The headquarter of our immune system resides in the gut and modulates autoimmune disease activation in multiple sclerosis.
Abstract
T helper 17 (TH17) cells are key players in multiple sclerosis (MS), and studies in animal models demonstrated that effector TH17 cells that trigger brain autoimmunity originate in the intestine. We validate in humans the crucial role of the intestinal environment in promoting TH17 cell expansion in MS patients. We found that increased frequency of TH17 cells correlates with high disease activity and with specific alterations of the gut mucosa-associated microbiota in MS patients. By using 16S ribosomal RNA sequencing, we analyzed the microbiota isolated from small intestinal tissues and found that MS patients with high disease activity and increased intestinal TH17 cell frequency showed a higher Firmicutes/Bacteroidetes ratio, increased relative abundance of Streptococcus, and decreased Prevotella strains compared to healthy controls and MS patients with no disease activity. We demonstrated that the intestinal TH17 cell frequency is inversely related to the relative abundance of Prevotella strains in the human small intestine. Our data demonstrate that brain autoimmunity is associated with specific microbiota modifications and excessive TH17 cell expansion in the human intestine.
INTRODUCTION
Multiple sclerosis (MS) is a chronic inflammatory demyelinating disease of the central nervous system (CNS) caused by myelin-specific, self-reactive T lymphocytes (1). T cells with high affinity for CNS autoantigens are present in healthy individuals (2), but they normally remain dormant and do not cross the blood-brain barrier to trigger autoimmune diseases in the brain unless they are activated in peripheral tissues, such as the lung and the intestine (3, 4). Relapsing-remitting MS (RRMS), the most common disease course, is characterized by clearly defined attacks of new or increasing neurologic symptoms followed by periods of partial or complete recovery (remissions). RRMS can be classified as either active [with clinical relapses and/or evidence of new magnetic resonance imaging (MRI) activity] or inactive. Disease activity in RRMS is correlated with periodic activation of myelin-specific T cells, but the mechanisms that regulate the aggressiveness of those self-reactive T cells are still largely unknown.
T helper 17 (TH17) cells are key players in MS pathogenesis, and acquisition of interleukin-17 (IL-17)–secreting phenotype by myelin-specific T cells enhances their pathogenicity. In experimental autoimmune encephalomyelitis (EAE), the preclinical model of MS, brain autoimmunity is driven by a proinflammatory T cell response that promotes infiltration of immune cells in the CNS. Although several B and T cell subsets orchestrate disease pathogenesis, effector TH17 cells represent the first wave of pathogenic T cells infiltrating the CNS (5) because of their ability to efficiently breach the blood-brain barrier (6). In humans, IL-17 was found in MS lesions, and whole-genome transcriptomic analysis revealed that IL-17 is the highest-ranking gene expressed in the active plaques (7).
Although TH17 cells are believed to be central in MS pathogenesis, the geography of their peripheral activation and expansion in humans is still unclear. In mice, activation of effector TH17 cells occurs mostly in the small intestine (8, 9), and acquisition of a TH17 cell phenotype by myelin-reactive T cells at the intestinal level enhances their pathogenicity and capacity to trigger brain autoimmunity (4, 10, 11). Commensal microbes residing in the small intestine, such as the segmented filamentous bacteria (SFB), induce brain autoimmunity in mice by selectively promoting TH17 cell differentiation (10). In humans, previous studies identified alterations of the microbiota composition in fecal samples of RRMS patients (12–16); however, it is still unclear how these modifications impinge on the immune system. Although two of these studies found a correlation between the microbiome modifications and specific immune markers in peripheral blood lymphocytes of RRMS patients compared to healthy controls (HCs) (14, 15), a specific analysis on how the microbiota alter immunity at the intestinal level in RRMS patients was never performed. Here, we performed analysis of immune cell subsets together with 16S ribosomal RNA (rRNA) sequencing of mucosa-associated microbiota in the small intestine of RRMS patients and HCs. Our data show that in RRMS patients, there is a selective expansion of effector TH17 cells in the small intestinal mucosa that is linked with specific microbiota modifications.
RESULTS
Study design
Small intestinal tissue and peripheral blood samples were collected from 36 individuals, including 19 RRMS patients and 17 HC subjects; details of the study population are illustrated in Table 1. Total lymphocytes were freshly isolated from intestinal tissue collected during esophagogastroduodenoscopy (EGD) and from peripheral blood, stained with fluorescent monoclonal antibodies against surface markers and intracellular cytokines, and analyzed by FACS (fluorescence-activated cell sorting) (Fig. 1). Microbial DNA was extracted from small intestinal samples obtained during EGD and analyzed through 16S rRNA gene sequencing on the Roche 454 platform. All patients enrolled in this study had a diagnosis of RRMS defined according to Poser’s diagnostic criteria, had different degrees of disability [Expanded Disability Status Scale (EDSS) score from 1 to 5.5], and were receiving treatment with immunomodulatory drugs [interferon-β (IFN-β), glatiramer acetate (GA), or fingolimod] (Table 2). Disease activity in a 2-year follow-up was evaluated by periodic neurological examinations and evaluations of clinical relapses and disability progression (EDSS), and by measurement of new lesions by MRI (Fig. 1). NEDA (no evidence of disease activity) is a new concept related to the absence of disease activity in the context of MS that takes into account the following clinical and neuroradiological parameters: clinical relapses, disability progression, and active lesions evaluated by MRI (17). Our RRMS patients were classified into two subgroups on the basis of the presence or absence of disease activity at a 2-year follow-up: patients with no evidence of disease activity (RRMS/NEDA, n = 9) and patients with evidence of disease activity (RRMS/EDA, n = 10).
Table 1. Demographics of study population.
All individuals recruited for the study underwent EGD for diagnostic purposes. Exclusion criteria for both HCs and RRMS patients were treatment with antibiotics or corticosteroids in the 3 months before EGD and history of gastroenteritis, gastric ulcer, irritable bowel disease, celiac disease, inflammatory bowel disease, and gastric and colorectal cancers. All patients recruited in this study have a diagnosis of RRMS defined according to Poser’s diagnostic criteria. None of the RRMS patients received corticosteroids in the 3 months before the EGD. The mean age of healthy individuals was slightly higher than that of RRMS patients, but our correlative analysis did not reveal any association between age and TH17 cell percentages and Prevotella/Streptococcus relative abundance. All samples analyzed were donated for research purposes, and each subject signed an informed consent. Data are means ± SD, unless otherwise indicated. RR-NEDA, RRMS/NEDA; RR-EDA, RRMS/EDA.
| Cohort | n | Age (years) | Age (mean) | Sex (women/men) | Other autoimmune diseases |
| Healthy | 17 | 27–74 | 48 ± 3 | 10/7 | NA |
| RRMS | 19 | 25–57 | 41 ± 2 | 11/8 | Autoimmune thyroiditis (1/19) |
| RR-NEDA | 9 | 26–56 | 40 ± 4 | 7/2 | — |
| RR-EDA | 10 | 25–57 | 41 ± 3 | 4/6 | — |
Fig. 1. Study design.
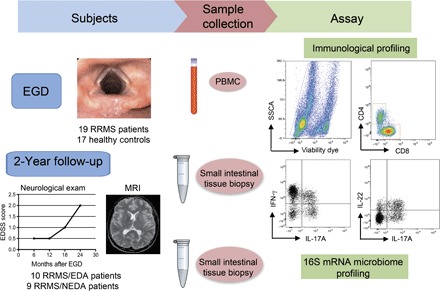
Tissue samples from the duodenal mucosa were collected during EGD from RRMS patients (n = 19) and HCs (n = 17). Simultaneously, we collected 6 to 7 ml of peripheral blood from the same individuals. Total lymphocytes were isolated from one to two fragments of intestinal tissue by collagenase digestion and from peripheral blood by Ficoll gradient. Mucosal immune cells and peripheral blood mononuclear cells (PBMCs) were FACS-analyzed for immunological profiling of the following TH cell subsets: IL-17A+CD4+ TH17 cells, IFN-γ+CD4+ TH1 cells, and IL-22+ TH22 cells. Microbial DNA was extracted from frozen intestinal tissue samples (seven RRMS and seven HC samples), and 16S rRNA sequencing was performed on the Roche 454 platform. At the 2-year end point, we stratified RRMS patients in two cohorts on the basis of evidence of disease activity by clinical and MRI criteria. RRMS patients with no evidence of disease activity in the 2-year follow-up were classified as RRMS/NEDA (n = 9), whereas RRMS patients with evidence of disease activity were classified as RRMS/EDA (n = 10). SSCA, side scatter FACS parameter.
Table 2. Clinical data.
All patients recruited in this study have a diagnosis of RRMS defined according to Poser’s diagnostic criteria. None of the RRMS patients received corticosteroids in the 3 months before the EGDs (time of intestinal biopsy collection). RRMS patients were divided into two subgroups on the basis of evidence of disease activity at 2-year follow-up: RRMS patients with no evidence of disease activity in the 2-year follow-up were classified as NEDA (n = 9), whereas RRMS patients with evidence of disease activity were classified as EDA (n = 10). All RRMS patients recruited in this study were undergoing different therapeutic immunomodulatory regimens (IFN-β, GA, and fingolimod) at the time of EGD and intestinal tissue sample analysis.
| MS patient ID | Sex | Age (years) | Disease activity (2-year follow-up) | EDSS | Current therapy |
| MS001 | F | 38 | NEDA | 3 | Fingolimod |
| MS002 | M | 49 | EDA | 3 | IFN-β |
| MS003 | F | 27 | EDA | 2 | IFN-β |
| MS004 | F | 49 | NEDA | 5.5 | GA |
| MS005 | F | 38 | EDA | 1.5 | IFN-β |
| MS006 | M | 42 | NEDA | 2 | GA |
| MS007 | M | 43 | EDA | 1.5 | GA |
| MS008 | M | 37 | EDA | 2 | Fingolimod |
| MS009 | M | 35 | EDA | 4 | GA |
| MS010 | M | 57 | EDA | 4 | GA |
| MS011 | F | 55 | NEDA | 1.5 | GA |
| MS012 | F | 26 | NEDA | 5 | GA |
| MS013 | F | 29 | NEDA | 1 | IFN-β |
| MS016 | M | 27 | NEDA | 1.5 | IFN-β |
| MS017 | F | 42 | NEDA | 1.5 | IFN-β |
| MS018 | F | 57 | EDA | 3.5 | Fingolimod |
| MS020 | F | 25 | EDA | 1 | GA |
| MS021 | F | 56 | NEDA | 2 | IFN-β |
| MS022 | M | 40 | EDA | 1 | GA |
Gut mucosal immunity in MS
Our multiparametric FACS analysis revealed a statistically significant increase in the frequencies of intestinal CD4+ T cells in RRMS patients compared to those in HCs (P < 0.01), whereas the frequency of total CD3+ T cells and that of CD8+ T cells were comparable between the two groups (Fig. 2A). Next, we examined the cytokine secretion profile of CD4+ TH cell subsets. Our data showed a statistically significant increase in the frequency of intestinal TH17 cells in RRMS patients compared to that of HCs (P < 0.01) (Fig. 2B). The higher frequency of TH17 cells in RRMS patients was detected in the small intestinal mucosa but not in the peripheral blood, where TH17 cells were poorly represented in both RRMS patients and HCs (Fig. 2B). The increase was limited exclusively to the TH17 cell subset, and the frequencies of other effector TH cell subsets, such as TH1 cells and TH22 cells, were comparable between the RRMS and HC groups (Fig. 2B).
Fig. 2. Intestinal T cell subsets in RRMS patients and HCs.
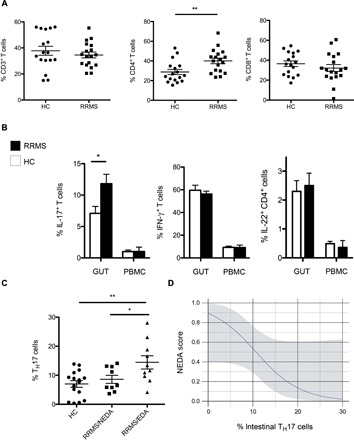
(A) Relative percentages of CD3+, CD4+, and CD8+ cells out of total lymphocytes isolated from the small intestinal mucosa of individuals with RRMS and of HCs. (B) Frequencies of different TH cell subsets (TH17, TH1, and TH22) in the intestinal mucosa (GUT) and PBMCs of HCs and RRMS patients. Blood samples were collected from the same individuals at the time of intestinal tissue sample collection. Data are means ± SEM. (C) RRMS patients were divided into two cohorts on the basis of disease activity. RRMS patients with evidence of disease activity at 2-year follow-up were classified as EDA, whereas patients with no disease activity were classified as NEDA. Percentages of intestinal TH17 cells out of total CD4+ T cells in HCs and RRMS/NEDA and RRMS/EDA patients are shown. (A to C) **P < 0.01 and *P < 0.05 by unpaired t test. (D) Logistic regression analysis showing correlation between TH17 cell percentages in the gut mucosa and predicted probability of showing disease activity at 2-year follow-up (NEDA score: RRMS/NEDA, 1; RRMS/EDA, 0). Odds ratio, 0.96; 95% CI, 0.02 to 0.98; P = 0.01.
Because our data revealed a bimodal trend in TH17 cell percentages in our RRMS patient cohort, we hypothesized that the TH17 cell frequency correlates with disease activity. To test this hypothesis, we divided our cohort of RRMS patients into two subgroups on the basis of the presence of disease activity at a 2-year follow-up, and we found that RRMS/NEDA patients with inactive disease had low percentages of intestinal TH17 cells, compared to those of HCs (Fig. 2C). Conversely, RRMS/EDA patients with active disease showed TH17 cell frequencies significantly augmented compared not only to HCs (P < 0.01) but also to RRMS/NEDA patients (P < 0.05) (Fig. 2C). A one standard deviation increase in the frequencies of TH17 in the intestine of RRMS patients was associated with a significant reduction in the predicted probability of being disease-free at 2 years [odds ratio, 0.21; 95% confidence interval (CI), 0.02 to 0.98; P = 0.01] (Fig. 2D). We also analyzed the percentages of double-positive IL-17+IFN-γ+ and IL-17+IL-22+ and found a statistically significant increase in the percentage of IL-17+IL-22+ T cells in the small intestine of RRMS/EDA patients with active disease compared to those in HCs, whereas the percentages of double-positive IL-17+IFN-γ+ were similar between the different groups (fig. S1).
At the time of EGD and intestinal immune profiling, all RRMS patients enrolled in this study were undergoing treatment with drugs that have strong immunomodulatory properties, such as interferon-β (IFN-β), GA, and fingolimod (Table 2). These immunomodulatory drugs can differentially modulate gut mucosal immunity. For example, recent studies demonstrated that type 1 interferons (T1IFNs), such as IFN-β play a beneficial anti-inflammatory role in the intestinal mucosa (18, 19). Hence, we determined whether administration of these immunomodulatory therapies specifically affects intestinal TH17 cell differentiation in our RRMS patients. We stratified our cohort of RRMS patients in three subgroups according to the type of treatment (IFN-β, GA, or fingolimod). Our analysis did not reveal statistically significant differences in the percentages of intestinal immune cell subsets between the three subgroups. However, we observed that the increase of intestinal TH17 cells was particularly evident in the GA-treated subgroup in comparison with HCs (fig. S2). We also conducted a correlative statistical analysis linking intestinal TH17 cell percentage to age, EDSS score (disability score), and disease duration but found no statistically significant association between these parameters (fig. S3).
Correlation between microbiota modifications, increased TH17 cell frequency and disease activity in RRMS
Next, we wanted to highlight the mechanism responsible for increased TH17 cell frequency in the small intestine of RRMS patients. Commensal microbiota play a crucial role in modulating intestinal TH17 cell expansion, and in EAE (the animal model of MS), gut colonization with SFB promotes brain autoimmunity by increasing intestinal TH17 cell differentiation (10). Hence, we determined whether enhanced TH17 cell differentiation in the small intestine of RRMS patients is linked with alterations in microbiota composition. TH17 cell–inducing microbes, such as SFB, are tightly associated with the epithelium and are scarce in the intestinal lumen and in stool samples (20, 21). Therefore, we analyzed the microbiota isolated from the small intestinal mucosa rather than from luminal contents (stool samples) to characterize bacterial strains that directly regulate TH17 cell differentiation (8, 9, 22, 23). Our taxonomic, functional, and diversity microbiome profiling did not show statistically significant differences in α-diversity either between HC and RRMS patients or between RRMS/EDA patients with disease activity and RRMS/NEDA patients with inactive disease (Fig. 3A). At the phylum level, again, we did not detect differences between HC and RRMS patients; but we did find a statistically significant higher abundance of Firmicutes and reduction of Bacteroidetes species in RRMS/EDA with disease activity compared to RRMS/NEDA patients (Fig. 3B). Genus-level analysis revealed that RRMS/EDA patients had higher representation of Streptococcus strains (Fig. 3C).
Fig. 3. Differences of gut mucosa-associated microbiota composition between RRMS patients and HCs.
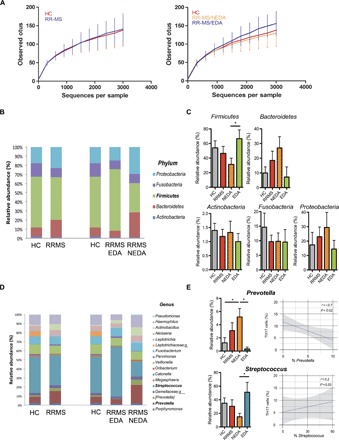
(A) α-Diversity of gut microbiota profiles assessed by 16S amplicon sequencing of DNA purified from human small intestinal tissue of HCs and RRMS patients with active (RRMS/EDA) and nonactive (RRMS/NEDA) disease. (B) Average phylum-level composition of microbiota isolated from small intestinal samples of HC and total RRMS, RRMS/NEDA, and RRMS/EDA patients. (C) Means ± SEM of the relative abundance of different phyla. *P < 0.05 by unpaired t test. (D) Average genus-level composition in four cohorts of individuals. (E) Left: Means ± SEM of the relative abundance of Prevotella and Streptococcus strains. *P < 0.05 by unpaired t test. Right: Regression linear plot in all individuals (RRMS + HC) showing inverse relation between percentages of intestinal TH17 cells and relative abundance of Prevotella strains.
A further characterization of the Streptococcus strains by amplification and sequencing revealed that they were mostly Streptococcus mitis and Streptococcus oralis, two bacterial strains that normally reside in the oral cavity (fig. S4). Notably, previous studies demonstrated that S. mitis is capable of inducing TH17 cell differentiation in humans (24). In addition, we found that RRMS/EDA patients had a statistically significant reduction in the relative abundance of Prevotella, commensal bacterial strains that have beneficial anti-inflammatory properties (25), in comparison with RRMS/NEDA patients (Fig. 3C). In RRMS/NEDA patients with inactive disease, the relative abundance of the anti-inflammatory Prevotella strains was higher in comparison not only with RRMS/EDA patients but also with HCs. A correlative statistical analysis confirmed a negative linear relationship between the frequency of intestinal TH17 cells and the relative abundance of Prevotella strains in all human small intestinal samples (r = −0.7, P ≤ 0.01), whereas no direct correlation between percentages of intestinal TH17 cells and relative abundance of Streptococcus strains was found (Fig. 3C). As for intestinal TH17 cell percentages, we also performed a correlative statistical analysis between the relative abundance of Prevotella and Streptococcus strains and age, EDSS score (disability score), and disease duration but found no statistically significant association among these parameters (fig. S5).
Treatment with immunomodulatory drugs in our RRMS cohort could also affect gut microbiota composition. In line with this idea, a recent study showed that treatment with GA modifies the microbiota composition in the stools of RRMS patients (12). Hence, we determined whether administration of different immunomodulatory drugs (IFN-β or GA) specifically modifies gut mucosa-associated microbiota in our RRMS patient cohort. Although our sample size was relatively small, our analysis revealed a statistically significant increase in the relative abundance of Prevotella strains in the IFN-β–treated subgroup compared with RRMS patients treated with GA (we could not perform a comparative analysis with fingolimod-treated patients because of the too-limited number of samples) (fig. S6). We detected no statistically significant difference in the relative abundance of Streptococcus strains (fig. S6) and no other differences at the family and genus levels between subgroups of RRMS patients stratified according to immunomodulatory treatment.
DISCUSSION
Modifications of the intestinal environment induced by either diet or microbiota composition have been linked to the pathogenesis of MS (12–16), but the mechanisms underlying this association are still largely unknown. In mice, a proinflammatory intestinal environment promotes brain autoimmunity by enhancing TH17 cell expansion and self-reactive T cell pathogenicity (4, 10). In these models, myelin-reactive T cells are driven toward a pathogenic TH17 phenotype in the small intestinal mucosa (11) and then migrate to the CNS where they mediate autoimmune damage of the myelin sheath. Our data confirm that brain autoimmunity is also associated in humans with intestinal expansion of TH17 cells. Although further investigation is needed to formally demonstrate that the TH17 cells residing in the small intestine of RRMS patients are myelin-specific, the selective TH17 cell expansion in RRMS/EDA patients with disease activity suggests that, similar to what is observed in mice with EAE, the gut environment enhances pathogenicity of self-reactive T cells by driving them toward a TH17 cell phenotype. In anti–MOG (myelin oligodendrocyte glycoprotein) T cell receptor transgenic mice that develop spontaneous EAE, the expansion of myelin reactive TH17 cells was detected in T cells residing in the intestinal wall (lamina propria T cells and in Peyer’s patches) but not in peripheral organs, such as spleen and lymph nodes (4). Our observation that increased TH17 cell frequency is present in the small intestinal mucosa but not in peripheral blood of RRMS patients further underlies the similarities between pathogenic TH17 cell differentiation in humans and in animal models of MS.
The acquisition of an IL-22+ phenotype by TH17 cells increased their effector function and proinflammatory properties (26, 27), and a recent study demonstrated that the presence of IL-22+ TH17 cells correlates with high disease activity in rheumatoid arthritis patients (28). Our data show that RRMS/EDA patients with active disease also have an increased percentage of double-positive IL-22+ TH17 cells. These results further suggest that increased differentiation and acquisition of a strong effector phenotype by TH17 cells in the small intestine correlate with disease activity in RRMS patients.
TH17 cell differentiation in the small intestine is regulated by the whole microbial community composition and also by some specific bacteria (29, 30). In rodents, a single bacterial type, the SFB, is sufficient to drive the accumulation of TH17 cells in the small intestinal lamina propria (8, 9). Studies in transgenic models of autoimmune arthritis (31) and EAE (10) demonstrated that SFB play a key role in the activation and expansion of self-reactive TH17 cells in the small intestine. Similarly, the TH17 cell expansion that we found in RRMS patients could be ascribed to overrepresentation of TH17-inducing commensal microbes that are present in the human small intestine. SFB have been found in multiple vertebrate species (32) and also in the human intestine (33). In a recent study, a functional analog of SFB, Bifidobacterium adolescentis, was isolated from the human intestine (21). In our human small intestinal samples, we did not detect SFB, and we found a limited amount of Bifidobacterium only in one intestinal tissue sample isolated from an RRMS patient. Hence, we exclude the possibility that increased TH17 cell frequency in RRMS was linked to excessive SFB or Bifidobacteria strain representation. However, TH17-promoting symbionts can be different in human populations residing in different geographical areas because of the effect of the environment on microbiota composition. In our human cohort, increased TH17 cell frequency in RRMS/EDA patients was associated with higher relative abundance of Streptococcus strains, in particular S. mitis and S. oralis. Previous studies showed that Streptococcus strains belonging to the S. mitis group (such as Streptococcus pneumoniae, S. mitis, and S. oralis) are capable of inducing TH17 cell differentiation in humans (24). S. mitis and S. oralis, the two Streptococcus strains that we found selectively increased in the small intestine of RRMS/EDA patients, are normally resident in the oral cavity, but both can cause local or invasive opportunistic infections (34). On the basis of our findings, we speculate that, under certain conditions, or because of still unknown virulence factors, these Streptococcus strains can colonize the small intestine and favor TH17 cell differentiation in the human gut mucosa. Notably, both Streptococcus strains can also grow under low-pH conditions, which are a prerequisite for colonizing the duodenum (35).
Enhanced TH17 cell differentiation in the intestine can be induced not only by overrepresentation of TH17-inducing bacteria but also by decreased abundance of beneficial microbial strains that reduce TH17 cell polarization. For example, Prevotella is capable of producing the anti-inflammatory metabolite propionate (36) that limits intestinal TH17 cell expansion in mice (37). So far, three of five studies that analyzed microbiome composition in RRMS patients from different geographical areas (Japan and United States) found a reduction in the relative abundance of Prevotella strains in MS patients compared to HC subjects (13, 14, 16). We confirmed these findings in our Italian cohort of RRMS patients and also found that the Prevotella decrease is selectively present in RRMS patients with active disease. In our human cohort, which includes not only RRMS patients but also HC subjects, we demonstrated a statistically significant negative correlation between the relative abundance of Prevotella and the TH17 cell frequency in the human small intestine. These data indicate that reduced Prevotella representation in RRMS patients is linked to TH17 cell expansion and disease activity. In RRMS/NEDA patients with inactive disease, the Prevotella strains were more abundant in comparison not only with RRMS/EDA patients with active disease but also with the HC group, as if overrepresentation of this beneficial anti-inflammatory strain represents a protective mechanism to counterbalance the effect of TH17 cell–inducing microbes and MS pathogenesis in these patients.
The current standard of practice in neurology is to treat RRMS patients with immunomodulatory drugs (IFN-β, GA, or fingolimod). Previous studies indicate that these immunomodulatory drugs can modulate gut mucosal immunity and microbiota composition. For example, IFN-β was reported to play a beneficial anti-inflammatory role in the murine intestine (18, 19), whereas GA can influence the microbiota profile in treated MS patients (12). Here, we demonstrate that the percentage of intestinal TH17 cells was particularly high in the GA-treated group that contained a high number of RRMS/EDA patients with active disease (5 out of 9). This finding further supports the notion that the percentage of intestinal TH17 cells correlates with disease activity in RRMS patients. We also analyzed the microbiota composition in RRMS patients stratified according to immunomodulatory therapy and found a higher increase in the relative abundance of Prevotella strains in the IFN-β–treated RRMS patients compared to the GA-treated cohort. Although our sample size was relatively small and we cannot make definitive conclusions on the capacity of IFN-β treatment to augment Prevotella strains in the human intestine, our preliminary data do suggest that the beneficial effect of IFN-β in RRMS may be related to modification of microbiota composition. Although a previous report showed several alterations in the microbiota composition in stool samples of GA-treated patients compared to untreated RRMS patients (12), our analysis on mucosa-associated microbiota did not reveal other significant differences in this subgroup. However, it must be noted that in our study, we compared GA-treated individuals only with RRMS patients treated with other immunomodulatory drugs (IFN-β) and not with untreated RRMS patients.
Our results point to a role of gut microbiota in regulating TH17 cell differentiation in RRMS patients; however, we cannot exclude the possibility that other environmental factors, such as diet composition, are also involved either directly or indirectly through microbiota modulation. High concentration of sodium chloride drives human and murine T cells toward a biased TH17 cell phenotype (38, 39), thus leading to the hypothesis that TH17 cell differentiation in the human intestine is regulated by dietary consumption of salt. We administered a dietary questionnaire to our RRMS patients and HC group; although we did not detect relevant differences in the salt intake between patients and controls, we cannot rule out the possibility that the quantity of dietary salt influences TH17 cell differentiation in RRMS patients.
Although modifications of the dietary habits have been held responsible for the marked spike in incidence of autoimmune diseases, the so-called “autoimmune epidemic” that has occurred in the last half-century (40, 41), it is yet to be clarified how diet modulates autoimmune diseases such as MS (42). Dietary factors can affect brain autoimmunity by favoring TH17 cell differentiation in the intestine acting directly on myelin-reactive T cells (38, 39) or through microbiota modifications (4). Our results show that increased TH17 cell frequency and microbiota modifications are not present in all RRMS patients but only in those patients with active disease. This strongly suggests that TH17 cell expansion and high-Streptococcus/low-Prevotella strain representation in the small intestine are associated with recurrence of myelin-reactive T cell activation and RRMS pathogenesis. Beneficial Prevotella strains are enriched in the intestine of children fed with a high-fiber/low-fat diet (25), thus indicating that modifications of the dietary habits in RRMS patients could promote a beneficial Prevotella-enriched gut microbiota that reduces TH17 cell differentiation and disease activity. Our data provide the first evidence that alterations of gut mucosal immunity induced by specific microbiota composition are associated with brain autoimmune disease in humans and could pave the way to new therapeutic strategies in RRMS aimed at regulating intestinal TH17 cell expansion through dietary approaches and/or probiotic administration.
MATERIALS AND METHODS
Study population
RRMS patients were recruited from the Center for Diagnosis and Treatment of Multiple Sclerosis of the San Raffaele Hospital (IRCCS San Raffaele Scientific Institute, Milan, Italy). RRMS patients and HC subjects (sex- and age-matched with RRMS patients) included in this study were adult male and female individuals who performed EGD for diagnostic purposes (dyspepsia, gastroesophageal reflux, altered bowel habits, etc.). Exclusion criteria for both RRMS patients and HCs were as follows: no antibiotic and corticosteroid treatment in the 3 months before EGD; no history of gastroenteritis, gastric ulcer, irritable bowel disease, celiac disease, inflammatory bowel disease, and gastric and colorectal cancers. None of the RRMS patients had an active clinical relapse at the time of tissue sampling and were all treated with an immunomodulatory drug (IFN-β, GA, or fingolimod) but not with immunosuppressive medications, such as axathioprine, mitoxantrone, and methotrexate. All individuals signed a written informed consent, complied with the study procedure, and were aware that they donated intestinal tissue fragments for research purpose. The study was approved by the Institutional Review Board of the IRCCS San Raffaele Scientific Institute (Ethical Committee Board), and all participants signed an informed consent before any data collection or study procedure.
Intestinal tissue sample collection
All EGDs were performed at the Gastroenterology Unit of the San Raffaele Hospital. Tissue fragments of duodenal mucosa (one to two from each individual) obtained during EGD were collected in 10% RPMI [RPMI 1640 containing 10% fetal bovine serum, penicillin/streptomycin (100 μg/ml), 2 mM glutamine, 10 mM Hepes, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, and 0.05 mM β-mercaptoethanol) and rapidly processed. Mucus and epithelial cells were removed from the tissue by incubation with 1 mM dithiothreitol and 5 mM EDTA in calcium- and magnesium-free Hanks’ balanced salt solution (HBSS) for 15 min at 37°C. Tissue samples were washed with HBSS; digested with collagenase A (1 mg/ml) (Roche Diagnostics Ltd.) in HBSS supplemented with calcium, magnesium, and deoxyribonuclease I (5 U/ml) (Roche Diagnostics Ltd.) for 1 hour at 37°C; and mechanically disrupted by gentle pipetting until dissociation was complete. One intestinal tissue sample was collected in RNAlater and immediately frozen at −80°C until DNA extraction for microbiota analysis.
Multiparametric FACS analysis
Single-cell suspensions isolated from intestinal tissue samples were passed through a 70-μm cell strainer and washed with HBSS. The number of total cells isolated from intestinal tissue samples ranged from 0.5 × 106 to 3.7 × 106, with a number of T cells (from 1 × 105 to 11 × 105) that were comparable between RRMS patients (5.7 × 105 ± 0.8 × 105) and HC (5.2 × 105 ± 0.7 × 105). PBMCs were isolated by Ficoll gradient and washed in complete RPMI 10. Intestinal cells and PBMCs were assessed for viability by Fixable Viability Dye eFluor 506 (eBioscience) and stained with APC-Cy7 (allophycocyanin-Cy7)–conjugated anti-human CD3 (clone SP34-2, BD Biosciences), PE (phycoerythrin)/Texas Red–conjugated anti-human CD4 (clone S3.5, Thermo Fisher), and Alexa Fluor 700–conjugated anti-human CD8 (clone OKT8, eBioscience) monoclonal antibodies. To stain for intracellular cytokines, cells were first stained with the monoclonal antibodies against surface molecules, fixed and permeabilized (BD Cytofix/Cytoperm solution kit, BD Biosciences), and then stained with eFluor 450–conjugated anti-human IFN-γ (clone 4S.B3, eBioscience), APC-conjugated anti-human IL-17A (clone eBio64DEC17, eBioscience), and PE-conjugated anti-human IL-22 (clone 142928, R&D Systems). Data were acquired using a four-laser cell analyzer, LSRFortessa (BD Biosciences), and analyzed by using FACSDiva software (BD Biosciences).
Microbiome analysis
The human microbiome analysis was performed by 16S amplicon sequencing on the 454-GS Junior platform (Roche). Total DNA was purified from human intestinal tissue biopsies of RRMS patients and HCs using QIAamp DNA Micro Kit (Qiagen), following the manufacturer’s instructions. The V3-V5 region of the 16S rRNA gene was amplified starting from 500 ng of extracted DNA using the FastStart High Fidelity PCR System (Roche), barcoded sample-specific primers [16S-F331: ACTCCTACGGGAGGCAGC and 16S-R920: CCGTCAATTCMTTTGAGTTT] under cycling conditions (95°C for 3 min; 45 cycles of 95°C for 30 s, 55°C for 45 s, and 72°C for 1 min; and 72°C for 8 min), and then stored at 4°C until usage. Amplicons were loaded on 1% agarose gel and purified with QIAquick Gel Extraction Kit (Qiagen). Extracted amplicons were purified with AMPure XP beads (Beckman Coulter) and used for emulsion polymerase chain reaction and ultradeep pyrosequencing, following the 454-GS Junior manufacturer’s instruction. Sequences with a high-quality score and a length of >250 bp were used for the taxonomic analysis with QIIME (Quantitative Insights Into Microbial Ecology version 1.9.0) software. The number of observed operational taxonomic units (OTUs) was calculated to determine the α-diversity. For all samples, at least 3000 sequences and a coverage rate of >99% were obtained.
The OTUs were identified using the UCLUST clustering method (43). Taxonomy to each sequence was assigned using the RDP Classifier Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy, which assigns taxonomies using Naïve Bayes classification (44). The chimeric sequences were identified with ChimeraSlayer (http://microbiomeutil.sourceforge.net/) and filtered from the sequences of an OTU file. The identification of Streptococcus species was performed, as previously described with minor modifications, by amplification and sequencing of the bacterial tuf gene from small intestinal samples (45, 46).
Statistical analyses
Statistical differences were calculated with PRISM 5.0 software (GraphPad) using unpaired, two-tailed t test. Logistic regression models adjusted for age, sex, ongoing MS therapy at the EGD, and previous immunosuppressant use were used to assess the association between gut TH17 cell percentages and subsequent disease activity at 2 years. Data were considered statistically significant when P values were ≤0.05.
Supplementary Material
Acknowledgments
We thank R. Furlan (Clinical Neuroimmunology Unit of the IRCCS San Raffaele Scientific Institute) for critical reading of the manuscript and all the participants who agreed to donate intestinal mucosa samples for research purposes. Funding: This work was supported by a research grant from the Italian Foundation for Multiple Sclerosis (Fondazione Italiana Sclerosi Multipla) to M.F. Author contributions: I.C., C.S., and J.D. performed all cellular immunology experiments. G.D.-C., M.J.M., V.M., and G.C. designed the cohort study, recruited MS patients, and performed neurological and MRI evaluations. G.D.-C. performed statistical analysis. E.R., A.M., and P.A.T. performed EGD and collected intestinal tissue samples. R.F. and F.C. performed microbiome analysis. V.M. and M.F. served as principal investigators. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/7/e1700492/DC1
fig. S1. Percentages of double-positive IL-17+IFN-γ+ and IL-17+IL-22+ T cells in the intestinal mucosa of HCs and RRMS patients with active disease (RRMS/EDA) or inactive disease (RRMS/NEDA).
fig. S2. Percentages of intestinal TH cell subsets in RRMS patients stratified according to immunomodulatory treatment.
fig. S3. Correlative statistical analysis between percentages of intestinal TH17 cells and age, EDSS score, and disease duration.
fig. S4. Characterization of Streptococcus species by amplification and sequencing.
fig. S5. Correlative analysis of relative abundance of Prevotella and Streptococcus strains in mucosa-associated microbiota and age, EDSS score, and disease duration.
fig. S6. Relative abundance of Prevotella and Streptococcus strains in HCs and RRMS patients stratified according to immunomodulatory treatment.
REFERENCES AND NOTES
- 1.Nylander A., Hafler D. A., Multiple sclerosis. J. Clin. Invest. 122, 1180–1188 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pette M., Fujita K., Wilkinson D., Altmann D. M., Trowsdale J., Giegerich G., Hinkkanen A., Epplen J. T., Kappos L., Wekerle H., Myelin autoreactivity in multiple sclerosis: Recognition of myelin basic protein in the context of HLA-DR2 products by T lymphocytes of multiple-sclerosis patients and healthy donors. Proc. Natl. Acad. Sci. U.S.A. 87, 7968–7972 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Odoardi F., Sie C., Streyl K., Ulaganathan V. K., Schläger C., Lodygin D., Heckelsmiller K., Nietfeld W., Ellwart J., Klinkert W. E. F., Lottaz C., Nosov M., Brinkmann V., Spang R., Lehrach H., Vingron M., Wekerle H., Flügel-Koch C., Flügel A., T cells become licensed in the lung to enter the central nervous system. Nature 488, 675–679 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Berer K., Mues M., Koutrolos M., Al Rasbi Z., Boziki M., Johner C., Wekerle H., Krishnamoorthy G., Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 479, 538–541 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Korn T., Anderson A. C., Bettelli E., Oukka M., The dynamics of effector T cells and Foxp3+ regulatory T cells in the promotion and regulation of autoimmune encephalomyelitis. J. Neuroimmunol. 191, 51–60 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kebir H., Kreymborg K., Ifergan I., Dodelet-Devillers A., Cayrol R., Bernard M., Giuliani F., Arbour N., Becher B., Prat A., Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat. Med. 13, 1173–1175 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lock C., Hermans G., Pedotti R., Brendolan A., Schadt E., Garren H., Langer-Gould A., Strober S., Cannella B., Allard J., Klonowski P., Austin A., Lad N., Kaminski N., Galli S. J., Oksenberg J. R., Raine C. S., Heller R., Steinman L., Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat. Med. 8, 500–508 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Ivanov I. I., Atarashi K., Manel N., Brodie E. L., Shima T., Karaoz U., Wei D., Goldfarb K. C., Santee C. A., Lynch S. V., Tanoue T., Imaoka A., Itoh K., Takeda K., Umesaki Y., Honda K., Littman D. R., Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaboriau-Routhiau V., Rakotobe S., Lécuyer E., Mulder I., Lan A., Bridonneau C., Rochet V., Pisi A., De Paepe M., Brandi G., Eberl G., Snel J., Kelly D., Cerf-Bensussan N., The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Lee Y. K., Menezes J. S., Umesaki Y., Mazmanian S. K., Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U.S.A. 108 (suppl. 1), 4615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esplugues E., Huber S., Gagliani N., Hauser A. E., Town T., Wan Y. Y., O’Connor W., Rongvaux A., Van Rooijen N., Haberman A. M., Iwakura Y., Kuchroo V. K., Kolls J. K., Bluestone J. A., Herold K. C., Flavell R. A., Control of TH17 cells occurs in the small intestine. Nature 475, 514–518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantarel B. L., Waubant E., Chehoud C., Kuczynski J., DeSantis T. Z., Warrington J., Venkatesan A., Fraser C. M., Mowry E. M., Gut microbiota in multiple sclerosis: Possible influence of immunomodulators. J. Invest. Med. 63, 729–734 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyake S., Kim S., Suda W., Oshima K., Nakamura M., Matsuoka T., Chihara N., Tomita A., Sato W., Kim S.-W., Morita H., Hattori M., Yamamura T., Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV clusters. PLOS ONE 10, e0137429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jangi S., Gandhi R., Cox L. M., Li N., von Glehn F., Yan R., Patel B., Antonietta Mazzola M., Liu S., Glanz B. L., Cook S., Tankou S., Stuart F., Melo K., Nejad P., Smith K., Topçuolu B. D., Holden J., Kivisäkk P., Chitnis T., De Jager P. L., Quintana F. J., Gerber G. K., Bry L., Weiner H. L., Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 7, 12015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tremlett H., Fadrosh D. W., Faruqi A. A., Hart J., Roalstad S., Graves J., Spencer C. M., Lynch S. V., Zamvil S. S., Waubant E. US Network of Pediatric MS Centers , Associations between the gut microbiota and host immune markers in pediatric multiple sclerosis and controls. BMC Neurol. 16, 182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J., Chia N., Kalari K. R., Yao J. Z., Novotna M., Mateo Paz Soldan M., Luckey D. H., Marietta E. V., Jeraldo P. R., Chen X., Weinshenker B. G., Rodriguez M., Kantarci O. H., Nelson H., Murray J. A., Mangalam A. K., Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci. Rep. 6, 28484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Havrdova E., Galetta S., Stefoski D., Comi G., Freedom from disease activity in multiple sclerosis. Neurology 74 (suppl. 3), S3–S7 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Katakura K., Lee J., Rachmilewitz D., Li G., Eckmann L., Raz E., Toll-like receptor 9-induced type I IFN protects mice from experimental colitis. J. Clin. Invest. 115, 695–702 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J.-Y., Kim M.-S., Kim E., Hee Cheon J., Lee Y.-S., Kim Y., Lee S.-H., Seo S.-U., Shin S.-H., Shim Choi S., Kim B., Chang S.-Y., Ko H.-J., Bae J.-W., Kweon M.-N., Enteric viruses ameliorate gut inflammation via toll-like receptor 3 and toll-like receptor 7-mediated interferon-β production. Immunity 44, 889–900 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Klaasen H. L. B. M., Koopman J. P., Poelma F. G. J., Beynen A. C., Intestinal, segmented, filamentous bacteria. FEMS Microbiol. Rev. 8, 165–180 (1992). [DOI] [PubMed] [Google Scholar]
- 21.Guan Tan T., Sefik E., Geva-Zatorsky N., Kua L., Naskar D., Teng F., Pasman L., Ortiz-Lopez A., Jupp R., Wu H.-J. J., Kasper D. L., Benoist C., Mathis D., Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. U.S.A. 113, E8141–E8150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atarashi K., Tanoue T., Ando M., Kamada N., Nagano Y., Narushima S., Suda W., Imaoka A., Setoyama H., Nagamori T., Ishikawa E., Shima T., Hara T., Kado S., Jinnohara T., Ohno H., Kondo T., Toyooka K., Watanabe E., Yokoyama S.-i., Tokoro S., Mori H., Noguchi Y., Morita H., Ivanov I. I., Sugiyama T., Nuñez G., Camp J. G., Hattori M., Umesaki Y., Honda K., Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Assa A., Butcher J., Li J., Elkadri A., Sherman P. M., Muise A. M., Stintzi A., Mack D., Mucosa-associated ileal microbiota in new-onset pediatric Crohn’s disease. Inflamm. Bowel Dis. 22, 1533–1539 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Engen S. A., Rukke H. V., Becattini S., Jarrossay D., Blix I. J., Petersen F. C., Sallusto F., Schenck K., The oral commensal Streptococcus mitis shows a mixed memory Th cell signature that is similar to and cross-reactive with Streptococcus pneumoniae. PLOS ONE 9, e104306 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Filippo C., Cavalieri D., Di Paola M., Ramazzotti M., Poullet J. B., Massart S., Collini S., Pieraccini G., Lionetti P., Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U.S.A. 107, 14691–14696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y., Awasthi A., Yosef N., Quintana F. J., Xiao S., Peters A., Wu C., Kleinewietfeld M., Kunder S., Hafler D. A., Sobel R. A., Regev A., Kuchroo V. K., Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13, 991–999 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghoreschi K., Laurence A., Yang X.-P., Tato C. M., McGeachy M. J., Konkel J. E., Ramos H. L., Wei L., Davidson T. S., Bouladoux N., Grainger J. R., Chen Q., Kanno Y., Watford W. T., Sun H.-W., Eberl G., Shevach E. M., Belkaid Y., Cua D. J., Chen W. J., O’Shea J. J., Generation of pathogenic TH17 cells in the absence of TGF-β signalling. Nature 467, 967–971 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jimeno R., Leceta J., Garín M., Ortiz A. M., Mellado M., Rodríguez-Frade J. M., Martínez C., Pérez-García S., Gomariz R. P., Juarranz Y., Th17 polarization of memory Th cells in early arthritis: The vasoactive intestinal peptide effect. J. Leukoc. Biol. 98, 257–269 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Ivanov I. I., de Llanos Frutos R., Manel N., Yoshinaga K., Rifkin D. B., Sartor R. B., Finlay B. B., Littman D. R., Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanabe S., The effect of probiotics and gut microbiota on Th17 cells. Int. Rev. Immunol. 32, 511–525 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Kriegel M. A., Sefik E., Hill J. A., Wu H.-J., Benoist C., Mathis D., Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci. U.S.A. 108, 11548–11553 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klaasen H. L. B. M., Koopman J. P., Van Den Brink M. E., Bakker M. H., Poelma F. G. J., Beynen A. C., Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Lab. Anim. 27, 141–150 (1993). [DOI] [PubMed] [Google Scholar]
- 33.Yin Y., Wang Y., Zhu L., Liu W., Liao N., Jiang M., Zhu B., Yu H. D., Xiang C., Wang X., Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J. 7, 615–621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willenborg J., Goethe R., Metabolic traits of pathogenic streptococci. FEBS Lett. 590, 3905–3919 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Wilkins J. C., Beighton D., Homer K. A., Effect of acidic pH on expression of surface-associated proteins of Streptococcus oralis. Appl. Environ. Microbiol. 69, 5290–5296 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwiertz A., Taras D., Schäfer K., Beijer S., Bos N. A., Donus C., Hardt P. D., Microbiota and SCFA in lean and overweight healthy subjects. Obesity 18, 190–195 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Li J., Sung C. Y. J., Lee N., Ni Y., Pihlajamäki J., Panagiotou G., El-Nezami H., Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc. Natl. Acad. Sci. U.S.A. 113, E1306–E1315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleinewietfeld M., Manzel A., Titze J., Kvakan H., Yosef N., Linker R. A., Muller D. N., Hafler D. A., Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature 496, 518–522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu C., Yosef N., Thalhamer T., Zhu C., Xiao S., Kishi Y., Regev A., Kuchroo V. K., Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature 496, 513–517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bach J.-F., The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 347, 911–920 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Lauer K., Diet and multiple sclerosis. Neurology 49 (suppl. 2), S55–S61 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Brenton J. N., Goldman M. D., A study of dietary modification: Perceptions and attitudes of patients with multiple sclerosis. Mult. Scler. Relat. Disord. 8, 54–57 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Kuczynski J., Stombaugh J., Walters W. A., González A., Caporaso J. G., Knight R., Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinformatics 10.1002/0471250953.bi1007s36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q., Garrity G. M., Tiedje J. M., Cole J. R., Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Picard F. J., Ke D., Boudreau D. K., Boissinot M., Huletsky A., Richard D., Ouellette M., Roy P. H., Bergeron M. G., Use of tuf sequences for genus-specific PCR detection and phylogenetic analysis of 28 streptococcal species. J. Clin. Microbiol. 42, 3686–3695 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X., Xing J., Li B., Wang P., Liu J., Use of tuf as a target for sequence-based identification of Gram-positive cocci of the genus Enterococcus, Streptococcus, coagulase-negative Staphylococcus, and Lactococcus. Ann. Clin. Microbiol. Antimicrob. 11, 31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/7/e1700492/DC1
fig. S1. Percentages of double-positive IL-17+IFN-γ+ and IL-17+IL-22+ T cells in the intestinal mucosa of HCs and RRMS patients with active disease (RRMS/EDA) or inactive disease (RRMS/NEDA).
fig. S2. Percentages of intestinal TH cell subsets in RRMS patients stratified according to immunomodulatory treatment.
fig. S3. Correlative statistical analysis between percentages of intestinal TH17 cells and age, EDSS score, and disease duration.
fig. S4. Characterization of Streptococcus species by amplification and sequencing.
fig. S5. Correlative analysis of relative abundance of Prevotella and Streptococcus strains in mucosa-associated microbiota and age, EDSS score, and disease duration.
fig. S6. Relative abundance of Prevotella and Streptococcus strains in HCs and RRMS patients stratified according to immunomodulatory treatment.


