Abstract
Sex differences and circadian variation are two major factors that affect the expression of drug-processing genes. This study aimed to examine sex differences in the circadian variation of hepatic cytochrome P450 (Cyp) genes and corresponding nuclear receptors. Adult mice were acclimated to environmentally controlled facilities for 2 wks, and livers were collected every 4 h during a 24-h period. Total RNA and protein were isolated and subjected to real-time reverse transcriptase–polymerase chain reaction (RT-PCR) and Western blot analysis. The mRNA expression of the aryl hydrocarbon receptor (AhR) and AhR-regulated Cyp1a1 and Cyp1a2 were higher in females and higher during the light phase. The mRNA expression of constitutive and rostane receptor (CAR) and CYP2B10 protein was female-predominant and higher in the dark phase. Pregnane X receptor (PXR) peaked around 18:00 h, but PXR-regulated Cyp3a11 and Cyp3a25 were higher at 10:00 h, without apparent sex dimorphism at protein levels. Peroxisome proliferator-activated receptor-a (PPARα), Cyp4a10, and Cyp4a14 were higher in females and peaked between 14:00 and 18:00 h. The mRNA levels of farnesoid X receptor (FXR), Cyp7a1, and Cyp27a1 peaked around 18:00 h and CYP7A1 protein was higher during the dark phase and higher in females. Cyp7b1(male-predominant) and Cyp2a4 (female-predominant) both showed circadian variation. Circadian variation of hepatic clock genes such as nuclear receptor Rev-erbα, cryptochrome 1 (Cry1), and brain muscle ARNT-like protein 1 (Bmal1) showed distinct patterns. Sex differences and circadian rhythmicity of Cyp genes and corresponding nuclear receptors exist in mouse liver that could impact xenobiotic metabolism and toxicity at different times of the day.
INTRODUCTION
Cytochrome P450 (Cyp) is a diverse superfamily of hemoproteins responsible for the metabolism of xeno-biotics such as drugs and endogenous molecules such as hormones and bile acids (Froy, 2009, 2011; Renaud et al., 2011). The first four Cyp families (Cyp1–4) play a major role in metabolizing xenobiotics, affecting drug pharmacokinetics and chemical-induced toxicity (Petrick & Klaassen, 2007; Renaud et al., 2011). Aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR), pregnane X receptor (PXR), and peroxisome proliferator-activated receptor-a (PPARα) are transcription factors that mediate xenobiotic inductionof Cyp genes (Petrick & Klaassen, 2007). In general, AhR mediates the induction of the Cyp1 family genes (e.g., Cyp1a1, Cyp1a2), CAR is responsible for the regulation of the Cyp2 family (e.g., Cyp2b10), PXR for the Cyp3 family (e.g., Cyp3a11, Cyp3a25), and PPARα for the Cyp4 family (Cyp4a10, Cyp4a14) (Petrick & Klaassen, 2007). Farnesoid X receptor (FXR) regulates bile acid synthesis genes such as Cyp7a1 (Chiang, 2009). Sex differences in rodent Cyp genes is a unique phenomenon (Hernandez et al., 2009; Hirao et al., 2011; Renaud et al., 2011; Sekimoto, 2011; Waxman & O’Connor, 2006).
Sex differences and circadian variation are two major sources of baseline gene expression variance in rodent livers that affect xenobiotic metabolism and toxicity (Corton et al., 2012). The biological clock in the liver is under the control of the master clock, the suprachiasmatic nucleus located in the brain, and controls Cyp expression to adapt to daily changes that the anticipates sleep and activity periods (Duez et al., 2008; Froy, 2009, 2011; Lim et al., 2006; Paschos et al., 2010). Many transcriptional regulators are involved in the regulation of tissue and daytime clock-controlled genes (Bozek et al., 2010). Disruption of the circadian clock is associated with various disorders, including metabolic syndrome, diabetes, and inflammatory diseases (Sukumaran et al., 2010), as well as accelerating liver carcinogenesis (Filipski et al., 2009). The hepatic circadian clock genes Cry1 and Cry2 play important roles in sex dimorphism of Cyp genes in the rodent liver (Bur et al., 2009).
The circadian variation of drug-processing genes in normal male C57 mice, including Cyp2b10, Cyp2e1, Cyp3a11, and Cyp4a14 (Zhang et al., 2009), and the bile acid synthesis gene Cyp7a1 (Zhang et al., 2011) was investigated by our laboratory. However, little is known about the sex differences in the circadian variation of Cyp genes. We have shown circadian and sex dimorphism of hepatic antioxidant components, including the Nrf2 and glutathione systems, the enzymatic and non-enzymatic antioxidant genes in normal mouse liver (Xu et al., 2012), as well as diurnal and sex variations of metallothionein-1 and -2 in mouse liver, kidney, and blood (Zhang et al., 2012). The present study further aimed to examine the sex differences in the circadian variation of major xenobiotic-metabolizing CYP enzymes (Cyp1–4 families) and CYP enzymes responsible for bile acid synthesis (Cyp7a1 and Cyp27a1). The corresponding nuclear receptors were also examined in normal mouse liver at both mRNA and protein levels.
MATERIALS AND METHODS
Animals
Adult 8-wk-old female and male outbred Kunming (KM) mice were obtained from the Animal Breeding Center (Chongqing, China), and maintained in the specific pathogen-free (SPF)-grade animal facilities (certificate no. SYXK2011-004). Mice were acclimatized for 2 wks in a temperature- and humidity-controlled facility with a standard 12-h light schedule (8:00–20:00 h). Mice had free access to SPF-grade rodent chow (SCXK2012-0002, Chongqing, China) and purified drinking water. In Western blot analysis, adult male and female C57BL/6 (C57) mice from Charles River Laboratories (Wilmington, MA, USA) were used. The circadian variations of these C57 male mice were reported in previous studies (Zhang et al., 2009, 2011). Mice (n 1/4 4/time point/sex) were anesthetized and livers were harvested at 6:00, 10:00, 14:00, 18:00, 22:00, and 2:00 h. Liver samples were frozen in liquid nitrogen and stored at 70 C prior to analysis.
All animal procedures follow the National Institutes of Health (NIH) Guide for Care and Use of Laboratory Animals, and were approved by the Institutional Animal Use and Care Committees of Zunyi Medical College and University of Kansas Medical Center. This research also adheres to the updated ethics policy of Chronobiology International (Portaluppi et al., 2010).
RNA isolation and real-time RT-PCR analysis
Approximately 50–100 mg of liver tissue was homogenized in 1 ml TRIzol (Invitrogen, Carlsbad, CA, USA). Total RNA was extracted according to the manufacturer’s instructions, followed by purification with RNeasy columns (Qiagen, Valencia, CA, USA). The quality of RNA was determined by the A260/A280 ratios. Purified RNA was reverse transcribed with the High Capacity Reverse Transcriptase Kit (Applied Biosystems, Foster City, CA, USA). The primers were designed with the Primer3 software and listed in Table S1. The Power SYBR Green Master Mix (Applied Biosystems) was used for real-time reverse transcriptase–polymerase chain reaction (RT-PCR) analysis. The expression of genes was first normalized with the housekeeping genes (b-actin and glyceraldehyde-3-phosphate dehydrogenase [G3PDH]) of the same sample, and the relative transcript levels were calculated using cycle threshold (ΔΔCt) methods and expressed as percentage of housekeeping genes.
Western blot analysis
Liver protein was extracted with T-PER tissue protein extraction kit (Thermo Scientific, Rockford, IL, USA) with freshly prepared proteinase inhibitors (Sigma, St. Louis, MO, USA). Protein concentrations were determined using the bicinchoninic acid (BCA) protein assay according to the manufacturer’s instructions (Thermo Scientific). Approximately 40 mg of cytosolic protein were used for immunoblotting of proteins of interest. The primary antibodies used in this study include CYP2B10 (equivalent to rat CYP2B1 (Renaud et al., 2011) (sc-53242), and FXR (sc-13063) from Santa Cruz Biotechnology (Santa Cruz, CA, USA); CYP7A1 (Ab2346) and β-actin (Ab8227) were from Abcam (Cambridge, MA, USA); and PPARα was from Norus Biochemicals (Littleton, CO, USA). Protein-antibody complexes were detected using an enhanced chemiluminescent kit (Thermo Scientific) and exposed to HyBlot CL autoradiography film (Denville Scientific, Metuchen, NJ, USA).
Statistics
The data were presented as mean and SEM. Special analysis of circadian rhythms followed the method of Refinetti et al. (2007), as described in our recent publications (Xu et al., 2012; Zhang et al., 2012). Sex difference at the same time point was analyzed by Student’s t test. P < 0.05 was considered statistically significant.
RESULTS
Circadian variation of the mRNA of clock genes in mouse liver
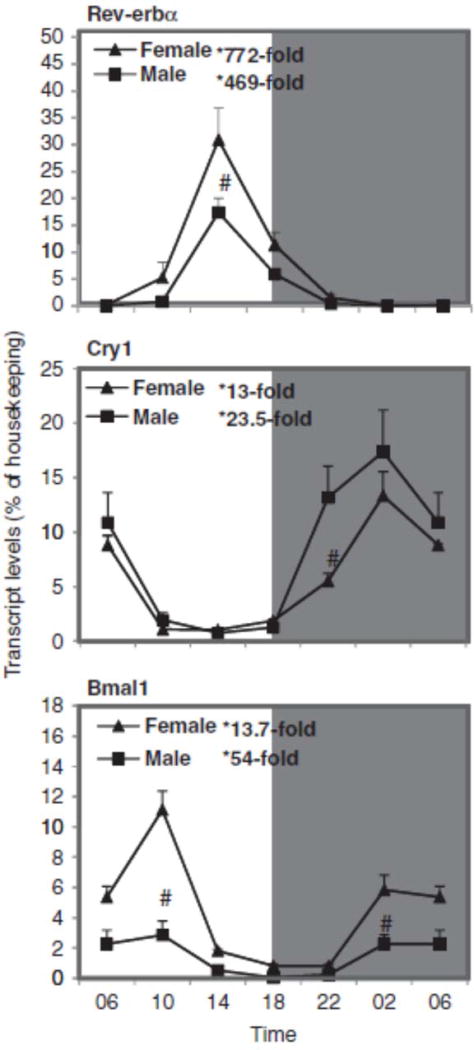
The clock genes nuclear receptor Rev-erbα, cryptochrome 1 (photolyase-like) (Cry1), and brain muscle ARNT-like protein 1 (Bmal1) were firstly examined in mouse liver. Figure 1 illustrates the relative mRNA levels of Rev-erbα, Cry1, and Bmal1 during a 24-h period at 4-h intervals. The peak/nadir differences for Rev-erbα were 772-fold for females (30.9 vs. 0.040) and 470-fold (17.4 vs. 0.037) for males. Although the peaks of females and males were about the same, the nadir of males is lower, resulting in higher fold changes. There was no sex difference in the circadian variation of Rev-erbα. Cry1 showed a 13-fold difference between nadir and peak in females and 23-fold in males. There was basically no sex difference in circadian variation of Cry1, except for slightly higher expression in males at 22:00 h. For the antiphase clock gene Bmal1, there was a 14-fold difference in females and 54-fold in males, and females had higher mRNA levels than males at 10:00 and 2:00 h. We have examined the expression of six clock genes in our recent publications (Xu et al., 2012; Zhang et al., 2009, 2011, 2012), and the oscillation patterns observed in C57 mice are similar in KM mice and represent typical circadian rhythms, thus validating further analysis of the Cyp genes.
FIGURE 1.
Circadian variations of mRNA levels of the clock genes such as nuclear receptor Rev-erbα, cryptochrome 1 (photolyase-like) (Cry1), and brain muscle ARNT-like protein 1 (Bmal1) in adult female and male mouse livers (n=4). *Significant circadian variation (p<0.05, the peak-to-valley ratio of each sex). #Significant sex variation (p<0.05, at the same time point).
Sex differences in the circadian variation of AhR, Cyp1a1, and Cyp1a2 mRNAs in mouse liver
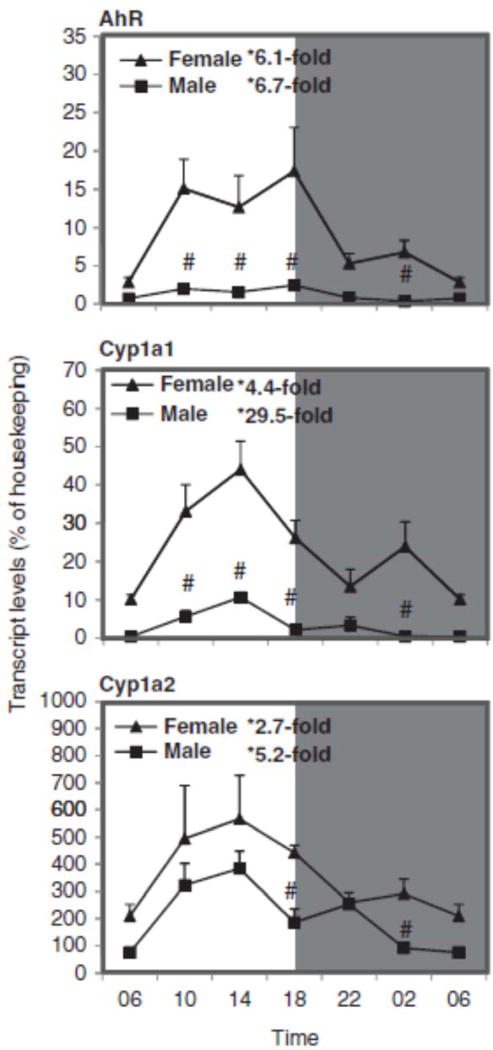
The circadian variations of the AhR-regulated genes, namely Cyp1a1 and Cyp1a2, are illustrated in Figure 2. The relative mRNA levels of these genes were higher in females than in males, and the circadian rhythms were evident in both females and males during a 24-h period at 4-h intervals. The expressions of AhR and AhR-regulated Cyp1a1 and Cyp1a2 peaked between 14:00 and 18:00 h. The nadir and peak differences for AhR (6.1-fold for females and 6.7-fold for males), Cyp1a1 (5-fold for females and 29-fold for males), and Cyp1a2 (3-fold for females and 5-fold for males) are also evident. Female mice had higher expression of AhR, Cyp1a1, and Cyp1a2 than males. Compared with the literature, the circadian variation of AhR and Cyp1a1 in C57 mice was also higher during the night than during daytime (Qu et al., 2009, 2010; Xu et al., 2010), and females had higher expression of Cyp1a than males in rats and pigs (Hirao et al., 2011; Sekimoto, 2011).
FIGURE 2.
Circadian variations of mRNA levels for the AhR and AhR-regulated Cyp1a1 and Cyp1a2 in adult female and male mouse livers (n=4). *Significant circadian variation (p<0.05, the peak-to-valley ratio of each sex). #Significant sex variation (p<0.05, at the same time point).
Sex differences in the circadian variation of CAR and Cyp2b10 in mouse liver
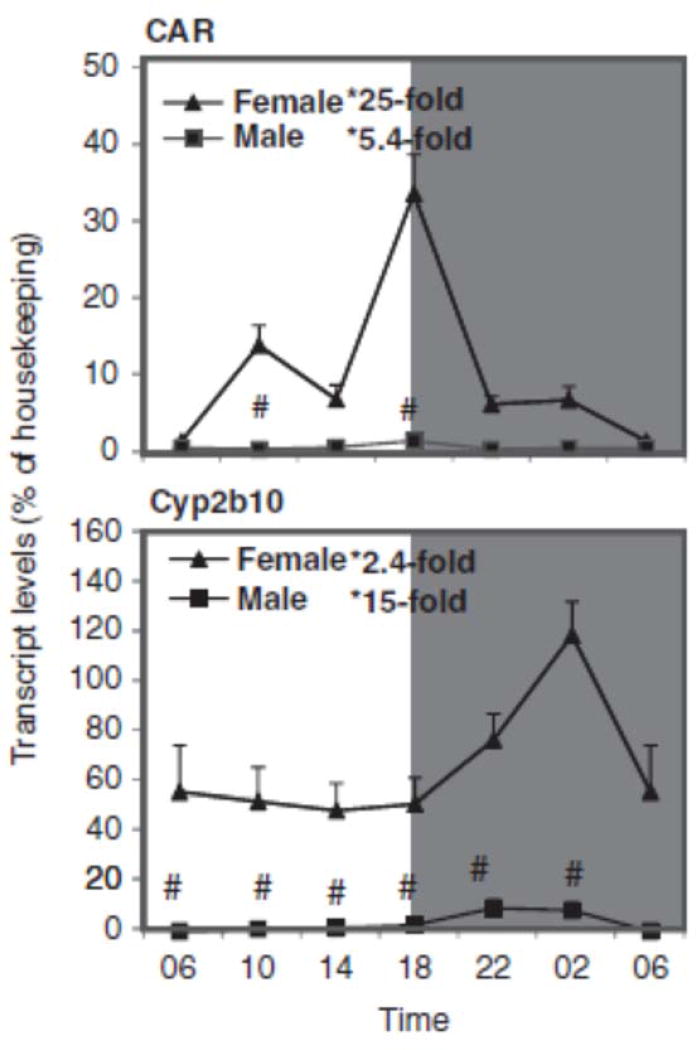
The expression of the CAR-regulated gene Cyp2b10 showed circadian and sex variation patterns, with higher expression in females (Figure 3). The relative mRNA levels of these genes vary during the 24-h period. The expression of CAR peaked at 18:00 h, but for Cyp2b10 at 22:00 h. Cyp2b10 expression pattern of KM mice was similar to that seen in C57 mice (Zhang et al., 2009). The nadir and peak differences for CAR (25-fold in females and 5-fold in males), and Cyp2b10 (2.4-fold in females and 15-fold in males) were also significant. Female mice had higher mRNA levels of CAR and Cyp2b10 than male mice.
FIGURE 3.
Circadian variations of mRNA levels for the CAR and CAR-regulated Cyp2b9 and Cyp2b10 in adult female and male mouse livers (n=4). *Significant circadian variation (p<0.05, the peak-to-valley ratio of each sex). #Significant sex variation (p<0.05, at the same time point).
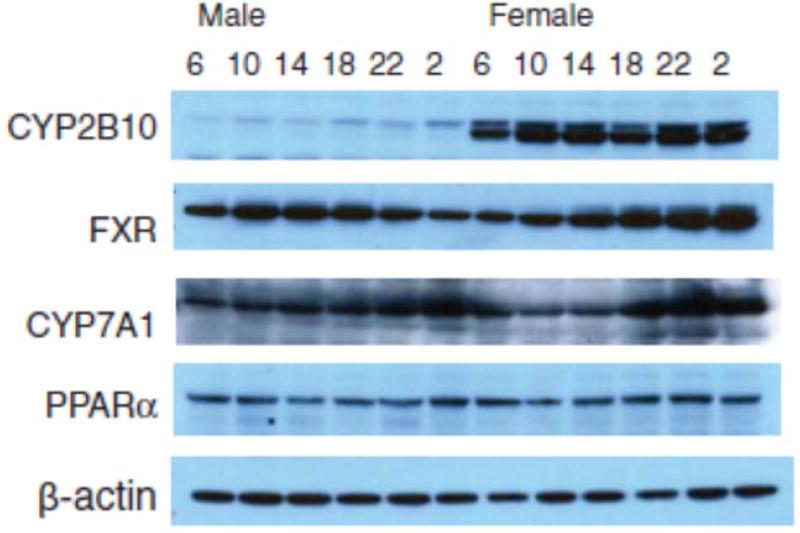
Western blot analysis of C57 mouse liver proteins confirmed higher expression of CYP2B10 in females (see Figure 8). In mice, CAR and CAR-regulated CYP2B10 are female-predominant (Renaud et al., 2011; Sekimoto, 2011).
FIGURE 8.
Representative immunoblots of CYP2B10, CYP3A, FXR, CYP7A1, PPARα, and b-actin in male and female mouse livers at the 6:00, 10:00, 14:00, 18:00, 22:00, and 2:00 h time points.
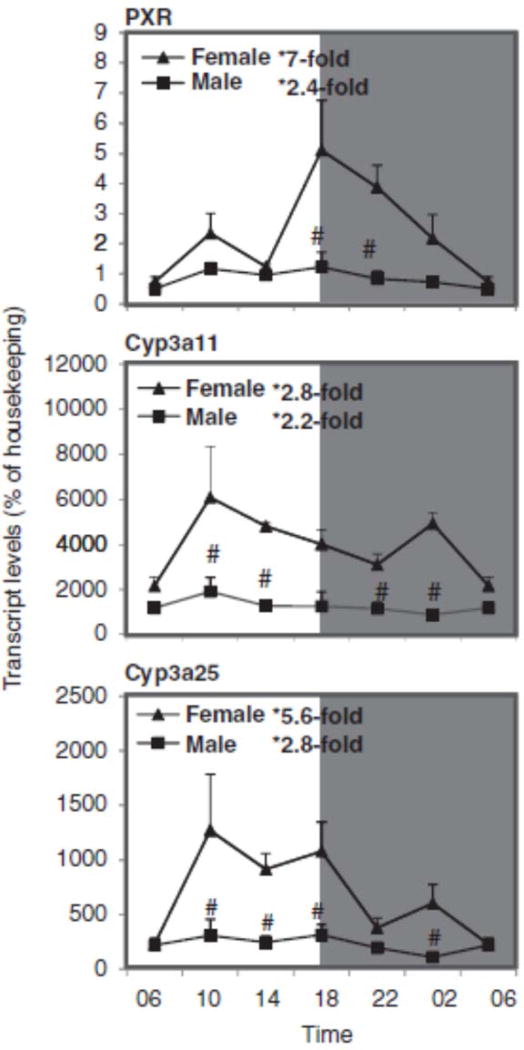
Sex differences in the circadian variation of PXR, Cyp3a11, and Cyp3a25 in mouse liver
Figure 4 shows the relative mRNA levels of PXR, Cyp3a11, and Cyp3a25 genes in mouse liver during the 24-h period. PXR expression peaked at 18:00 h, and both Cyp3a11 and Cyp3a25 peaked around 10:00 h, similar to that in C57 mice (Zhang et al., 2009). The nadir and peak differences for PXR were 7-fold for females and 2.4-fold for males, for Cyp3a11 they were 2.8-fold for females and 2.2-fold for males, and for Cyp3a25 they were 5.6-fold for females and 2.8-fold for males. In mice, Cyp3a11 and Cyp3a44 mRNAs are higher in females than males (Hernandez et al., 2009; Renaud et al., 2011), whereas in rats Cyp3a9 is female-predominant but Cyp3a2 is male-predominant (Hirao et al., 2011). The higher expression of CYP3A4 in women is thought to be under the control of growth hormones (Waxman & O’Connor, 2006).
FIGURE 4.
Circadian variations of mRNA levels for the PXR and PXR-regulated Cyp3a11 and Cyp3a25 in adult female and male mouse livers (n=4). *Significant circadian variation (p<0.05, the peak-to-valley ratio of each sex). #Significant sex variation (p<0.05, at the same time point).
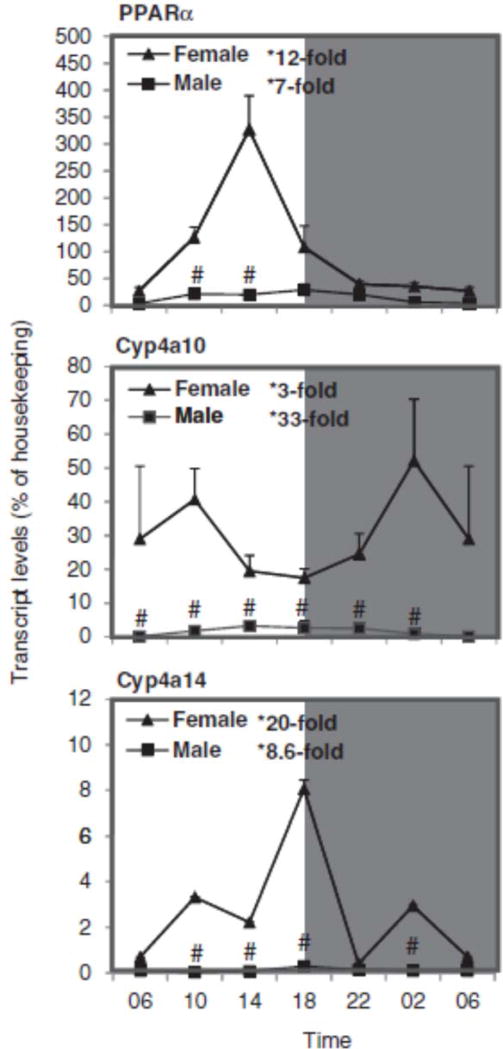
Sex differences in the circadian variation of PPARα, Cyp4a10, and Cyp4a14 in mouse liver
Figure 5 illustrates the relative mRNA levels of PPARα and PPARα-regulated Cyp4a10 and Cyp4a14 during the 24-h period. PPARα reached a peak around 18:00 h, similar to that in C57 mice (Zhang et al., 2009). Western blot analysis of PPARα showed higher expression of PPARα protein in females, and PPARα appeared to peak during the dark phase (22:00–2:00 h). Cyp4a10 reached a peak at 2:00 h. The nadir and peak differences for PPARα were 12-fold in females and 7-fold in males, and for Cyp4a10 they were 3-fold in females and 33-fold in males. Cyp4a14 in KM mice peaked at 18:00 h, also similar to that in C57 mice (Zhang et al., 2009). The nadir and peak differences for Cyp4a14 were 20-fold in females and 9-fold in males. Although males showed more apparent circadian variation of Cyp4a10 than females, females had higher basal levels of Cyp4a10 mRNA. In comparison, mRNA levels of Cyp4a14 in female C57 mice are also 5–8-fold higher than males (Renaud et al., 2011), and Cyp4a10 is also higher in female rats (Hirao et al., 2011).
FIGURE 5.
Circadian variations of mRNA levels for the PPARα and PPARα-regulated Cyp4a10 and Cyp4a14 in adult female and male mouse livers (n=4). *Significant circadian variation (p<0.05, the peak-to-valley ratio of each sex). #Significant sex variation (p<0.05, at the same time point).
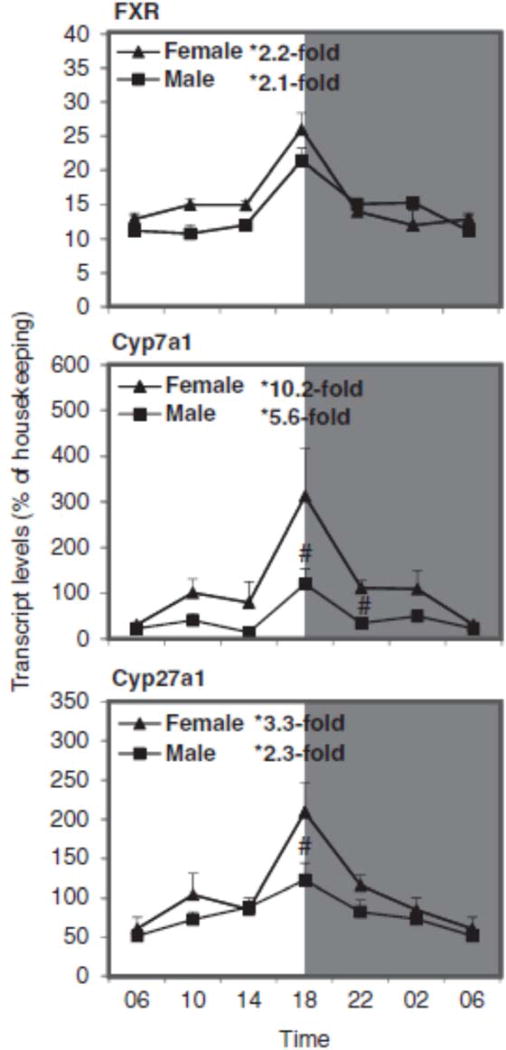
Sex differences in circadian variation of FXR, Cyp7a1, and Cyp27a1 in mouse liver
Figure 6 illustrates the relative mRNA levels of FXR, Cyp7a1, and Cyp27a1 during the 24-h period. For FXR and Cyp7a1, peak mRNA level was seen around 18:00 h, similar to that in C57 mice (Zhang et al., 2011). The nadir and peak differences for FXR were 2-fold in both females and males; for Cyp7a1 there were 10-fold differences in females and 5.6-fold in males. The nadir and peak differences for Cyp27a1 were 3.3-fold in females and 2.3-fold in males. Although mRNA levels of FXR were similar between males and females, the mRNA levels of Cyp7a1 and Cyp27a1 were higher in females at 18:00 h, similar to those reported in a recent publication in C57 mice (Fu et al., 2012). Western blot analysis showed that expression of FXR and CYP7A1 proteins were higher during the dark phase, i.e., 18:00–2:00 h, and females had slightly higher CYP7A1 protein levels (Figure 8).
FIGURE 6.
Circadian variations of mRNA levels for the FXR and FXR-regulated Cyp7a1 and Cyp27a1 in adult female and male mouse livers (n=4). *Significant circadian variation (p<0.05, the peak-to-valley ratio of each sex). #Significant sex variation (p<0.05, at the same time point).
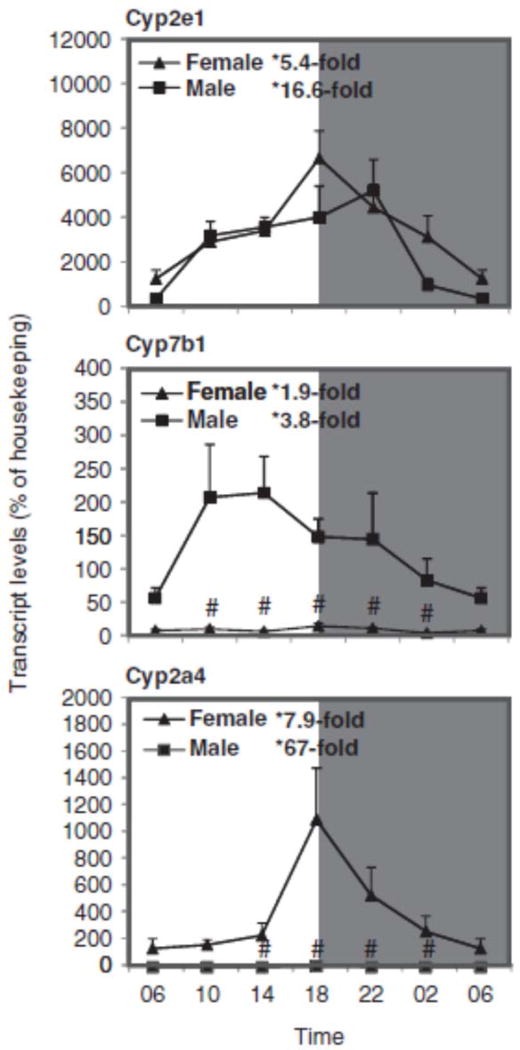
Sex differences in the circadian variation of Cyp2e1, Cyp2a4, and Cyp7b1 in the mouse liver
Figure 7 illustrates the relative mRNA levels of Cyp2e1, Cyp2a4, and Cyp7b1 during the 24-h period. The basal expression of Cyp2e1 was very high, peaked between 18:00 and 22:00 h, similar to that observed in C57 mice (Zhang et al., 2009) and no sex difference was evident. The nadir and peak differences for Cyp2e1 mRNA were 5.4-fold in females and 16.6-fold in males. In comparison, Cyp7b1 is a male-predominant gene, with male/female ratio up to 30-fold, whereas Cyp2a4 is a female-predominant gene, with female/male ratio of 180-fold. The nadir and peak differences for Cyp7b1 were 2-fold in females and 4-fold in males, and for Cyp2a4 they were 8-fold in females and 67-fold in males, as Cyp2a4 expression level in males was very low. In comparison, the sex dimorphism observed in KM mice is similar to that in C57 mice for Cyp7b1 (Bur et al., 2009; Fu et al., 2012) and Cyp2a4 (Renaud et al., 2011).
FIGURE 7.
Circadian variations of mRNA levels for the major metabolism gene Cyp2e1 and examples for female-predominant, Cyp2a4, and male-predominant, Cyp7b1, genes in adult female and male mouse livers (n=4). *Significant circadian variation (p<0.05, the peak-to-valley ratio of each sex). #Significant sex variation (p<0.05, at the same time point).
DISCUSSION
The present work clearly demonstrates sex differences in the circadian variation of Cyp genes, including AhR and AhR-regulated Cyp1a1 and Cyp1a2, CAR and CAR-regulated Cyp2b10, PXR and PXR-regulated Cyp3a11 and Cyp3a25, PPARα and PPARα-regulated Cyp4a10 and Cyp4a14, and FXR and FXR-regulated Cyp7a1 and Cyp27a1. The most abundant Cyp2e1 and the typical female-predominant Cyp2a4 and male-predominant Cyp7b1 also showed circadian rhythmicity. In general, females had higher CYP transcript levels, consistent with our recent publications (Fu et al., 2012; Renaud et al., 2011). This is a systematic investigation on the circadian and sex variations of Cyp genes in the outbred KM mice, the most commonly used mouse strain in China for pharmacology and toxicology studies. The results obtained are generally comparable to those of inbred C57 mice (Fu et al., 2012; Renaud et al., 2011; Zhang et al., 2009, 2011).
Mammals have developed an endogenous circadian clock located in the suprachiasmatic nucleus of the anterior hypothalamus that responds to circadian changes (Dibner et al., 2010). Similar clocks have been found in peripheral tissues, such as the liver (Dibner et al., 2010; Duez et al., 2008; Froy, 2009, 2011; Lim et al., 2006; Paschos et al., 2010). The circadian clock is composed of the cyclical antiphase variation in the expression of Bmal1 and multiple Cry and Per gene isoforms. The clock genes Per, Cry, and Rev-erbα are the regulatory targets of the CLOCK:BMAL1 heterodimer. PER and CRY heterodimerize and function as negative regulators of Clock:Bmal1 activity (Dibner et al., 2010; Lim et al., 2006; Paschos et al., 2010). The typical rhythmicities of these three major clock genes in the livers of KM mice (Figure 1) are similar to those in C57 mice (Zhang et al., 2009, 2011), which is essential for valid comparison of sex differences in the circadian variation of Cyp genes and the corresponding nuclear receptors between the two mouse strains.
Cyp1 enzymes are important in the metabolic activation of some carcinogens such as polycyclic aromatic hydrocarbons and heterocyclic amines, and are induced through AhR activation (Froy, 2009; Hernandez et al., 2009; Huang et al., 2002). The circadian oscillation patterns of AhR and AhR-regulated Cyp1a1 and Cyp1a2 in KM mice were similar to those in C57 mice (Qu et al., 2009, 2010; Xu et al., 2010). Disruption of the clock gene Per1 and Per2 reduced AhR-mediated induction of CYP1A1 by dioxin (Qu et al., 2009). The Clock-mutant (Clk/Clk) mice failed to show significant oscillations in the expression of AhR and induction of CYP1A1 in response to a single intraperitoneal injection of benzo[α]pyrene, a ligand of AhR (Tanimura et al., 2011). The present data and the literature indicate that rhythmicity of AhR and the Cyp1 family genes is under the control of the biological clock. Because AhR and Cyp1a1 are important in carcinogen bioactivation, sex differences in the circadian variation of AhR and CYP1A induction are implicated in altering the carcinogenicity of some chemicals (Levi & Schibler, 2007).
CAR regulates the Cyp2 family of genes (Petrick & Klaassen, 2007). The expression of CAR in KM mice reached a peak at 18:00 h, a pattern similar to that in C57 mice (Zhang et al., 2009). The circadian rhythms of CAR-regulated Cyp2b10 are evident. The female-predominant expression of Cyp2b10 was evident not only at the mRNA level (Figure 3), but also at the CYP2B10 protein level (Figure 8), consistent with that in C57 mice (Renaud et al., 2011). It is proposed that the PAR-domain basic leucine zipper (PAR bZip) transcription factors (DBP, TEF, and HLF) regulate diurnal variation of CAR and CAR-related Cyp genes in mouse liver (Levi & Schibler, 2007). CAR activation increases the CYP2 family of enzymes that metabolize xenobiotics and drugs, and their circadian variations are an important consideration for the timing of drug administration (Levi & Schibler, 2007; Zmrzljak & Rozman, 2012).
CYP3A is the most abundant liver CYP enzyme. It accounts for up to 50% of the oxidative drug metabolism in human liver and also contributes to the oxidation of many steroids and bile acids (Froy, 2009; Waxman & O’Connor, 2006). In humans, CYP3A4 activity is sexually dimorphic, as indicated by the more rapid, CYP3A4- dependent clearance of certain drugs in women compared with men. The observed sex difference in CYP3A4 metabolic activity is thought to be due to differences in growth hormone patterns (Waxman & O’Connor, 2006). PXR and PXR-regulated Cyp3 gene family in rodents are also responsible for the metabolism of a majority of xenobiotics (Petrick & Klaassen, 2007). Urinary 6β-hydroxycortisol-to-cortisol ratio reflects CYP3A activity in the liver, and shows circadian variation (Ohno et al., 2000). In mice, Cyp3a11 and Cyp3a25 are homologues to human CYP3A4 (Renaud et al., 2011). In the present study, diurnal variations of PXR, Cyp3a11, and Cyp3a25 were evident. The expression of PXR peaked around 18:00 h in the C57 mouse liver (Zhang et al., 2009). Sex and circadian variations of Cyp3 genes are also evident in rat livers (Hirao et al., 2011). Because the CYP3 family genes are important in drug metabolism, understanding their circadian variation is helpful in chronopharmacology.
PPARα and PPARα-regulated Cyp4a10 and Cyp4a14 are important in lipid and energy metabolism (Petrick & Klaassen, 2007). Circadian expression of mouse PPARα requires the basic helix-helix PAS protein CLOCK (Oishi et al., 2005), and the rhythms of the clock gene Bmal1, Rev-erbα (Nr1d1), and Dbp are disrupted in PPARα-null mice fed a ketogenic diet (Oishi et al., 2010). In the present study, the circadian expression of PPARα in KM mice is similar to that in BALB/c and C57 mice (Oishi et al., 2010). The expression of PPARα and PPARα-regulated Cyp4a10 and Cyp4a14 showed an apparent sex dimorphism, as higher expression was seen in females, similar to the sex dimorphism pattern of these genes in C57 mice (Renaud et al., 2011) and in rats (Hirao et al., 2011).
The clock gene Rev-erbα participates in the circadian regulation of Cyp genes for bile acid homeostasis (Le et al., 2009). FXR and FXR-regulated Cyp7a1 and Cyp27a1 are important in bile acid synthesis and homeostasis. Cyp7a1 is the rate-limiting enzyme in converting cholesterol to bile acids (Chiang, 2009). The clock gene Rev-erbα alters Cyp7a1 circadian pattern and its basal expression (Duez et al., 2008; Le et al., 2009). In Per1(−/−)/Per2(−/−) mice, serum bile acid levels were elevated and the circadian expressions of key bile acid synthesis and transport genes, including Cyp7a1 and Ntcp, were lost (Ma et al., 2009). CLOCK is also essential for the robust circadian expression of hepatic Cyp7a1 and Cyp8b1, the classic pathway of bile acid synthesis. Clock-controlled genes such as Rev-erbα and Dbp are direct regulators required for the robust circadian rhythm of Cyp7a1 (Ma et al., 2009). Multiple transcriptional factors, including Rev-erbα, Dbp, FXR, PPARα, and HNF4α, participate in circadian rhythmicity of Cyp7a1 and bile acid synthesis genes (Noshiro et al., 2007). In C57 mice, the expression of Cyp7a1 in females was higher than males at different ages, from 3 to 27 months (Fu et al., 2012). Female C57 mice also had higher expression of Cyp27a1 and BA transporters Ntcp and Oatp1b2 than males, but lower expression of Oatp1a1 and Cyp8b1 (Fu et al., 2012). Thus, sex and circadian variations of bile acid homeostasis involve bile acid signaling, synthesis, and transport.
CYP2E1 is one of the most abundant CYP enzymes in the liver, and plays an important role in alcohol metabolism and the metabolism of a variety of chemicals (Froy, 2009, 2011). The diurnal rhythm of CYP2E1 is responsible for circadian variation of the hepatotoxicity of carbon tetrachloride (Bruckner et al., 2002), and for the disposition kinetics of chlorzoxazone (Khemawoot et al., 2007). In the present study, the expression of Cyp2e1 was highest among all Cyp genes under investigation, and clearly follows the pattern of circadian variation (Matsunaga et al., 2008). However, no sex dimorphism for Cyp2e1 was observed in KM mouse liver. In comparison, the female-predominant Cyp2a4 and male-predominant Cyp7b1 were evident and both showed circadian variation. Cyp2a4 is known to be regulated by the clock gene Dbp (Lavery et al., 1999), whereas Cyp7b1 is known to be regulated by the clock gene Cry (Bur et al., 2009), suggesting that the biological clock also controls the sex dimorphism of CYP enzyme genes.
Accumulating evidence indicates that disruption of clock gene expression alters responses of nuclear receptor signaling pathways and alters the circadian expression of Cyp genes. For example, the diurnal variation of TCDD (2,3,7,8-tetrachlorodibenzo-p-dioxin) induction of Cyp1a1 was abolished in Period circadian clock gene nullizygous mice (Qu et al., 2009); in the Clock-mutant Clk/Clk mice, the basal expression of AhR and Cyp1a1 was low, and the AhR ligand benzo[α]pyrene induction of Cyp1a1 was attenuated (Tanimura et al., 2011); in Period circadian clock gene nullizygous mice (Per(/) mice), dark-phase expression of Cyp1a2 was disrupted, resulting in less hepatotoxicity when acetaminophen was dosed at 20:00 h (Kakan et al., 2011). On the other hand, the dark-phase high expression level of Cyp2e1 and low levels of glutathione make animals susceptible to the hepatotoxicity of acetaminophen (Xu et al., 2012) and carbontetrachloride (Bruckner et al., 2002). In Per1(−/−)/Per2(−/−) mice, the circadian rhythmicity of Cyp7a1, Cyp8b1, and Cyp27a1, the major CYP enzymes for bile acid metabolism, was disrupted, resulting in hepatic cholestasis (Ma et al., 2009). Taken together, the biological clock clearly controls Cyp expression, resulting in altered drug metabolism and toxicity.
The influence of the time of day on drug efficacy and toxicity is receiving more and more attention (Froy, 2009, 2011; Lim et al., 2006; Paschos et al., 2010; Zmrzljak & Rozman, 2012). Drugs and toxins can influence the circadian rhythm of the liver; drug administration timing could also influence pharmacokinetics and pharmacodynamics of drugs (Gachon & Firsov 2011; Zmrzljak & Rozman, 2012). Disruption of circadian rhythms may lead to manifestations of the metabolic syndrome (Froy, 2011). In essence, the time matters in the control of xenobiotic detoxification (Levi & Schibler, 2007; Zmrzljak & Rozman, 2012), and rhythmicity of CYP enzyme genes provides the molecular basis for the dosing-time dependence of drug toxicities and efficacy, and the knowledge of circadian variation of Cyp genes can in turn be used in improving or designing chronotherapeutics for the patients (Froy, 2009, 2011; Gachon & Firsov 2011).
In the present study, expression of most Cyp genes were female-predominant except of CYP7b1, which is male-predominant. A few Cyp genes did not show sex dimorphism (i.e., Cyp2e1). We did not measure estrous cycles of these mice, but the female-predominant Cyp expression seems to be a unique phenomenon (Renaud et al., 2011), and may not be under the estrous cycle control. Nullizygous for the circadian regulator gene Cry1 and Cry2 changed the pattern of sex dimorphism in mouse liver, such as Cyp7b1 (Bur et al., 2009). Sex differences in Cyp expression are most striking in rodents, but are also seen, albeit to a much smaller degree, in humans. Females have a higher incidence of drug-induced liver toxicity (Maddrey, 2005), and this could be due, at least in part, to pharmacokinetic differences in the expression of Cyp genes (Corton et al., 2012).
In summary, this study clearly demonstrated the circadian variation in hepatic drug metabolism genes in mice, including the Cyp1, Cyp2, Cyp3, and Cyp4 gene families and bile acid synthesis genes (Cyp7a1 and Cyp27a1) and their corresponding nuclear receptors AhR, CAR, PXR, PPARα, and FXR. In addition, sex dimorphism was also evident, and females had higher expression of most Cyp genes. Sex differences in the circadian variation of Cyp genes are important factors in drug metabolism affecting the efficacy and toxicity.
Supplementary Material
Acknowledgments
This study was supported by National Natural Science Foundation of China (grant no. 81160415), and supported in part by National Institutes of Health grants ES-009649, ES-019487, and DK-081461. The authors also acknowledge the support from Science and Technology Foundation of Guizhou Province (grant nos. TZJF2009-41, 2010-5, and C-453) and Foundation of Zunyi Medical College.
The authors alone are responsible for the content and writing of the paper.
Footnotes
DECLARATION OF INTEREST
The authors report no conflicts of interest.
References
- Bozek K, Rosahl AL, Gaub S, et al. Circadian transcription in liver. Biosystems. 2010;102:61–9. doi: 10.1016/j.biosystems.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Bruckner JV, Ramanathan R, Lee KM, Muralidhara S. Mechanisms of circadian rhythmicity of carbon tetrachloride hepatotoxicity. J Pharmacol Exp Ther. 2002;300:273–81. doi: 10.1124/jpet.300.1.273. [DOI] [PubMed] [Google Scholar]
- Bur IM, Cohen-Solal AM, Carmignac D, et al. The circadian clock components CRY1 and CRY2 are necessary to sustain sex dimorphism in mouse liver metabolism. J Biol Chem. 2009;284:9066–73. doi: 10.1074/jbc.M808360200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–66. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corton JC, Bushel PR, Fostel J, O’Lone RB. Sources of variance in baseline gene expression in the rodent liver. Mutat Res. 2012;746:104–12. doi: 10.1016/j.mrgentox.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–49. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Duez H, van der Veen JN, Duhem C, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erb alpha. Gastroenterology. 2008;135:689–98. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- Filipski E, Subramanian P, Carrière J, et al. Circadian disruption accelerates liver carcinogenesis in mice. Mutat Res. 2009;680:95–105. doi: 10.1016/j.mrgentox.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Froy O. Cytochrome P450 and the biological clock in mammals. Curr Drug Metab. 2009;10:104–15. doi: 10.2174/138920009787522179. [DOI] [PubMed] [Google Scholar]
- Froy O. The circadian clock and metabolism. Clin Sci. 2011;120:65–72. doi: 10.1042/CS20100327. [DOI] [PubMed] [Google Scholar]
- Fu ZD, Csanaky IL, Klaassen CD. Gender-divergent profile of bile acid homeostasis during aging of mice. PLoS ONE. 2012;7:e32551. doi: 10.1371/journal.pone.0032551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Firsov D. The role of circadian timing system on drug metabolism and detoxification. Expert Opin Drug Metab Toxicol. 2011;7:147–58. doi: 10.1517/17425255.2011.544251. [DOI] [PubMed] [Google Scholar]
- Hernandez JP, Mota LC, Huang W, et al. Sexually dimorphic regulation and induction of P450s by the constitutive androstane receptor (CAR) Toxicology. 2009;256:53–64. doi: 10.1016/j.tox.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao J, Nishimura M, Arakawa S, et al. Sex and circadian modulatory effects on rat liver as assessed by transcriptome analyses. J Toxicol Sci. 2011;36:9–22. doi: 10.2131/jts.36.9. [DOI] [PubMed] [Google Scholar]
- Huang P, Ceccatelli S, Rannug A. A study on diurnal mRNA expression of CYP1A1, AHR, ARNT, and PER2 in rat pituitary and liver. Environ Toxicol Pharmacol. 2002;11:119–26. doi: 10.1016/s1382-6689(01)00111-9. [DOI] [PubMed] [Google Scholar]
- Kakan X, Chen P, Zhang J. Clock gene mPer2 functions in diurnal variation of acetaminophen induced hepatotoxicity in mice. Exp Toxicol Pathol. 2011;63:581–5. doi: 10.1016/j.etp.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Khemawoot P, Nishino K, Ishizaki J, et al. Circadian rhythm of cytochrome P4502E1 and its effect on disposition kinetics of chlorzoxazone in rats. Eur J Pharmacol. 2007;574:71–6. doi: 10.1016/j.ejphar.2007.06.032. [DOI] [PubMed] [Google Scholar]
- Lavery DJ, Lopez-Molina L, Margueron R, et al. Circadian expression of the steroid 15 alpha-hydroxylase (Cyp2a4) and coumarin 7-hydroxylase (Cyp2a5) genes in mouse liver is regulated by the PAR leucine zipper transcription factor DBP. Mol Cell Biol. 1999;19:6488–99. doi: 10.1128/mcb.19.10.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G, Claudel T, Gatfield D, et al. REV-ERB alpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi F, Schibler U. Circadian rhythms:mechanisms andtherapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- Lim FL, Currie RA, Orphanides G, Moggs JG. Emerging evidence for the interrelationship of xenobiotic exposure and circadian rhythms: a review. Xenobiotica. 2006;36:1140–51. doi: 10.1080/00498250600861819. [DOI] [PubMed] [Google Scholar]
- Ma K, Xiao R, Tseng HT, et al. Circadian dysregulation disrupts bile acid homeostasis. PLoS ONE. 2009;4:e6843. doi: 10.1371/journal.pone.0006843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddrey WC. Drug-induced hepatotoxicity. J Clin Gastroenterol. 2005;39:S83–9. doi: 10.1097/01.mcg.0000155548.91524.6e. [DOI] [PubMed] [Google Scholar]
- Matsunaga N, Ikeda M, Takiguchi T, et al. The molecular mechanism regulating 24-hour rhythm of CYP2E1 expression in the mouse liver. Hepatology. 2008;48:240–51. doi: 10.1002/hep.22304. [DOI] [PubMed] [Google Scholar]
- Noshiro M, Usui E, Kawamoto T, et al. Multiple mechanisms regulate circadian expression of the gene for cholesterol 7alpha-hydroxylase (Cyp7a), a key enzyme in hepatic bile acid biosynthesis. J Biol Rhythms. 2007;22:299–311. doi: 10.1177/0748730407302461. [DOI] [PubMed] [Google Scholar]
- Ohno M, Yamaguchi I, Ito T, et al. Circadian variation of the urinary 6beta-hydroxycortisol to cortisol ratio that would reflect hepatic CYP3A activity. Eur J Clin Pharmacol. 2000;55:861–5. doi: 10.1007/s002280050708. [DOI] [PubMed] [Google Scholar]
- Oishi K, Shirai H, Ishida N. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor alpha (PPARαlpha) in mice. Biochem J. 2005;386:575–81. doi: 10.1042/BJ20041150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Uchida D, Ohkura N, Horie S. PPARα deficiency augments a ketogenic diet-induced circadian PAI-1 expression possibly through PPARg activation in the liver. Biochem Biophys Res Commun. 2010;401:313–18. doi: 10.1016/j.bbrc.2010.09.060. [DOI] [PubMed] [Google Scholar]
- Paschos GK, Baggs JE, Hogenesch JB, FitzGerald GA. The role of clock genes in pharmacology. Annu Rev Pharmacol Toxicol. 2010;50:87–214. doi: 10.1146/annurev.pharmtox.010909.105621. [DOI] [PubMed] [Google Scholar]
- Petrick JS, Klaassen CD. Importance of hepatic induction of constitutive androstane receptor and other transcription factors that regulate xenobiotic metabolism and transport. Drug Metab Dispos. 2007;35:1806–15. doi: 10.1124/dmd.107.015974. [DOI] [PubMed] [Google Scholar]
- Portaluppi F, Smolensky MH, Touitou Y. Ethics and methods for biological rhythm research on animals and human beings. Chronobiol Int. 2010;27:1911–29. doi: 10.3109/07420528.2010.516381. [DOI] [PubMed] [Google Scholar]
- Qu X, Metz RP, Porter WW, et al. Disruption of period gene expression alters the inductive effects of dioxin on the AhR signaling pathway in the mouse liver. Toxicol Appl Pharmacol. 2009;34:370–7. doi: 10.1016/j.taap.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Metz RP, Porter WW, et al. The clock genes period 1 and period 2 mediate diurnal rhythms in dioxin-induced Cyp1A1 expression in the mouse mammary gland and liver. Toxicol Lett. 2010;196:28–32. doi: 10.1016/j.toxlet.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refinetti R, Cornélissen G, Halberg F. Procedures for numerical analysis of circadian rhythms. Biol Rhythm Res. 2007;38:235–325. doi: 10.1080/09291010600903692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud HJ, Cui JY, Khan M, Klaassen CD. Tissue distribution and gender-divergent expression of 78 cytochrome P450 mRNAs in mice. Toxicol Sci. 2011;124:261–277. doi: 10.1093/toxsci/kfr240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto M. Sex- and species-differences on xenobiotic-induced toxicity: differences in constitutive and xenobiotic-mediated expression of cytochrome P450 1A subfamily enzymes (CYP1As) Yakugaku Zasshi. 2011;131:415–22. doi: 10.1248/yakushi.131.415. [DOI] [PubMed] [Google Scholar]
- Sukumaran S, Almon RR, DuBois DC, Jusko WJ. Circadian rhythms in gene expression: relationship to physiology, disease, drug disposition and drug action. Adv Drug Deliv Rev. 2010;62:904–17. doi: 10.1016/j.addr.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura N, Kusunose N, Matsunaga N, et al. Aryl hydrocarbon receptor-mediated Cyp1a1 expression is modulated in a CLOCK-dependent circadian manner. Toxicology. 2011;290:203–7. doi: 10.1016/j.tox.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Waxman DJ, O’Connor C. Growth hormone regulation of sex-dependent liver gene expression. Mol Endocrinol. 2006;11:2613–29. doi: 10.1210/me.2006-0007. [DOI] [PubMed] [Google Scholar]
- Xu CX, Krager SL, Liao DF, Tischkau SA. Disruption of CLOCK-BMAL1 transcriptional activity is responsible for aryl hydrocarbon receptor-mediated regulation of Period1 gene. Toxicol Sci. 2010;115:98–108. doi: 10.1093/toxsci/kfq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YQ, Zhang D, Jin T, et al. Diurnal variation of hepatic antioxidant gene expression in mice. PLoS ONE. 2012;7:e44237. doi: 10.1371/journal.pone.0044237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Jin T, Xu YQ, et al. Diurnal-and sex-related difference of metallothionein expression in mice. J Circadian Rhythms. 2012;10:5. doi: 10.1186/1740-3391-10-5. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YK, Yeager RL, Klaassen CD. Circadian expression profiles of drug-processing genes and transcription factors in mouse liver. Drug Metab Dispos. 2009;37:106–15. doi: 10.1124/dmd.108.024174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YK, Guo GL, Klaassen CD. Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS ONE. 2011;6:e16683. doi: 10.1371/journal.pone.0016683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmrzljak UP, Rozman D. Circadian regulation of the hepatic endobiotic and xenobitoic detoxification pathways: the time matters. Chem Res Toxicol. 2012;25:811–24. doi: 10.1021/tx200538r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.