Abstract
Cells interact with the extracellular environment through molecules expressed on the membrane. Disruption of these membrane-bound interactions (or encounters) can result in disease progression. Advances in super-resolution microscopy have allowed membrane encounters to be examined, however, these methods cannot image entire membranes and cannot provide information on the dynamic interactions between membrane-bound molecules. Here, we show a novel DNA probe that can transduce transient membrane encounter events into readable cumulative fluorescence signals. The probe, which translocates from one anchor site to another, such as motor proteins, is realized through a toehold-mediated DNA strand displacement reaction. Using this probe, we successfully monitored rapid encounter events of membrane lipid domains using flow cytometry and fluorescence microscopy. Our results show a preference for encounters within different lipid domains.
Cellular communication and signalling depends on molecules expressed on the membrane, especially proteins and lipids. During the resultant cell signalling, these protein and lipid molecules interact and regulate key functions such as signal transduction1. Consequently, disruption of such membrane-bound interactions, or encounters, can result in disease progression, such as that seen in cancer or inflammatory and metabolic disorders2. Indeed, understanding interaction patterns on live cell membranes is critical to biological studies. Powerful imaging techniques have been developed to track individual receptors on cell membrane surfaces3,4 and to examine cluster formation following up/downstream encounters between interacting membrane-bound cellular components, for example, proteins or lipids5,6. However, such imaging techniques may often be confounded by the absence of cluster formation. Moreover, the duration of these encounters, estimated to range from microseconds to milliseconds3, is generally too fast to be analysed by single-molecule tracking and co-localization techniques. Thus, signalling transient molecular encounter events on live cell membrane remains a technical challenge. Here, we report a DNA probe to monitor such membrane encounters. The probe functions by transducing transient encounter events into readable cumulative fluorescence signals.
Lipid domain theory holds that the cell membrane is not passive diffusing environment, but rather exerts preferential association among lipids, sterols and specific proteins7. These diffusion-restricting membrane structures are believed to play critical functions in membrane signalling and trafficking. However, the understanding of lipid domains has been limited by indirect methods, such as detergent extraction or mechanical disruption8. Recent progress in Förster resonance energy transfer (FRET) microscopy and super-resolution microscopy has begun to allow the direct measurement of lipid domains on live cell membranes. However, FRET microscopy is currently limited to the study of membrane interactions mainly between proteins, rather than lipids6. Furthermore, super-resolution methods, such as stimulated emission depletion or fluorescence photo-activation localization microscopy imaging, require costly instrumentation and provide lateral distribution of each individual membrane molecule, but no dynamic interaction information among membrane-bound molecules9. Stimulated emission depletion-fluorescence correlation spectroscopy, a new type of technique to study membrane encounters, normally cannot be used spatially across the entire membrane since it only allows the acquisition of data at one single small observation spot at a diameter of around 40 nm (ref. 10). In comparison, our method can confirm the ‘encounter’ of studied membrane molecules within <10 nm, that is, the length of DNA probe.
Characterization of DNA probe
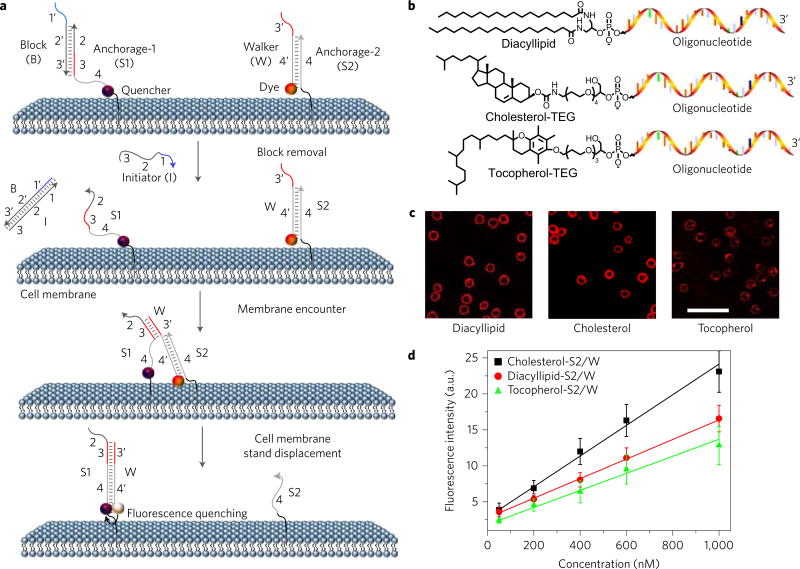
This DNA probe is inspired by the tiny motor proteins that power locomotion in the cellular world11. Artificial DNA probes have been produced to mimic motor proteins, step onto anchor sites (S) and move along a track (T)12,13. Here, we prove that live cell membranes and membrane compounds (for example, lipids or proteins) can be employed as the track and anchor sites, respectively, to construct a well-regulated dynamic DNA system (Fig. 1a). The translocation of the DNA probe (W) from one anchor site (S2) to another (S1) is realized through a toehold-mediated DNA strand displacement reaction12, in which two complementary DNA strands (W and S1) hybridize to each other, displacing, in this example, prehybridized strand S2 from the S2/W conjugate. Under our experimental conditions, the strand displacement reaction rate linearly correlates with the rate of anchor site encounter on the membrane (see Methods, equation (5)). As a result, the encounter dynamics of two anchor sites, or two target membrane components, can be measured by calculating the time-dependent variation of DNA probe translocation.
Figure 1. Anchoring and operation scheme of DNA probe on live cell membrane.
a, Schematic illustration of the operation of DNA probe on a live cell membrane. Here, the initiator (I) strand removes the block strand (B) by a strand displacement reaction. In this way, the translocation of the DNA probe (W) from one anchor site (S2) to another (S1) is triggered. b, Chemical structures of diacyllipid-, cholesterol- and tocopherol-conjugated oligonucleotides for membrane lipid domain studies. c, Confocal fluorescence microscopy images of carboxytetramethylrhodamine-labelled diacyllipid-, cholesterol- and tocopherol-conjugated S2/W conjugates (representative of six images). 500 nM of each conjugate was incubated with 5 × 105 Ramos cells at room temperature for 2 h, and unbound conjugates were removed. Scale bar, 50 µm. d, Flow cytometry evaluation of the anchoring efficiency of DNA probe on Ramos cell membranes. 0–1,000 nM initial concentrations of each conjugate were incubated with 5 × 105 Ramos cells at room temperature for 2 h, and unbound conjugates were removed. The error bars stand for the standard deviation from 5,000 cell events at each conjugate concentration.
We first characterized the kinetics of DNA probe locomotion between two anchor sites in buffer solution. In this experiment, a 6-carboxyfluorescein (FAM) dye was labelled onto the 3′-end of the DNA probe, and a corresponding quencher, Dabcyl, was attached to the 5′-end of anchor site S1. The locomotion of the DNA probe from anchor site S2 to S1 was monitored by the quenched fluorescence after mixing 10–50 nM S1 strand with 50 nM S2/W conjugate. Similar to previous reports14,15, this strand displacement reaction follows a second-order reaction model (rate constant = 5.1 × 105 M−1 s−1), and the reaction rate linearly correlates with the anchor site encounter rate (Supplementary Fig. 1).
To fine tune the locomotion of the DNA probe, we further introduced a block strand (B) and an initiator strand (I). The block (B) strand prevents strand displacement before the designated measurement and functions by blocking the DNA probe from recognizing the toehold region of anchor site S1, that is, domain 3, as shown in Fig. 1a. On the other hand, the initiator (I) strand removes the block strand by a strand displacement reaction (Fig. 1a). In this way, DNA probe locomotion can be triggered only after the addition of I, thus allowing precise regulation over displacement reaction initiation and resultant accurate measurement of strand displacement rate, which, to restate, is linearly correlated with anchor site encounter rate (Supplementary Figs 1 and 2).
Next, to anchor the DNA probe system onto a live cell membrane, DNA anchor strands S1 and S2 were chemically synthesized with several natural membrane components, including phospholipid, cholesterol and tocopherol (Fig. 1b)16,17. After incubating carboxytetramethylrhodamine (TMR)-labelled probe/anchor conjugates (500 nM at room temperature for 2 h) with a model B-cell Burkitt’s lymphoma cell line (Ramos) and removing unbound probes, the membrane anchoring of DNA probes could be clearly observed with fluorescence microscopy (Fig. 1c). Flow cytometry was further applied to calculate the degree of cell membrane anchoring. Efficient labelling of phospholipid, cholesterol and tocopherol anchor/DNA probe conjugates was observed on the membrane of both Ramos cells and CCRF-CEM cells, another human T lymphoblast cell line (Supplementary Fig. 3). Similar to a previous report18, membrane density of DNA anchoring could be precisely regulated over a large concentration range.
To investigate whether the DNA anchoring would remain bound to the membrane or be internalized after a series of cellular incubation times, we used a FAM-labelled reporter DNA to exclusively label membrane-attached oligonucleotides19. The cell membrane-anchored oligonucleotides, but not the internalized ones, were hybridized and labelled with the reporter DNA, which was not taken up by the cells based on the negatively charged phosphate groups of the oligonucleotides. As shown in Supplementary Fig. 4, during the 4 h of incubation, 85–96% of DNA anchoring remained bound to the cell membrane, as determined by the cellular fluorescence intensity, thereby allowing long-term measurement of DNA translocations.
To study the efficiency of anchoring different oligonucleotide conjugates on the same cell membrane, Cy5.5-labelled phospholipid-S1/B and FAM-labelled phospholipid-S2/W conjugates were simultaneously incubated with Ramos cells. Using flow cytometry, controlled amounts of both conjugates co-localized on the same cell membrane, as observed in different fluorescence channels (Supplementary Fig. 5), indicating successful anchor of the DNA probe onto a live cell membrane.
Locomotion of DNA probe
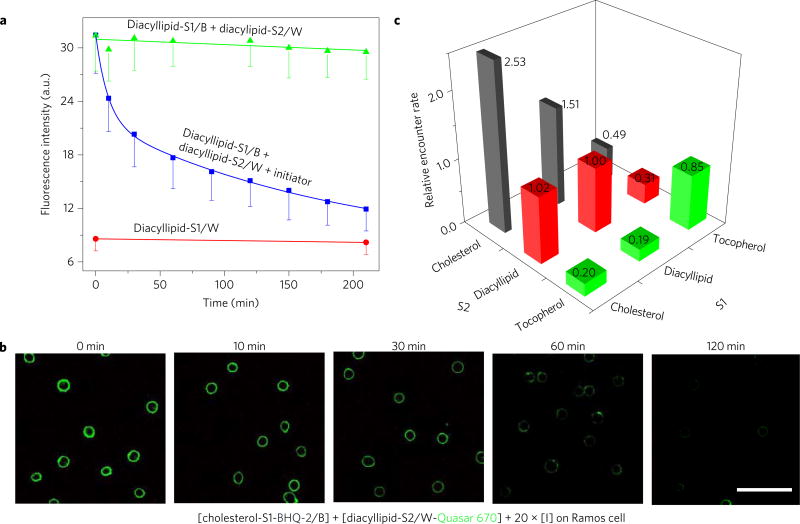
DNA probe translocation only requires initial encounters of several microseconds (Supplementary Discussion 1)14,20. Therefore, we asked if our DNA probe could be used to study previously untraceable membrane encounters in the microsecond range, such as those between membrane lipids3. To test this possibility, we examined the encounter rate of two diacyllipids. To accomplish this, we anchored a FAM-labelled diacyllipid-S2/W conjugate and a Dabcyl-labelled diacyllipid-S1/B conjugate onto a Ramos cell membrane. To avoid disruption of natural membrane interactions, the overall cell membrane modification percentage was estimated to be less than 1%. After adding I strand, the flow cytometry signal was monitored, and, as shown in Fig. 2a and Supplementary Fig. 6, a classic second-order exponential decay of cellular fluorescence indicated that a diacyllipid encounter rate-dependent DNA translocation had occurred on the cell membrane. This same phenomenon, as explained more fully in the Methods, was confirmed by fluorescence microscopy when studying membrane encounter dynamics between cholesterol and diacyllipid anchors (Fig. 2b). In another control experiment, the effect of temperature on the DNA probe system was further investigated (Supplementary Figs 7 and 11). As temperature dropped, the diffusion coefficient of diacyllipids correspondingly decreased. Indeed, a slower fluorescence quenching was observed at lower temperatures, which may at least partially indicate such changes in the lipid diffusion coefficient. All together, these experiments prove that changes in fluorescence intensity of the DNA probe provide an efficient mechanism by which to measure the encounter dynamics of cell membrane components.
Figure 2. Locomotion of DNA probe on live cell membrane.
a, Locomotion of DNA probe between two diacyllipid-conjugated anchor sites as monitored with flow cytometry. Initially 500 nM of each DNA conjugate was incubated with 5 × 105 Ramos cells ml−1. After washing the non-binding probes away, 10 µM of initiator strand I was added to initiate the strand displacement reactions at time 0. The solid lines in the figures are the fitting curves based on the bimolecular interaction model. All experiments were repeated three times. The error bars stand for the standard deviation from 5,000 cell events at each time point. b, Confocal fluorescence microscopy images for the diacyllipid–cholesterol encounter study. Quasar 670 fluorescent decay was monitored after incubating 150 nM BHQ-2-labelled cholesterol conjugate (cholesterol-S1-BHQ-2/B) and 300 nM Quasar 670-labelled diacyllipid conjugate (diacyllipid-S2/ W-Quasar 670) with 5 × 105 Ramos cells ml−1. Scale bar, 50 µm. The disappearance of Quasar 670 fluorescence (green) over time indicates membrane encounter dynamics between cholesterol and diacyllipid anchors. c, Graph showing relative Ramos cell membrane encounter rates among diacyllipid, cholesterol and tocopherol. Different encounter rates were normalized to that of diacyllipid–diacyllipid. The number of these relative encounter rates are also displayed. Original flow cytometry data are shown in Supplementary Fig. 8 with three repeated measurements. The relative encounter rates were calculated based on equation (6) in the Methods.
Next, we examined the preferential encounter dynamics among different lipid domains. Based on different viscosity and composition, lipid domains are classified as liquid-ordered (lo) or liquid-disordered (ld) domains. Therefore, to perform this study, we synthesized three types of DNA-linked membrane anchor: diacyllipids (L, lo partition based on saturated lipid composition, Supplementary Fig. 14), tocopherols (T, exclusive ld partition) and cholesterols (C, partitioning depends on membrane lipid composition)21–23. We named each DNA probe system with two letters. Taking LC as an example, the first letter, L, corresponds to diacyllipid-linked S1, and the second letter, C, corresponds to cholesterol-linked S2. Thus LC stands for the locomotion of dye-conjugated probe from cholesterol-S2 to quencher-linked diacyllipid-S1. After immobilizing approximately the same amount of anchor/oligonucleotide conjugates on the Ramos cell membrane (using 150 nM C, 300 nM L or 400 nM T based on Supplementary Figs 3 and 6), the fluorescence quenching rate of each DNA probe was measured with flow cytometry. At this point, it should be recalled that the strand displacement reaction follows a second-order reaction model and that the reaction rate linearly correlates with the lipid encounter rate.
The encounter rate of each anchor pair, for example, diacyllipid–cholesterol pair based on LC (and CL) probe measurement, was then calculated and compared (Fig. 2c, Supplementary Discussion 2 and Methods). The encounter rate among studied lipid pairs on Ramos cell membranes was found to be in the range of 8 to 84 times per millisecond, with differences up to 10-fold in the order . Here, stands for the averaged encounter rate of LC and CL probe measurements. It is worth mentioning that the observed encounter rates between heterogeneous lipid pairs will depend on the sequence of anchoring DNA strands, for example, relative encounter rate of LC was slightly larger than that of CL (Fig. 2c). This fact could be explained by different orientation and spatial availability of the DNA strand after lipid anchoring. As a result, the probability of strand displacement in a given collision can be slightly varied. To attenuate such orientation effect on strand displacement, a longer and more flexible linker between DNA and lipid/protein could be preferred (Supplementary Fig. 6). Based on the result, encounter rates for anchors of the same type (for example, CC) were found to be generally higher compared with those for heterogeneous encounters (for example, or ). This result can be expected based on the lipid domain theory, which holds that the same lipid tends to be confined in the same type of lipid domain, thus interacting more frequently.
It was also observed that faster diffusion does not necessarily equate with more frequent encounters. For example, although tocopherol has a diffusion coefficient that is 2.0-fold over that of diacyllipid (Supplementary Figs 12 and 13), TT encounters are less frequent compared with LL encounters (0.85-fold). Since lipid domains confine the membrane distribution of lipid molecules, their local concentration is increased.
DNA probe competition game to study encounter preference
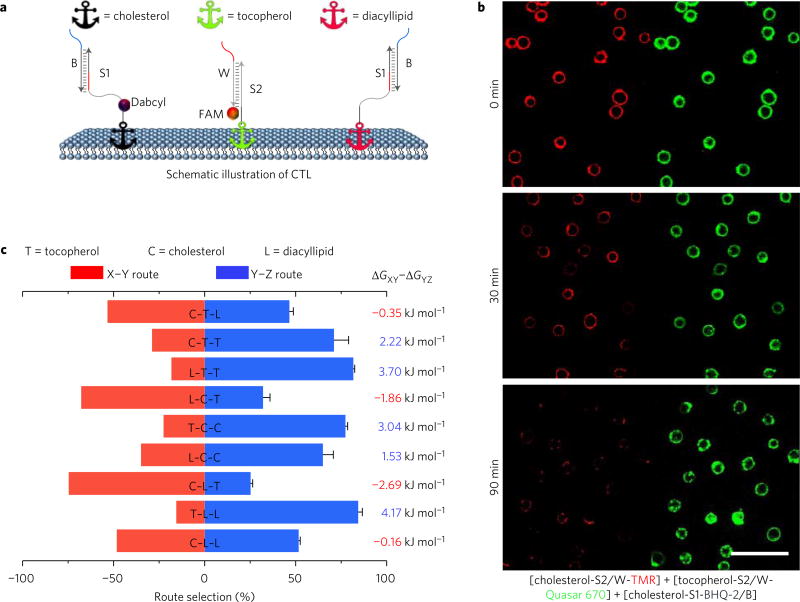
To study the preference of one type of membrane anchor (Y) towards potential encounter anchors (X and Z) on the membrane, we further designed a DNA probe competition game, termed XYZ. Taking CTL as an example, (Fig. 3a), in this game, dye-labelled probe strand initially conjugated with tocopherol anchor T is given two possible destinations, quencher-labelled cholesterol C or unlabelled diacyllipid L anchor sites. If tocopherol anchor T prefers an encounter with cholesterol anchor C, a fast and complete quenching will be observed. In contrast, a slower and less complete event indicates that tocopherol–diacyllipid encounter T–L was preferred, essentially because the translocation of probe to diacyllipid L anchor does not result in fluorescence intensity change. The same sequence (S1) for both destinations guarantees that the choice will be based on encounter preference, but not oligonucleotide hybridization rate differences. As shown in Fig. 3, diacyllipid preferred to interact with another saturated phospholipid (84%) instead of tocopherol (16%), with a nearly equal chance of interacting with another diacyllipid (52%) or cholesterol (48%). If given only the choice between cholesterol and tocopherol, the diacyllipid-conjugated probe showed a threefold preference for cholesterol. The encounter preference of each anchor can be summarized as L ≈ C > T for encounter with diacyllipid, C > L > T for encounter with cholesterol, and T > C > L for encounter with tocopherol. Therefore, this competition game provides a single-step approach to simultaneously compare two encounter pairs on the same cell surface. Compared with individual encounter rate measurement, the competition game reduces experiment-to-experiment and cell-to-cell errors.
Figure 3. DNA probe competition game to study encounter preference.
a, Schematic showing DNA probe competition game results among diacyllipid, cholesterol and tocopherol anchors on the Ramos cell membrane. Initially, 150 nM cholesterol, 300 nM diacyllipid and 400 nM tocopherol conjugates were incubated with 5 × 105 Ramos cells ml−1. b, Fluorescence microscopy study of the TCC competition game on the Ramos cell membrane. In this game, BHQ-2 quencher-labelled cholesterol (cholesterol-S1-BHQ-2/B) was given two possible destinations, TMR-labelled cholesterol (red, cholesterol-S2/W-TMR) or Quasar 670-labelled tocopherol (green, tocopherol-S2/W-Quasar 670) anchor sites. The quenching of TMR fluorescence (‘winning’ site) was apparently faster than that of Quasar 670 fluorescence (‘losing’ site), indicating that cholesterol–cholesterol encounters are preferred than those of cholesterol– tocopherol. Scale bar, 50 µm. c, Route selection was between Dabcyl-labelled anchor (red bar) and non-labelled anchor (blue bar). In the divergent stacked bar chart, all the bar length represents 100%. The left-shifted bar indicates a preferred X–Y encounter, while right-shifted bar shows that a Y–Z encounter was preferred. Taking C–T–L as an example, in this game, dye-labelled probe strand initially conjugated with tocopherol anchor T is given two possible destinations, quencher-labelled cholesterol C or unlabelled diacyllipid L anchor sites. Here, the divergent stacked bar chart indicates 54% tocopherol anchor will encounter preferably with a cholesterol anchor (C–T route), while 46% encounters with a diacyllipid anchor (T–L route). Original flow cytometry data are shown in Supplementary Fig. 8 for three repeated measurements. The relative route choice was calculated based on equation (10) in the Methods. Gibbs free energy changes (ΔG) were calculated based on the corresponding competition results.
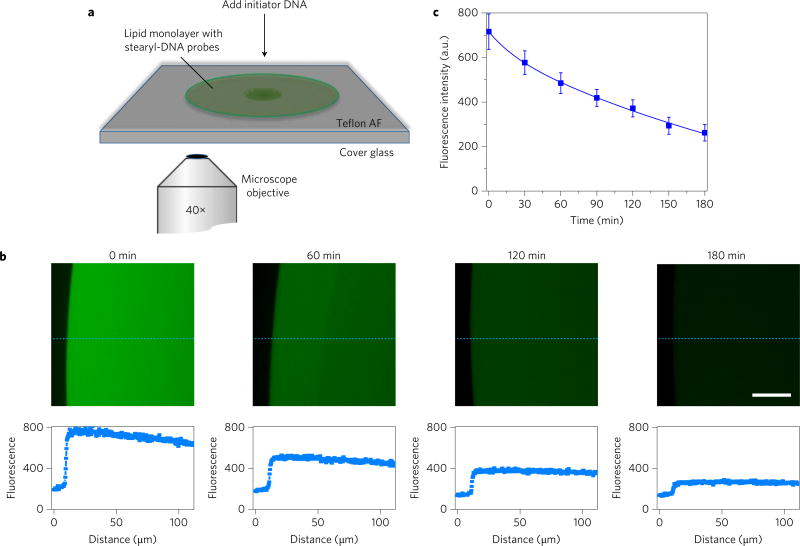
To calculate the membrane encounter rates among these cholesterol, diacyllipid and tocopherol molecules, we need to identify the membrane strand displacement reaction probability after encounter. To study this reaction probability, a stearyl-DNA-incorporated lipid monolayer film was prepared (Fig. 4a). The encounter rate of DNA probe in this film follows a free-diffusing two-dimensional Smoluchowski model15,24. Based on this model and effective fluorescence signal change (Fig. 4), membrane strand displacement reaction probability after a lipid-S1 and lipid-S2/W encounter was calculated to be 0.032. Further based on this encounter reaction probability, in this study, the lipid encounter rates on individual Ramos cell membrane are calculated to be in the range of 8.5 to 89/ms (Supplementary Discussion 2).
Figure 4. Locomotion of DNA probe on model lipid monolayer film.
a, Illustration of experimental set-up at the fluorescence microscope. Soybean polar extract lipid solution was spiked with stearyl-S2/W-FAM conjugate and stearyl-S1-Dabyl/B conjugate separately at a DNA:lipid ratio of 1:10,000. After dropping and mixing these two DNA–lipid mixtures on a Teflon AF-coated cover glass, a circularly spreading lipid patch was formed. Fluorescence signal of lipid biofilm was monitored for 3 h after adding excess amount of initiator DNA strand. b, Fluorescence microscopy images of stearyl–DNA encounter on model lipid monolayer film. Scale bar, 30 µm. The representative fluorescence intensity curves along the dashed blue line are indicated below each panel. Two other positions were also analysed for each image to calculate the average fluorescence intensity of the lipid monolayer film. c, Locomotion of DNA probe between two stearyl-conjugated anchor sites on model lipid monolayer biofilm as monitored with fluorescence microscopy. At each time point, the fluorescence signal from the edge of the lipid film to 100 µm towards the centre was averaged. The error bars stand for the standard deviation from these data. All experiments were repeated three times.
In addition, these encounter rates can be further applied to estimate the effective cell membrane diffusion area of each lipid. The existence of lipid domains confines the membrane distribution of lipid molecules, thus lipid effective membrane concentration increased. Based on the increased effective encounter rate compared with that in the free diffusion mode, the percentage of lipid diffusion area across cell membrane can be calculated. In our study, the diffusion areas of diacyllipid, cholesterol and tocopherol were estimated to cover 52, 39 and 77%, respectively, of the Ramos cell membrane (Supplementary Discussion 3). This information can be potentially important to estimate the sizes of lipid domains on live cell membranes.
DNA probe to study membrane protein encounter rates
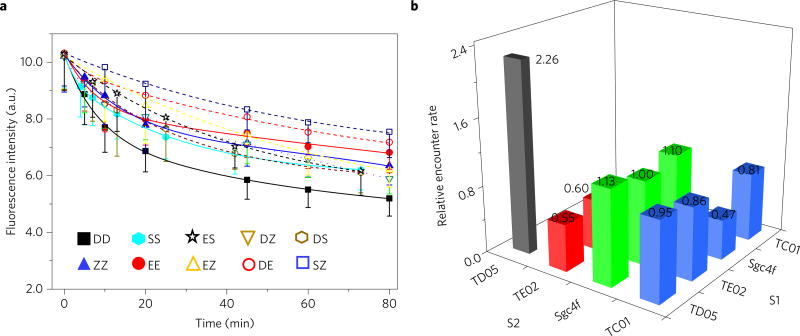
Finally, to test whether the method can be applied to various anchors, such as membrane proteins, we further tethered the DNA probe to membrane protein-binding ligands, for example, aptamers. Aptamers are single-stranded DNAs or RNAs, which can selectively recognize a wide range of targets, including cell membrane proteins25. Four proteins co-expressed on the Ramos cell surface were chosen and designated D, E, S and Z, because some of their identities were unknown. D, E, S and Z were respectively targeted by DNA aptamers TD05, TE02, Sgc4f and TC01 (Fig. 5a and Supplementary Fig. 9)26. The target-binding properties of these aptamers were tested and confirmed to be preserved after conjugation with S1 or S2 oligonucleotides (Supplementary Fig. 10). Applying the same method as shown in Fig. 1a, the encounter rate for the same type of protein was found to increase in the order of , with differences up to 3.8-fold (Fig. 5b). The accurate calculation of membrane protein encounter rates can be difficult here. The orientation of protein-linked DNA probes during encounters will influence the efficiency of the strand displacement reaction. A careful design of the linker region and binding site on the protein is important (Supplementary Fig. 6). In our case, it is interesting to note that the most frequent interaction was between two TD05 aptamers, whose target, immunoglobulin heavy mu chain (IgM), is known to aggregate as a type of B-cell receptor for signal transduction on the membrane27. The fast initial quenching of DD may stem from DNA probe locomotion within the same IgM pentamer. These results indicate that the DNA probe can be used to study the encounter dynamics of various membrane components.
Figure 5. DNA probe–aptamer conjugates to study membrane protein encounter rates.
a, Locomotion of DNA probe between two aptamer-conjugated anchor sites as monitored with flow cytometry. Initially, 100 nM TC01 (Z)-, 400 nM TD05 (D)-, 600 nM TE02 (E)- or 1 µM Sgc4f (S)-conjugated DNA probe conjugates were incubated with 5 × 105 Ramos cells ml−1. After washing the non-binding probes away, 1 µM initiator strand I was added to initiate the strand displacement reactions at time 0. All experiments were repeated at least three times. The error bars stand for the standard deviation from 5,000 cell events at each time point. The fitting curves are based on second-order reaction, Exponential Decay 2 function. The solid lines are used for fitting data between the same type of protein (that is, DD, ZZ, SS and EE), whereas the dashed lines are used between heterogeneous pairs (that is, ES, DZ, DS, EZ, DE and SZ). b, Summarized relative encounter rates among the four types of aptamer–oligonucleotide conjugates studied. Different encounter rates were normalized to that of ES, that is, the locomotion of dye-conjugated probe from Sgc4f aptamer-conjugated S2 to quencher-linked TE02 aptamerconjugated S1. The number of these relative encounter rates are also displayed. The relative encounter rates were calculated based on equation (6) in the Methods.
Conclusions
A major goal in biology aims to understand the underlying structure of cell signalling networks. By applying the most recent advancements of DNA nanotechnology, our approach offers a practical mechanism to study such structures with the ultimate aim of detecting downstream pathophysiological dysregulation induced by abnormal lipid and protein interactions. This novel membrane DNA probe can be easily conjugated with different cell anchors to perform cellular measurement of rapid membrane encounters. Compared with other imaging probes based on cluster formation after membrane encounters, including fluorescent protein-based FRET assays3–6, our method can transduce transient membrane encounter events into cumulative cell surface signals, thus transient membrane lipid encounters can be studied. With standard fluorescence microscopy and flow cytometry, this DNA probe can detect and calculate encounter rates and preferences during various live cell membrane signalling events.
Methods
Manipulation of the membrane DNA probe
The ligand-conjugated DNA duplexes X-S1/B and Y-S2/W were incubated separately in 1× phosphate-buffered saline (PBS) buffer (pH = 7.4 with 137 mM NaCl and 2.7 mM KCl) for 1 h before use. Each conjugate was then incubated at a concentration of 200 nM with 5 × 105 cells ml−1 in 200 µl binding buffer (containing 4.5 g l−1 glucose, 5 mM MgCl2, 0.1 mg ml−1 yeast tRNA and 1 mg ml−1 bovine serum albumin (BSA) in Dulbecco’s PBS with calcium chloride and magnesium chloride) and shaken every 20 min. X-S1/B conjugates were generally mixed with cells for 20 min prior to the addition of Y-S2/W conjugate to ensure that no X-S1/W conjugates were formed before initiating the strand displacement reactions. Cells were then washed three times with PBS to remove free probes and resuspended in binding buffer. After washing and discarding the non-binding probes, 20-fold (compared with the initial concentration of S1/B conjugate) initiator strand I was typically added to initiate the strand displacement reaction. During each experiment, the initial fluorescence signal was examined before adding the initiator strand to demonstrate the proper concentration of DNA probes; meanwhile, the same batch of cells was used for the control experiment by adding the same amount of DNA probes and wrapping the dye/quencher labelling (X-S1-quencher/Y-S2-dye versus X-S2-dye/Y-S1-quencher) in order to confirm that a similar concentration of both ligands was labelled. Each experiment was repeated three times.
Measurement of membrane strand displacement efficiencies
The cellular fluorescence signal was monitored with a FACScan cytometer (Becton Dickinson Immunocytometry Systems) by counting 5,000 events at each time point, using channel #3 for the 6-carboxyfluorescein dye and channel #5 for the PE-Cy5.5 dye. The confocal microscope images were acquired by an Olympus FV500-IX81 with a 488 nm argon laser and a 543/633 nm helium/neon laser (Olympus America) for fluorescence signals from TMR dye- or Quasar 670 dye-modified DNA probes.
Kinetics of cell surface locomotion
The dynamic toehold-mediated strand displacement reaction has been proven to follow a second-order reaction model14,15. In our system, the locomotion of DNA probe in buffer solution can be written as:
| (1) |
Based on previous simulations, it should be noted that the contribution of the reverse rate constant k−1 will be negligible15; as a result, k1 functions as an apparent displacement process rate constant. Based on the Stern–Volmer equation, the fraction of the maximum fluorescence change indicates the reaction efficiency at any specific time. Since the toehold binding process is mediated by the effective concentration of the incoming strand (that is, the encounter rate), k1 is a diffusion rate-influenced constant. As a proof, two studies using biophysical models have reported that tethered hybridization, instead of free diffusion, will effectively increase the rates of strand displacement reactions28,29. The operation of the DNA probe on the cell membrane can be written as a two-step process:
| (2) |
| (3) |
Since a large excess of I strand is introduced to remove block strand B, the first step appears as a pseudo-first-order reaction, which is almost finished within a minute Supplementary Fig. 2). As a result, the second step, that is, probe locomotion, is the rate-limiting step in this process, and the entire rate law can be written as:
| (4) |
Note that CCS1 and CWS2 represent DNA probe density on the cell membrane in units of mol cm–2 and that the overall process is second-order, depending on the original immobilization amounts of both B/S1 and W/S2 conjugates. After solving the apparent locomotion rate constant k″, the encounter rate (F) of the two oligonucleotide-tethered ligands on the cell membrane can be calculated by
| (5) |
where P represents the reaction probability after an S1 and W/S2 encounter, which can be obtained based on the Smoluchowski equation24. The diffusion coefficients of free oligonucleotide strands and that of oligonucleotide-modified membrane ligands can be obtained based on fluorescence recovery after photobleaching30. Since F is a factor that depends on the membrane density of each immobilized strand, a relative encounter rate between two encounter pairs with similar surface concentration can be a more meaningful indicator for comparison. In our case, since (1) the same strand sequences are used to study different ligand interactions and (2) all reaction directions are assumed to occur along the surface of the cell membrane, PAA and PBB can be considered the same. Thus, the relative encounter rate between ligand A–A and ligand B–B can be obtained as
| (6) |
In equation (6), only the relative concentrations of DNA probes are needed, and these can be determined from the fluorescence enhancement by flow cytometry or fluorescence microscopy. Therefore, information about the absolute density of oligonucleotides on the cell membrane is not necessary. As an example, the relative encounter rate between two diacyllipid-conjugated anchors was studied under different initial concentrations of DNA probes (Supplementary Fig. 3). It has been demonstrated that a direct correlation exists between the encounter rate and the relative membrane density of DNA probe. Moreover, by comparing the experimental data with the theoretical fitting curve, at the low membrane density condition (≤ 300 nM), which is employed later in this study, the apparent locomotion rate constant k″ can be viewed as independent of the initial concentration of the probes, thus validating our approach that extracts the inherent encounter rate differences among various surface ligands.
In our study, experimental fluorescence data were implemented with OriginPro 8. Based on the nature of second-order reaction, the built-in Exponential Decay 2 function “y = A1 *exp(−x/t1) + A2 *exp(−x/t2 ) + y0″ was used for fitting the data. In equation (4), the simplest case occurred when initial membrane concentrations of S1 and WS2 were almost the same. Then k″ could be obtained through half-time measurements, which is the time it takes for a fluorescence signal to decrease to half of the original value after subtracting the background signal. Then, , where stands for initial S1 concentration at time 0, which was determined from the fluorescence calibration curve. However, in the case of different concentrations between S1 andWS2, a plot of versus time would be plotted, and the slope of the linear curve would be .
Kinetics of cell membrane competition game
Cell membrane fluorescence change was monitored during the first 80 min after adding I strand, and the pathway selection was studied by comparing the kinetic decay results of the XYZ system with that of the ZYX system. ZYX is the system for which the two possible final destinations will be reversely labelled, that is, unlabelled X anchor site and quencher-labelled Z anchor site. A second fluorescence kinetic decay curve will be measured in such condition. For two second-order competition reactions
| (7) |
| (8) |
the reaction rate ratio can be expressed as
| (9) |
At a specific time t when the product strand concentration (CXW and CZW) is still small compared with that of the initial strand (CX0 and CZ0), the relative choice of towards X and Z can be calculated based on percentage fluorescence decrease at time t, fXY (=(CXW/CX0)) and fYZ (=(CZW/CZ0)), as
| (10) |
Preparation of lipid monolayer film spiked with DNA probe
The lipid monolayer film was prepared following ref. 15. Teflon AF-coated microscope glass coverslides were prepared by spin-coating. In short, after thoroughly cleaning and blow-drying the coverslides, 1.2% Teflon AF solution (diluted from 6% with Fluorinert FC-770) was added. Spin-coating was performed at 2,000 r.p.m. for 1 min. The coverslides were then baked at 180 °C for 5 min to finish coating. To prepare DNA–lipid mixtures, soybean polar extract lipid solutions were spiked with stearyl-S2/W-FAM conjugate and stearyl-S1-Dabcyl/B conjugate, respectively, at a DNA:lipid ratio of 1:10,000. After equilibration at 8 °C overnight for DNA incorporation into the lipid layer, 10 µl DNA–lipid mixture was dried under reduced pressure to remove chloroform for 1 h, and then rehydrated into 10 µl 1× PBS buffer. 1 µl of both stearyl-S2/W-FAM and stearyl-S1-Dabcyl/B lipid solution were added and mixed on the above-prepared coverslides that were coated with Teflon AF. After adding excess amount of initiator DNA strand to start the encounter measurement, fluorescence signal of the lipid biofilm was monitored for 3 h. At each time point, the averaged fluorescence signal from the edge of the lipid film to 100 µm towards the centre was used to plot and calculate the DNA strand displacement reaction efficiency.
Manipulation of DNA probe to study membrane protein encounter rates
Locomotion of DNA probe between two aptamer-conjugated anchor sites was manipulated similarly as other membrane DNA probes. The DNA probe–aptamer duplexes X-S1/B and Y-S2/W were incubated separately in 1× PBS buffer for 1 h before use. 100 nM TC01-, 400 nM TD05-, 600 nM TE02- or 1 µM Sgc4f-conjugated DNA probe conjugates were then incubated with 5 × 105 cells ml−1 in 200 µl binding buffer and shaken every 20 min. Cells were then washed three times with PBS to remove free probes and resuspended in binding buffer. After washing and discarding the non-binding probes, 1 µM initiator strand I was added to initiate the strand displacement reaction. Here, to measure the heterogeneous encounters, , that is, the averaged encounter rate of XY and YX probe measurements, the same batch of cells was used for the control experiment by adding the same amount of DNA probes and wrapping the dye/quencher labelling (X-S1-quencher/Y-S2-dye versus X-S2-dye/Y-S1-quencher). All experiments were repeated at least three times.
Data availability
All relevant data are available from the authors, requests should be addressed to M.Y. and/or W.T.
Supplementary Material
Acknowledgments
The authors thank X. Fang, G. Fanucci, V.D. Kleiman, K. Salaita and Y. Tseng for helpful discussions and suggestions. This work is supported by NSFC grants (NSFC 21521063 and NSFC 21327009), and the US National Institutes of Health (GM079359 and CA133086). The work of G.B. was supported in part by a Key Project of the Major Research Plan of NSFC (no. 91130004), a NSFC A3 Project (no.11421110002), NSFC Tianyuan Projects (no. 11426235; no. 11526211), a NSFC Innovative Group Fund (no.11621101) and US NSF FRG DMS-0968360.
Footnotes
Author contributions
M.Y., Y.L. and D.H. contributed equally to this work. M.Y., D.H. and W.T. conceived and designed the experiments. M.Y. and Y.L. performed the experiments. M.Y., D.H., L.P. and G.B. analysed the data. M.Y., D.H., Y.L., T.C., C.S.W. and L.Z. synthesized lipid-DNA and other reagents. Y.L., M.Y., L.Q. and Q.L. prepared giant unilamellar vesicles and lipid monolayer. M.Y. and W.T. co-wrote the manuscript. W.T. supervised the project. All authors discussed the results and commented on the manuscript.
Supplementary information is available in the online version of the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Kholodenko BN. Cell signaling dynamics in time and space. Nat. Rev. Mol. Cell. Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wymann MP, Schneiter R. Lipid signaling in disease. Nat. Rev. Mol. Cell. Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 3.Groves JT, Parthasarathy R, Forstner MB. Fluorescence imaging of membrane dynamics. Annu. Rev. Biomed. Eng. 2008;10:311–338. doi: 10.1146/annurev.bioeng.10.061807.160431. [DOI] [PubMed] [Google Scholar]
- 4.Chung I, et al. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- 5.Endres NF, et al. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152:543–556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loura LMS, Prieto M. FRET in membrane biophysics: an overview. Front. Physiol. 2011;2:1–11. doi: 10.3389/fphys.2011.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 8.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 9.Hess ST, et al. Dynamic clustered distribution of hemagglutinin resolved at 40nm in living cell membranes discriminates between raft theories. Proc. Natl Acad. Sci. USA. 2007;104:17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eggeling C, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 11.Schliwa M, Woehlke G. Molecular motors. Nature. 2003;422:759–765. doi: 10.1038/nature01601. [DOI] [PubMed] [Google Scholar]
- 12.Zhang DY, Seelig G. Dynamic DNA nanotechonology using strand-displacement reactions. Nat. Chem. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 13.Rudchenko M, et al. Autonomous molecular cascades for evaluation of cell surfaces. Nat. Nanotech. 2013;8:580–586. doi: 10.1038/nnano.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang DY, Winfree E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 2009;131:17303–17314. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- 15.Hannestad JK, et al. Kinetics of diffusion-mediated DNA hybridization in lipid monolayer films determined by single-molecule fluorescence spectroscopy. ACS Nano. 2013;7:308–315. doi: 10.1021/nn304010p. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, Kwong B, Irvine DJ. Membrane anchored immunostimulatory oligonucleotides for in vivo cell modification and localized immunotherapy. Angew. Chem. Int. Ed. 2011;50:7052–7055. doi: 10.1002/anie.201101266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gartner ZJ, Bertozzi CR. Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc. Natl Acad. Sci. USA. 2009;106:4606–4610. doi: 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borisenko GG, Zaitseva MA, Chuvilin AN, Pozmogova GE. DNA modification of live cell surface. Nucleic Acids Res. 2009;37:e28. doi: 10.1093/nar/gkn1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H, Johnston AP. A programmable sensor to probe the internalization of proteins and nanoparticles in live cells. Angew. Chem. Int. Ed. 2013;52:5744–5748. doi: 10.1002/anie.201301243. [DOI] [PubMed] [Google Scholar]
- 20.Yin Y, Zhao X. Kinetics and dynamics of DNA hybridization. Acc. Chem. Res. 2011;44:1172–1181. doi: 10.1021/ar200068j. [DOI] [PubMed] [Google Scholar]
- 21.Beales PA, Vanderlick TK. Application of nucleic acid-lipid conjugates for the programmable organisation of liposomal modules. Adv. Colloid. Interface Sci. 2014;207:290–305. doi: 10.1016/j.cis.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Loew M, et al. Lipid domain specific recruitment of lipophilic nucleic acids. J. Am. Chem. Soc. 2010;132:16066–16072. doi: 10.1021/ja105714r. [DOI] [PubMed] [Google Scholar]
- 23.Beales PA, Vanderlick TK. Partitioning of membrane-anchored DNA between coexisting lipid phases. J. Phys. Chem. B. 2009;113:13678–13686. doi: 10.1021/jp9006735. [DOI] [PubMed] [Google Scholar]
- 24.Hardt SL. Rates of diffusion controlled reactions in one, two and three dimensions. Biophys. Chem. 1979;10:239–243. doi: 10.1016/0301-4622(79)85012-7. [DOI] [PubMed] [Google Scholar]
- 25.Tan W, Donovan MJ, Jiang J. Aptamers from cell-based selection for bioanalytical applications. Chem. Rev. 2013;113:2842–2862. doi: 10.1021/cr300468w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Z, et al. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 27.Mallikaratchy P, et al. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt’s lymphoma cells. Mol. Cell. Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Genot AJ, Zhang DY, Bath J, Turberfield AJ. Remote toehold: a mechanism of flexible control of DNA hybridization kinetics. J. Am. Chem. Soc. 2011;133:2177–2182. doi: 10.1021/ja1073239. [DOI] [PubMed] [Google Scholar]
- 29.Chandran H, Gopalkrishnan N, Phillips A, Reif J. DNA. Vol. 17. Springer Verlag; 2011. Localized hybridization circuits; pp. 64–83. [Google Scholar]
- 30.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available from the authors, requests should be addressed to M.Y. and/or W.T.