS. Solutions of MNS were prepared gravimetrically in ethanol and diluted into water. The absorbance of triplicate serial dilution was recorded at 378 nm yielding an extinction coefficient of 10,380 ± 300 M−1 cm−1.
PDI preparation and handling
A His-tagged PDI construct was expressed and purified as described previously with minor modifications [45]. An additional four elutions of 10, 25, 50 and 100 mM imidazole in 50 mM phosphate, pH 8.0, containing 300 mM NaCl were utilized. Fractions with PDI of the highest purity were combined and dialyzed against 2 x 4 L of same buffer, and then subject to a second affinity purification step as above. PDI was then dialyzed against 50 mM phosphate containing 1 mM EDTA, pH 7.5, and concentrated using an EMD Millipore Amicon Ultra 30K device. With this extended purification protocol, the CxxC motifs of purified PDI were shown to be essentially completely oxidized by DTNB titration. Reduced PDI was obtained by first incubating the protein for 15 min with a 20-fold molar excess of DTT in phosphate buffer at room temperature. Incubation volumes of 0.7 mL or less were applied to a PD-10 column equilibrated with the same buffer to ensure baseline separation between PDIred and excess DTT (confirmed by sampling small aliquots of each 0.5 mL fraction for –SH content using DTNB). Fractions containing PDIred were combined and distributed for storage at −80 °C. PDI mutants of the CxxC motifs in both the a and a' domains were obtained using the QuikChange II Mutagenesis kit (Stratagene) with appropriate primers, sequenced to verify mutations, and handled as above.
Multiple Turnover Assay
A BMG POLARstar OMEGA plate reader with 96-well black flat-bottomed polystyrene plates from COSTAR (Corning) was used to monitor the increase in fluorescence accompanying the reduction of BD-SS. Wells contained 200 μL of 0–75 nM oxidized PDI and either 5 mM GSH or 5 μM DTT in buffer. Routinely, assays were started using the instrument’s reagent injector capability to deliver 10 μL of 15 μM BD-SS in phosphate buffer followed by 3 s of orbital shaking (resulting in a 4.8% dilution of PDI, thiol reagents, and BD-SS). Measurements were then taken in fluorescence intensity well-mode (excitation filter 485-12 and emission filter 520) for durations of 250 1 s cycles. All experiments were conducted in triplicate. When evaluating inhibitors, compounds were added in the following order to generate 200 μL of solution: PDI, reductant, inhibitor. After 15 min incubation at room temperature, the increase in relative fluorescence was plotted as a function of time after the addition of BD-SS. Experiments were corrected for background thiol reduction of BD-SS.
Reoxidation Assay
Wells initially contained 200 μL of 75 nM PDIred in phosphate buffer. The assay was initiated by injecting 10 μL of 15 μM BD-SS followed by 3 s of orbital shaking to give final concentrations of 71.4 nM rPDI and 714 nM BD-SS. Fluorescence measurements were taken with 1 s cycles over 250 s, and fluorescence at t=0 was subtracted. The sensitivity of the instrument was set using 150 nM BD-SS that had been treated with 10 mM DTT for 30 min. When evaluating inhibitors, reagents were pre-incubated for 15 min with PDIred before the addition of BD-SS to the well, as above.
rRNase Titrations
Reduced RNase was prepared as described previously [46]. Solutions of increasing rRNase concentrations were added to 40 μM MNS in buffer and the absorbance at 378 nm was monitored over time, applying a 530–730 nm scatter correction. The resulting absorbance values at 35 s were plotted against the concentration of rRNase thiols.
Calculations
Kd values for the reaction R – L ↔ R + L (where R represents a thiol-containing moiety, L is MNS, and R-L the corresponding MNS adduct) were determined by non-linear least squares fitting of absorbance changes, Aobs, to the concentration of added ligand using the equation:
using Prism 6 (GraphPad Software). Here, A0 and Af represent initial and final absorbance readings, Rt = R + R-L and Lt = L + R-L. Plots of the kinetics of MNS bleaching by glutathione were fit using non-linear regression to a two-phase decay using Prism 6. The amplitudes for the total absorbance changes were then plotted as a function of thiol (Rt) concentration as above.
RESULTS
A sensitive PDI assay using a commercially-available fluorogenic disulfide
Two broad categories of PDI assays have been employed [9,12,32,43,47,48]. The first involves the ability of PDI to catalyze the isomerization of mispaired disulfides in peptides and proteins. The second follows the PDI-mediated reduction of insulin, or a range of disulfide containing dye conjugates including the eosin self-quenched disulfide dimer developed by Raturi and Mutus (Fig. 3A) [43]. Here, we first describe a variation of this assay replacing the dieosin reagent with a commercially-available disulfide based on the BODIPY-FL fluorophore (BD-SS) [49]. BD-SS is almost non-fluorescent in buffer but becomes ~55-fold more fluorescent upon reduction (exciting at 490 nm, emitting at 513 nm; Fig. S1). Reduction of BD-SS may also be followed, with lower sensitivity, using the sizable absorbance changes at 505 nm (Fig. S2) yielding an extinction coefficient of 101,700 ± 1,800 M−1cm−1 (Fig. S3).
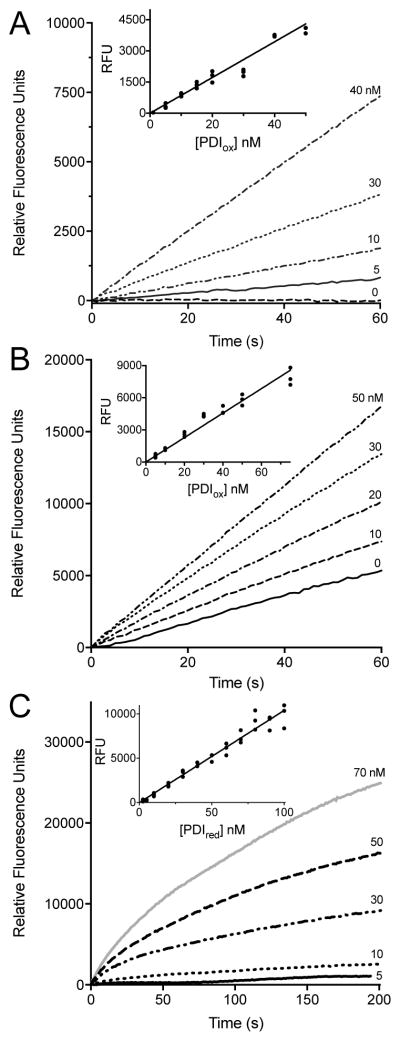
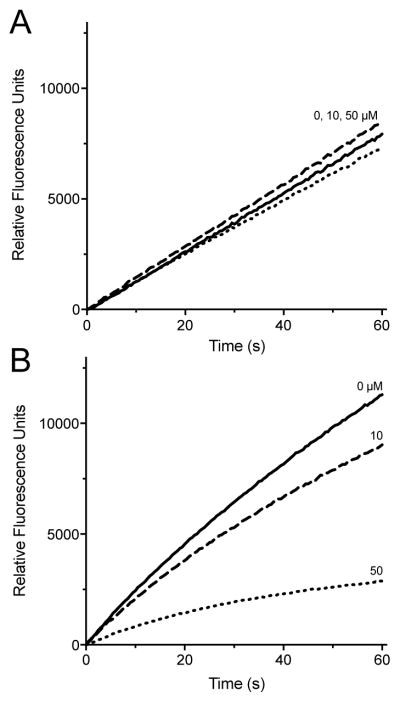
The fluorescence assay is conveniently conducted in a plate reader. Fig. 4A shows progress curves following the PDI mediated-reduction of BD-SS using 5 μM DTT at pH 7.5. Here, the non-enzymatic background for the reduction of BD-SS is minimal, and the increase in fluorescence at each PDI concentration is almost linear over 60 s. The inset in Fig. 4A shows the fluorescence at 30 s is linearly dependent on the PDI concentration. Concentrations of PDI down to 5 nM can be comfortably assessed under these conditions.
Fig. 4.
Multiple turnover and reoxidation PDI assays. Multiple turnover assays in buffer with (A) 5 μM DTT or (B) 5 mM GSH using increasing concentrations of oxidized PDI. The inset demonstrates the linearity of absorbance readings taken at 30 s as a function PDIox concentration, corrected for background thiol reduction of BD-SS. Assays were performed in triplicate, see Materials and Methods. (C) Reoxidation assays in buffer performed in triplicate using increasing concentrations of PDIred added to a final concentration of 714 nM BD-SS. Inset depicts absorbance readings at 30 s plotted against PDIred concentration.
The concentration of 5 μM DTT used in Fig. 4A was chosen to replicate conditions used in the dieosin assay [43]. However, using such a low concentration of thiol substrate, while it minimizes the non-enzymatic background rate of dye reduction, increases the possibility that thiol-reactive inhibitors might also deplete the substrate and thus contribute to an artifactually lowered rate of PDI under these conditions. We thus explored the possibility of assaying PDI in the presence of 5 mM GSH as a more realistic approximation of the high concentrations of thiols that exist intracellularly [27]. While the background rate with this 500-fold increased substrate thiol concentration is now significant, the stimulation on the addition of nanomolar levels of PDI remains clear (Fig. 4B). Background correction of the corresponding rates yields a linear response to PDI with a detection limit of about 5 nM (see inset Fig. 4B). While Di-E-GSSG remains a highly sensitive assay reagent for PDI, the BD-SS disulfide introduced here shows an ~8-fold lower background rate with 5 mM GSH (see Fig. S4).
As a complementary approach, we wanted to evaluate the ability of thiol-reactive inhibitors to impact the reactivity of reduced CxxC motifs of PDI in the complete absence of additional potential competing small molecule thiols. While replacing thiol substrates in multiple turnover assays with the phosphine reducing agent, TCEP, is a potentially workable strategy (see Fig. S5), TCEP was found to react directly with the enone functionality of nitrostyrenes (not shown). A more general and robust approach is to exploit reoxidation experiments in which reduced PDI is mixed with the fluorogenic reagent in the absence of additional reductant. Here BD-SS, like the dieosin reagent [43], provides a sensitive response when mixed with PDIred (Fig. 4C; see Materials and Methods). The inset to Fig. 4C plots the appearance of fluorescence at 30 s from 5–50 nM PDIred.
The BD-SS multiple turnover assay can utilize a range of reducing agents including 5 - 500 μM DTT, 5 – 5000 μM GSH and 5 – 250 μM TCEP, as well as 10 – 50 μM reduced RNase thiols (Fig. S6). The assay can be performed from pH 6 – 9.5 (see Methods) and can be extended to other redox proteins including human thioredoxin and ERp57 (Fig. S7–10; for preparation of ERp57, see Fig. S8). In subsequent sections we utilize both reoxidation and conventional steady-state reductase assays to characterize the potency of representative inhibitors of PDI in the absence and presence of competing thiols.
MNS as an inhibitor of PDI
Steady state assays of PDI in the presence of 4.75 mM GSH is shown in Fig. 5A. All wells were pre-incubated for 15 min with 5 mM GSH and the indicated concentration of the nitrostyrene, MNS. The assays were then initiated by the addition of a small volume of BD-SS reagent so that the concentration of inhibitor and competing thiol would be lowered only slightly (by 4.8%). Thus conditions between pre-incubation and assay are comparable; hence the outcome is not subject to potential bias that may accompany a major dilution of a reversible inhibitor prior to assay. These data show that MNS concentrations of up to 50 μM are ineffective at inhibiting PDI turnover in the presence of 5 mM GSH (for clarity, hereafter nominal concentrations are those prior to the small dilution accompanying BD-SS addition; see Materials and Methods). As a control, assays using pre-reduced PDI, under otherwise identical conditions, were similarly unaffected by MNS in the presence of 5 mM GSH (Fig. S11). In contrast, reoxidation experiments reveal marked inhibition by 50 μM MNS when PDIred is pre-incubated for 15 min with MNS in the absence of competing thiols (Fig. 5B; see Materials and Methods). The differences between Fig. 5A and 5B prompted a closer examination of the reaction between MNS and the sulfhydryl groups of GSH and PDIred.
Fig. 5.
Effect of MNS on PDI activity in multiple turnover and reoxidation assays. (A) Activity of 75 nM oxidized PDI with 0 (——), 10 (– – –) or 50 (. . .) μM MNS in the presence of 5 mM GSH (corrected for non-enzymatic background, see Materials and Methods). (B) The development of fluorescence upon mixing 714 nM BD-SS to 75 nM PDIred preincubated for 15 min with 0 (——), 10 (– – –) or 50 μM (. . .) MNS. Curves represent the average of traces recorded in triplicate.
MNS as a Michael acceptor for thiol nucleophiles
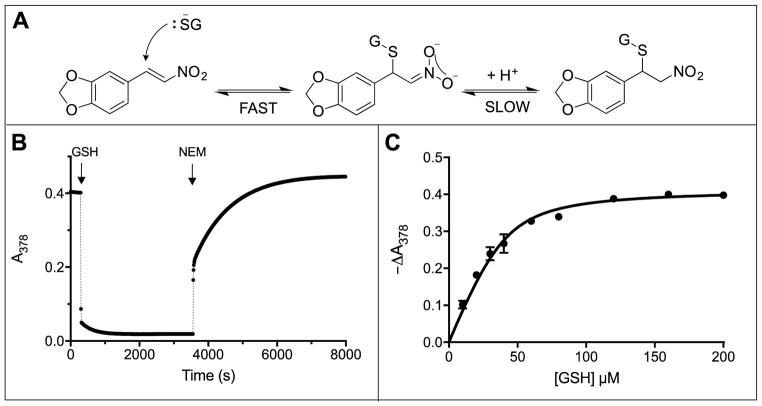
Several thiol-containing reagents have previously been shown to form Michael adducts with nitrostyrenes [50–52]. The reaction involves two steps (Fig. 6A). For example, attack of the glutathione thiolate on MNS leads to discharge of the enone chromophore and is followed by a slower rehybridization of the nitronate anion [53] coupled with protonation at C1. Fig. 6B shows that the nitrostyrene chromophore (40 μM) is rapidly and almost entirely bleached by 400 μM GSH (see also Fig. S12); the subsequent addition of 5 mM NEM completely reverses adduct formation. Titration experiments at pH 7.5 (Fig. 6C) yield a Kd of 6.7 ± 1.4 μM for the overall reaction shown in Fig. 6A. Thus a typical intracellular concentration of GSH (e.g. 5 mM) would be expected to capture >99.9% of MNS in the form of the Michael adducts depicted in Fig. 6A (see Methods). Furthermore, adduct formation is rapid; Fig. 6B shows that this reaction is half-complete in < 6 seconds with 0.4 mM GSH.
Fig. 6.
Reaction between MNS and glutathione. (A) Michael adduct formation to the enone moiety of MNS followed by a slower rehybridization at C1. (B) Absorbance traces at 378 nm following the addition of 400 μM GSH to 40 μM MNS dissolved in phosphate buffer, pH 7.5. Subsequent addition of 5 mM NEM (50 μL to a final volume of 1.05 mL) reversed the bleaching (see also Fig. S12). The increase in the final absorbance after 2 h compared to the initial reading before GSH addition reflects a small contribution of NEM to the total absorbance at 378 nm. (C) Absorbance changes accompanying the addition of GSH to 40 μM MNS. Data in triplicate are fit to a Kd of 6.7 ± 1.4 μM (see Methods).
Using the bleaching of MNS absorbance as observed above, we then titrated MNS with reduced PDI (Fig. S13). The absorbance of MNS significantly decreased upon addition of PDIred in solution, demonstrating binding to exposed thiols. Addition of excess NEM restores the absorbance loss, supporting protein thiol conjugation (data not shown). Since the binding avidity between GSH and rPDI for MNS are both in the low micromolar range (Fig. 6C and Fig. S13), this nitrostyrene would be expected to be an ineffective inhibitor of PDI in the presence of 5 mM GSH (Fig. 5A). The problem of MNS mediated thiol sequestration is potentially more acute due to the plethora of additional thiols found intracellularly [27]. In the next section we briefly examine the ability of MNS to conjugate to thiol groups in folded and unfolded proteins.
MNS reacts with protein thiols
We first examined two folded cytosolic proteins to see if MNS could react with their cysteine residues. Aldolase, containing 2 reactive surface accessible cysteine residues out of 8 total [54], is able to conjugate to MNS (Fig. S14). Pyruvate kinase (1 out of 9 cysteine residues are DTNB-reactive in the absence of denaturing agents (data not shown)) is also slowly labeled with MNS (Fig. S14). While cysteine residues at the surface of folded proteins likely contribute to the intracellular inventory of MNS thiol adducts, we now demonstrate that unfolded proteins present a more dramatic instance of nitrostyrene sequestration.
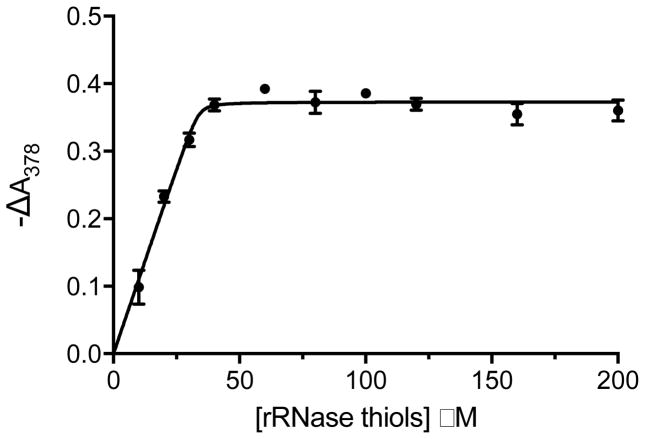
We chose reduced RNase as a well-studied example of the molten globular state [55,56] and a commonly-employed substrate of PDI. Fig. 7 depicts a spectrophotometric titration showing that MNS binds stoichiometrically to the 8 cysteines of reduced RNase with tight affinity. In addition, mixtures of reduced RNase and MNS show ladders of masses using MALDI-TOF analyses corresponding to multiple nitrostyrene labels attached to the polypeptide chain (Fig. S15). This surprisingly tight binding extends the range of potential targets for MNS and again suggests that nitrostyrenes are likely to exert pleiotropic effects intracellularly (see later).
Fig. 7.
Titration of reduced RNase with MNS. MNS, 40 μM, was mixed with 10–200 μM RNase thiols and the absorbance at 378 nm was followed with time. The reaction was rapid, with a half life of less than 2 s at each concentration. The change in absorbance at 378 nm was recorded 30 s after mixing (see Materials and Methods). Titrations were performed in triplicate. The solid line represents a Kd of 0.1 μM.
Evaluating APAO and 16F16 as inhibitors of PDI
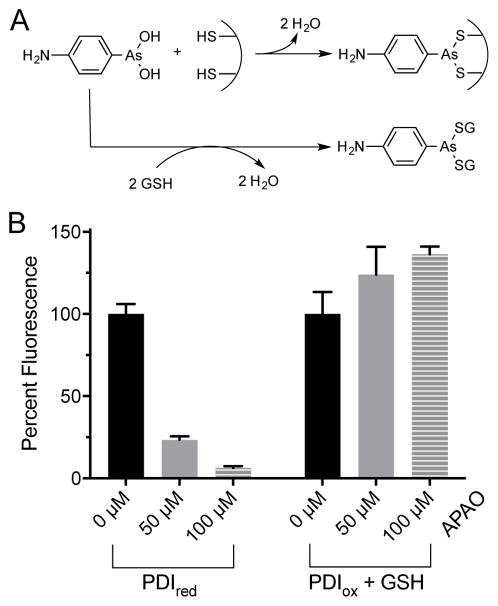
We next examined the model arsenical APAO (Fig. 1) as an inhibitor of PDI using the BD-SS assay. Phenylarsine oxides have been widely used as inhibitors in cell biology [57–59] and can coordinate reduced CxxC motifs and related redox-active dithiols in a variety of cellular oxidoreductases (Fig. 8A) [60]. When we tested the competency of PDIred as a reductant of BD-SS in the reoxidation format in the absence of competing thiols, 50 and 100 μM APAO were found to strongly inhibit the appearance of fluorescence (by approximately 75 and 90%, respectively; Fig. 8B). However, in the turnover assay with 5 mM competing GSH thiols (Fig. 8B and S11) inhibition of PDI was abolished, as we observed with other arsenicals [61,62].
Fig. 8.
Effects of APAO on PDI activity. (A) Schematic of APAO interaction with thiols. (B) Left: following incubation of 75 nM PDIred with 0 (black), 50 (gray), and 100 (gray stripe) μM APAO for 15 min, assays were initiated by BD-SS addition. Fluorescence after 30 sec was plotted for triplicate experiments normalized to the corresponding value in the absence of APAO. Right: multiple turnover assay incubating PDIox with 5 mM GSH and 0 (black), 50 (gray), or 100 (gray stripe) μM APAO. Percent fluorescence was plotted for triplicate experiments 30 s after BD-SS injection as before.
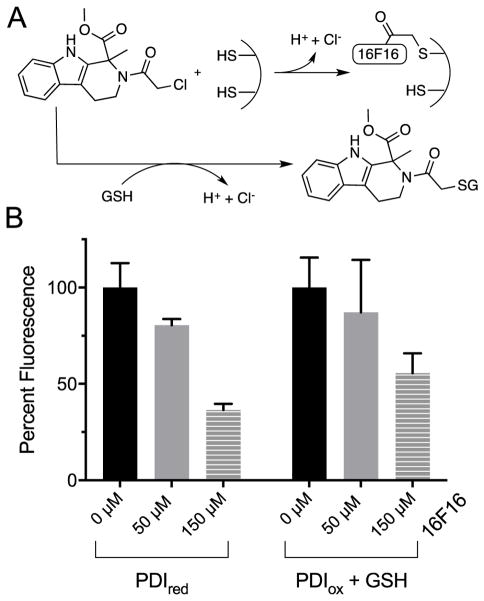
The nitrostyrene and arsenical derivatives considered above form reversible complexes with target thiols and are strongly susceptible to sequestration by competing non-PDI thiol groups. We next consider the competition between PDIred and GSH for an alkylating agent which forms an essentially irreversible thioether linkage with target thiols (Fig. 9A). First described by Stockwell and coworkers [24], the chloroacetamide derivative 16F16 was isolated through high throughput screening as an inhibitor of cell apoptosis. PDI was later identified as a target of this alkylating agent by treating cell lysates with a fluorescent derivative of 16F16 [24]. Fig. 9B shows that a 15 min preincubation of PDIred with 16F16 followed by the addition of BD-SS in the absence of competing thiols showed significant inhibition of BD-SS reduction in this non-turnover assessment of activity. The additional inclusion of 5 mM GSH in the preincubation step attenuated inhibition but loss of activity remained significant (Fig. 9B and S11). Here, intracellular concentrations of GSH was unable to provide strong protection against inactivation as the nucleophilic outer cysteine of each CxxC pair of PDIred is likely more intrinsically reactive towards 16F16 than GSH under these conditions [63,64].
Fig. 9.
16F16 inhibits PDI in the presence of competing thiols. (A) Schematic of 16F16 thiol interaction. (B) Left: solutions 75 nM PDIred with 0 (black), 50 (gray) and 150 (gray stripe) μM 16F16 were incubated for 15 min before the addition of 714 nM BD-SS. The percent fluorescence after 30 s, normalized to PDI activity in the absence of inhibitor was plotted for triplicate experiments. Right: multiple turnover assay with 75 nM PDIox, 5 mM GSH and 0 (black), 50 (gray) and 150 (gray stripe) μM 16F16 were preincubated before addition of BD-SS. Plots depict triplicate experiments, normalized to inhibitor-free PDI activity.
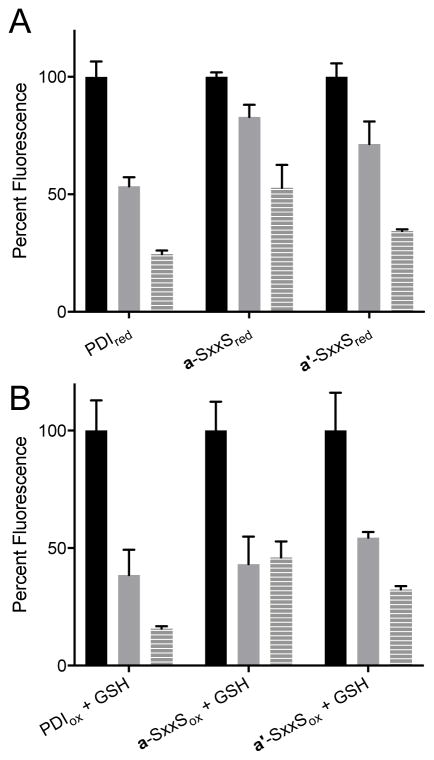
Analyzing rutin with PDI mutants
Recently, high throughput screening led to the identification of quercetin-3-rutinoside (rutin; Fig. 1D) as a reversible inhibitor of PDI [25]. Inhibition reflects binding of the flavonoid to the b'x domain of PDI rather than a covalent interaction with the CxxC motif of the a and a' domains of PDIred [26]. Since these studies used the less sensitive insulin reductase assay, we were interested in applying the BD-SS reoxidation and turnover assays to assess their suitability for high throughput applications. As before, the inhibitor was incubated with PDIred for 15 min prior to the addition of BD-SS. Fig. 10 shows the effect of 1 and 10 μM rutin on the activity of PDIred, demonstrating attenuated reoxidation after 30 s. When challenged with 5 mM GSH, rutin succeeded in inhibiting PDI turnover activity, supporting a non-thiol based method of inhibition [26].
Fig. 10.
Rutin as an inhibitor of wild-type and CxxC mutant PDIs. (A) Following a 15 min incubation of 75 nM PDIred or 150 nM mutant PDIred with 0 (black), 1 (gray) or 10 (gray stripe) μM rutin, 714 nM BD-SS was added and the increase in fluorescence was monitored. Percent fluorescence after 30 s was normalized to PDIred without inhibitor. (B) Multiple turnover assays were completed including 5 mM in GSH in the preincubation as above.
Lin et al. generated a series of truncations of the full-length wild-type PDI and demonstrated that constructs containing either a or a' redox active domains were inhibited by rutin providing they contained the b'x domain (e.g. abb'x or bb'xa') [26]. We sought to extend this observation by exploring the effect of rutin on full-length mutants of PDI in which either the a or a' domains were rendered inactive by replacing the catalytic disulfide CxxC with an SxxS sequence (See Materials and Methods). Fig. 10 shows that rutin is able to inhibit the redox activity of both a and a' CxxC motifs in the context of the full-length multi-domain oxidoreductase, in the absence or presence of 5 mM GSH.
DISCUSSION
In this work we have applied a convenient PDI assay variant, based on a commercially-available fluorogenic disulfide, to examine the potency of thiol-reactive inhibitors intended to target the CxxC motifs of the isomerase. The issue is of particular significance in view of the increasing numbers of thiol-reactive reagents that have been introduced as inhibitors of PDI. We first address intracellular forms of PDI because they present particular difficulties in terms of the multi-millimolar concentrations of potentially competing thiols. The protocols introduced here, where PDI is preincubated with 5 mM GSH in the presence of the inhibitor and then assayed with essentially the same thiol concentration, show that two inhibitors of PDI, MNS and APAO, are unlikely to be effective intracellularly because they are so effectively sequestered by endogenous thiols. However, exposing cells to these membrane-permeant reagents would be expected to result in the internalization of high concentrations of these compounds [65,66] as they are progressively trapped as intracellular thiol adducts with GSH and proteins. The cellular outcomes of such accumulations are likely to add to the difficulties in ascribing a singular target of these non-specific thiol reagents.
Our examination of the irreversible alkylating agent 16F16 (Fig. 1) as an inhibitor of PDI shows that its impact would be somewhat attenuated by typical intracellular concentrations of GSH. Additionally, of course, there are a plethora of protein-derived thiol groups which may further contribute to the sequestration of this chloroacetamide derivative. Some of these targeted protein species may be cellularly critical and yet be present at intracellular concentrations that are far below that of the highly-abundant PDI. As such, these rarer proteins may escape notice on gels in favor of the most prominent bands. A related issue of specificity is that there are many protein disulfide isomerases with a range of abundances that may also be susceptible to alkylation at their redox-active CxxC motifs. It would be interesting to learn what percentage of the most abundant intracellular PDI is derivatized by 16F16 and whether this extent rationalizes the biological effects of this reagent.
In contrast to the intracellular cases, the presence of a number of protein disulfide isomerases at the cell surface [8,67] provides an opportunity to utilize thiol-reactive inhibitors in a milieu of relatively low competing thiols. In terms of the arsenicals, a number of mono- and bis-As(III) species are pronounced inhibitors of reduced PDI in the absence of competing small molecule thiols [61,68,69]. Although exofacial oxidoreductases may be more effectively inhibited by thiol-directed reagents, complications in the interpretation of biological data may still remain. We illustrate them with a brief consideration of the interaction between MNS and PDI.
In an interesting report Chen et al. attributed the suppression of cell adhesion, spreading, migration and invasion of MDA-MB-231 human breast cancer cells in part to the inhibition of surface bound PDI by MNS [19]. They demonstrated significant (~50%) inhibition of surface PDI by 20 μM MNS using the dieosin GSSG assay in the presence of 5 μM DTT as the exogenous reductant. The abrogation of the effect of MNS on cell adhesion in the presence of 5 mM N-acetyl-cysteine or 2-mercaptoethanol likely reflects the ability of these thiols to essentially completely sequester the nitrostyrene as their corresponding Michael adducts [19]. With regards to the site of inhibition, reversal of the cellular effects of MNS by treating cells with a membrane-impermeant dithiol such as 2,3-dimercapto-1-propanesulfonate (DMPS) does not provide strong evidence that a target is itself extracellular. In principle, the presence of a high concentration of extracellular thiol-reactive reagent could drive efflux of the permeant MNS by reversing intracellular complexes and relieving inhibition from the true intracellular target of nitrostyrene action (Fig. 2A and 6B).
In summary, this work outlines some of the challenges associated with tracing a cellular effect induced by a thiol-reactive reagent to the cognate protein target. This problem is acute when inhibition is reversible, because this condition generally precludes the application of conventional analytical techniques to establish the extent and specificity of labeling. As part of the characterization of potential targets, it is important to confirm in vitro that candidate proteins are inhibited under realistic concentrations of endogenous thiols. We illustrate this here with PDI; the BD-SS assay protocols provide a convenient way to evaluate the likely efficacy of the growing number thiol and non-thiol-directed inhibitors of this medically-important class of isomerases.
Supplementary Material
Highlights.
thiol-directed inhibitors of protein disulfide isomerase are examined
a sensitive PDI assay using a commercially-available BODIPY disulfide is introduced
the assay is compatible with physiologically-relevant levels of reduced glutathione
inclusion of competing free thiols can drastically attenuate the inhibition of PDI
the significant challenges for the intracellular targeting of PDI are discussed
Acknowledgments
FUNDING
This work was funded in part by National Institutes of Health [GM26643]. Mass spectrometry experiments were supported by the Delaware COBRE program, with a grant from the National Institute of General Medical Sciences [5 P30 GM110758-02].
We thank Dr. Sharon Rozovsky and Mr. Devin Hudson for discussions, Ms. Qingqing Chen for a gift of human thioredoxin, and anonymous reviewers for helpful comments.
Abbreviations
- APAO
4-amino-phenylarsine oxide
- BD-SH
reduced BODIPY FL-L Cystine
- BD-SS
BODIPY FL-L Cystine
- DTNB
5,5-dithio-bis-(2-nitrobenzoic acid)
- GSH
glutathione
- MNS
3,4-methylenedioxy-β-nitrostyrene
- NEM
N-ethylmaleimide
- PDI
protein disulfide isomerase
- PDIox
oxidized protein disulfide isomerase
- PDIred
reduced protein disulfide isomerase
- RNase
ribonuclease A
- -SH and -SS-
generic thiol and disulfide
- TCEP
tris(2-carboxyethyl)phosphine
- THP
tris(hydroxymethyl)phosphine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goldberger RF, Epstein CJ, Anfinsen CB. Acceleration of reactivation of reduced bovine pancreatic ribonuclease by a microsomal system from rat liver. J Biol Chem. 1963;238:628–35. http://www.jbc.org/content/238/2/628.long. [PubMed] [Google Scholar]
- 2.Venetianer P, Straub FB. The enzymic reactivation of reduced ribonuclease. Biochim Biophys Acta. 1963;67:166–68. doi: 10.1016/0006-3002(63)91812-2. http://www.ncbi.nlm.nih.gov/pubmed/13996655. [DOI] [PubMed] [Google Scholar]
- 3.Galligan JJ, Petersen DR. The human protein disulfide isomerase gene family. Hum Genomics. 2012;6:6. doi: 10.1186/1479-7364-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rigobello MP, Donella-Deana A, Cesaro L, Bindoli A. Distribution of protein disulphide isomerase in rat liver mitochondria. Biochem J. 2001;356:567–70. doi: 10.1042/bj3560567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turano C, Coppari S, Altieri F, Ferraro A. Proteins of the PDI family: Unpredicted non-ER locations and functions. J Cell Physiol. 2002;193:154–63. doi: 10.1002/jcp.10172. [DOI] [PubMed] [Google Scholar]
- 7.Gerner C, Holzmann K, Meissner M, Gotzmann J, Grimm R, Sauermann G. Reassembling proteins and chaperones in human nuclear matrix protein fractions. J Cell Biochem. 1999;74:145–51. http://www.ncbi.nlm.nih.gov/pubmed/10404385. [PubMed] [Google Scholar]
- 8.Araujo TLS, Zeidler JD, Oliveira PVS, Dias MH, Armelin HA, Laurindo FRM. Protein disulfide isomerase externalization in endothelial cells follows classical and unconventional routes. Free Radic Biol Med. 2016;103:199–208. doi: 10.1016/j.freeradbiomed.2016.12.021. [DOI] [PubMed] [Google Scholar]
- 9.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979;254:9627–32. http://www.ncbi.nlm.nih.gov/pubmed/385588. [PubMed] [Google Scholar]
- 10.Wang CC, Li W, Ren J, Fang J, Ke H, Gong W, Feng W, Wang CC. Structural Insights into the Redox-Regulated Dynamic Conformations of Human Protein Disulfide Isomerase. Antioxid Redox Signal. 2012;19:36–45. doi: 10.1089/ars.2012.4630. [DOI] [PubMed] [Google Scholar]
- 11.Darby NJ, Creighton TE. Functional properties of the individual thioredoxin-like domains of protein disulfide isomerase. Biochemistry. 1995;34:11725–35. doi: 10.1021/bi00037a009. [DOI] [PubMed] [Google Scholar]
- 12.Hatahet F, Ruddock LW. Protein Disulfide Isomerase: A Critical Evaluation of Its Function in Disulfide Bond Formation. Antioxid Redox Signal. 2009;11:2807–50. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 13.Puig A, Gilbert HF. Anti-chaperone behavior of BiP during the protein disulfide isomerase-catalyzed refolding of reduced denatured lysozyme. J Biol Chem. 1994;269:25889–96. http://www.ncbi.nlm.nih.gov/pubmed/7929293. [PubMed] [Google Scholar]
- 14.Cai H, Wang CC, Tsou CL. Chaperone-like activity of protein disulfide isomerase in the refolding of a protein with no disulfide bonds. J Biol Chem. 1994;269:24550–52. http://www.ncbi.nlm.nih.gov/pubmed/7929125. [PubMed] [Google Scholar]
- 15.Wilson R, Lees JF, Bulleid NJ. Protein disulfide isomerase acts as a molecular chaperone during the assembly of procollagen. J Biol Chem. 1998;273:9637–43. doi: 10.1074/JBC.273.16.9637. [DOI] [PubMed] [Google Scholar]
- 16.Mares RE, Meléndez-López SG, Ramos MA. Acid-denatured Green Fluorescent Protein (GFP) as model substrate to study the chaperone activity of protein disulfide isomerase. Int J Mol Sci. 2011;12:4625–36. doi: 10.3390/ijms12074625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho J, Furie BCB, Coughlin SR, Furie BCB. A critical role for extracellular protein disulfide isomerase during thrombus formation in mice. J Clin Invest. 2008;118:1123–31. doi: 10.1172/JCI34134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mor-Cohen R. Disulfide Bonds as Regulators of Integrin Function in Thrombosis and Hemostasis. Antioxid Redox Signal. 2016;24:16–31. doi: 10.1089/ars.2014.6149. [DOI] [PubMed] [Google Scholar]
- 19.Chen I-H, Chang F-R, Wu Y-C, Kung P-H, Wu C-C. 3,4-Methylenedioxy-β-nitrostyrene inhibits adhesion and migration of human triple-negative breast cancer cells by suppressing β1 integrin function and surface protein disulfide isomerase. Biochimie. 2015;110:81–92. doi: 10.1016/j.biochi.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Fenouillet E, Barbouche R, Courageot J, Miquelis R. The Catalytic Activity of Protein Disulfide Isomerase Is Involved in Human Immunodeficiency Virus Envelope Mediated Membrane Fusion after CD4 Cell Binding. J Infect Dis. 2001;183:744–52. doi: 10.1086/318823. [DOI] [PubMed] [Google Scholar]
- 21.Khan MMG, Simizu S, Kawatani M, Osada H. The potential of protein disulfide isomerase as a therapeutic drug target. Oncol Res. 2011;19:445–53. doi: 10.3727/096504011x13123323849717. http://www.ncbi.nlm.nih.gov/pubmed/22715587. [DOI] [PubMed] [Google Scholar]
- 22.Flaumenhaft R, Furie B, Zwicker JI. Therapeutic Implications of Protein Disulfide Isomerase Inhibition in Thrombotic Disease Significance. Arterioscler Thromb Vasc Biol. 2014;35:16–23. doi: 10.1161/ATVBAHA.114.303410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett TA, Edwards BS, Sklar LA, Rogelj S. Sulfhydryl regulation of L-selectin shedding: phenylarsine oxide promotes activation-independent L-selectin shedding from leukocytes. J Immunol. 2000;164:4120–29. doi: 10.4049/jimmunol.164.8.4120. http://www.ncbi.nlm.nih.gov/pubmed/10754306. [DOI] [PubMed] [Google Scholar]
- 24.Hoffstrom BG, Kaplan A, Letso R, Schmid RS, Turmel GJ, Lo DC, Stockwell BR. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat Chem Biol. 2010;6:900–6. doi: 10.1038/nchembio.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jasuja R, Passam FH, Kennedy DR, Kim SH, Van Hessem L, Lin L, Bowley SR, Joshi SS, Dilks JR, Furie B, Furie BC, Flaumenhaft R. Protein disulfide isomerase inhibitors constitute a new class of antithrombotic agents. J Clin Invest. 2012;122:2104–2113. doi: 10.1172/JCI61228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin L, Gopal S, Sharda A, Passam F, Bowley SR, Stopa J, Xue G, Yuan C, Furie BCB, Flaumenhaft R, Huang M, Furie BCB. Quercetin-3-rutinoside Inhibits Protein Disulfide Isomerase by Binding to Its b’x Domain. J Biol Chem. 2015;290:23543–52. doi: 10.1074/jbc.M115.666180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen RE, Roth D, Winther JR. Quantifying the global cellular thiol-disulfide status. Proc Natl Acad Sci U S A. 2009;106:422–7. doi: 10.1073/pnas.0812149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montero D, Tachibana C, Rahr Winther J, Appenzeller-Herzog C. Intracellular glutathione pools are heterogeneously concentrated. Redox Biol. 2013;1:508–513. doi: 10.1016/j.redox.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Couët J, de Bernard S, Loosfelt H, Saunier B, Milgrom E, Misrahi M. Cell Surface Protein Disulfide-Isomerase Is Involved in the Shedding of Human Thyrotropin Receptor Ectodomain. Biochemistry. 1996;35:14800–5. doi: 10.1021/bi961359w. [DOI] [PubMed] [Google Scholar]
- 30.Griffith OW. Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic Biol Med. 1999;27:922–35. doi: 10.1016/S0891-5849(99)00176-8. [DOI] [PubMed] [Google Scholar]
- 31.Wu J, Xu K, Landers JP, Weber SG. An in situ measurement of extracellular cysteamine, homocysteine, and cysteine concentrations in organotypic hippocampal slice cultures by integration of electroosmotic sampling and microfluidic analysis. Anal Chem. 2013;85:3095–103. doi: 10.1021/ac302676q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe MM, Laurindo FRM, Fernandes DC. Methods of measuring protein disulfide isomerase activity: a critical overview. Front Chem. 2014;2:73. doi: 10.3389/fchem.2014.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang XM, Fitzgerald M, Grant CM, Hogg PJ. Redox control of exofacial protein thiols/disulfides by protein disulfide isomerase. J Biol Chem. 1999;274:2416–23. doi: 10.1074/JBC.274.4.2416. [DOI] [PubMed] [Google Scholar]
- 34.Chen I-H, Shih H-C, Hsieh P-W, Chang F-R, Wu Y-C, Wu C-C. HPW-RX40 restores anoikis sensitivity of human breast cancer cells by inhibiting integrin/FAK signaling. Toxicol Appl Pharmacol. 2015;289:330–40. doi: 10.1016/j.taap.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 35.Park J, Pei D. trans-Beta-nitrostyrene derivatives as slow-binding inhibitors of protein tyrosine phosphatases. Biochemistry. 2004;43:15014–21. doi: 10.1021/bi0486233. [DOI] [PubMed] [Google Scholar]
- 36.Villar JA, Lima FT, Veber CL, Oliveira AR, Calgarotto AK, Marangoni S, da Silva SL. Synthesis and evaluation of nitrostyrene derivative compounds, new snake venom phospholipase A2 inhibitors. Toxicon. 2008;51:1467–1478. doi: 10.1016/j.toxicon.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 37.Zehavi U, Naim M. Inhibition of Escherichia coli β-galactosidase by 2-nitro-1-(4,5-dimethoxy-2-nitrophenyl)ethyl, a photoreversible thiol label. Biochim Biophys Acta. 1996;1293:238–42. doi: 10.1016/0167-4838(95)00254-5. [DOI] [PubMed] [Google Scholar]
- 38.Hsieh PW, Chang YT, Chuang WY, Shih HC, Chiang SZ, Wu CC. The synthesis and biologic evaluation of anti-platelet and cytotoxic β-nitrostyrenes. Bioorganic Med Chem. 2010;18:7621–27. doi: 10.1016/j.bmc.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 39.Wang WY, Hsieh PW, Wu YC, Wu CC. Synthesis and pharmacological evaluation of novel β-nitrostyrene derivatives as tyrosine kinase inhibitors with potent antiplatelet activity. Biochem Pharmacol. 2007;74:601–611. doi: 10.1016/j.bcp.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Reis J, Oliveira C, Milhazes N, Viña D, Borges F. Exploring Nitrostyrene as a Scaffold for a New Class a of Monoamine Oxidase Inhibitors. Lett Drug Des Disovery. 2013;9:958–61. [Google Scholar]
- 41.Kim JHJH, Kim JHJH, Lee GE, Lee JE, Chung IK. Potent inhibition of human telomerase by nitrostyrene derivatives. Mol Pharmacol. 2003;63:1117–24. doi: 10.1124/mol.63.5.1117. [DOI] [PubMed] [Google Scholar]
- 42.Messerschmitt PJ, Rettew AN, Schroeder NO, Brookover RE, Jakatdar AP, Getty PJ, Greenfield EM. Osteosarcoma phenotype is inhibited by 3,4-methylenedioxy-β-nitrostyrene. Sarcoma. 2012;2012:479712. doi: 10.1155/2012/479712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raturi A, Mutus B. Characterization of redox state and reductase activity of protein disulfide isomerase under different redox environments using a sensitive fluorescent assay. Free Radic Biol Med. 2007;43:62–70. doi: 10.1016/j.freeradbiomed.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 44.Stevenson KJ, Hale G, Perham RN. Inhibition of pyruvate dehydrogenase multienzyme complex from Escherichia coli with mono- and bifunctional arsenoxides. Biochemistry. 1978;17:2189–92. doi: 10.1021/bi00604a026. http://www.ncbi.nlm.nih.gov/pubmed/352396. [DOI] [PubMed] [Google Scholar]
- 45.Rancy PC, Thorpe C. Oxidative protein folding in vitro: a study of the cooperation between quiescin-sulfhydryl oxidase and protein disulfide isomerase. Biochemistry. 2008;47:12047–56. doi: 10.1021/bi801604x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hudson DA, Thorpe C. Mia40 is a facile oxidant of unfolded reduced proteins but shows minimal isomerase activity. Arch Biochem Biophys. 2015;579:1–7. doi: 10.1016/j.abb.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 48.Ali Khan H, Mutus B. Protein disulfide isomerase a multifunctional protein with multiple physiological roles. Front Chem. 2014;26:70. doi: 10.3389/fchem.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar D, Meenan BJ, Dixon D. Glutathione-mediated release of Bodipy® from PEG cofunctionalized gold nanoparticles. Int J Nanomedicine. 2012;7:4007–22. doi: 10.2147/IJN.S33726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim T-R, Choi S-Y. Synthesis of Nucleophilic Adducts of Thiols (IV). Addition of Glutathione to beta-Nitrostyrene Derivatives. Bull Korean Chem Soc. 1983;4:92–95. [Google Scholar]
- 51.Louis-Ferdinand RT, Fuller GC. Rate assay for estimation of thiol affinity of sulfhydryl-reactive agents: Estimation of SH-reactivity. J Pharm Sci. 1969;58:1155–57. doi: 10.1002/jps.2600580931. [DOI] [PubMed] [Google Scholar]
- 52.Bernasconi CF, Schuck DF. Kinetics of reversible thiolate ion addition to substituted β-nitrostyrenes in water. Radicaloid transition state or principle of nonperfect synchronization? J Org Chem. 1992;57:2365–73. doi: 10.1021/jo00034a032. [DOI] [Google Scholar]
- 53.Bernasconi CF. Intrinsic barriers of reactions and the principle of nonperfect synchronization. Acc Chem Res. 1987;20:301–8. doi: 10.1021/ar00140a006. [DOI] [Google Scholar]
- 54.Lai CY, Chen C, Smith JD, Horecker BL. The number, distribution and functional implication of sulfhydryl groups in rabbit muscle aldolase. Biochem Biophys Res Commun. 1971;45:1497–1505. doi: 10.1016/0006-291X(71)90189-6. [DOI] [PubMed] [Google Scholar]
- 55.Jacob J, Dothager RS, Thiyagarajan P, Sosnick TR. Fully reduced ribonuclease A does not expand at high denaturant concentration or temperature. J Mol Biol. 2007;367:609–15. doi: 10.1016/j.jmb.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Trewhella J, Goldenberg DP. Small-angle X-ray scattering of reduced ribonuclease A: effects of solution conditions and comparisons with a computational model of unfolded proteins. J Mol Biol. 2008;377:1576–92. doi: 10.1016/j.jmb.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerhard R, John H, Aktories K, Just I. Thiol-Modifying Phenylarsine Oxide Inhibits Guanine Nucleotide Binding of Rho but Not of Rac GTPases. Mol Pharmacol. 2003;63:1349–55. doi: 10.1124/mol.63.6.1349. [DOI] [PubMed] [Google Scholar]
- 58.Fletcher MC, Samelsons LE, June CH. Complex Effects of Phenylarsine Oxide in T Cells. Induction of Tyrosine Phosphorylation and Calcium Mobilization Independent of CD45 Expression. J Biol Chem. 1993;268:23697–703. [PubMed] [Google Scholar]
- 59.Doussiere J, Poinas A, Blais C, Vignais PV. Phenylarsine oxide as an inhibitor of the activation of the neutrophil NADPH oxidase. Eur J Biochem. 1998;251:649–58. doi: 10.1046/j.1432-1327.1998.2510649.x. [DOI] [PubMed] [Google Scholar]
- 60.Shen S, Li X-F, Cullen WR, Weinfeld M, Le XC. Arsenic binding to proteins. Chem Rev. 2013;113:7769–92. doi: 10.1021/cr300015c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sapra A, Ramadan D, Thorpe C. Multivalency in the Inhibition of Oxidative Protein Folding by Arsenic(III) Species. Biochemistry. 2015;54:612–21. doi: 10.1021/bi501360e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sapra A, Thorpe C. An Arsenical–Maleimide for the Generation of New Targeted Biochemical Reagents. J Am Chem Soc. 2013;135:2415–18. doi: 10.1021/ja310553h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mössner E, Iwai H, Glockshuber R. Influence of the pKa value of the buried, active-site cysteine on the redox properties of thioredoxin-like oxidoreductases. FEBS Lett. 2000;477:21–26. doi: 10.1016/S0014-5793(00)01738-5. [DOI] [PubMed] [Google Scholar]
- 64.Hawkins HC, Freedman RB. The reactivities and ionization properties of the active-site dithiol groups of mammalian protein disulphide-isomerase. Biochem J. 1991;275:335–39. doi: 10.1042/bj2750335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bernardo PH, Brasch N, Chai CLL, Waring P. A novel redox mechanism for the glutathione-dependent reversible uptake of a fungal toxin in cells. J Biol Chem. 2003;278:46549–55. doi: 10.1074/jbc.M304825200. [DOI] [PubMed] [Google Scholar]
- 66.Hirano S, Kobayashi Y, Cui X, Kanno S, Hayakawa T, Shraim A. The accumulation and toxicity of methylated arsenicals in endothelial cells: important roles of thiol compounds. Toxicol Appl Pharmacol. 2004;198:458–67. doi: 10.1016/j.taap.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 67.Essex DW, Chen K, Swiatkowska M. Localization of protein disulfide isomerase to the external surface of the platelet plasma membrane. Blood. 1995;86:2168–73. [PubMed] [Google Scholar]
- 68.Donoghue N, Yam PTW, Jiang XM, Hogg PJ. Presence of closely spaced protein thiols on the surface of mammalian cells. Protein Sci. 2000;9:2436–45. doi: 10.1110/ps.9.12.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramadan D, Rancy PC, Nagarkar RP, Schneider JP, Thorpe C. Arsenic (III) Species Inhibit Oxidative Protein Folding in Vitro. Biochemistry. 2009;20:424–32. doi: 10.1021/bi801988x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.