Abstract
A key goal of modernmedicine is target-specific therapeutic intervention.However, most drugs lackselectivity, resulting in“off-target” side effects.To address the requirements of “targeted therapy,”aptamers, which are artificial oligonucleotides,have beenusedas novel targeting ligands to construct aptamer drug conjugates (ApDC)that can specifically bind to a broad spectrum of targets, including diseased cells. Accordingly, the applicationof aptamers in targeted drug delivery has attracted broad interest due to their impressive selectivity and affinity, low immunogenicity, easy synthesis with high reproducibility, facile modification, and relatively rapid tissue penetration with no toxicity. Functionally, aptamers themselves can be used as macromolecular drugs, and they are also commonly used in biomarker discovery and targeted drug delivery. In this review,we will highlight the most recent advances in the development of aptamers and aptamer conjugates, and discuss their potential in targeted therapy.
Keywords: aptamer, ApDC, cell-SELEX, conjugate, nanomaterials, targeted drug delivery
Graphical abstract
1. Introduction
Off-target drug delivery, especially in the cases of anticancer drugs, can cause serious side effects to normal tissues and organs. Moreover, the resulting suboptimal dosage in diseased cells may lead to inefficient therapeutic efficacy and even cause drug resistance. Targeted drug delivery, which can selectively and efficiently address chemotherapeutics to diseased sites,isexpected to significantly reduce systematic toxicity and improve therapeutic effect.1. Indeed, thus far, hundreds of approaches for targeted drug delivery have been developed, and some have been approved by the U.S. Food and Drug Administration (FDA) for cancer treatment1, 2. For example, OKT3 (muromonab-CD3), an IgG2a CD3-specific transplant rejection drug, was the first FDA-approved therapeutic antibody (Hooks et al., 1991)3.Since then, antibodies have been widely used in targeted drug delivery. However, there are stilla number of limitations associated withantibody-based targeted therapy, includingtheirlarge size, high productioncost, high batch-to-batch variation, high immunogenicity, low thermal stability, and complicated modifications4-7.
Aptamers were first reported in 19908. They are artificialoligonucleotidesgenerated by an in vitroselection technique called Systematic Evolution of Ligands by Exponential Enrichment (SELEX). As demonstrated, their binding affinities arecomparable or superior tothose of most antibodies. In addition, aptamers have further advantages over antibodies, such as smallsize, low production cost, facile chemical modification, low immunogenicity, lowbatch-to-batch variation, high chemical stability, rapid tissue penetration, and no toxicity9-17. Because of these advantages, aptamers have attracted tremendous attention in the areas of biosensing, imaging and drug delivery.10, 18-22 To date, numerous high-affinity aptamers have been selected for a broad range of target molecules, including metal ions, small molecules, peptides, proteins, and even whole cells or viruses23-30. Recently,theTan group used whole cells as targetsanddeveloped a simple and efficient cell-based aptamer selection strategy called cell-SELEX31. This process enables selection of aptamersagainst native molecular signatures present on the membrane surface of target cells. Taken together, aptamers selected via cell-SELEX have greatpotential as highly specific ligandsfor targeted drug delivery. In this review, we will highlight the most recent advances in the development of aptamers and aptamer conjugates and discuss their potential in targetedtherapies.
2. Aptamerselectionand Cell-SELEX
2.1 Advantages of aptamers compared with antibodies
It has been nearly three decades since scientists first reported Systematic Evolution of Ligands by Exponential Enrichment (SELEX), a process forin vitro selection of aptamers against targets of interest. The term “aptamer” comes from the Latin aptus,orfit, and Greekmeros, or part, which suggests the lock-and-key relationship between target and aptamer. More specifically, aptamers areRNA or DNA oligomers that spontaneously fold into specific three-dimensional conformations that can bind defined targets with high affinity and specificity. Compared to antibodies, these nucleic acid moleculesare easily engineered by a fully controlled synthesis process and, hence, are much more cost-effective to produce. As such, aptamers have been introduced as core elements in research disciplines ranging from materials science to biomedicine, particularly, targeted drug delivery32-35.
2.1.1 Controllable and cost-effectivesynthesis
Because aptamers can be chemically producedin vitro, selection protocols can be controlled and conveniently adjusted in a test tube on demand.Because of their thermal and chemical stability, aptamers can be synthesized with no need to apply physiological temperature or buffers, whereas antibodies, as proteins, are prone to contamination and loss of biological activity under unstable experimental conditions. Moreover, antibodies often suffer from batch-to-batch variation and canbe denatured or easily degraded under poor storage or transport conditions. On the other hand, chemically synthesized aptamers are temperature-resistant and can tolerate transport with no restrictive requirements for cooling.This eliminates the need for a continuous cold chain and reduces the cost of long-term storage or transport of aptamers. In brief, aptamers can be readily synthesized in large quantities and atlow cost, whereas antibody production is relatively laborious and requires live cell screening.
2.1.2 Non-immunogenic and nontoxic withbroadtarget selection
Aptamers havelow immunogenicity and notoxicityin vivo. Compared with antibodies, aptamers with smaller size (∼30 kDa) cansurpassantigen-antibody recognition, and they have been found to recognize the IgG Fc region in mouse36. In contrast to antibodies, aptamers can bind to hundreds of molecules of different sizes and structures without compromising immune response. To date, many aptamer targets have been identified, including cocaine37, 38, growth factors39-41, peptides42, 43, toxins44, 45, viral proteins46, 47, even live cells and tissues48-51, most of whichwereperformed successfully under controlled aptamer selection and translated into clinical practice.
2.1.3 Effective penetration
Based on their small size, aptamers can penetrate tissue barriers smoothly and be internalized, thus facilitating their use in biosensing and drug delivery. For example, as an alternative to epithelial cell adhesion molecule (EpCAM) antibody, Pu etal.51developed a time- and cost-saving molecular tool for the diagnosis ofcancers of epithelial origin using a DNA-based EpCAM aptamer, SYL3C.Ristau et al.52demonstrated an EGFR aptamer that could recognize twice as many epitopes as its antibody counterpart, while aprostate-specific membrane antigen (PSMA) aptamer identifiedahundred-foldmore epitopes compared with the PSMA antibody.
2.2 SELEX and Cell-SELEX
Conventional SELEX is a well-controlled and efficient technology,involvingmultiple selection rounds of exponential amplification and enrichment for the screening of oligonucleotides with high affinities for their targets from random-sequence libraries. The total size of the library can range from 1014 to 1015, providing for a wide range of 3-dimensional folding structures 53. As shown in Figure 1, the SELEX procedure starts with the design of pools of nucleic acid sequences containing a randomized central core30-50nt long flanked by two conserved primer binding sites for enzymatic pool replication. Therandom sequences in the initial pool will fold into uniquesecondary and tertiary structures, some of whichcan form aptamer-target complexes. In the following step, the binding sequences are eluted and amplified by PCR (DNA aptamers) or RT-PCR (RNA aptamers), and reaction products are used as a new aptamer subpool for the next round of selection. These steps represent a single round of SELEX, but to obtain aptamers with high affinity and specificity, 7 to 20 iterative rounds are generally required.
Figure 1.
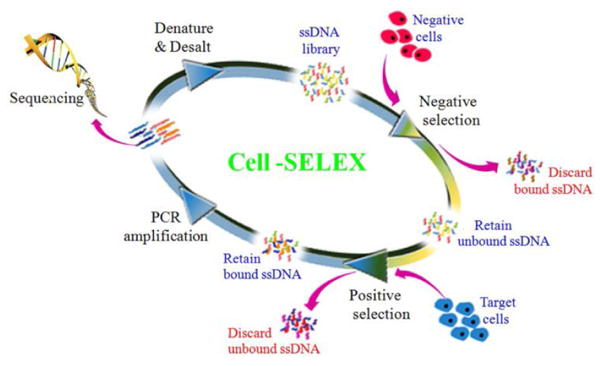
Schematic illustration of the cell-SELEX process (Reproduced with permission from Reference59).
Although the process has been developed over the course of many years, conventional SELEX still remainstime- and labor-intensive. In addition, some SELEX-selected aptamers fail to recognize their targets as expected. However,improvements have been made using classical techniques for aptamer selection, such as magnetic bead-based SELEX54, capillary electrophoresis-SELEX55,56, automated SELEX57,and cell-SELEX58, 59. Suchtechniques have been able to immobilize SELEX targets, facilitate the capture of the targets, or even manipulate time-consuming repetitive cycles automatically during selection.It has been a challenge to select aptamers able to recognize a cognate target ligand in its native conformation. To address this, the Tan grouprecently developed a modified SELEX technology termed Cell-SELEX, which uses whole living cells, including myeloid leukemia, lymphocytic leukemia, liver cancer, small-cell and non-small cell lung cancer, as targets60-62.This allows aptamer recognition of a target molecule in its native conformation with corresponding translational value and clinical application.
Moreover, cell-SELEX has brought the scientific community closer to realizing preferential binding to novel biomarkersand transport of drug payloads to disease cells. For instance,theTan group recently developed a truncated DNA aptamer termed XQ-2d59, with high affinity and specificity for pancreatic ductal adenocarcinoma (PDAC), and applied it to in vivo imaging and clinical tissue recognition. Previously, they identifiedthe target of aptamer TOV6 to be a cell-surface membrane receptor, stress-induced phosphoprotein 1 (STIP1)63,associated with poor survival outcome in epithelial ovarian cancer (EOC). They also found that aptamer TD05 targeted Immunoglobulin Heavy mu chain (IGHM) associated with Burkitt's lymphoma (American)(Ramos cells)64. In all, more and more aptamers selected through Cell-SELEX are advancing the potential for early diagnosis and imaging, as well as targeted therapy.
3. Aptamersasmacromoleculardrugs
The development of monoclonal antibodies is currently driving the targeted therapy revolution. However, aptamers have also been utilized as macromolecular drugs. Sullenger et al.65first found that nucleic acidscould prevent the activation of viral gene expression by overexpressinga trans-activation response decoy in host cells,resulting in the inhibition of viral replication. Since then, pegaptanib (Macugen, Pfizer)was approved by the FDA in 2004 as the first therapeutic aptamer for anti-VEGF treatment of neovascular age-related macular degeneration66. In addition to Macugen, abroad array of aptamers has beendesigned to inhibit or activate their targets in order to affect downstream signaling, thereby making them potentially useful as pharmaceutical or therapeutic agents in cancers. Notably, aptamer AS141167, which is currently in phase II clinical trials,can recognize a BCL-2 mRNA binding protein,nucleolin, associated with acute myelogenous leukemia (AML). Upon binding, AS1411 can beimmediately internalized,disruptingintracellular pathways andinhibiting cancer cell proliferation.Aptamersarealso considered good anticoagulant agents, as determined by Dobrovolsky et al. who developed DNA aptamers against thrombin to preventthrombin-induced clotting and platelet cell aggregation67. Anticoagulant aptamers are active against thrombin, prothrombin, coagulation factor VII, Factor IX, Factor X, and von Willebrand factor(vWF)68-74.So far, many of the aptamers usedasanticoagulants are in the first phase of clinical trials.
Apart from aptamers in clinical trials or in clinical treatment, as noted above,developmental work is underwayto perfect more aptamer-based drugs. For instance, a DNA aptamer, termed RA10-6, efficiently blocks IL-17 binding to IL-17RA in a dose-dependent mannerin vitro75. The injection of RA10-6 to an experimental mouse model of osteoarthritis resultedin thereduction ofIL-6 levels and synovial thickening, revealingRA10-6 as a potent adjunctive agent for the early treatment of osteoarthritis. Another study focused on advanced glycation end products (AGEs) and their receptor (RAGE), both of which play important roles in diabetic complications, such as nephropathy, retinopathy and neuropathy. According to the report, these diabetic complications result from inflammatory reactions activated by AGEs. Kaidaet al. established an animal model of type 2 diabetes with renal injury KKAy/Ta mice76. They detected increased urinary albumin and 8-hydroxy-2′-deoxy-guanosine levels inthese mice, as well as glomerular hypertrophy and enhanced extracellular matrix accumulation. However, the AGEs-aptamer was able to arrest experimental diabetic nephropathy in this mouse model.Thus, such aptamers with efficacy against cell proliferation, metabolic dysregulation,inflammation and coagulation are likely to make significant contributions to the treatment of various diseases in the near future.
4. Aptamer-drugconjugatesin targeted drug delivery
Aptamers linked toanticancer drugs have enabled the selective delivery of therapeutic compounds to diseased sites.In the section below, we cite representative examples of direct aptamer-drug conjugationbya chemical covalent linker or physical intercalation.
4.1 Covalent conjugation
Covalent conjugation between aptamers and chemotherapeutics dates back to an early experiment of Huang et al. using DNA aptamer sgc8, which specifically targetsprotein tyrosine kinase 7 (PTK7) overexpressed on human T-cell acute lymphoblastic leukemia (T-ALL) fromtheCCRF-CEM cell line77. Cleavage of a sgc8-Doxorubicin (Dox) conjugate was evoked in an acidic environment with pH 4.5–5.5 in order to control release of Dox. Tests of cell viability in vitro demonstrated that the sgc8c-Doxconjugate was potent in lowering toxicity towards nontarget cells compared with the unconjugated parent Dox(Figure 2). Nevertheless, this strategydid present some flaws,such as low copy number of drugs conjugated onto each aptamer. In response, Boyaciogul et al.78 synthesized a novel dimeric aptamer complex (DAC) for high-capacity targeted drug delivery. More recently,Wang et al.79proposed and synthesized a more efficient strategy that not only enhanced the drug payload capacity of aptamer-drug conjugates, but also provided spatiotemporal controllability of intracellular drug release.A frequently prescribed anticancer drug, 5-fluorouracil (5-FU) for the treatment of colorectal cancer and pancreatic cancer, has been incorporated into anApDCsgc8-(5-FU)5conjugate, in which one sgc8 aptamer carries 5 copies of 5-FU, thereby increasing drug payload capacity and decreasing cost. On the other hand, a photocleavable (PC) linker has been used to link a drug moiety with the backbone of phosphoramidite, which served as a modular buildingblock.Under light irradiation, the cleavage of the PC linker released the tethered 5-FU molecules from the aptamer backbone, inhibiting cancer cell proliferation. Meanwhile, the recently recommended combinational chemotherapy has led to correspondingly increased side effects. To address this,Zhu et al.80utilized a simple biocompatible reaction to construct ApDCs with multiple drug copies, including anthracycline drugs (e.g., Doxorubincin, Daunorubicin, Epirubicin, and cisplatin), which could inhibit cell proliferation by disrupting cell division and inducing cell apoptosis. In this conjugate, a crosslinker, formaldehyde, was used to form a methylene linker with the 2-NH2 on deoxyguanosine (dG) and the 3-NH2 group of Dox on each side. The 2-NH2 on the dG conjugate allowed one aptamer tocarrymultiple drugs in the ApDCs. Furthermore, temperature-dependent cleavage of the methylene linker offered gradual drug release at physiological temperature, thus enabling efficient production of ApDCs with high drug loading and drug release controllability.
Figure 2.
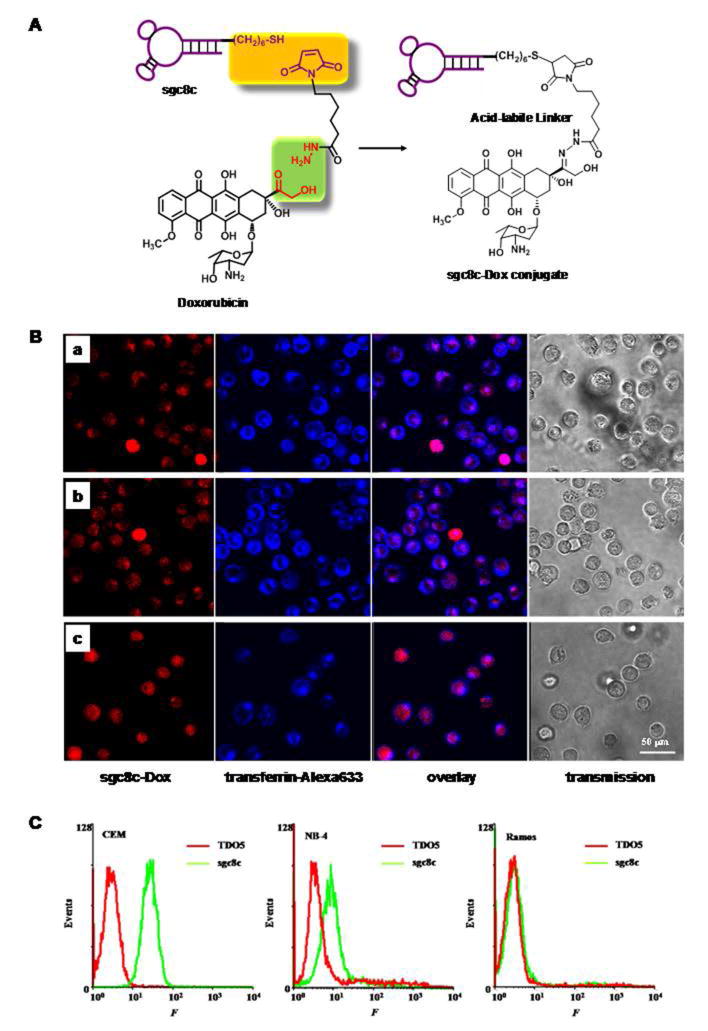
Sgc8c-Dox conjugates for targeted drug delivery. (A) Schematic diagrams depicting sgc8c-Dox covalent conjugation via acid-labile linkages. (B) Distribution of sgc8c-Dox conjugates inside CCRF-CEM cells after incubation with cells for (a) 30 min, (b) 1 h, and (c) 2 h. From left to right, the fluorescence confocal images were monitored for sgc8c-Dox, transferrin-Alexa633, overlay of these two channels, and bright field channel, respectively. (C) Flow cytometry assay for the binding of biotin-labeled TDO5 and sgc8c with three different cell lines: CCRF-CEM, NB-4, and Ramos. Cells (105/mL) were incubated with biotin-labeled TDO5 and sgc8c at 37 °C for 20 min in 100 μL culture medium without FBS. After washing twice, cells were mixed with streptavidin-(R-phycoerythrin) (20 min on ice), and the fluorescence was determined by flow cytometry(Reproduced with permission from Reference77).
4.2 Noncovalentaptamerconjugation through intercalation
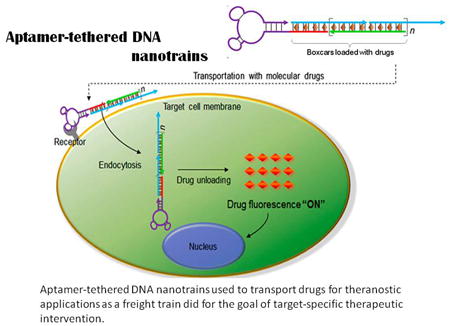
In addition to stablecovalent bonding, noncovalent aptamer-drug conjugation, with the simplicity of programmable nucleic acid engineering, represents another attractive drug delivery strategy fortargeted therapy. In 2006, a PSMA-targeting RNA aptamer, A10, synthesized by Farokhzad et al. was considered as the first physical complex with drug molecules requiring no covalent modifications81and having an intrinsic intercalating site for Dox. According to their report, approximately 1.2 times dose Dox wasintercalated into the A10 aptamer, leading to selective cytotoxicity and relatively abundantdrug loading against PSMA-positive cells. Another aptamer-Dox complexwas then developed by Liu et al. for targeteddrug delivery to breast cancer. However, in retrospect, neither strategy was able to meet full drug-carrying capacity. To achieve this requirement for in vivo cancer treatment, aptamer-tethered DNA nanotrains (aptNTrs) wereexplored by the Tan group82. In this study, modified sgc8 aptamer acted as a locomotive for targeting, while the remaining dsDNA nanoconstructs, which contained numerous Dox intercalation sites, acted as boxcars for drug delivery. The resultant sgc8c-NTrs displayed high cargo loading capacity with a Dox:sgc8-NTr molar ratio of 50:1. This special and efficient delivery platform holds promise for minimizing side effects and broadening applications for targeted drug delivery (Figure 3).
Figure 3.
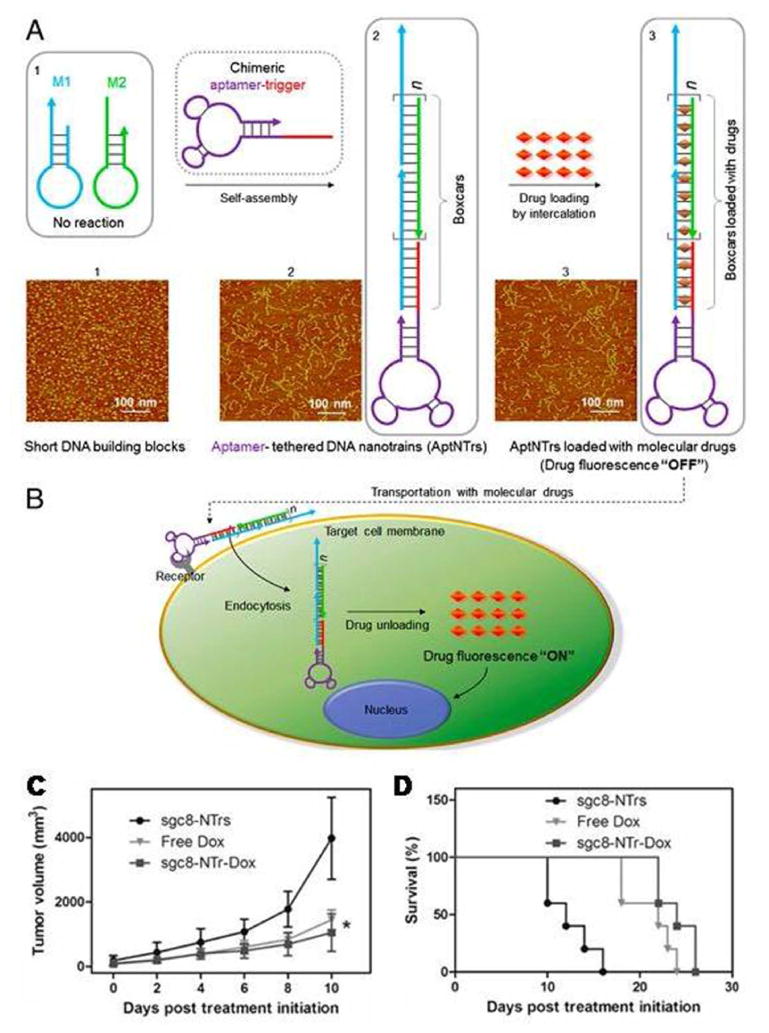
Aptamer-tetheredDNAnanotrains (aptNTrs) used to transport drugsfortheranostic applications. (A) Schematic diagram depicting the self-assembly of aptNTrs from two partially complementary short hairpin monomers upon initiation by an aptamer-tethered probe through hybridization chain reaction (HCR). AFM images (1–3) show the morphologies of the corresponding nanostructures.(B) Drugs were specificallytransported to target cancer cells by aptNTrs and unloaded to induce cytotoxicity to target cells. (C) Potent antitumor efficacy and reduced side effects of drugs transported by aptNTrs. Tumor volumes of subcutaneousCEM xenograft mouse tumors were measured after drug administration up to day 10 (n = 5). Asterisk on day 10 represents significant differences between tumor volumes of free Dox- and sgc8-NTr-Dox-treated mice (*P< 0.05, n = 5; Student's-t test). (D) Percentage of surviving mice after treatment initiation(Reproducedwith permission from Reference82).
Another drawback involved the limited recognition of single aptamers given the heterogeneity of clinical samples from different patients.Zhu et al.83 sought to solve this problem by developing a bi-specific aptamer-based drug delivery systemcontainingtwo aptamers, sgc8 and sgd5a, with drug-intercalating dsDNA as both linker and drug carrier, able to recognize two subtypes of a cancer with heterogeneous biomarkers, therebyovercomingmany diagnostic and therapeutic complications for future clinical applications.
5. Targeted delivery usingaptamer-functionalized nanomaterials
Combining size at the nanometer scale and unique structures, nanomaterials, such as nanoparticles, liposomes, and hydrogels,84-93 offer many physical, chemical and biological properties conducive to aptamer functionalization and targeted drug delivery. For example, nanomaterials offer large surface area-to-volume ratios, the ability to travel through the blood stream withoutblockage of the microvasculature,and the ability to be taken up by cells through endocytosisand penetrate tissues.Broadening the scope of both aptamers and nanomaterials, many of the advantages of aptamers are compatible with nanomaterials, resulting in conjugates able to minimize the drawbacks of each technology when applied separately to address the same biological issues, e.g., targeted drug delivery. Below we discuss some of these innovative bioconjugates.
5.1 Aptamer-conjugated gold nanoparticles for targeted therapy
Gold nanoparticles (AuNPs)are the most common and stable metallic nanomaterialsclinically appliedover the last few decades. AuNPs can form thiolated complexes by stable Au:S bonds and introduce diverse molecules enabling multiple functionalities for drug delivery. Moreover, the inert nature of AuNPs ensures nontoxicity to living cells, as demonstrated byin vitro studies, making AuNPs ideal candidates for drug transport. The size and shape of AuNPs, e.g. nanorods, nanospheres, nanoshells, and nanocages, can be precisely adjusted for a variety of applications.
Among a wide array of cancer treatments, photothermal therapy (PTT) is relatively noninvasive and benign.PTT simplyexposes biological tissues to higher temperatures to promote the destruction of abnormal cells. Gold nanorods (AuNRs), providing a higher absorption cross section per unit volume than other AuNPs, are more feasible for future clinical PTT.Huang et al.94conjugated aptamer sgc8c to Au-Ag NRs for selective PTT in vitro. The binding affinity of this sgc8c-NR conjugate for targeted CCRF-CEM cells was foundto be 26-fold greater than that ofsgc8c used alone.Alongwith itshigh absorption efficiency and superior photothermal transfer, this bioconjugate is highly promising for specific recognition and targeted PTT.Following this work,aptamer CSC1 targeting DU145 prostate cancer cellsand aptamer CSC13 targetingasubpopulationof DU145cancer stem cellswere linked to the surface of AuNRs. The bi-specific CSC13-AuNRcomplexes were able to kill both cancer cells and cancer stem cells using NIR laser irradiation95.
For more effective drug loading,the hollow interior of AuNPs can be utilized, or their surface can be coated with mesoporous materials to enlarge pore volume and surface area. As an example of the first type, gold nanospheres (HAuNS) are composed of an Au shell with a hollow interior96, which, compared to AuNPs, has similar size, surface charge, and equivalent Au concentration, but 3.5-fold greater drug loading capacity(Figure 4A).A delivery vehicle has also been explored using HAuNS together with a highly specific RNA aptamer to target CD30.TheresultantAptamer-HAuNS-Dox showed selective destruction of lymphoma tumor cells with minimal “off-target” effects. Coating AuNRs with mesoporous silica (AuMPs)has attracted attention as a potential drug delivery system owing to the high surface area for effective drug loading andmesoporous container for functional nucleic acids for controlled drug delivery and release97, 98.Aptamer AS1411 has been utilized as a molecular gate grafted onto the surface of AuMPs forming a dimeric G-quadruplex structure to cap guest molecules. Whenirradiated by NIR light, the photothermal effect leads to a dehybridization of the linked DNA duplex that anchors the capping molecules, therebyallowing the specific release of the loaded cargo99.Thus, by its functionalization as both targeting and capping agent, aptamer AS1411 makes this AuMP-based porous nanocarrier a good choice for remote-controlled targeted drug delivery, as itsin vivoapplicationis enabled by laser-induced thermal stimulus (Figure 4B).
Figure 4.
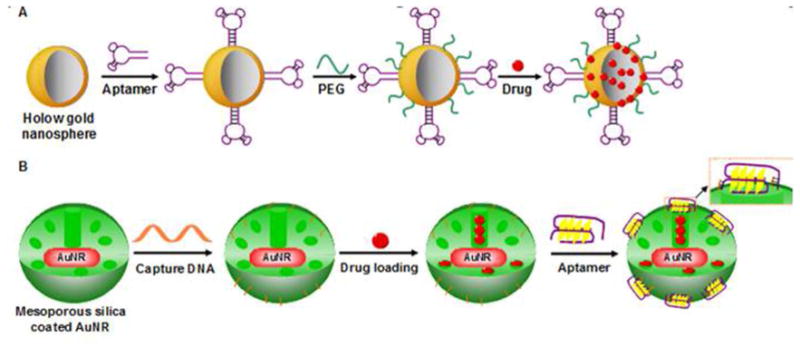
Schematic diagrams of nanoplatforms based on aptamer-conjugated gold nanoparticles for targeted drugdelivery. (A) Apt-HAuNS-Dox nanoscale drug carrier. (B) AuNR-based mesoporous silica nanocarrier(Reproduced with permission from Reference97).
5.2 Other aptamer-conjugated nanoparticles for targeted therapy
Softnanoparticles, such as liposomesand hydrogels, also exhibitunique advantages,including high water-solubility, enhancedaccumulation at the tumor cells, prolonged circulationtime in the blood andinherent biocompatibility.
Liposomes, which are artificially prepared vesicles composed of a lipid bilayer, are biocompatible and biodegradable. To form aptamer-liposome bioconjugates, cholesterol-modified aptamers can be spontaneously anchored on the outer shell during the formation of the NP conjugate.Thisadvantage has been utilized to engineer liposomesfor the delivery of toxic chemotherapeutic drugs, such as cisplatin and taxol. As reported, compared to nontargeting liposomes, the AS1411 aptamer-functionalized liposomes containing taxol have increased rates of cellular uptake and cytotoxicity for MCF-7 breast cancer cells100. Athymic nude mice bearing xenograft MCF-7 tumors treated with intratumoral injection of aptamer-functionalized liposomesexhibited earlier onset of tumor inhibition and improved anticancer efficacy.Kang et al. have successfully extended the application of aptamer-functionalized liposomes to the cellular level by simplification of the aptamer-modified liposome synthesis method101. Each liposome had approximately 250 aptamers tethered to its surface to facilitate target binding, and several thousand FITC-Dextran (FD) molecules (drug proxy)wereloaded inside. Within 30 minutes of incubation time, the aptamer-liposome conjugates could specifically bind with target cells and release the loaded model drug. Targeted drug delivery was guaranteed by sgc8 aptamer binding with its target CEM cells for breast cancer.
Hydrogels are crosslinked hydrophilic polymer structures that can hold large amounts of water or biological fluids in their pore spaces. As one of the newest classes of polymer-based systems, target-responsive hydrogels have found numerous biomedical and pharmaceutical applications102-105.Li et al.88recently reported DNA nanohydrogels created through a self-assembly process. These DNA nanohydrogels consist of three elements, respectively termed Y-shaped monomer A with three sticky ends (YMA), Y-shaped monomer B with one sticky end (YMB), and DNA linker (LK) with two sticky ends. DNA nanohydrogels are size-controllable by varying the ratio of YMA to YMB. By incorporating different functional elements, such as aptamers, disulfide linkages, and therapeutic genes into different building blocks, the synthesized aptamer-based nanohydrogels (Y-gel-Apt) can be used for targeted and stimuli-responsive gene therapy. Y-gel-Apt strongly inhibited cell proliferation and migration in target A549 cells, but not in control cells. By taking advantage of both aptamers and nanohydrogels, this Y-gel-Apt with facile self-assembly, effective cellular uptake, and superior biocompatibility holdspromise as a candidate for targeted gene or drug delivery in cancer therapy.
6. Conclusion
Extensive genome sequencing and proteomic analysis in past decades have generated vast quantities of information for disease biology, changing, as a result,some inherent concepts in treating disease.The conjugation of less selective therapeutics withtarget-specific ligands has emerged as a promising alternativeto the use of antibodies.Based on their unique advantages and properties, compared to antibodies,aptamershaveattracted increasing attentionfor application in biomedicine to achieve targeted therapy. Cell-SELEX, whichselects aptamers againstwhole live cells without prior knowledge of molecular signatures onthe cell surface, is an ideal toolfor the selection of aptamers able to preferentially bind to diseased cells.Over past decades, this technologyhas enabled the generation ofbulk aptamers targetinga number of proteins,including PSMA, PTK7, IGHM, and nucleolin. Aptamerscan also act as macromolecular drugs in a myriad of human diseases, includingcancers andmetabolic diseases,as well as disorders resulting in inflammatory conditions and coagulation abnormality. Moreover, bioconjugation between nanomaterials and aptamers takes advantage of both technologies, making aptamer-based NP conjugates ideal vehicles for drug delivery applications, especially for the controlled release of drugs. Taken together, aptamershavethus farsuccessfully performedas drugs or guidance systems for targeted drug delivery and will certainly become even more widespread clinical tools in biomedical applications.
Figure 5.
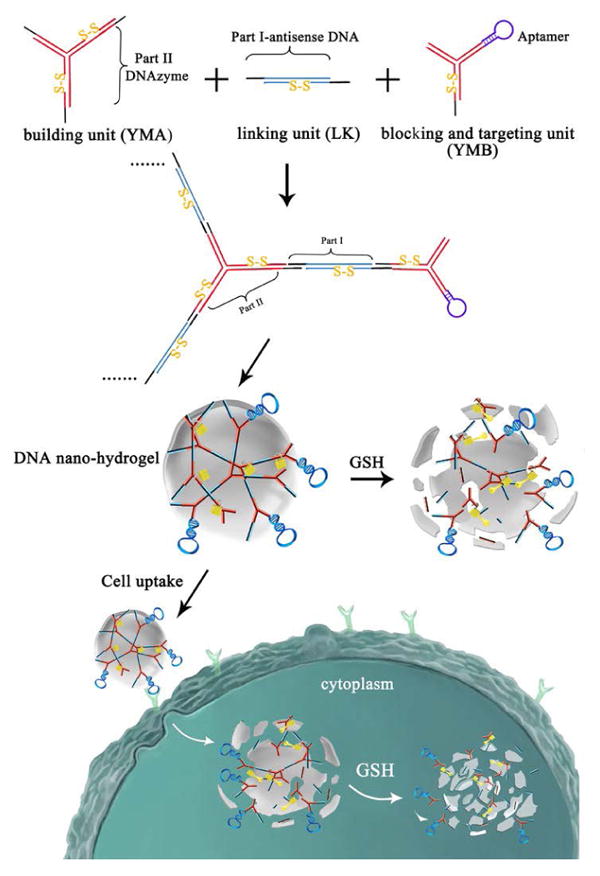
Schematic illustration of Stimuli-Responsive DNA Nanohydrogel formation (Reproduced with permission from Reference88).
Acknowledgments
This research was supported by NSFC grants (NSFC 21502050,NSFC 81370983,NSFC 8140086410, NSFC 81500692,NSFC 21521063 and NSFC 21327009), the Foundation of National Key Scientific Instrument and Equipment Development Projects (2011YQ0301241403), the Hunan Province Natural Science Key Fund Project (2014SK2003), the Foundation of China Hunan Provincial Science & Technology Department (2012FJ4371,S2014S2032,2010SK2003),the Science & Research Foundation of Hunan Health Department (B2010-007, 132013-010, 132011-014) Fundamental Research Funds for the Central Universities of Central South University (2013 zzts083) and the Youth Scientific Fund of Xiangya Hospital(2015Q03). This work is also supported by the National Institutes of Health (GM079359 , GM 111386 and CA133086).
References
- 1.Dosio F, Brusa P, Cattel L. Immunotoxins and anticancer drug conjugate assemblies: the role of the linkage between components. Toxins (Basel) 2011;3:848–883. doi: 10.3390/toxins3070848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farokhzad OC. Targeted polymeric therapeutic nanoparticles: design, development and clinical translation. Chem Soc Rev. 2012;41:2971–3010. doi: 10.1039/c2cs15344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooks MA, Wade CS, Millikan WJ., Jr Muromonab CD-3: a review of its pharmacology, pharmacokinetics, and clinical use in transplantation. Pharmacotherapy. 1991;11:26–37. [PubMed] [Google Scholar]
- 4.Brader ML, Estey T, Bai S, Alston RW, Lucas KK, Lantz S, Landsman P, Maloney KM. Examination of thermal unfolding and aggregation profiles of a series of developable therapeutic monoclonal antibodies. Mol Pharm. 2015;12:1005–1017. doi: 10.1021/mp400666b. [DOI] [PubMed] [Google Scholar]
- 5.de Figueiredo IR, Freire JM, Flores L, Veiga AS, Castanho MA. Cell-penetrating peptides: A tool for effective delivery in gene-targeted therapies. IUBMB Life. 2014 doi: 10.1002/iub.1257. [DOI] [PubMed] [Google Scholar]
- 6.Guijarro-Munoz I, Compte M, Alvarez-Vallina L, Sanz L. Antibody gene therapy: getting closer to clinical application? Curr Gene Ther. 2013;13:282–290. doi: 10.2174/15665232113139990025. [DOI] [PubMed] [Google Scholar]
- 7.Smaglo BG, Aldeghaither D, Weiner LM. The development of immunoconjugates for targeted cancer therapy. Nat Rev Clin Oncol. 2014;11:637–648. doi: 10.1038/nrclinonc.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 9.Gallina ME, Zhou Y, Johnson CJ, Harris-Birtill D, Singh M, Zhao H, Ma D, Cass T, Elson DS. Aptamer-conjugated, fluorescent gold nanorods as potential cancer theradiagnostic agents. Mater Sci Eng C Mater Biol Appl. 2016;59:324–332. doi: 10.1016/j.msec.2015.09.101. [DOI] [PubMed] [Google Scholar]
- 10.Kang L, Yang B, Zhang X, Cui L, Meng H, Mei L, Wu C, Ren S, Tan W. Enzymatic cleavage and mass amplification strategy for small molecule detection using aptamer-based fluorescence polarization biosensor. Anal Chim Acta. 2015;879:91–96. doi: 10.1016/j.aca.2015.03.030. [DOI] [PubMed] [Google Scholar]
- 11.Kanwar JR, Roy K, Maremanda NG, Subramanian K, Veedu RN, Bawa R, Kanwar RK. Nucleic acid-based aptamers: applications, development and clinical trials. Curr Med Chem. 2015;22:2539–2557. doi: 10.2174/0929867322666150227144909. [DOI] [PubMed] [Google Scholar]
- 12.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Lan X. Aptamer Oligonucleotides: Novel Potential Therapeutic Agents in Autoimmune Disease. Nucleic Acid Ther. 2015;25:173–179. doi: 10.1089/nat.2014.0529. [DOI] [PubMed] [Google Scholar]
- 14.Sun H, Tan W, Zu Y. Aptamers: versatile molecular recognition probes for cancer detection. Analyst. 2016;141:403–415. doi: 10.1039/c5an01995h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu X, Dickey DD, Chen SJ, Giangrande PH. Structural computational modeling of RNA aptamers. Methods. 2016 doi: 10.1016/j.ymeth.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu G, Ye M, Donovan MJ, Song E, Zhao Z, Tan W. Nucleic acid aptamers: an emerging frontier in cancer therapy. Chem Commun (Camb) 2012;48:10472–10480. doi: 10.1039/c2cc35042d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Z, Song Y, Li C, Zou Y, Zhu L, An Y, Yang CJ. Monoclonal surface display SELEX for simple, rapid, efficient, and cost-effective aptamer enrichment and identification. Anal Chem. 2014;86:5881–5888. doi: 10.1021/ac501423g. [DOI] [PubMed] [Google Scholar]
- 18.Alibolandi M, Ramezani M, Abnous K, Hadizadeh F. AS1411 Aptamer-Decorated Biodegradable Polyethylene Glycol-Poly(lactic-co-glycolic acid) Nanopolymersomes for the Targeted Delivery of Gemcitabine to Nonsmall Cell Lung Cancer In Vitro. J Pharm Sci. 2016 doi: 10.1016/j.xphs.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 19.Gijs M, Aerts A, Impens N, Baatout S, Luxen A. Aptamers as radiopharmaceuticals for nuclear imaging and therapy. Nucl Med Biol. 2015 doi: 10.1016/j.nucmedbio.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Meng HM, Zhang X, Lv Y, Zhao Z, Wang NN, Fu T, Fan H, Liang H, Qiu L, Zhu G, et al. DNA dendrimer: an efficient nanocarrier of functional nucleic acids for intracellular molecular sensing. ACS Nano. 2014;8:6171–6181. doi: 10.1021/nn5015962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xing H, Zhang CL, Ruan G, Zhang J, Hwang K, Lu Y. Multimodal Detection of a Small Molecule Target Using Stimuli-Responsive Liposome Triggered by Aptamer-Enzyme Conjugate. Anal Chem. 2016;88:1506–1510. doi: 10.1021/acs.analchem.5b04031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou C, Chen T, Wu C, Zhu G, Qiu L, Cui C, Hou W, Tan W. Aptamer CaCO3 nanostructures: a facile, pH-responsive, specific platform for targeted anticancer theranostics. Chem Asian J. 2015;10:166–171. doi: 10.1002/asia.201403115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang YC, Yang CY, Sun RL, Cheng YF, Kao WC, Yang PC. Rapid single cell detection of Staphylococcus aureus by aptamer-conjugated gold nanoparticles. Sci Rep. 2013;3:1863. doi: 10.1038/srep01863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng H, Beck J, Nassal M, Hu KH. A SELEX-screened aptamer of human hepatitis B virus RNA encapsidation signal suppresses viral replication. PLoS One. 2011;6:e27862. doi: 10.1371/journal.pone.0027862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gopinath SC, Hayashi K, Kumar PK. Aptamer that binds to the gD protein of herpes simplex virus 1 and efficiently inhibits viral entry. J Virol. 2012;86:6732–6744. doi: 10.1128/JVI.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong KL, Sooter LJ. Single-Stranded DNA Aptamers against Pathogens and Toxins: Identification and Biosensing Applications. Biomed Res Int. 2015;2015:419318. doi: 10.1155/2015/419318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowalska E, Bartnicki F, Pels K, Strzalka W. The impact of immobilized metal affinity chromatography (IMAC) resins on DNA aptamer selection. Anal Bioanal Chem. 2014;406:5495–5499. doi: 10.1007/s00216-014-7937-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urak KT, Shore S, Rockey WM, Chen SJ, McCaffrey AP, Giangrande PH. In vitro RNA SELEX for the generation of chemically-optimized therapeutic RNA drugs. Methods. 2016 doi: 10.1016/j.ymeth.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.You M, Zhu G, Chen T, Donovan MJ, Tan W. Programmable and multiparameter DNA-based logic platform for cancer recognition and targeted therapy. J Am Chem Soc. 2015;137:667–674. doi: 10.1021/ja509263k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P, Chen HW, Li Y, Tan W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 32.Liang H, Zhang XB, Lv Y, Gong L, Wang R, Zhu X, Yang R, Tan W. Functional DNA-containing nanomaterials: cellular applications in biosensing, imaging, and targeted therapy. Acc Chem Res. 2014;47:1891–1901. doi: 10.1021/ar500078f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shum KT, Zhou J, Rossi JJ. Aptamer-based therapeutics: new approaches to combat human viral diseases. Pharmaceuticals (Basel) 2013;6:1507–1542. doi: 10.3390/ph6121507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi M, Yang S, Peng Z, Liu C, Li J, Zhong W, Yang R, Tan W. Two-photon graphene oxide/aptamer nanosensing conjugate for in vitro or in vivo molecular probing. Anal Chem. 2014;86:3548–3554. doi: 10.1021/ac5000015. [DOI] [PubMed] [Google Scholar]
- 35.Zichel R, Chearwae W, Pandey GS, Golding B, Sauna ZE. Aptamers as a sensitive tool to detect subtle modifications in therapeutic proteins. PLoS One. 2012;7:e31948. doi: 10.1371/journal.pone.0031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma J, Wang MG, Mao AH, Zeng JY, Liu YQ, Wang XQ, Ma J, Tian YJ, Ma N, Yang N, et al. Target replacement strategy for selection of DNA aptamers against the Fc region of mouse IgG. Genet Mol Res. 2013;12:1399–1410. doi: 10.4238/2013.April.25.11. [DOI] [PubMed] [Google Scholar]
- 37.Reinstein O, Yoo M, Han C, Palmo T, Beckham SA, Wilce MC, Johnson PE. Quinine binding by the cocaine-binding aptamer. Thermodynamic and hydrodynamic analysis of high-affinity binding of an off-target ligand. Biochemistry. 2013;52:8652–8662. doi: 10.1021/bi4010039. [DOI] [PubMed] [Google Scholar]
- 38.Slavkovic S, Altunisik M, Reinstein O, Johnson PE. Structure-affinity relationship of the cocaine-binding aptamer with quinine derivatives. Bioorg Med Chem. 2015;23:2593–2597. doi: 10.1016/j.bmc.2015.02.052. [DOI] [PubMed] [Google Scholar]
- 39.Kalnins A, Thomas MN, Andrassy M, Muller S, Wagner A, Pratschke S, Rentsch M, Klussmann S, Kauke T, Angele MK, et al. Spiegelmer Inhibition of MCP-1/CCR2--Potential as an Adjunct Immunosuppressive Therapy in Transplantation. Scand J Immunol. 2015;82:102–109. doi: 10.1111/sji.12310. [DOI] [PubMed] [Google Scholar]
- 40.Ramaswamy V, Monsalve A, Sautina L, Segal MS, Dobson J, Allen JB. DNA Aptamer Assembly as a Vascular Endothelial Growth Factor Receptor Agonist. Nucleic Acid Ther. 2015;25:227–234. doi: 10.1089/nat.2014.0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravalli A, Rivas L, De la Escosura-Muniz A, Pons J, Merkoci A, Marrazza G. A DNA Aptasensor for Electrochemical Detection of Vascular Endothelial Growth Factor. J Nanosci Nanotechnol. 2015;15:3411–3416. doi: 10.1166/jnn.2015.10037. [DOI] [PubMed] [Google Scholar]
- 42.Rhinehardt KL, Srinivas G, Mohan RV. Molecular Dynamics Simulation Analysis of Anti-MUC1 Aptamer and Mucin 1 Peptide Binding. J Phys Chem B. 2015;119:6571–6583. doi: 10.1021/acs.jpcb.5b02483. [DOI] [PubMed] [Google Scholar]
- 43.Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang J, Xie J, Shao N, Yan Y. The DNA aptamers that specifically recognize ricin toxin are selected by two in vitro selection methods. Electrophoresis. 2006;27:1303–1311. doi: 10.1002/elps.200500489. [DOI] [PubMed] [Google Scholar]
- 45.Tang J, Yu T, Guo L, Xie J, Shao N, He Z. In vitro selection of DNA aptamer against abrin toxin and aptamer-based abrin direct detection. Biosens Bioelectron. 2007;22:2456–2463. doi: 10.1016/j.bios.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 46.Jeon SH, Kayhan B, Ben-Yedidia T, Arnon R. A DNA aptamer prevents influenza infection by blocking the receptor binding region of the viral hemagglutinin. J Biol Chem. 2004;279:48410–48419. doi: 10.1074/jbc.M409059200. [DOI] [PubMed] [Google Scholar]
- 47.Koch TH, Smith D, Tabacman E, Zichi DA. Kinetic analysis of site-specific photoaptamer-protein cross-linking. J Mol Biol. 2004;336:1159–1173. doi: 10.1016/j.jmb.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 48.Dai H, Ye M, Peng M, Zhou W, Bai H, Xiao X, Ma B, Zhou J, Tang S, Yao S, et al. Aptamer TY04 inhibits the growth of multiple myeloma cells via cell cycle arrest. Tumour Biol. 2014;35:7561–7568. doi: 10.1007/s13277-014-1920-2. [DOI] [PubMed] [Google Scholar]
- 49.Leach JC, Wang A, Ye K, Jin S. A RNA-DNA Hybrid Aptamer for Nanoparticle-Based Prostate Tumor Targeted Drug Delivery. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17030380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J, Liu H, Sefah K, Liu B, Pu Y, Van Simaeys D, Tan W. Selection of aptamers specific for adipose tissue. PLoS One. 2012;7:e37789. doi: 10.1371/journal.pone.0037789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pu Y, Liu Z, Lu Y, Yuan P, Liu J, Yu B, Wang G, Yang CJ, Liu H, Tan W. Using DNA aptamer probe for immunostaining of cancer frozen tissues. Anal Chem. 2015;87:1919–1924. doi: 10.1021/ac504175h. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hong-Min Meng, Ting Fu, Xiao-Bing Zhang, Weihong Tan. Cell-SELEX-based aptamer-conjugated nanomaterials for theranostic applications. National Science Review. 2015;2:71–84. [Google Scholar]
- 52.Ristau BT, O'Keefe DS, Bacich DJ. The prostate-specific membrane antigen: lessons and current clinical implications from 20 years of research. Urol Oncol. 2014;32:272–279. doi: 10.1016/j.urolonc.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo KT, Yan XR, Huang GJ, Xu CX, Chai YS, Zhang ZQ. Screening and characterization of DNA aptamers with hTNF-alpha binding and neutralizing activity. Sheng Wu Gong Cheng Xue Bao. 2003;19:730–733. [PubMed] [Google Scholar]
- 54.Lou X, Qian J, Xiao Y, Viel L, Gerdon AE, Lagally ET, Atzberger P, Tarasow TM, Heeger AJ, Soh HT. Micromagnetic selection of aptamers in microfluidic channels. Proc Natl Acad Sci U S A. 2009;106:2989–2994. doi: 10.1073/pnas.0813135106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mosing RK, Bowser MT. Isolating aptamers using capillary electrophoresis-SELEX (CE-SELEX) Methods Mol Biol. 2009;535:33–43. doi: 10.1007/978-1-59745-557-2_3. [DOI] [PubMed] [Google Scholar]
- 56.Mosing RK, Mendonsa SD, Bowser MT. Capillary electrophoresis-SELEX selection of aptamers with affinity for HIV-1 reverse transcriptase. Anal Chem. 2005;77:6107–6112. doi: 10.1021/ac050836q. [DOI] [PubMed] [Google Scholar]
- 57.Eulberg D, Buchner K, Maasch C, Klussmann S. Development of an automated in vitro selection protocol to obtain RNA-based aptamers: identification of a biostable substance P antagonist. Nucleic Acids Res. 2005;33:e45. doi: 10.1093/nar/gni044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin C, Zheng J, Li C, Qiu L, Zhang X, Tan W. Aptamers Selected by Cell-SELEX for Molecular Imaging. J Mol Evol. 2015;81:162–171. doi: 10.1007/s00239-015-9716-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu X, Zhao Z, Bai H, Fu T, Yang C, Hu X, Liu Q, Champanhac C, Teng IT, Ye M, et al. DNA Aptamer Selected against Pancreatic Ductal Adenocarcinoma for in vivo Imaging and Clinical Tissue Recognition. Theranostics. 2015;5:985–994. doi: 10.7150/thno.11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen HW, Medley CD, Sefah K, Shangguan D, Tang Z, Meng L, Smith JE, Tan W. Molecular recognition of small-cell lung cancer cells using aptamers. ChemMedChem. 2008;3:991–1001. doi: 10.1002/cmdc.200800030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shangguan D, Cao ZC, Li Y, Tan W. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clin Chem. 2007;53:1153–1155. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- 62.Shangguan D, Meng L, Cao ZC, Xiao Z, Fang X, Li Y, Cardona D, Witek RP, Liu C, Tan W. Identification of liver cancer-specific aptamers using whole live cells. Anal Chem. 2008;80:721–728. doi: 10.1021/ac701962v. [DOI] [PubMed] [Google Scholar]
- 63.Van Simaeys D, Turek D, Champanhac C, Vaizer J, Sefah K, Zhen J, Sutphen R, Tan W. Identification of cell membrane protein stress-induced phosphoprotein 1 as a potential ovarian cancer biomarker using aptamers selected by cell systematic evolution of ligands by exponential enrichment. Anal Chem. 2014;86:4521–4527. doi: 10.1021/ac500466x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi H, Tang Z, Kim Y, Nie H, Huang YF, He X, Deng K, Wang K, Tan W. In vivo fluorescence imaging of tumors using molecular aptamers generated by cell-SELEX. Chem Asian J. 2010;5:2209–2213. doi: 10.1002/asia.201000242. [DOI] [PubMed] [Google Scholar]
- 65.Sullenger BA, Gallardo HF, Ungers GE, Gilboa E. Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell. 1990;63:601–608. doi: 10.1016/0092-8674(90)90455-n. [DOI] [PubMed] [Google Scholar]
- 66.Zhou B, Wang B. Pegaptanib for the treatment of age-related macular degeneration. Exp Eye Res. 2006;83:615–619. doi: 10.1016/j.exer.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 67.Dobrovolsky AB, Titaeva EV, Khaspekova SG, Spiridonova VA, Kopylov AM, Mazurov AV. Inhibition of thrombin activity with DNA-aptamers. Bull Exp Biol Med. 2009;148:33–36. doi: 10.1007/s10517-009-0627-7. [DOI] [PubMed] [Google Scholar]
- 68.Chang JY, Chantrathammachart P, Monroe DM, Key NS. Studies on the mechanism of action of the aptamer BAX499, an inhibitor of tissue factor pathway inhibitor. Thromb Res. 2012;130:e151–157. doi: 10.1016/j.thromres.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 69.Gilbert JC, DeFeo-Fraulini T, Hutabarat RM, Horvath CJ, Merlino PG, Marsh HN, Healy JM, Boufakhreddine S, Holohan TV, Schaub RG. First-in-human evaluation of anti von Willebrand factor therapeutic aptamer ARC1779 in healthy volunteers. Circulation. 2007;116:2678–2686. doi: 10.1161/CIRCULATIONAHA.107.724864. [DOI] [PubMed] [Google Scholar]
- 70.Krishnan A, Vogler EA, Sullenger BA, Becker RC. The effect of surface contact activation and temperature on plasma coagulation with an RNA aptamer directed against factor IXa. J Thromb Thrombolysis. 2013;35:48–56. doi: 10.1007/s11239-012-0778-7. [DOI] [PubMed] [Google Scholar]
- 71.Reshetnikov RV, Sponer J, Rassokhina OI, Kopylov AM, Tsvetkov PO, Makarov AA, Golovin AV. Cation binding to 15-TBA quadruplex DNA is a multiple-pathway cation-dependent process. Nucleic Acids Res. 2011;39:9789–9802. doi: 10.1093/nar/gkr639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soule EE, Bompiani KM, Woodruff RS, Sullenger BA. Targeting Two Coagulation Cascade Proteases with a Bivalent Aptamer Yields a Potent and Antidote-Controllable Anticoagulant. Nucleic Acid Ther. 2016;26:1–9. doi: 10.1089/nat.2015.0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zavyalova E, Golovin A, Pavlova G, Kopylov A. Module-activity relationship of G-quadruplex based DNA aptamers for human thrombin. Curr Med Chem. 2013;20:4836–4843. doi: 10.2174/09298673113206660283. [DOI] [PubMed] [Google Scholar]
- 74.Zavyalova E, Golovin A, Reshetnikov R, Mudrik N, Panteleyev D, Pavlova G, Kopylov A. Novel modular DNA aptamer for human thrombin with high anticoagulant activity. Curr Med Chem. 2011;18:3343–3350. doi: 10.2174/092986711796504727. [DOI] [PubMed] [Google Scholar]
- 75.Chen L, Li DQ, Zhong J, Wu XL, Chen Q, Peng H, Liu SQ. IL-17RA aptamer-mediated repression of IL-6 inhibits synovium inflammation in a murine model of osteoarthritis. Osteoarthritis Cartilage. 2011;19:711–718. doi: 10.1016/j.joca.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 76.Kaida Y, Fukami K, Matsui T, Higashimoto Y, Nishino Y, Obara N, Nakayama Y, Ando R, Toyonaga M, Ueda S, et al. DNA aptamer raised against AGEs blocks the progression of experimental diabetic nephropathy. Diabetes. 2013;62:3241–3250. doi: 10.2337/db12-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang YF, Shangguan D, Liu H, Phillips JA, Zhang X, Chen Y, Tan W. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. Chembiochem. 2009;10:862–868. doi: 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boyacioglu O, Stuart CH, Kulik G, Gmeiner WH. Dimeric DNA Aptamer Complexes for High-capacity-targeted Drug Delivery Using pH-sensitive Covalent Linkages. Mol Ther Nucleic Acids. 2013;2:e107. doi: 10.1038/mtna.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang R, Zhu G, Mei L, Xie Y, Ma H, Ye M, Qing FL, Tan W. Automated modular synthesis of aptamer-drug conjugates for targeted drug delivery. J Am Chem Soc. 2014;136:2731–2734. doi: 10.1021/ja4117395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu G, Niu G, Chen X. Aptamer-Drug Conjugates. Bioconjug Chem. 2015;26:2186–2197. doi: 10.1021/acs.bioconjchem.5b00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci U S A. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu G, Zheng J, Song E, Donovan M, Zhang K, Liu C, Tan W. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc Natl Acad Sci U S A. 2013;110:7998–8003. doi: 10.1073/pnas.1220817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu G, Meng L, Ye M, Yang L, Sefah K, O'Donoghue MB, Chen Y, Xiong X, Huang J, Song E, et al. Self-assembled aptamer-based drug carriers for bispecific cytotoxicity to cancer cells. Chem Asian J. 2012;7:1630–1636. doi: 10.1002/asia.201101060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ara MN, Matsuda T, Hyodo M, Sakurai Y, Ohga N, Hida K, Harashima H. Construction of an aptamer modified liposomal system targeted to tumor endothelial cells. Biol Pharm Bull. 2014;37:1742–1749. doi: 10.1248/bpb.b14-00338. [DOI] [PubMed] [Google Scholar]
- 85.Cai R, Yang D, Peng S, Chen X, Huang Y, Liu Y, Hou W, Yang S, Liu Z, Tan W. Single Nanoparticle to 3D Supercage: Framing for an Artificial Enzyme System. J Am Chem Soc. 2015;137:13957–13963. doi: 10.1021/jacs.5b09337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li J, Hong CY, Wu SX, Liang H, Wang LP, Huang G, Chen X, Yang HH, Shangguan D, Tan W. Facile Phase Transfer and Surface Biofunctionalization of Hydrophobic Nanoparticles Using Janus DNA Tetrahedron Nanostructures. J Am Chem Soc. 2015;137:11210–11213. doi: 10.1021/jacs.5b05650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li J, Mo L, Lu CH, Fu T, Yang HH, Tan W. Functional nucleic acid-based hydrogels for bioanalytical and biomedical applications. Chem Soc Rev. 2016;45:1410–1431. doi: 10.1039/c5cs00586h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li J, Zheng C, Cansiz S, Wu C, Xu J, Cui C, Liu Y, Hou W, Wang Y, Zhang L, et al. Self-assembly of DNA nanohydrogels with controllable size and stimuli-responsive property for targeted gene regulation therapy. J Am Chem Soc. 2015;137:1412–1415. doi: 10.1021/ja512293f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu Y, Purich DL, Wu C, Wu Y, Chen T, Cui C, Zhang L, Cansiz S, Hou W, Wang Y, et al. Ionic Functionalization of Hydrophobic Colloidal Nanoparticles To Form Ionic Nanoparticles with Enzymelike Properties. J Am Chem Soc. 2015;137:14952–14958. doi: 10.1021/jacs.5b08533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Song ZL, Chen Z, Bian X, Zhou LY, Ding D, Liang H, Zou YX, Wang SS, Chen L, Yang C, et al. Alkyne-functionalized superstable graphitic silver nanoparticles for Raman imaging. J Am Chem Soc. 2014;136:13558–13561. doi: 10.1021/ja507368z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sung TC, Chen WY, Shah P, Chen CS. A replaceable liposomal aptamer for the ultrasensitive and rapid detection of biotin. Sci Rep. 2016;6:21369. doi: 10.1038/srep21369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiong X, Wu C, Zhou C, Zhu G, Chen Z, Tan W. Responsive DNA-based hydrogels and their applications. Macromol Rapid Commun. 2013;34:1271–1283. doi: 10.1002/marc.201300411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Z, Liu C, Bai J, Wu C, Xiao Y, Li Y, Zheng J, Yang R, Tan W. Silver nanoparticle gated, mesoporous silica coated gold nanorods (AuNR@MS@AgNPs): low premature release and multifunctional cancer theranostic platform. ACS Appl Mater Interfaces. 2015;7:6211–6219. doi: 10.1021/acsami.5b00368. [DOI] [PubMed] [Google Scholar]
- 94.Huang YF, Sefah K, Bamrungsap S, Chang HT, Tan W. Selective photothermal therapy for mixed cancer cells using aptamer-conjugated nanorods. Langmuir. 2008;24:11860–11865. doi: 10.1021/la801969c. [DOI] [PubMed] [Google Scholar]
- 95.Wang J, Sefah K, Altman MB, Chen T, You M, Zhao Z, Huang CZ, Tan W. Aptamer-conjugated nanorods for targeted photothermal therapy of prostate cancer stem cells. Chem Asian J. 2013;8:2417–2422. doi: 10.1002/asia.201300375. [DOI] [PubMed] [Google Scholar]
- 96.Zhao N, You J, Zeng Z, Li C, Zu Y. An ultra pH-sensitive and aptamer-equipped nanoscale drug-delivery system for selective killing of tumor cells. Small. 2013;9:3477–3484. doi: 10.1002/smll.201202694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Qin C, Fei J, Wang A, Yang Y, Li J. Rational assembly of a biointerfaced core@shell nanocomplex towards selective and highly efficient synergistic photothermal/photodynamic therapy. Nanoscale. 2015;7:20197–20210. doi: 10.1039/c5nr06501a. [DOI] [PubMed] [Google Scholar]
- 98.Wang G, Chen Z, Wang W, Yan B, Chen L. Chemical redox-regulated mesoporous silica-coated gold nanorods for colorimetric probing of Hg2+ and S2. Analyst. 2011;136:174–178. doi: 10.1039/c0an00597e. [DOI] [PubMed] [Google Scholar]
- 99.Borghei YS, Hosseini M, Dadmehr M, Hosseinkhani S, Ganjali MR, Sheikhnejad R. Visual detection of cancer cells by colorimetric aptasensor based on aggregation of gold nanoparticles induced by DNA hybridization. Anal Chim Acta. 2016;904:92–97. doi: 10.1016/j.aca.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 100.Xing H, Tang L, Yang X, Hwang K, Wang W, Yin Q, Wong NY, Dobrucki LW, Yasui N, Katzenellenbogen JA, et al. Selective Delivery of an Anticancer Drug with Aptamer-Functionalized Liposomes to Breast Cancer Cells and. J Mater Chem B Mater Biol Med. 2013;1:5288–5297. doi: 10.1039/C3TB20412J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kang H, Trondoli AC, Zhu G, Chen Y, Chang YJ, Liu H, Huang YF, Zhang X, Tan W. Near-infrared light-responsive core-shell nanogels for targeted drug delivery. ACS Nano. 2011;5:5094–5099. doi: 10.1021/nn201171r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wei X, Tian T, Jia S, Zhu Z, Ma Y, Sun J, Lin Z, Yang CJ. Target-responsive DNA hydrogel mediated “stop-flow” microfluidic paper-based analytic device for rapid, portable and visual detection of multiple targets. Anal Chem. 2015;87:4275–4282. doi: 10.1021/acs.analchem.5b00532. [DOI] [PubMed] [Google Scholar]
- 103.Yan L, Zhu Z, Zou Y, Huang Y, Liu D, Jia S, Xu D, Wu M, Zhou Y, Zhou S, et al. Target-responsive “sweet” hydrogel with glucometer readout for portable and quantitative detection of non-glucose targets. J Am Chem Soc. 2013;135:3748–3751. doi: 10.1021/ja3114714. [DOI] [PubMed] [Google Scholar]
- 104.Yang H, Liu H, Kang H, Tan W. Engineering target-responsive hydrogels based on aptamer-target interactions. J Am Chem Soc. 2008;130:6320–6321. doi: 10.1021/ja801339w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhu Z, Guan Z, Jia S, Lei Z, Lin S, Zhang H, Ma Y, Tian ZQ, Yang CJ. Au@Pt nanoparticle encapsulated target-responsive hydrogel with volumetric bar-chart chip readout for quantitative point-of-care testing. Angew Chem Int Ed Engl. 2014;53:12503–12507. doi: 10.1002/anie.201405995. [DOI] [PubMed] [Google Scholar]


