Abstract
Impedance spectroscopy has been applied in prokaryotic and eukaryotic cytometry as a label-free method for the investigation of adherent cells. In this paper, its use for characterizing the growth dynamics of P. aeruginosa biofilms is described and compared to crystal violet staining and confocal microscopy. The method allows monitoring the growth of biofilm-forming P. aeruginosa in a continuous and label-free manner over a period of 72 h in a 96 well plate format. Impedance curves obtained for P. aeruginosa PA14 wild type and mutant strains with a transposon insertion in pqsA and pelA genes exhibited distinct phases. We propose that the slope of the declining curve following a maximum at ca. 35–40 h is a measure of biofilm formation. Transplant experiments with P. aeruginosa biofilms and paraffin suggest that the impedance also reflects pellicle formation at the liquid-air interface, a barely considered contributor to impedance. Finally, the impairment of biofilm formation upon treatment of cultures with L-arginine and with ciprofloxacin, tobramycin and meropenem was studied by single frequency impedance spectroscopy. We suggest that these findings qualify impedance spectroscopy as an additional technique to characterize biofilm formation and its modulation by small molecule drugs.
Introduction
Infections caused by pathogenic bacteria represent a major health threat that is expected to rise further in the future1. The formation of biofilms is in many cases a prerequisite for the development of chronic bacterial infections that can persist for months or years2, because they increase the resistance of bacterial pathogens to drug therapies3–8. Biofilms not only constitute a physical barrier that impedes the penetration of antibiotics, but also exhibit an altered metabolism compared to planktonic bacteria9. In a biofilm, microbes are embedded in a self-produced matrix of extracellular polymeric substances (EPS), which consist of DNA, proteins and polysaccharides.
P. aeruginosa forms various types of biofilms, which differ in the composition of their EPS, depending on its genetic background and environmental cues10. Important polysaccharides for the three-dimensional biofilm are pel, psl and alginate10–15. In case of the commonly used laboratory strain PA14, the polysaccharide pel is mandatory for biofilm formation14, 16. Concordant, the pelA::Tn mutant is not capable of forming a pellicle biofilm at the air liquid interface. It is also known that the P. aeruginosa quinolone signal (PQS) plays a significant role as a signal molecule for the PQS quorum sensing system. The latter system regulates the production of extracellular DNA, an important biofilm matrix component11. Accordingly, a psqA::Tn mutant was found to form less biofilm in a microscopic biofilm screen as well as in a murine animal model17–19.
Biofilm structures are induced by nutrition cues and/or non-lethal concentrations of antibiotics20. Pseudomonas aeruginosa has become an important model organism to study biofilms, as this opportunistic Gram-negative pathogen is the frequent cause of devastating chronic, biofilm-associated infections, for example in cystic fibrosis or wound patients21, 22.
To develop new therapeutic solutions for such infections, there is a high demand for diagnostic methods that also characterize the biofilm phenotype of pathogens. Common techniques mostly rely on colorimetric measurements of biofilm formation23–36. A frequently applied method is the staining of biofilms by crystal violet; however, this technique suffers from high standard deviations, cannot determine viability and is an endpoint measurement, reflecting only a single time point. Two recent, commercially available techniques are the Calgary Biofilm Device and the Biofilm Ring Test. Both of them define antimicrobial efficacies in relation to biofilm formation in a 96 well-plate format30, 37. Detailed three-dimensional information on biofilms of P. aeruginosa was obtained by automated confocal laser scanning microscopy (CLSM) following a live and death staining of cells33. This 96 well-plate-based optical method could determine antibiotic efficacy in biofilms in a quantitative, spatially resolved manner. Its utility was also proven in a genetic screen for mutants affecting the formation of biofilms18 that allowed deducing specific biofilm phenotypes.
‘Single-frequency impedance analysis’ measures the opposition that an electrochemical system exhibits upon application of an alternating voltage. Also biological matrices constitute such an electrochemical system, featured by energy dissipater (resistor) and energy storage (capacitor) elements. The use of EIS to characterize changes in the density and morphology of eukaryotic cells adherent to an electrode surface has been pioneered by Giaever and Keese38–40. As the method provides a label free, non-invasive and quantitative assessment of cellular processes like protrusion, proliferation, viability, migration, trans-endothelial invasion, wound healing, apoptosis, cytotoxicity, or cell-cell adhesion, and as it is simple, prone to miniaturization and to low-cost formats, EIS has found widespread applications in cell biology, bioanalysis, and medicine40–51. Impedance spectroscopy was also applied since the 1970’s52 for the detection of bacteria in food and manufacturing industries53. A variety of impedance studies on bacterial biofilm formation have been reported54–56. Among those, Paredes et al. characterized the biofilm formation of Staphylococcus epidermis in four different devices57. It was shown that the change in impedance during biofilm formation in an unstirred device with a sensor on the bottom was primarily driven by the capacitance. This effect was explained by an increased charge transfer resistance by the coating of the sensor material. Gottschamel et al. found that contactless bioimpedance measurements are particularly sensitive toward capacitive changes during biofilm formation and maturation58. Kim et al. observed a decrease in the double-layer capacitance during adhesion and biofilm maturation of P. aeruginosa at the electrode surface, and speculated that bacteria obstructed double-layer charging by blocking the surface59. A microfluidic sensor platform was constructed by Bruchmann et al. to investigate the growth and the viability of P. aeruginosa strains in a time-dependent manner60. The impact of disinfectants and detergents on the mostly continuously increasing impedance curves (at a starting OD of 0.5) was also reported on that system. Ward et al. constructed a low-cost printed carbon sensor and used frequency sweeps at distinct time points to reveal a distinct impedance signature of P. aeruginosa that is shaped by the production of phenazines61. The authors summarize that the impedance signal is influenced by six mechanisms: (i) the production of redox compounds; (ii) the deposition of biofilm material on the electrode surface; (iii) charge transfer through the attachment of cells and microbial nanowires; (iv) the presence of microbial cells in close proximity to the electrode surface; (v) breakdown of nutrients within the electrolyte; and (vi) the adsorption of proteins to the electrode surface. Of course, the relative contributions of these mechanisms to impedance may change over the growth period of the microorganism.
In this paper, we highlight differences in the dynamics of biofilm formation of three P. aeruginosa strains by following the single frequency impedance spectroscopy over 72 h in a label-free manner. The setup was successfully applied to study the effects of three standard antibiotics on biofilm formation over time. Finally, we describe another contributing factor to impedance that, to the best of our knowledge, has not been considered so far, i.e. the pellicle biofilm that forms at the air-liquid interface.
All studies were performed by single-frequency impedance spectroscopy using a commercial instrument, the xCELLigence Real Time Cell Analyzer (RTCA) from Acea Biosciences that measures impedance in 96 well plates equipped with gold microelectrodes. The instrument expresses impedance in form of a standardized, dimensionless parameter termed cell-index. We suggest that the slope of the cell index at specific detection time periods can characterize pellicle biofilm formation with performance features (standard deviations and effect sizes) superior to crystal violet staining. A part of the results has been included in a patent application by us (see conflict of interest statement)62. Due to the automated setup using microtiter plates, impedance spectroscopy may have additional applications, e.g. for the screening of compound libraries to discover inhibitors and/or enhancers of biofilm formation.
Results
Characterization of Pseudomonas aeruginosa biofilms by inverted confocal microscopy, crystal violet staining and single-frequency impedance spectroscopy
Three different strains of Pseudomonas aeruginosa that were well-characterized with respect to their biofilm-forming ability, i.e. the PA14 wild type (wt) and the pelA::Tn and pqsA::Tn transposon mutants, were selected for analysis by single-frequency impedance spectroscopy. The results were compared to those obtained by confocal microscopy and crystal violet staining as reference techniques. In crystal violet staining experiments, the biofilm was visible as a ring at the air-liquid interface (pellicle), while confocal microscopy measured structures on the bottom of microtiter plates at the solid-liquid interphase (Fig. 1A,B). The pellicle staining was robust, tolerating rigorous washing steps. In contrast, the solid-liquid biofilm was very fragile and strongly dependent on the formed pellicle as well as on nutrient and oxygen levels. The pellicle biofilm of PA14 was increasingly formed over a period of 72 h in half-area µclear 96 well plates as well as in the 96 well E-plates that were used for impedance measurements (Fig. 1B,C). The pelA::Tn mutant did not form any pellicle in both plate types, confirming preceding reports in similar experimental setups. In contrast, the staining of the pqsA::Tn mutant’s pellicle was even stronger than that of the wild type. The data obtained by inverted confocal microscopy at the solid-liquid interface gave a different picture (Fig. 1D,E). Here, the pqsA::Tn mutant showed reduced levels of biofilm, as the calculated biovolume of the 3D biofilm stack (E) was lower compared to the wild type. In contrast, the calculated biovolumes of the pelA::Tn mutant were as high as (24 h, 72 h) or higher than (48 h) those of the wild type. However, the microscopic images in Fig. 1D show that the pelA::Tn mutant did not form a three-dimensional biofilm structure, as opposed to the wild type and the pqsA::Tn mutant. Visual microscopic observations revealed that the pelA::Tn biovolume was a result of a massive accumulation of planktonic cells, which biased the biovolume calculation. This was also visible in the time series presented and holds true for the 24 h values of the wild type and in part the pqsA::Tn mutant. Microtiter plate biofilms usually change over time due to changes in nutrient and oxygen availability. While the induced dispersal can be seen for the wild type and pqsA::Tn mutant, the appearance of pelA::Tn mutant was not changed. Based on crystal violet staining, the pelA::Tn mutant was impaired in biofilm formation at the air-liquid interface, while inverted confocal microscopy showed a reduction in biovolume for the pqsA::Tn mutant at the solid-liquid interface.
Figure 1.

Comparison of biofilms measured with inverted confocal laser scanning microscopy (CLSM) and with crystal violet staining. (A) Schematic visualization of biofilm localization in a microtiter plate well. (B) Example of the biofilm stained with crystal violet in an E-plate 96 (ACEA, USA). (C) Biofilm biomass determined with crystal violet staining in a half-area, µclear microtiter plate (n = 12) (Greiner Bio-One). The crystal violet background of the medium was subtracted. (D) Easy 3D projections of biofilm structures stained with the BacLight Viability Kit. (E) Biofilm biovolume determined from biofilm structures as depicted in Fig. 1D with a customized solution of the image analysis software Definiens (n = 6). Red bars: PA14 Wild type; olive bars: pqsA::Tn mutant; light green bars: pelA::Tn mutant. n indicates the number of technical replicates.
As most studies of impedance spectroscopy measure biofilms at distinct time points, we wanted to investigate whether the technique could capture the dynamics of biofilm formation through continuous measurements. In addition, the ability of the technique to discern differences between the strains was probed. The cell index values of the single-frequency impedance measurements over 72 h are shown in Fig. 2A. The cell index of the negative control (LB medium alone) was not constant, but decreased slowly over time. In all experiments with P. aeruginosa, a pronounced decay of the cell index over approximately 8 hours occurred, whose magnitude was variable between experiments. For the pelA::Tn mutant, a structureless, constantly increasing cell index was observed after that time period. In contrast, the curves obtained for the PA14 wt and the pqsA::Tn mutants increased to a maximum after ca. 33 h, and then showed a linear decay over time for a period of ca. 10 h. After a second minimum, a signal increase was observed that lasted to the end of the measurement time (see also Table S1 and Figures S2–S4). All three strains produced an approximately equal number of colony forming units per well after 72 h, demonstrating that the overall growth was equal. This finding indicates that the differences in single-frequency impedance curves did not reflect differences in growth, and that bacteria were alive after a 72 h cultivation period in the 96 well E-plate without further nutrient additions. The impedance dynamics were referenced to crystal violet staining intensities of the strains under equivalent conditions (Fig. 2B). Crystal violet staining demonstrated that the majority of the pellicle biofilm was formed in the time between 24 and 48 h. Shorter time points (<20 h) could not be captured by crystal violet, as a visible, physical biofilm was not formed in that period. Thus, in the period of biofilm formation (ca. 24–48 h), the cell index curves were featured by a local maximum and a linear decline (Fig. 2A).
Figure 2.
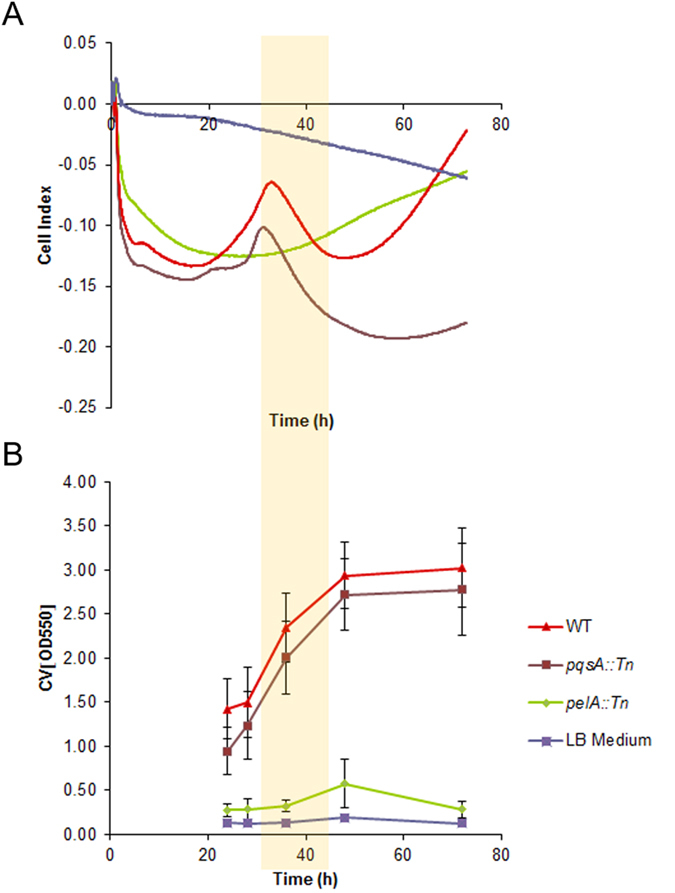
(A) Changes in the standardized single-frequency impedance (expressed as cell indices) and (B) crystal violet staining measurements of pellicle biofilm over 72 h, during biofilm growth in LB culture medium of PA14 wt, pqsA::Tn mutant, and pelA::Tn mutant (n = 24 for each mutant) and LB Medium (n = 8). The PA14 wild type and pqsA::Tn are biofilm formers; pelA::Tn does not form biofilm. n indicates the number of technical replicates. The period of linear decline of the cell index is highlighted by a yellow bar.
Single-frequency impedance spectroscopy detects biofilm changes at the air-liquid interface
In order to probe the specific effect of the pellicle biofilm on the cell index, we performed two experiments: First, we added paraffin (10–100 µl) as an artificial substrate to LB medium (Fig. 3A). Paraffin was chosen to mimic the addition of an inert, liquid material on the top of the broth solution, as it is immiscible with water and therefore floats on top of the medium due to its lower density. Because its chemical composition consists of alkanes, paraffin has a low dielectric constant, in contrast to the pellicle biofilm of P. aeruginosa. Secondly, a pellicle biofilm was transplanted with a metal loop into wells filled with LB medium (Figs 3B and S6). The pellicle biofilm was obtained by growing PA14 in a 96 well plate in LB medium for 72 h. We observed that the cell index was reduced by the addition of pellicle biofilm (Fig. 3B); the decrease was steeper than that obtained for the control (added PBS buffer in varying amounts of 10–80 µl). This effect cannot be explained by the addition of an immiscible layer on top of the LB medium, as the addition of paraffin led to unchanged slopes of cell index (Fig. 3A). We concluded from this experiment that the electrical single frequency impedance is strongly influenced by a bacterial biofilm at the air-liquid interface. In addition, we concluded that the declining cell index curve is a characteristic of pellicle biofilm formation.
Figure 3.
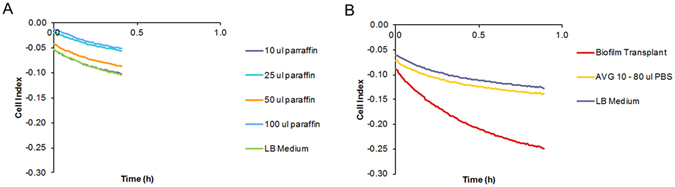
Effect of the addition of biofilm and control substances on top of the bacterial broth on single-frequency impedance. (A) Impedance curves following addition of paraffin in different amounts (n = 4). (B) Impedance curves following addition of PA14 pellicle biofilm (n = 4) or different amounts of PBS buffer (n = 1). As the curves for PBS buffer additions had identical shapes, an average curve is shown. n indicates the number of technical replicates.
Pharmacological interference of biofilm formation can be monitored by single-frequency impedance spectroscopy
The cell index curves depicted in Fig. 2 demonstrate that the growth of biofilm is a process with distinct phases. We next wanted to probe whether cellular single-frequency impedance spectroscopy can capture the effects of low molecular weight compounds that interfere with biofilm formation. First, the effect was tested through the addition of 0.4% and 0.8% (w/w) L-arginine. L-Arginine is a model compound for biofilm suppression that acts by stimulating anaerobic metabolism in P. aeruginosa and other biofilm-forming bacteria63, 64.
Because the decline of the single-frequency impedance curve following the maximum after ca. 30–35 h coincided with the period of biofilm growth (yellow bar in Fig. 2A), and as it captured the effect of an added pellicle biofilm, the slope of the linear phase of the declining cell index, determined by fitting the data to a local linear model, was used as a parameter for pellicle biofilm formation. The decline of the cell index became less steep upon the addition of L-arginine in a concentration dependent manner; the effect occurred in both PA14 wt as well as the pqsA::Tn mutant (Fig. 4A, Table S2 and Figure S3). Furthermore, the biofilm formation started earlier with pqsA::Tn (28.4 h ± 0) than with the wt (30.5 h ± 0.7). Also, 0.4% and 0.8% (w/w) L-arginine shortened the induction time for the biofilm formation of pqsA::Tn 29.2 h ± 1.5, 26.7 h ± 0.7 and the wt 25.0 h ± 0.7, 24.2 h ± 0.7, respectively. In parallel, the biofilm growth under these conditions was monitored by crystal violet staining after 43 hours as a reference, showing reduced intensities for both strains upon addition of L-arginine, as expected (Fig. 4B). For the end point measurement with crystal violet, maximal fold changes of 0.40 and 0.13 in OD620 values with the wt and the pqsA::Tn mutant were measured with maximal relative standard deviations of 46% and 17%, respectively. Single-frequency impedance measurements showed fold changes of 0.48 and 0.47 in slope for the two strains, respectively, with corresponding maximal relative standard deviations of 17% and 14%. Thus, the higher dynamic range and the smaller standard deviation of the impedance measurements suggest that the method may be better suited to track subtle changes in biofilm growth than crystal violet measurements. In fact, recording multiple time points by the CV method requires is experimentally tedious, associated with large error bars between measurements, and the frequent removal of plates from the incubator constitutes a perturbation of microbial growth (Figure S4).
Figure 4.

Impact of L-arginine (n=3) (A) on the cell index slope measured by single-frequency impedance spectroscopy over 41 h and (B) on the crystal violet staining intensity of PA14 wt and the pqsA::Tn mutant strains. The crystal violet background of the medium was subtracted. n indicates the number of technical replicates.
The method has also been used to study the effect of antibiotics on biofilm formation of the PA14 wt over 70 h. For this purpose, ciprofloxacin, tobramycin and meropenem have been tested, as they represent current standard treatments of P. aeruginosa infections. All three antibiotics changed the impedance curves in a concentration-dependent manner, indicating effects on biofilm formation (Fig. 5, Table S3, Figures S5 and S7). Interestingly, single-frequency impedance spectroscopy could monitor effects at antibiotic concentrations that did not impact planktonic growth in the same period of time. For example, the addition of 0.125 µg/ml ciprofloxacin clearly changed the impedance curve, although growth, determined by OD measurements of bacterial cultures in the presence of antibiotics, was not impaired at this concentration (Fig. 5C). At a concentration of 0.25 µg/ml, steady biofilm formation was not observed anymore, but planktonic growth was still possible. The effect on impedance was quantified by three parameters at varying antibiotic concentrations: The first parameter is the time of onset of a linear decrease of the cell index (Fig. 5A). The second parameter is the duration of a linear decrease of the cell index (Fig. 5A). The third parameter is the cell index slope during that time period (Fig. 5B). As outlined above, both parameters were determined by a linear trendline that was fitted to the data (Figures S5 and S7). For ciprofloxacin and tobramycin, a less steep decline could be observed at increasing concentrations, indicating an impaired biofilm formation. For meropenem on the other hand, the cell index slope was rather constant. Especially at the highest tested concentrations, all antibiotics led to later onsets of linear declines of impedance; at the same time, the period of linear decline was longer compared to untreated samples, indicating a delayed and attenuated biofilm formation. When antibiotic concentrations are sufficient to inhibit bacterial growth, changes in impedance diminish, and a constant curve is obtained (see for example Scheme S5, Tobramycin concentrations from 7.5 to 30 µg/ml).
Figure 5.
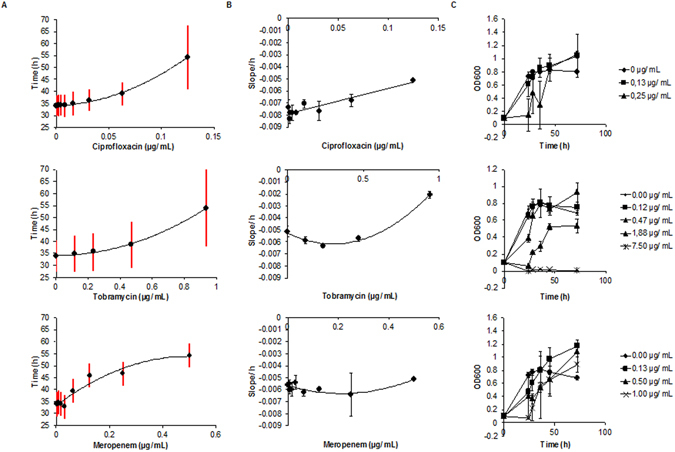
Impact of ciprofloxacin, tobramycin, and meropenem on biofilm formation of the PA14 wt strain over 70 h. (A) Impact of antibiotics on the time period of biofilm formation. The red vertical bars cover the time period of linear decline of the cell index value at a given antibiotic concentration, as determined by the fitting model. The lower end of the red bar marks the onset of CI decline, the upper end marks the end of the decline (n = 3 or 4). (B) The slope of the cell index is plotted against the antibiotic concentration (n = 3 or 4). (C) Growth curves of PA14 at various antibiotic concentrations over time (n = 4). The starting OD600 at t = 0 was 0.1. n indicates the number of technical replicates.
Discussion
The single-frequency impedance of microbial broth solutions is modulated by a variety of biological, chemical and electrochemical processes. As these processes are dynamic along the bacterial life cycle, the deconvolution of the main contributors to impedance is a subject of current research59, 61. By following the single-frequency impedance of P. aeruginosa PA14 over a time period of 72 h, we observed curve shapes that were featured by a maximum at ca. 30–35 h followed by a period of decline of ca. 10 h. These features could be abrogated by a pelA::Tn mutant that was not able of forming a biofilm, although it was not impaired in growth, which implies that the impedance curve is indeed shaped by biofilm formation. Bruchmann et al. reported a continuous rise of impedance over days at slightly higher starting OD’s (0.5 vs. 0.1 in our study) of P. aeruginosa cultures using a microfluidic device60, implying that the dynamics of the impedance signal is strongly influenced by the instrumental setup, i.e. by flow vs. static biofilm growth conditions. One important difference is that pellicle formation is hampered by the flow. This suggests that the pellicle may account for differences in signal shape between the studies. In our study, the effect of the pellicle biofilm was separated from that of the liquid broth by a transplant experiment, demonstrating that the pellicle alone changed single-frequency impedance. This effect cannot be explained by the addition of an immiscible layer on top of the LB medium, as the addition of paraffin, an inert material with low dielectric constant, led to unchanged slopes of the cell index. The finding highlights the pellicle biofilm formed at the air-liquid interface as another contributing factor to impedance. Ward et al. recently noted the pellicle effect as well when putting the electrode in contact with the liquid air interface61. The electrode in the present study is located at the bottom of the well distant from the pellicle, demonstrating that the pellicle can exert a remote effect at a distance of ca. 0.8 cm to the electron surface. We suggest that the pellicle may increase the capacitance of the material, thereby contributing to an overall decline of impedance. However, the molecular mechanism behind the effect remains unclear at this stage. This suggestion is in line with the fact that the linear decrease of the impedance curve coincides with the growth period of the pellicle, as monitored by crystal violet staining. The overall shape of the single-frequency impedance curve over 73 h is complex and influenced by various electrochemical mediators and processes along the growth phase of P. aeruginosa. Therefore, we do not imply that the cell index at any time is specific for biofilm formation. However, the comparison between biofilm formers and the respective isogenic controls cultivated under identical conditions suggests that the signal in a specific time period, which coincides with the formation of a pellicle biofilm, indeed reflects biofilm growth. We therefore suggest the length and the slope of the linear part of the cell index as simple descriptors of biofilm formation in P. aeruginosa.
In our studies with adherent eukaryotic cells, CI values up to +5 were observed (unpublished data). In contrast, the CI values detected for P. aeruginosa biofilms in this study were about 20fold smaller. As eukaryotic cells are larger and adhere to the electrode surface in dense (mono)layers, their impact on the resistance to impedance is a lot larger than that of Pseudomonas cells, whose attachment to the electrode surface was limited.
Using L-arginine as a suppressor of biofilm formation, a head-to-head comparison with crystal violet staining demonstrated that quantification by impedance curves was associated with higher dynamic ranges and smaller standard deviations than the notoriously variable crystal violet staining method, thereby enabling to disclose also subtle perturbances of a biofilm. This was further explored by monitoring the effects of three standard antibiotics on single-frequency impedance and its descriptors slope, onset and duration of linear decline. It is well-documented that antibiotics induce significant alterations of gene expression even at low doses65–68. We observed that sublethal concentrations that did not impair bacterial growth also altered the impedance of P. aeruginosa PA14 cultures, and that the substance effects were different. At higher concentrations, all antibiotics delayed the onset and extended the period of linear curve decline, which we interpret as a consequence of impaired biofilm formation.
In summary, single-frequency impedance spectroscopy with a commercially available instrument has been applied to characterize the growth dynamics of biofilm-forming P. aeruginosa strains in a label-free manner. The technique revealed distinct phases of biofilm formation and characterized the effects of antibiotics and other biofilm-modulating compounds with a precision that was superior to crystal violet staining. Using mutant strains and transplant experiments, we demonstrated that the pellicle biofilm formed at the air-liquid interface is an important contributor to impedance.
As the measurements are done in 96 well plates in an automated fashion, and as a 384 well plate format is supported by the instrument, single-frequency impedance spectroscopy may find utility for the medium throughput screening of compound libraries to discover modulators of biofilm formation. Future work will be devoted to extending the method to other biofilm-forming pathogens, and to gaining an enhanced understanding of impedance curve dynamics, for example to disclose the molecular processes that lead to the curve maximum. More detailed descriptor functions that also capture the earlier part of impedance curves may be conceived in order to obtain compound signatures that reflect a specific mode of action. While such an application of single-frequency impedance spectroscopy has been reported for eukaryotic cells69, 70, the current work suggests that it is also viable for biofilm-forming bacteria. A diagnostic utility for clinical purposes may be derived following miniaturization of the assays to a low-cost chip format, as demonstrated for other applications of impedance microbiology.
Material and Methods
Strains and cultivation conditions
The reference strain Pseudomonas aeruginosa PA14 (DSM 50071) and two mutant strains with a transposon insertion within the pqsA::Tn or the pelA::Tn gene were applied in this study10, 17, 71. Both mutants have been shown to result in a low biofilm morphotype16, 18. The mutant strains were obtained from the PA14 transposon mutant library of the F.M. Ausubel lab. And the strains were stored in glycerol stocks at −70 °C. The pre-cultures (4 ml lysogeny broth (LB) medium) were incubated overnight at 37 °C and 180 rpm. Chemicals were obtained from commercial sources (ciprofloxacin hydrochloride from Biomedicals no. 02199020, tobramycin sulfate salt from Serva no. 36562.01, meropenem trihydate from Sigma Aldrich no. M2574, L-arginine from AppliChem no. A3709). The compounds were diluted with LB media and distributed to the test plate just before the incubation with bacteria. The concentration ranges for the antibiotics were selected according to MIC values reported on the EUCAST website, while literature data were adapted for arginine concentrations72.
Biofilm growth assay
The cultivation of static biofilms at the bottom of 96 well microtiter plates was performed as previously described33. Overnight cultures were adjusted with LB to an OD600 of 0.02. From these cultures, 100 µl were transferred into a half-area, μclear microtiter plate (Greiner Bio-One), covered with an air-permeable foil and incubated at 37 °C in an incubator with humid atmosphere for 24 h, 48 h and 72 h. To stain the bacteria, 60 µl of staining solution containing Syto9 and propidium iodide with final concentrations of 2.1 µM and 12.5 µM, respectively (LIVE/DEAD BacLight Bacterial Viability kit, Molecular Probes/Life Technologies), was added per well. Stacks of the biofilms were acquired with an automated confocal laser-scanning microscope (TCS SP8, Leica Microsystems) equipped with an x40/1.10 water objective and a z-step size of 3 μm between the image layers. The acquired image stacks were analysed with a customized solution from the software developer XD (Definiens) to calculate biofilm-relevant parameters and visualized with the software IMARIS (Bitplane).
Crystal violet staining
To quantify biofilm formation, a 96 well plate was inoculated from a pre-culture73. LB start cultures were prepared with an OD600 of 0.1 (200 µl/well). To measure the attached ring of the pellicle biofilm, cultures were withdrawn by suction, and the wells were washed with 200 µl H2O to remove media and unattached cell material. To stain the biofilm, 150 µl crystal violet staining solution (0.1% m/v in H2O) was added per well and incubated for 30 min at room temperature. The staining solution was removed, and the wells were washed with 200 µl H2O. After the plates were dried at room temperature, 200 µl ethanol (95%) was added to each well to destain the pellicle biofilm ring for 30 min at room temperature. 125 µl of the violet ethanol solution was transferred into a fresh 96-well plate, and the absorbance was measured at 550, 580 and 620 nm.
Single-frequency impedance measurement
Before inoculation with pre-cultures, the background impedance of the culture media was measured from 100 µl LB medium per well in an E-plate 96 (ACEA, USA) with the xCELLigence RTCA SP (single plate) instrument (ACEA, USA). The analyzer with the E-plate was placed in a 37 °C incubator with 5% CO2. For each well, the background was subtracted from the measurement points by setting the cell index to zero at t = 0. Thereby, the cell index is a self-calibrated value. After pausing the system, the E-plate was reinstalled in the tissue culture incubator after inoculating of the wells with 200 µl of a LB-diluted overnight culture (start OD600 of 0.1). In a 5 minute interval for each well the single-frequency impedance signal was monitored for at least 41 h with default settings. The xCELLigence instrument displays the impedance as a standardized dimensionless parameter termed cell index (CI). Because the change in the electrical impedance at a certain frequency is divided by the nominal impedance value, the CI represents a relative value. The CI at a given time point t (CI(t)) is calculated as follows (Eq. 1).
| 1 |
where ƒn is the frequency at which the impedance measurement is carried out; R(ƒn, t) is the measured impedance at frequency ƒn at time point t; R(ƒn, t0) is the measured impedance at frequency ƒn at time point t0 (usually t0 is the time when the background is measured) and Zn is the corresponding nominal impedance value of ƒn. All experiments have been performed at a frequency of 10 kHz and a nominal impedance value of 15 Ohm.
The observed CI values fell into the measurement range of the XCellingence instrument. The lower limit of CI for a reproducible measurement is 0.01–0.02, translating to an impedance change of 0.15 ohm–0.3 ohm. Whilst the impedance analyzer could certainly reproducibly resolve 0.15–0.3 ohm impedance changes, the RTCA system measurement could be affected by other factors such electrical contact resistance between the E-Plate and E-Plate station and other parts of measurement system. The upper limit of CI is experted to be >100, since the system could reproducibly measure the impedance up to many thousands of ohms, or even higher74.
The electrical potential at the electrodes is about 10 mV. The Alternating Current (AC) voltage is from an impedance analyzer where the voltage signal is digitally synthesized to a voltage amplitude (about 20 mV) and sinusoidal waveform of several variable frequencies (e.g. 10 kHz, 25 kHz, 50 kHz). There is no reference electrode in the system. The AC voltage signal from the analyzer is applied to the electrodes through electronic switching circuits. The system measured AC electrical current by monitoring the voltage drop across of an internal resistance. The impedance is calculated from the division of the voltage by the current74. All experiments in the study have been conducted at a frequency of 10 kHz. The open circuit potential remains constant during the experiment. The change in the electrical current going through the system leads to the derivation of the change in the monitored impedance.
The gold electrodes are of interdigitated, circle-on-line electrode geometry. The circle is of 90 μm diameter. The separation between the neighboring electrode lines is 20 μm. The electrodes are fabricated on glass substrate with lithographic techniques75.
The equivalent circuit of impedance (Z) can be described as a serial combination of the resistance (Z R = R) and reactance (Z C = 1/jwc) (Eq. 2).
| 2 |
For identification of the linear range of cell index decay, reflecting the biofilm growth period, a 5 h sliding window was moved in 1 hour steps along the time axis, and a local linear model was fitted. Only data from 20 h onwards was considered, based on empirical data on the lag phase of P. aeruginosa biofilm growth. Next, local linear models were removed if r2 was below the cut-off of 0.97 (Figure S1). In the presence of tobramycin, the r2 was adapted to 0.89 based on a higher noise ratio at a concentration of 0.94 µg/ml. This however did not lead to an inaccurate measurement, given the calculated variances of slopes of sliding windows in the presence of tobramycin did not increase (Figure S7). The range of biofilm growth was defined as beginning from the start of the first to the end of the last of a series of consecutive sliding windows fulfilling the defined criteria, while allowing for a maximum of one local linear model with r2 below the cutoff.
To mimic the presence of pellicle biofilm in a point measurement, paraffin, which is a hydrophobic, electrically inactive compound, was added in different amounts (10–100 µL) (n = 4) to separate wells that contained LB medium. Paraffin is immiscible with water and floats on top of the medium due to its lower density. In a second set of experiments, biofilm grown in a 96 well plate in LB medium for 72 h was transplanted with a metal loop into four wells filled with LB medium. To transplant the pellicle biofilm, it was first carefully loosened by adding 30 µl PBS under the pellicle biofilm structure with a pipette. In control wells, varying amounts of PBS buffer (10, 20, 40, and 80 µl) (n = 1) were added to capture the inpact of PBS on single-frequency impedance. CI values were compared to the LB medium (n = 8). In both sets of experiments, the xCELLigence system was paused before addition.
Electronic supplementary material
Acknowledgements
We would like to thank Dr. Xiaobo Wang and Dr. Silke Schwengberg, Acea Biosciences, for helpful discussions and information on the XCelligence instrument.
Author Contributions
B.K. and J.v.D. designed and performed biofilm experiments using single-frequency impedance spectroscopy and crystal violet staining. M.M. designed and performed biofilm experiments using confocal microscopy. J.T. analyzed the cell index data by fitting to a linear model. J.v.D., M.B., S.H. and C.W. analyzed the data and wrote the manuscript.
Competing Interests
Jozef B.J.H. van Duuren, Bianka Karge, Mark Brönstrup, and Christoph Wittmann have filed a patent application (PCT/EP2015/080540) on ‘Impedance spectroscopy for characterizing biofilms’.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05273-5
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.European Centre for Disease prevention and Control/European Medicines Agencies (ECDPC/EMA) ECDC/EMEA Joint Technical report. Stockholm: ECDPC/EMA. The bacterial challenge: time to react. A call to narrow the gap between multidrug-resistant bacteria in the EU and the development of new antibacterial agents (2009).
- 2.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science (New York, N.Y.) 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 3.Davies D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug. Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert P, McBain AJ. Biofilms: their impact on health and their recalcitrance toward biocides. Am. J. Infect Control. 2001;29:252–255. doi: 10.1067/mic.2001.115673. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert P, Maira-Litran T, McBain AJ, Rickard AH, Whyte FW. The physiology and collective recalcitrance of microbial biofilm communities. Adv. Microb. Physiol. 2002;46:202–256. [PubMed] [Google Scholar]
- 6.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 7.Bjarnsholt T, Ciofu O, Molin S, Givskov M, Hoiby N. Applying insights from biofilm biology to drug development - can a new approach be developed? Nat. Rev. Drug Discov. 2013;12:791–808. doi: 10.1038/nrd4000. [DOI] [PubMed] [Google Scholar]
- 8.Coenye T, Van Dijck P, Bjarnsholt T, Forsberg A. Microbial biofilms–the coming of age of a research field. Pathog. Dis. 2014;70:203–204. doi: 10.1111/2049-632X.12169. [DOI] [PubMed] [Google Scholar]
- 9.Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 2009;73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 2004;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 11.Allesen-Holm M, et al. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 12.Diggle SP, et al. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 2003;50:29–43. doi: 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- 13.Davies DG, Chakrabarty AM, Geesey GG. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 1993;59:1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colvin KM, et al. PelA deacetylase activity is required for Pel polysaccharide synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2013;195:2329–2339. doi: 10.1128/JB.02150-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jennings LK, et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA. 2015;112:11353–11358. doi: 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colvin KM, et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 2012;14:1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haussler S, Becker T. The pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog. 2008;4:e1000166. doi: 10.1371/journal.ppat.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musken M, Di Fiore S, Dotsch A, Fischer R, Haussler S. Genetic determinants of Pseudomonas aeruginosa biofilm establishment. Microbiol. Mol. Biol. Rev. 2010;156:431–441. doi: 10.1099/mic.0.033290-0. [DOI] [PubMed] [Google Scholar]
- 19.Komor U, et al. Biofilm formation by Pseudomonas aeruginosa in solid murine tumors - a novel model system. Microbes Infect. 2012;14:951–958. doi: 10.1016/j.micinf.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman LR, et al. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 21.Lyczak JB, Cannon CL, Pier GB. Lung Infections Associated with Cystic Fibrosis. Clin. Microbiol. Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/S1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 23.Pierce CG, et al. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 2008;3:1494–1500. doi: 10.1038/nprot.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Repp KK, Menor SA, Pettit RK. Microplate Alamar blue assay for susceptibility testing of Candida albicans biofilms. Med. Mycol. 2007;45:603–607. doi: 10.1080/13693780701581458. [DOI] [PubMed] [Google Scholar]
- 25.Huang CT, Yu FP, McFeters GA, Stewart PS. Nonuniform spatial patterns of respiratory activity within biofilms during disinfection. Appl. Environ. Microbiol. 1995;61:2252–2256. doi: 10.1128/aem.61.6.2252-2256.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramage G, Vande Walle K, Wickes BL, López-Ribot JL. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob. Agents Chemother. 2001;45:2475–2479. doi: 10.1128/AAC.45.9.2475-2479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitts B, Hamilton MA, Zelver N, Stewart PS. A microtiter-plate screening method for biofilm disinfection and removal. J. Microbiol. Meth. 2003;54:269–276. doi: 10.1016/S0167-7012(03)00034-4. [DOI] [PubMed] [Google Scholar]
- 28.Ahn SJ, Ahn SJ, Wen ZT, Brady J, Burne RA. Characteristics of biofilm formation by Streptococcus mutans in the presence of saliva. Infect. Immun. 2008;76:4259–4268. doi: 10.1128/IAI.00422-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 30.Ceri H, et al. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 1999;37:1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooun A, Liu S, Lewis K. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2000;44:640–646. doi: 10.1128/AAC.44.3.640-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson ME, Ceri H, Morck DW, Buret AG, Read RR. Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can. J. Vet. Res. 2002;66:86–92. [PMC free article] [PubMed] [Google Scholar]
- 33.Müsken M, Di Fiore S, Römling U, Häussler S. A 96-well-plateg-based optical method for the quantitative and qualitative evaluation of Pseudomonas aeruginosa biofilm formation and its application to susceptibility testing. Nat. Protoc. 2010;5:1460–1469. doi: 10.1038/nprot.2010.110. [DOI] [PubMed] [Google Scholar]
- 34.Sandoe JAT, Wysome J, West AP, Heritage J, Wilcox MH. Measurement of ampicillin, vancomycin, linezolid and gentamicin activity against enterococcal biofilms. J. Antimicrob. Chemother. 2006;57:767–770. doi: 10.1093/jac/dkl013. [DOI] [PubMed] [Google Scholar]
- 35.De Kievit TR, et al. Multidrug efflux pumps: Expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2001;45:1761–1770. doi: 10.1128/AAC.45.6.1761-1770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tré-Hardy M, Vanderbist F, Traore H, Devleeschouwer MJ. In vitro activity of antibiotic combinations against Pseudomonas aeruginosa biofilm and planktonic cultures. Int. J. Antimicrob. Agents. 2008;31:329–336. doi: 10.1016/j.ijantimicag.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 37.Chavant P, Gaillard-Martinie B, Talon R, Hebraud M, Bernardi T. A new device for rapid evaluation of biofilm formation potential by bacteria. J. Microbiol. Methods. 2007;68:605–612. doi: 10.1016/j.mimet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 38.Giaever I, Keese CR. A morphological biosensor for mammalian cells. Nature. 1993;366:591–592. doi: 10.1038/366591a0. [DOI] [PubMed] [Google Scholar]
- 39.Giaever I, Keese CR. Micromotion of mammalian cells measured electrically. Proc. Natl. Acad. Sci. USA. 1991;88:7896–7900. doi: 10.1073/pnas.88.17.7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giaever I, Keese CR. Monitoring fibroblast behavior in tissue culture with an applied electric field. Proc. Natl. Acad. Sci. USA. 1984;81:3761–3764. doi: 10.1073/pnas.81.12.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Armando Gagliardi P, et al. Real-time monitoring of cell protrusion dynamics by impedance responses. Sci. Rep. 2015;5:10206. doi: 10.1038/srep10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ke N, Wang X, Xu X, Abassi YA. The xCELLigence system for real-time and label-free monitoring of cell viability. Methods Mol. Biol. 2011;740:33–43. doi: 10.1007/978-1-61779-108-6_6. [DOI] [PubMed] [Google Scholar]
- 43.Pietrosimone, K. M., Yin, X., Knecht, D. A. & Lynes, M. A. Measurement of cellular chemotaxis with ECIS/Taxis. J. Vis. Exp., doi:10.3791/3840 (2012). [DOI] [PMC free article] [PubMed]
- 44.Rahim, S. & Uren, A. A real-time electrical impedance based technique to measure invasion of endothelial cell monolayer by cancer cells. J. Vis. Exp., doi:10.3791/2792 (2011). [DOI] [PMC free article] [PubMed]
- 45.Keese CR, Wegener J, Walker SR, Giaever I. Electrical wound-healing assay for cells in vitro. Proc. Natl. Acad. Sci. USA. 2004;101:1554–1559. doi: 10.1073/pnas.0307588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arndt S, Seebach J, Psathaki K, Galla HJ, Wegener J. Bioelectrical impedance assay to monitor changes in cell shape during apoptosis. Biosens. Bioelectron. 2004;19:583–594. doi: 10.1016/S0956-5663(03)00269-0. [DOI] [PubMed] [Google Scholar]
- 47.Tiruppathi C, Malik AB, Del Vecchio PJ, Keese CR, Giaever I. Electrical method for detection of endothelial cell shape change in real time: assessment of endothelial barrier function. Proc. Natl. Acad. Sci. USA. 1992;89:7919–7923. doi: 10.1073/pnas.89.17.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atienza JM, Yu N, Wang X, Xu X, Abassi Y. Label-free and real-time cell-based kinase assay for screening selective and potent receptor tyrosine kinase inhibitors using microelectronic sensor array. J. Biomol. Screen. 2006;11:634–643. doi: 10.1177/1087057106289334. [DOI] [PubMed] [Google Scholar]
- 49.Atienza JM, Zhu J, Wang X, Xu X, Abassi Y. Dynamic monitoring of cell adhesion and spreading on microelectronic sensor arrays. J. Biomol. Screen. 2005;10:795–805. doi: 10.1177/1087057105279635. [DOI] [PubMed] [Google Scholar]
- 50.Benson K, Cramer S, Galla HJ. Impedance-based cell monitoring: barrier properties and beyond. Fluids and barriers of the CNS. 2013;10:5. doi: 10.1186/2045-8118-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.di Blasio L, et al. PDK1 regulates focal adhesion disassembly by modulating endocytosis of alphavbeta3 integrin. J. Cell Sci. 2015;128:863–877. doi: 10.1242/jcs.149294. [DOI] [PubMed] [Google Scholar]
- 52.Cady, P. In New approaches to the identification of microorganisms (ed Illeni T Heden CG) 74–79 (Wiley, 1975).
- 53.Firstenberg-Eden, R. & Eden, G. Impedance microbiology. (Research Studies Press, 1984).
- 54.Varshney M, Li Y. Interdigitated array microelectrodes based impedance biosensors for detection of bacterial cells. Biosens. Bioelectron. 2009;24:2951–2960. doi: 10.1016/j.bios.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 55.Dheilly A, et al. Monitoring of microbial adhesion and biofilm growth using electrochemical impedancemetry. Appl. Microbiol. Biotechnol. 2008;79:157–164. doi: 10.1007/s00253-008-1404-7. [DOI] [PubMed] [Google Scholar]
- 56.Kim S, Yu G, Kim T, Shin K, Yoon J. Rapid bacterial detection with an interdigitated array electrode by electrochemical impedance spectroscopy. Electrochim. Acta. 2012;82:126–131. doi: 10.1016/j.electacta.2012.05.131. [DOI] [Google Scholar]
- 57.Paredes J, Becerro S, Arana S. Comparison of real time impedance monitoring of bacterial biofilm cultures in different experimental setups mimicking real field environments. Sensors Actuators, B: Chem. 2014;195:667–676. doi: 10.1016/j.snb.2014.01.098. [DOI] [Google Scholar]
- 58.Gottschamel J, et al. Development of a disposable microfluidic biochip for multiparameter cell population measurements. Anal. Chem. 2009;81:8503–8512. doi: 10.1021/ac901420u. [DOI] [PubMed] [Google Scholar]
- 59.Kim T, Kang J, Lee JH, Yoon J. Influence of attached bacteria and biofilm on double-layer capacitance during biofilm monitoring by electrochemical impedance spectroscopy. Water Res. 2011;45:4615–4622. doi: 10.1016/j.watres.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 60.Bruchmann J, Sachsenheimer K, Rapp BE, Schwartz T. Multi-channel microfluidic biosensor platform applied for online monitoring and screening of biofilm formation and activity. PloS one. 2015;10:e0117300. doi: 10.1371/journal.pone.0117300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ward AC, Connolly P, Tucker NP. Pseudomonas aeruginosa can be detected in a polymicrobial competition model using impedance spectroscopy with a novel biosensor. PloS one. 2014;9:e91732. doi: 10.1371/journal.pone.0091732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brönstrup, M., Wittmann, C., van Duuren, J. & Karge, B. Impedance spectroscopy for characterizing biofilms. PCT/EP2015/080540 (2015).
- 63.Kolderman E, et al. L-arginine destabilizes oral multi-species biofilm communities developed in human saliva. PloS one. 2015;10:e0121835. doi: 10.1371/journal.pone.0121835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Borriello G, Richards L, Ehrlich GD, Stewart PS. Arginine or nitrate enhances antibiotic susceptibility of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 2006;50:382–384. doi: 10.1128/AAC.50.1.382-384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blazquez J, Couce A, Rodriguez-Beltran J, Rodriguez-Rojas A. Antimicrobials as promoters of genetic variation. Curr. Opin. Microbiol. 2012;15:561–569. doi: 10.1016/j.mib.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 66.Dorr T, Lewis K, Vulic M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 2009;5:e1000760. doi: 10.1371/journal.pgen.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morita Y, Tomida J, Kawamura Y. Responses of Pseudomonas aeruginosa to antimicrobials. Front. Microbiol. 2014;4:422. doi: 10.3389/fmicb.2013.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chow L, Waldron L, Gillings MR. Potential impacts of aquatic pollutants: sub-clinical antibiotic concentrations induce genome changes and promote antibiotic resistance. Front. Microbiol. 2015;6:803. doi: 10.3389/fmicb.2015.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abassi YA, et al. Kinetic cell-based morphological screening: prediction of mechanism of compound action and off-target effects. Chem. Biol. 2009;16:712–723. doi: 10.1016/j.chembiol.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xi Z, et al. Mode of action classification of chemicals using multi-concentration time-dependent cellular response profiles. Comput. Biol. Chem. 2014;49:23–35. doi: 10.1016/j.compbiolchem.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 71.Liberati NT, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang L, Hinz AJ, Nadeau JP, Mah TF. Pseudomonas aeruginosa tssC1 links type VI secretion and biofilm-specific antibiotic resistance. J. Bacteriol. 2011;193:5510–5513. doi: 10.1128/JB.00268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ahn SJ, Ahn SJ, Wen ZT, Brady LJ, Burne RA. Characteristics of biofilm formation by Streptococcus mutans in the presence of saliva. Infect. Immun. 2008;76:4259–4268. doi: 10.1128/IAI.00422-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dr. Xiaobo Wang co-founder and CTO of Acea Biosciences personal communication (2017).
- 75.Solly K, Wang X, Xu X, Strulovici B, Zheng W. Application of real-time cell electronic sensing (RT-CES) technology to cell-based assays. Assay and drug development technologies. 2004;2:363–372. doi: 10.1089/adt.2004.2.363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


