Abstract
Background
Studies have shown a familial predisposition for anterior cruciate ligament (ACL) rupture and have been followed by genetic-association studies on polymorphisms in candidate genes in recent years. To date, no systematic review with a best-evidence synthesis has evaluated the influence of genetics on this devastating knee injury.
Objective
Our objective was to evaluate the association between genetic variants and ACL rupture.
Methods
We performed an extensive search in Embase, MEDLINE, Web of Science, Scopus, PubMed Publisher, Cochrane Register of Clinical Trials, and Google scholar up to 24 August 2015. Studies were eligible if they met the following inclusion criteria: (1) design was a case–control study, retrospective or prospective follow-up study, or a randomized controlled trial (RCT); (2) the study examined the association between a genetic variant and ACL rupture in both an ACL and a control group. We determined the risk of bias for all included studies.
Results
We included a total of 16 studies (eight at high risk of bias and eight with an unclear risk) that examined 33 different DNA variants. Conflicting evidence was found for the COL1A1 rs1800012 and COL3A1 rs1800255 variants, whereas limited evidence was found for no association of the COL5A1 rs12722 and rs13946 and COL12A1 rs970547 variants (all encoding collagen). Evidence was insufficient to draw conclusions as to whether any other genetic variant identified in this review had any association with ACL rupture.
Conclusions
More research is needed to support a clear association between ACL rupture and genetic variants. Genome-wide studies are recommended for exploring more potential genetic variants. Moreover, large prospective studies are needed to draw robust conclusions.
Key Points
| Anterior cruciate ligament (ACL) rupture is a very common and severe knee injury that predominantly occurs while participating in sports. It incurs high costs and has disastrous clinical consequences. Studies in recent years have suggested that genetic predisposition is an important factor in its etiology. |
| This is the first systematic review with a best-evidence synthesis regarding associations between genetic variants and ACL rupture. We found some potential genetic variants that require further investigation, especially since we identified large heterogeneity in the broad genetic variants studied and outcome definitions. |
| More research with large samples, phenotype homogeneity, and less bias is needed for a better understanding of the etiology of ACL rupture. This would allow us to take appropriate measures to screen for and prevent this injury and its clinical consequences. |
Introduction
An anterior cruciate ligament (ACL) rupture is a very common and severe knee injury that predominantly occurs during sports participation, primarily via a non-contact mechanism [1, 2]. An ACL rupture is often accompanied by meniscal tears (approximately 50%), medial collateral ligament injuries (22%), and chondral lesions (16–46%) and results in a tenfold increased risk of knee osteoarthritis [3–6]. As a result, an ACL rupture is referred to as ‘the stroke of the knee’ or ‘an old knee in a young patient’ [7]. ACL rupture reconstruction is one of the most commonly performed orthopedic procedures, with an increasing incidence across the globe: England (13.5 per 100,000 person-years), Scandinavian countries (32–38 per 100,000 person-years), Australia (52.0 per 100,000 person-years), and USA (43.5 per 100,000 person-years) [8–13]. In absolute numbers, this means between 100,000 and 200,000 ACL ruptures are reconstructed annually in the USA alone [13, 14]. The high incidence, high costs, and disastrous clinical consequences of ACL rupture mean it is important to be aware of the cause and mechanism behind this injury. A better understanding regarding the risk factors, etiology, and mechanism is an important step in screening for and preventing ACL rupture.
ACL rupture risk is determined by intrinsic and extrinsic factors. Extrinsic factors include the intensity of the physical activity and the type of playing surface [15–17]. Intrinsic factors include differences in anatomy, sex, neuromuscular control, and hormonal constitution [18–20]. For example, the incidence of ACL rupture is 3–6 times higher in women than in men [15, 21], which could be partially explained by the smaller intercondylar notch, higher estrogen concentration, and a movement pattern with an increased hip adductor moment and knee valgus found in women [18, 20]. Previous studies have indicated a familial predisposition for ACL rupture. An individual with an ACL rupture was twice as likely to have a relative with an ACL rupture [22]. Hewett et al. [23] pointed out that twins with an ACL rupture shared the same multiple risk factors. This might be explained by an active lifestyle, since athletes tend to injure their ACL more often than non-athletes do. However, genetics or other intrinsic variations could also be of influence.
A number of studies have suggested associations between ACL rupture and various genetic variants, possibly suggesting that genetic predisposition is a factor of importance in ACL rupture. John et al. [24] recently published a systematic review on a topic similar to ours, albeit with some notable methodological differences between the two reviews. In an attempt to conduct more sensitive research, we searched more databases. We also used a different risk-of-bias assessment tool and a best-evidence approach to synthesizing the data, which allowed us to weigh results for potential risk of bias and to grade evidence. We believe these methodological differences enabled us to generate more accurate conclusions.
To date, no systematic review with a best-evidence synthesis has been performed concerning genetics and ACL rupture. The objective of this systematic review was to summarize the current evidence for an association between genetic variants and ACL rupture.
Methods and Materials
Protocol
The reporting in this systematic review was conducted according to the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) statement [25].
Eligibility Criteria
Studies were included in the systematic review if they met the following inclusion criteria: (1) design was a case–control, retrospective or prospective follow-up study, or a randomized controlled trial; (2) the study examined the association between a genetic variant and an ACL rupture in both an ACL and a control group; (3) the study was written in English, Dutch, German, French, Spanish, Turkish, or Swedish. We excluded studies for which no full text was available, animal studies, and reviews.
Information Sources and Search
We conducted a systematic search of the following databases up to 24 August 2015: Embase, MEDLINE, Web of Science, Scopus, PubMed Publisher, Cochrane Register of Clinical Trials, and Google scholar. The following search strategy was used in Embase: (‘anterior cruciate ligament’/de OR ‘anterior cruciate ligament injury’/de OR ‘anterior cruciate ligament rupture’/de OR (‘knee injury’/de AND (‘sports and sport related phenomena’/exp OR ‘ligament injury’/exp)) OR (‘sport injury’/de AND (knee/exp OR ‘knee ligament’/exp OR ‘ligament injury’/exp)) OR (‘anterior cruciate’ OR acl OR ((ligament*) NEAR/6 (injur* OR rupture* OR trauma* OR tear*))):ab,ti) AND (genetics/exp OR ‘genetic parameters’/exp OR (genetic* OR genom* OR gene OR genes OR (famil* NEAR/3 predispos*)):ab,ti) NOT ([animals]/lim NOT [humans]/lim). This search strategy was transferred into similar search strategies in the databases described above. References in reviews and full-text articles were screened to retrieve more studies that could be eligible for this systematic review.
Study Selection
The results of the seven different search strategies were combined and duplicates removed using EndNoteX5. Three authors screened the results of these database searches independently by title and abstract. The final selection for inclusion of the remaining full-text articles was made by the same independent authors. Discrepancies were resolved by consensus.
Data-Collection Process
One author extracted the general information, study design, sample size, gene, corresponding variant, and product of each study.
Risk-of-Bias Assessment
Three reviewers, independent of each other, assessed the risk of bias of the studies using the Cochrane Centre ‘case–control tool’ [26]. Any disagreements were resolved by consensus. This risk-of-bias tool included six questions, four of which addressed bias (see Table 1). Selection bias scoring was based on the source of recruiting for cases and controls. Ideally, cases were compared with population-based controls. Confounding was scored based on age and sex. Ideally, both groups were matched or adjusted for age and sex. In addition, the studies included were scored for information bias. Ideally, the methods used to extract DNA and to genotype the genetic variant were the same. The risk of bias was divided into three ranks: low, high, and unclear risk of bias. A study was labelled ‘high risk’ if at least one bias question was answered with ‘no’ and ‘low risk’ of bias when all other questions were answered with ‘yes’. A study was labelled ‘unclear risk’ of bias if all questions were answered ‘doubtful’ or a mix of ‘doubtful’ and ‘yes’.
Table 1.
List of questions used to assess risk of bias
| # | Criterion | Question |
|---|---|---|
| 1 | Case | Are the cases defined clearly and adequately? |
| 2 | Control | Are the controls defined clearly and adequately? |
| 3 | Selection bias | Is selection bias excluded sufficiently? |
| 4 | Defined exposure | Is the exposure defined clearly, and is the method used to assess this exposure appropriate? |
| 5 | Determination | Was blinding to exposure status maintained before determination of disease? |
| 6 | Confounding | Are the main confounders identified and taken into account adequately for the design and analysis? |
Information bias comprises questions 4 and 5
Summary Measures
An overview with odds ratios (ORs) was given of various genetic variants and their associations with ACL rupture. The ACL group consisted of individuals who experienced an ACL rupture. The control group consisted of controls with no history of ACL rupture. When possible, the association with ACL rupture was examined, with subgroups being stratified according to sex and non-contact versus contact mechanism, since these factors are known to influence the risk of an ACL rupture.
Synthesis of Results
We refrained from statistically pooling the data because of the different genetic variants and the heterogeneity of the risk of bias between studies, providing a narrative summary of the results as an alternative. Therefore, we performed a ‘best-evidence’ synthesis based on the study of van Tulder et al. [27]. Evidence was defined as generally consistent if ≥75% of the studies/cohorts reported consistent findings. Strong evidence was defined as two or more studies with a low risk of bias and generally consistent findings in all studies/cohorts. Moderate evidence was defined as one study with low risk of bias and two or more studies/cohorts with a high risk of bias and generally consistent findings. Limited evidence was defined as generally consistent findings in one study with a low risk of bias or two or more studies with a high risk of bias. Insufficient evidence was defined as a finding in one study with a high risk of bias. Conflicting evidence was defined as <75% of the studies reporting consistent findings.
Results
Study Selection
Combining the search results of all databases retrieved a total of 2559 studies. After removing duplicates, 1433 studies remained. A further 1412 studies were excluded after screening title and abstract. Five studies were excluded after accessing the full text: two because they did not examine the association between genetic variants and ACL ruptures and three because they lacked full text. Ultimately, 16 articles fell within the scope of this systematic review. A flowchart of this process is shown in Fig. 1.
Fig. 1.
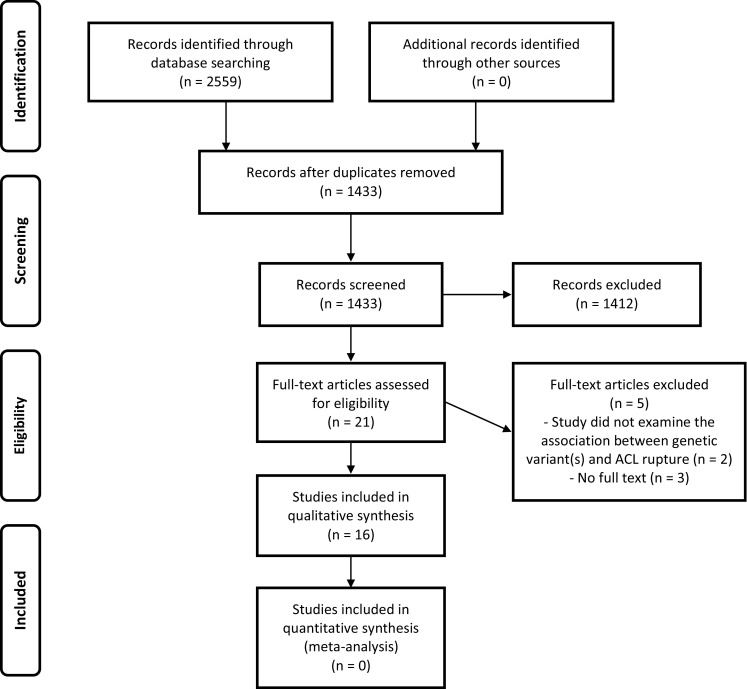
PRISMA flowchart showing the study-selection process [25]. ACL anterior cruciate ligament
Study Characteristics
A summary of the 16 included studies is shown in Table 2; all were case–control studies. Ten examined genetic variants in or near genes encoding collagens (collagen type I, alpha 1 [COL1A1]; collagen type III, alpha 1 [COL3A1]; collagen type V, alpha 1 [COL5A1]; collagen type VI, alpha 1 [COL6A1]; collagen type XII, alpha 1 [COL12A1]), one study examined proteoglycans (aggrecan [ACAN], biglycan [BCN], decorin [DCN], fibromodulin [FMOD], lumican [LUM]), two examined matrix metalloproteinases (matrix metalloproteinase 1 [MMP1], matrix metalloproteinase 3 [MMP3], matrix metalloproteinase 10 [MMP10], matrix metalloproteinase 12 [MMP12]), one study examined a variant near growth-differentiation factor (growth differentiation factor 5 [GDF5]), one investigated variants in genes involved in the angiogenesis-associated signaling cascade (vascular endothelial growth factor A [VEGFA], kinase insert domain receptor [KDR], nerve growth factor beta [NGFB], hypoxia-inducible factor 1-alpha [HIF1A]), and, finally, one study focused on elastin (ELN) and fibrillin (fibrillin 2 [FBN2]). A total of 33 different genetic variants were examined. O’Connell et al. [34] examined two different case–control cohorts in one study: South African and Polish. O’Connell et al. [34] and Ficek et al. [29] examined the COL12A1 gene in the same population; however, O’Connell et al. [34] only performed stratified analyses, and Ficek et al. [29] analysed only the overall results. Therefore, these two studies were considered independent of each other.
Table 2.
Study characteristics of the included studies
| Study | Design | Patients with ACL rupture (n) | Controls (n) | Gene | Product | Variant |
|---|---|---|---|---|---|---|
| Ficek et al. [28] 2013 | Case–control | 91 | 143 | COL1A1 | Collagen | rs1800012 |
| rs1107946 | ||||||
| Ficek et al. [29] 2014 | Case–control | 91 | 143 | COL12A1 | Collagen | rs970547 |
| Khoschnau et al. [30] 2008 | Case–control | 233 | 325 | COL1A1 | Collagen | rs1800012 |
| Khoury et al. [31] 2015 | Case–control | 141 | 219 | ELN | Elastin | rs2071307 |
| FBN2 | Fibrillin | rs331079 | ||||
| Malila et al. [32] 2011 | Case–control | 86 | 100 | MMP3 | Matrix metalloproteinase | –1612 |
| Mannion et al. [33] 2014 | Case–control | 227 | 234 | ACAN | Proteoglycans | rs2351491 |
| rs1042631 | ||||||
| rs1516797 | ||||||
| BGN | rs1126499 | |||||
| rs1042103 | ||||||
| DCN | rs13312816 | |||||
| rs516115 | ||||||
| FMOD | rs7543148 | |||||
| rs10800912 | ||||||
| LUM | rs2268578 | |||||
| O’Connell et al. [34]a 2015 | Case–control | 242b | 235b | COL3A1 | Collagen | rs1800255 |
| 91c | 91c | COL6A1 | rs35796750 | |||
| Posthumus et al. [35] 2009 | Case–control | 117 | 130 | COL1A1 | Collagen | rs1800012 |
| Posthumus et al. [36] 2009 | Case–control | 129 | 216 | COL5A1 | Collagen | rs13946 |
| rs12722 | ||||||
| Posthumus et al. [37] 2010 | Case–control | 129 | 216 | COL12A1 | Collagen | rs240736 |
| rs970547 | ||||||
| Posthumus et al. [38] 2012 | Case–control | 129 | 216 | MMP1 | Matrix metalloproteinase | rs1799750 |
| MMP3 | rs679620 | |||||
| MMP10 | rs486055 | |||||
| MMP12 | rs2276109 | |||||
| Rahim et al. [39] 2014 | Case–control | 227 | 227 | VEGFA | Angiogenesis-associated signaling cascade genes | rs699947 |
| rs1570360 | ||||||
| rs2010963 | ||||||
| KDR | rs1870377 | |||||
| rs2071559 | ||||||
| NGFB | rs6678788 | |||||
| HIF1A | rs11549465 | |||||
| Raleigh et al. [40] 2013 | Case–control | 126 | 216 | GDF5 | Growth differentiation factor | rs143383 |
| Stępień-Słodkowska et al. [41] 2013 | Case–control | 138 | 183 | COL1A1 | Collagen | rs1800012 |
| Stępień-Słodkowska et al. [42] 2015 | Case–control | 138 | 183 | COL3A1 | Collagen | rs1800255 |
| Stępień-Słodkowska et al. [43] 2015 | Case–control | 138 | 183 | COL5A1 | Collagen | rs13946 |
| rs12722 |
ACAN aggrecan, BCN biglycan, COL12A1 collagen type XII, alpha 1, COL1A1 collagen type I, alpha 1, COL3A1 collagen type III, alpha 1, COL5A1 collagen type V, alpha 1, COL6A1 collagen type VI, alpha 1, DCN decorin, ELN elastin, FBN2 fibrillin 2, FMOD fibromodulin, GDF5 growth differentiation factor 5, HIF1A hypoxia-inducible factor 1-alpha, KDR kinase insert domain receptor, LUM lumican, MMP1 matrix metalloproteinase 1, MMP10 matrix metalloproteinase 10, MMP12 matrix metalloproteinase 12, MMP3 matrix metalloproteinase 3, NGFB nerve growth factor beta, VEGFA vascular endothelial growth factor A
aTwo different cohorts were analyzed in this study, as indicated by footnote ‘b’ or ‘c’
bSouth African population
cPolish population
Risk of Bias
An overview of the risk of bias is shown in Table 3. Eight studies were considered unclear risk of bias [28, 29, 36, 38, 40–43] and eight were labelled high risk of bias [30–35, 37, 39].
Table 3.
Risk-of-bias summary: review authors’ judgements of each risk-of-bias item for each included studya
| Study | Case (1) | Control (2) | Selection bias (3) | Defined exposure (4) | Determination exposure (5) | Confounding (6) | Overallb |
|---|---|---|---|---|---|---|---|
| Ficek et al. [28] 2013 | + | + | ? | + | ? | + | ? |
| Ficek et al. [29] 2014 | + | + | ? | + | ? | + | ? |
| Khoschnau et al. [30] 2008 | + | ? | + | + | + | − | − |
| Khoury et al. [31] 2015 | + | + | ? | + | ? | − | − |
| Malila et al. [32] 2011 | + | + | ? | + | ? | − | − |
| Mannion et al. [33] 2014 | + | + | ? | + | ? | − | − |
| O’Connell et al. [34] 2015 | + | ? | ? | + | + | − | − |
| Posthumus et al. [35] 2009 | + | + | ? | + | ? | − | − |
| Posthumus et al. [36] 2009 | + | + | ? | + | ? | + | ? |
| Posthumus et al. [37] 2010 | + | + | ? | + | ? | − | − |
| Posthumus et al. [38] 2012 | + | + | ? | + | ? | + | ? |
| Rahim et al. [39] 2014 | + | ? | ? | + | ? | - | − |
| Raleigh et al. [40] 2013 | + | + | ? | + | ? | + | ? |
| Stępień-Słodkowska et al. [41] 2013 | + | + | ? | + | ? | ? | ? |
| Stępień-Słodkowska et al. [42] 2015 | + | + | ? | + | ? | ? | ? |
| Stępień-Słodkowska et al. [43] 2015 | + | + | ? | + | ? | ? | ? |
aNumbers 1–6 in the column headings correspond to the questions listed in Table 1; − indicates the risk of bias question was answered ‘no’, + indicates the risk of bias question was answered ‘yes’, ? indicates the risk of bias question could not be answered either ‘yes’ or ‘no’ and was answered with a ‘doubtful’ or ‘unknown’
b− indicates a high risk of bias, ? indicates an unclear risk of bias
Results of Association Studies in the Complete Populations
Results of the association studies are shown in Table 4.
Table 4.
Results of genetic studies examining associations between genetic variants and anterior cruciate ligament rupture
| Study | Gene | Protein | Variant | Genetic analysis | OR (95% CI) | p-Value | Risk of bias |
|---|---|---|---|---|---|---|---|
| Ficek et al. [28] 2013 | COL1A1 | Collagen type I | rs1800012 | GG vs. GT + TT | Not shown | >0.05 | Unclear |
| GT vs. GG + TT | Not shown | >0.05 | |||||
| TT vs. GG + GT | Not shown | >0.05 | |||||
| rs1107946 | GG vs. GT + TT | Not shown | >0.05 | ||||
| GT vs. GG + TT | Not shown | >0.05 | |||||
| TT vs. GT + GG | Not shown | >0.05 | |||||
| Ficek et al. [29] 2014 | COL12A1 | Collagen type XII | rs970547 | GG vs. GA + AA | Not shown | >0.05 | Unclear |
| GA vs. GG + AA | Not shown | >0.05 | |||||
| AA vs. GG + GA | Not shown | >0.05 | |||||
| Khoschnau et al. [30] 2008 | COL1A1 | Collagen type I | rs1800012 | GG vs. GG | 1 | >0.05 | High |
| GT vs. GG | 1.19 (0.82–1.75) | >0.05 | |||||
| TT vs. GG | 0.12 (0.02–0.92) | <0.05 d | |||||
| Khoury et al. [31] 2015 | ELN | Elastin | rs2071307 | GG vs. GA + AA | Not shown | >0.05 | High |
| GA vs. GG + AA | Not shown | >0.05 | |||||
| AA vs. GG + GA | Not shown | >0.05 | |||||
| FBN2 | Fibrillin-2 | rs331079 | GG vs. GC + CC | Not shown | >0.05 | ||
| GC vs. GG + CC | Not shown | >0.05 | |||||
| CC vs. GG + GC | Not shown | >0.05 | |||||
| Malila et al. [32] 2011 | MMP3 | Matrix metalloproteinase type 3 | –1612 | 5A+ vs. 5A− | 1.39 (0.72–2.67) | >0.05 | High |
| 5A− vs. 5A+ | 0.72 (0.37–1.38) | >0.05 | |||||
| Mannion et al. [33] 2014 | DCN | Decorin | rs516115 | GG vs. GA + AA | 9.23 (1.17–73.01)e | 0.015 | High |
| GA vs. GG + AA | Not shown | >0.05 | |||||
| AA vs. GG + GA | Not shown | >0.05 | |||||
| rs13312816 | AA vs. AT + TT | Not shown | >0.05 | ||||
| AT vs. AA + TT | Not shown | >0.05 | |||||
| TT vs. AA + AT | Not shown | >0.05 | |||||
| ACAN | Aggrecan | rs2351491 | CT vs. CC + TT | Not shown | >0.05 | ||
| CC vs. CT + TT | Not shown | >0.05 | |||||
| TT vs. CT + CC | Not shown | >0.05 | |||||
| rs1042631 | TT vs. CT + CC | Not shown | >0.05 | ||||
| CT vs. TT + CC | Not shown | >0.05 | |||||
| CC vs. CT + TT | Not shown | >0.05 | |||||
| rs1516797 | TT vs. GT + GG | Not shown | >0.05 | ||||
| GT vs. GG + TT | Not shown | >0.05 | |||||
| GG vs. GT + TT | Not shown | >0.05 | |||||
| BGN | Biglycan | rs1126499 | CC vs. CT + TT | Not shown | >0.05 | ||
| CT vs. CC + TT | Not shown | >0.05 | |||||
| TT vs. CC + CT | Not shown | >0.05 | |||||
| rs1042103 | GG vs. GA + AA | Not shown | >0.05 | ||||
| GA vs. GG + AA | Not shown | >0.05 | |||||
| AA vs. GG + GA | Not shown | >0.05 | |||||
| FMOD | Fibromodulin | rs7543148 | GG vs. GA + AA | Not shown | >0.05 | ||
| GA vs. GG + AA | Not shown | >0.05 | |||||
| AA vs. GG + GA | Not shown | >0.05 | |||||
| rs10800912 | CC vs. CT + TT | Not shown | >0.05 | ||||
| CT vs. CC + CT | Not shown | >0.05 | |||||
| TT vs. CC + CT | Not shown | >0.05 | |||||
| LUM | Lumican | rs2268578 | TT vs. TC + CC | Not shown | >0.05 | ||
| TC vs. TT + CC | Not shown | >0.05 | |||||
| CC vs. CT + TT | Not shown | >0.05 | |||||
| O’Connell et al. [34]a 2015 | COL3A1 | Collagen type IIIb | rs1800255 | AA vs. GA + GG | Not shown | >0.05 | High |
| GA vs. GG + AA | Not shown | >0.05 | |||||
| GG vs. AA + GA | Not shown | >0.05 | |||||
| Collagen type IIIc | AA vs. GA + GG | 3.8 (1.1–12.8) | 0.036 | ||||
| GA vs. AA + GG | Not shown | >0.05 | |||||
| GG vs. AA + GA | Not shown | >0.05 | |||||
| Collagen type VIb | rs35796750 | TT vs. TC + CC | Not shown | >0.05 | |||
| TC vs. CC + TC | Not shown | >0.05 | |||||
| CC vs. TT + CT | Not shown | >0.05 | |||||
| Posthumus et al. [35] 2009 | COL1A1 | Collagen type I | rs1800012 | TT vs. GT + GG | 0.08 (<0.01–1.46) | 0.031 | High |
| GT vs. TT + GG | Not shown | >0.05 | |||||
| GG vs. GG + GT | Not shown | >0.05 | |||||
| Posthumus et al. [36] 2009 | COL5A1 | Collagen type V | rs12722 | TT vs. CT + CC | Not shown | >0.05 | Unclear |
| CT vs. TT + CC | Not shown | >0.05 | |||||
| CC vs. TT + CT | Not shown | >0.05 | |||||
| rs13946 | TT vs. CT + CC | Not shown | >0.05 | ||||
| CT vs. TT + CC | Not shown | >0.05 | |||||
| CC vs. TT + CT | Not shown | >0.05 | |||||
| Posthumus et al. [37] 2010 | COL12A1 | Collagen type XII | rs970547 | GG vs. GA + AA | Not shown | >0.05 | High |
| GA Vs. AA + GG | Not shown | >0.05 | |||||
| AA VS. GG + GA | Not shown | >0.05 | |||||
| rs240736 | TT vs. CT + CC | Not shown | >0.05 | ||||
| CT vs. TT + CC | Not shown | >0.05 | |||||
| CC vs. TT + CT | Not shown | >0.05 | |||||
| Posthumus et al. [38] 2012 | MMP1 | Matrix metalloproteinase type 1 | rs1799750 | CC vs. CT + TT | Not shown | >0.05 | Unclear |
| CT vs. CC + TT | Not shown | >0.05 | |||||
| TT vs. CC + CT | Not shown | >0.05 | |||||
| MMP3 | Matrix metalloproteinase type 3 | rs679620 | 1G1G vs. 1G2G + 2G2G | Not shown | >0.05 | ||
| 1G2G vs. 1G1G + 2G2G | Not shown | >0.05 | |||||
| 2G2G vs. 1G1G + 1G2G | Not shown | >0.05 | |||||
| MMP10 | Matrix metalloproteinase type 10 | rs486055 | GG vs. GA + AA | Not shown | >0.05 | ||
| GA vs. GG + AA | Not shown | >0.05 | |||||
| AA vs. GG + GA | Not shown | >0.05 | |||||
| MMP12 | Matrix metalloproteinase type 12 | rs2276109 | AA vs. AG + GG | Not shown | >0.05 | ||
| AG vs. AA + GG | Not shown | >0.05 | |||||
| GG vs. AA + AG | Not shown | >0.05 | |||||
| Rahim et al. [39] 2014 | VEGFA | Vascular endothelial growth factor A | rs699947 | CC vs. CA + AA | Not shown | >0.05 | High |
| CA vs. CC + AA | Not shown | >0.05 | |||||
| AA vs. CC + CA | Not shown | >0.05 | |||||
| rs1570360 | GG vs. GA + AA | Not shown | >0.05 | ||||
| GA vs. GG + AA | 1.70 (1.16–2.50) | 0.007 | |||||
| AA vs. GG + GA | Not shown | >0.05 | |||||
| rs2010963 | GG vs. GC + CC | Not shown | >0.05 | ||||
| GC vs. GG + CC | Not shown | >0.05 | |||||
| CC vs. GC + GG | Not shown | >0.05 | |||||
| KDR | Kinase insert domain receptor | rs1870377 | TT vs. TA + AA | Not shown | >0.05 | ||
| TA vs. TT + AA | Not shown | >0.05 | |||||
| AA vs. TT + AT | Not shown | >0.05 | |||||
| rs2071559 | GG vs. GA + AA | Not shown | >0.05 | ||||
| GA vs. GG + AA | Not shown | >0.05 | |||||
| AA vs. GA + GG | Not shown | >0.05 | |||||
| NGFB | Nerve growth factor beta | rs6678788 | CC vs. CT + TT | Not shown | >0.05 | ||
| CT vs. GG + TT | Not shown | >0.05 | |||||
| TT vs. GT + CC | Not shown | >0.05 | |||||
| HIF1A | Hypoxia-inducible factor 1-alpha | rs11549465 | CC vs. CT + CC | Not shown | >0.05 | ||
| CT vs. CC + TT | Not shown | >0.05 | |||||
| TT vs. CT + CC | Not shown | >0.05 | |||||
| Raleigh et al. [40] 2013 | GDF5 | Growth-differentiation hormone factor | rs143383 | TT vs. CT + CC | Not shown | >0.05 | Unclear |
| CT vs. TT + CC | Not shown | >0.05 | |||||
| CC vs. TT + CT | Not shown | >0.05 | |||||
| Stępień-Słodkowska et al. [41] 2013 | COL1A1 | Collagen type I | rs1800012 | GG vs. GT + TT | Not shown | 0.046 | Unclear |
| GT vs. GG + TT | Not shown | >0.05 | |||||
| TT vs. GT + GG | Not shown | >0.05 | |||||
| Stępień-Słodkowska et al. [42] 2015 | COL3A1 | Collagen type III | rs1800255 | GG vs. GA + AA | 0.78 (0.49–1.24) | >0.05 | Unclear |
| GA vs. GG + AA | Not shown | >0.05 | |||||
| AA vs. GG + GA | 5.05 (1.62–15.78) | 0.003 | |||||
| Stępień-Słodkowska et al. [43] 2015 | COL5A1 | Collagen type V | rs13946 | CC vs. CT + TT | Not shown | >0.05 | Unclear |
| CT vs. CC + TT | Not shown | >0.05 | |||||
| TT vs. CC + CT | Not shown | >0.05 | |||||
| rs12722 | CC vs. CT + TT | Not shown | >0.05 | ||||
| CT vs. CC + TT | Not shown | >0.05 | |||||
| TT vs. CC + CT | Not shown | >0.05 |
Bold type indicates statistical significance (p < 0.05)
ACAN aggrecan, ACL anterior cruciate ligament, BCN biglycan, COL12A1 collagen type XII, alpha 1, COL1A1 collagen type I, alpha 1, COL3A1 collagen type III, alpha 1, COL5A1 collagen type V, alpha 1, COL6A1 collagen type VI, alpha 1, DCN decorin, ELN elastin, FBN2 fibrillin 2, FMOD fibromodulin, GDF5 growth differentiation factor 5, HIF1A hypoxia-inducible factor 1-alpha, KDR kinase insert domain receptor, LUM lumican, MMP1 matrix metalloproteinase 1, MMP10 matrix metalloproteinase 10, MMP12 matrix metalloproteinase 12, MMP3 matrix metalloproteinase 3, NGFB nerve growth factor beta, OR odds ratio, VEGFA vascular endothelial growth factor A
aTwo different cohorts were analysed in this study, as indicated by footnote ‘b’ or ‘c’
bSouth African group
cPolish group
dNo exact p-value was reported
eThis genotype was over-represented (OR = 9.23) in the control group compared with the ACL group, which is consistent with a protective effect in the ACL group (OR = 1/9.23 = 0.11)
Collagen
The most frequently studied gene was COL1A1. Conflicting evidence was found for an association between TT and GG genotype of the COL1A1 rs1800012 variant and ACL rupture. Conflicting evidence was found for an association between AA genotype of the COL3A1 rs1800255 variant and ACL rupture. Limited evidence was found for no association between COL5A1 rs12722, COL5A1 rs13946, and COL12A1 rs970547 variants and ACL rupture. Insufficient evidence was found for no association between COL1A1 rs1107946, COL6A1 rs35796750, and COL12A1 rs240736 variants and ACL rupture.
Proteoglycans
Insufficient evidence was found for an association between GG (protective) genotype of the DCN rs516115 variant and ACL rupture. Insufficient evidence was found for no association between DCN rs13312816, ACAN rs2351491, ACAN rs1042631, ACAN rs1516797, BGN rs1126499, BGN rs1042103, FMOD rs7543148, FMOD rs10800912, and LUM rs2268578 variants and ACL rupture.
Matrix Metalloproteinases
Insufficient evidence was found for no association between MMP1 rs1799750, MMP3 rs679620, MMP3-1612, MMP10 rs486055, and MMP12 rs2276109 variants and ACL rupture.
Angiogenesis-Associated Signaling Cascade and Growth Differentiation Hormone Factor
Insufficient evidence was found for an association between GA genotype (harmful) of the VEGFA rs1570360 variant and ACL rupture. Insufficient evidence was found for no association between VEFGA rs699947, VEFGA rs2010963, KDR 1870377, KDR rs2071559, NGFB rs6678788, HIF1A rs11549465, and GDF5 rs143383 variants and ACL rupture.
Elastin and Fibrillin
Insufficient evidence was found for no association between ELN rs2071307 variant and ACL rupture or for no association between FBN2 rs331079 variant and ACL rupture.
Stratified Analysis
In addition to the overall analyses, studies investigated genetic variants in sex and/or (non-) contact stratified analyses. However, because of the small sample sizes, insufficient data were available to report sufficient evidence regarding those analyses. Therefore, they were not included in this review.
Discussion
In this systematic review, we summarized the current literature on genetic variants predicting the risk of ACL rupture. We found conflicting evidence for the COL1A1 rs1800012 GG and TT genotype and COL3A1 rs1800255 AA genotype and limited evidence for no association between COL5A1 rs13946, COL5A1 rs12722, and COL12A1 rs970547 variants and ACL rupture. We also found associations, albeit with insufficient evidence, regarding the DCN rs516115 GG genotype (protective) and VEGFA rs1570360 GA genotype (harmful) and ACL rupture. Moreover, a large number of genetic variants were found not to have an association. However, those genetic variants were studied only once; therefore, evidence for those DNA variants was also considered insufficient. We included 16 studies in this review, with a total of 33 different genetic variants. Many studies were found to have an unclear or high risk of bias and confounding. In addition, we identified large heterogeneity in the genetic variants studied, outcome definition, and the genetic contrast studied, which made it impossible to conduct a formal meta-analysis of these studies. Therefore, we performed a best-evidence synthesis. Overall, we found some potential genetic variants that could influence the risk of ACL rupture. However, more data are needed to support a clear association between genetic variants and ACL rupture. Larger and more genetic studies are required to obtain a better understanding of these possible associations.
John et al. [24] recently published a similar systematic review. However, there are some notable differences between the two studies. First, John et al. [24] presented the results as a narrative review and concluded that, of the 20 genes examined, ten were positively associated with an ACL rupture. Their review does not appear to fully justify their finding that 50% (COL1A1, COL12A1, COL5A1, COL3A1, MMP3, MMP12, and various ECM) of the genes examined so far are positively associated with an ACL tear, especially when mentioning contradictory results for some specific genetic variants such as COL1A1 or COL3A1. The current analysis presented the findings using a best-evidence synthesis by van Tulder et al. [27]. A best-evidence synthesis provides stronger evidence and takes a different approach to presenting the results than does a simple narrative summary. Consequently, the results and conclusions of the current analysis concerning the associations between genetic variants and ACL injury differ from and have greater methodological power than those of John et al. [24]. Second, this review included two additional studies [30, 40]. John et al. [24] excluded one of these [30] because both the ACL and the posterior cruciate ligaments were included and analysed together in one population group. They also excluded a different study [40], likely because of differences between our search strategies and because this review searched more databases. Third, in contrast to John et al. [24], we did not account for subgroups such as sex and injury mechanism because of small sample sizes. Fourth, this review concentrated on polymorphisms rather than less well-studied haplotypes or alleles. Fifth, John et al. [24] did not report the results of genetic variants for which no association was found with ACL rupture. This review represents a survey of all investigated genetic variants and genotypes with their ORs and p-values, even if no association was found at all. This approach was taken in the interests of maximum transparency and clarity, allowing readers and researchers to decide which genetic variants to investigate in the future, taking into account confidence intervals and statistical significance data.
Variants of the investigated genes, displayed in Table 4, are involved in the synthesis, strength, and homeostasis of the ligament. COL1A1 encodes for collagen type I, which provides mechanical strength to several tissues, including ligaments [44]. COL3A1 encodes for collagen type III and is involved in collagen type I fibrillogenesis [45]. COL5A1 encodes collagen type V, which is engaged with collagen type I in constructing heterotypic fibrils and also regulates the diameter of those fibrils [46]. Collagen type XII, encoded by COL12A1, is the largest member of the fibril-associated collagens and regulates the organization and mechanical properties of collagen fibril bundles [47]. Decorin, encoded by the DCN gene, belongs to the small group of proteoglycans and is engaged in limiting the diameter of collagen fibrils during fibrillogenesis [48]. VEGFA encodes vascular endothelial growth factor A, is a regulator of angiogenesis, and increases the expression of the matrix metalloproteinases [49]. The consequences of those genetic variants are not exactly known. To date, a limited number of the genetic variants involved in the synthesis, strength, and homeostasis of the ligament have been investigated. Moreover, most of those genetic variants were only studied once. Only some genetic variants in or near COL1A1 (4x), COL3A1 (3x), COL5A1 (2x), and COL12A1 (2x) were studied in more than one independent study/cohort.
Our included studies had some limitations. No limit was set on minimum sample size to enable us to include all possible studies because genetic studies require more participants than most of our included studies had. Sample size became very small when groups were stratified according to sex or mechanism of injury and, therefore, we did not report any stratified analyses. In total, 14 published studies used, partially or fully, the same population for cases and controls [28, 29, 31, 33–43], which increased the risk of bias. Most likely, every gene variant has its own potential, influencing the risk of an ACL rupture. For example, the VEGFA rs1570360 and DCN rs516115 variants were examined in nearly the same population group, which was overrepresented in the ACL rupture group [33, 39]. While both of these genetic variants could have increased the risk of ACL rupture, the possibility remains that only one of them was the actual risk contributor while the association of the other genetic variant was modified (confounding or effect modification) by the actual risk variant. Thus, another limitation is possible: confounding by ethnicity (population stratification). If the risk of an ACL rupture differs between different ethnicities but there is also a variation in the statistical distribution of a genetic variant between ethnic groups, the association between the ACL rupture and genetic variant may be confounded by the ethnic background of the studied population [50].
The heterogeneity of several study aspects, such as differences in population and type of genetic variants, meant that a meta-analysis was not possible. Therefore, we performed a best-evidence synthesis as an alternative. All of our studies were case-controlled as it is nearly impossible, and unnecessary, to conduct a randomized controlled trial given the research question. However, prospective cohort studies would provide stronger evidence.
Another major issue remains the possible underlying heterogenetic etiology in genetic studies, for example in patients with osteoarthritis [51]. Therefore, future research should also focus on evaluating larger samples and resolving phenotype heterogeneity to facilitate more comprehensive study of the genetics of ACL rupture, for example by investigating genetic variants in established genome-wide studies conducted to assess other factors such as osteoarthritis [51–54].
In the risk-of-bias assessment, eight studies were considered at high risk of bias. Eight studies were labelled at unclear risk of bias, which exemplifies the quality of the research and the conclusions that can be distilled.
Research has already been conducted for various genes involved in production, strength, or homeostasis of the ACL. However, as mentioned, larger and more genetic studies are required to provide a better understanding of the possible associations with ACL rupture. Every genetic variant examined should be re-examined to obtain a better understanding of the influence of these genes. Furthermore, research is encouraged for collagen and matrix metalloproteinase genes other than those already studied [55, 56]. Tendons and ligaments largely share the same components and both belong to the soft tissues. A systematic review by Claessen et al. [57] indicated no association between the COL1A1 rs1800012 variant and Achilles tendon ruptures, which does not clarify whether ACL and Achilles tendon ruptures might share the same genetic risk factors. More variants found in tendon ruptures, such as the TIMP gene, should also be investigated in relation to ACL ruptures [57]. This also applies to genetic variants found in osteoarthritis, since ACL rupture is a major risk factor for osteoarthritis [52, 53].
Some studies addressed the interaction between two gene variants on one chromosome, called haplotypes, which were found to modify the risk of an ACL rupture [28, 33, 34, 38, 39]. If genetic variants were not found to influence the risk, an association was still found due to the haplotype of those two gene variants. More research would be needed to clarify the exact role of haplotypes.
The examination of ACL rupture genetics is valuable, since knowledge of the genetic variants involved could contribute to an understanding of the etiology and risk factors in ACL tears. In addition, genetic variants could help in screening and prevention. Each person has a unique genetic profile. Some studies already suggest using genetic profiles to enhance athletic performance [58, 59]. Taking appropriate preventive measures might decrease the risk of an ACL rupture, as well as its costs [60, 61].
Screening for genetic variants could be implemented in different scenarios. Professional sports organizations might take appropriate measures regarding players at high risk of an ACL rupture. High-risk players could decide at a young age not to start a career in high-risk sports. Furthermore, screening could be implemented in families with an active lifestyle if a first-degree relative has a history of an ACL rupture. However, in reality, screening for risk of ACL rupture would be a complex task. It should be noted that none of the genetic variants solely influence the risk and thus could not be used as a diagnostic tool in isolation. As previously stated, an ACL rupture is determined by various intrinsic and extrinsic factors in which each factor contributes a small amount to the risk. Therefore, multifactorial and comprehensive models are designed to predict the risk of ACL rupture [62]. In future, when an association with genetics is found, genetic risk factors might be included in the current multifactorial models predicting the risk of ACL rupture [63]. Appropriate screening and prevention programs might be implemented based on those models. Individuals understand, and are interested in, the benefits of genomic testing in psychological and medical terms [64]. Unfortunately, genetic testing remains a source of moral and ethical controversy [64, 65].
Conclusion
More evidence is needed to draw significant conclusions regarding the association between genetic variants and ACL rupture. We did find some genetic variants that potentially contribute. Conflicting evidence was found for COL1A1 rs1800012 and COL3A1 rs1800255, whereas limited evidence was found for no association with COL5A1 rs13946, COL5A1 rs12722, and COL12A1 rs970547. Finally, we found insufficient evidence for an association between ACL rupture and DCN rs516115 GG genotype (protective) or VEGFA rs1570360 GA genotype (harmful). The genetic variants included in this systematic review account for only a small number of the genes involved in the biology of the ligament. ACL rupture has a high incidence and incurs extreme consequences. Therefore, more high-quality and homogenous data are needed to provide a better understanding of the etiology, which in future might improve screening and prevention programs. Research should primarily focus on the components and homeostasis of the ligament. These future genetic investigations should be performed in large (collaborative) genome-wide association studies with large sample sizes and phenotype homogeneity to explore for more potential genetic variants. However, and more importantly, future research should focus on (large) prospective studies so that clinically significant conclusions can be drawn.
Acknowledgements
The authors thank the medical librarian, WM Bramer, for his assistance with the systematic literature search.
Compliance with Ethical Standards
Funding
No sources of funding were used to assist in the preparation of this article.
Conflicts of interest
Mustafa Kaynak, Frank Nijman, Joyce van Meurs, Max Reijman, and Duncan Meuffels declare that they have no conflicts of interest relevant to the content of this review.
References
- 1.Gianotti SM, Marshall SW, Hume PA, et al. Incidence of anterior cruciate ligament injury and other knee ligament injuries: a national population-based study. J Sci Med Sport. 2009;12(6):622–627. doi: 10.1016/j.jsams.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Griffin LY, Agel J, Albohm MJ, et al. Noncontact anterior cruciate ligament injuries: risk factors and prevention strategies. J Am Acad Orthop Surg. 2000;8(3):141–150. doi: 10.5435/00124635-200005000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Brophy RH, Zeltser D, Wright RW, et al. Anterior cruciate ligament reconstruction and concomitant articular cartilage injury: incidence and treatment. Arthroscopy. 2010;26(1):112–120. doi: 10.1016/j.arthro.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Meuffels DE, Favejee MM, Vissers MM, et al. Ten year follow-up study comparing conservative versus operative treatment of anterior cruciate ligament ruptures. A matched-pair analysis of high level athletes. Br J Sports Med. 2009;43(5):347–351. doi: 10.1136/bjsm.2008.049403. [DOI] [PubMed] [Google Scholar]
- 5.Oiestad BE, Engebretsen L, Storheim K, et al. Knee osteoarthritis after anterior cruciate ligament injury: a systematic review. Am J Sports Med. 2009;37(7):1434–1443. doi: 10.1177/0363546509338827. [DOI] [PubMed] [Google Scholar]
- 6.Yoon KH, Yoo JH, Kim KI. Bone contusion and associated meniscal and medial collateral ligament injury in patients with anterior cruciate ligament rupture. J Bone Jt Surg Am. 2011;93(16):1510–1518. doi: 10.2106/JBJS.J.01320. [DOI] [PubMed] [Google Scholar]
- 7.Meuffels DE, Poldervaart MT, Diercks RL, et al. Guideline on anterior cruciate ligament injury. Acta Orthop. 2012;83(4):379–386. doi: 10.3109/17453674.2012.704563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jameson SS, Dowen D, James P, et al. Complications following anterior cruciate ligament reconstruction in the English NHS. Knee. 2012;19(1):14–19. doi: 10.1016/j.knee.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Granan LP, Forssblad M, Lind M, et al. The Scandinavian ACL registries 2004–2007: baseline epidemiology. Acta Orthop. 2009;80(5):563–567. doi: 10.3109/17453670903350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granan LP, Bahr R, Steindal K, et al. Development of a national cruciate ligament surgery registry: the Norwegian National Knee Ligament Registry. Am J Sports Med. 2008;36(2):308–315. doi: 10.1177/0363546507308939. [DOI] [PubMed] [Google Scholar]
- 11.Lind M, Menhert F, Pedersen AB. The first results from the Danish ACL reconstruction registry: epidemiologic and 2 year follow-up results from 5,818 knee ligament reconstructions. Knee Surg Sports Traumatol Arthrosc. 2009;17(2):117–124. doi: 10.1007/s00167-008-0654-3. [DOI] [PubMed] [Google Scholar]
- 12.Janssen KW, Orchard JW, Driscoll TR, et al. High incidence and costs for anterior cruciate ligament reconstructions performed in Australia from 2003–2004 to 2007–2008: time for an anterior cruciate ligament register by Scandinavian model? Scand J Med Sci Sports. 2012;22(4):495–501. doi: 10.1111/j.1600-0838.2010.01253.x. [DOI] [PubMed] [Google Scholar]
- 13.Buller LT, Best MJ, Baraga MG, et al. Trends in anterior cruciate ligament reconstruction in the United States. Orthop J Sports Med. 2015;3(1):2325967114563664. doi: 10.1177/2325967114563664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mather RC, 3rd, Koenig L, Kocher MS, et al. Societal and economic impact of anterior cruciate ligament tears. J Bone Jt Surg Am. 2013;95(19):1751–1759. doi: 10.2106/JBJS.L.01705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph AM, Collins CL, Henke NM, et al. A multisport epidemiologic comparison of anterior cruciate ligament injuries in high school athletics. J Athl Train. 2013;48(6):810–817. doi: 10.4085/1062-6050-48.6.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hewett TE, Myer GD, Ford KR, et al. Biomechanical measures of neuromuscular control and valgus loading of the knee predict anterior cruciate ligament injury risk in female athletes: a prospective study. Am J Sports Med. 2005;33(4):492–501. doi: 10.1177/0363546504269591. [DOI] [PubMed] [Google Scholar]
- 17.Orchard J, Seward H, McGivern J, et al. Intrinsic and extrinsic risk factors for anterior cruciate ligament injury in Australian footballers. Am J Sports Med. 2001;29(2):196–200. doi: 10.1177/03635465010290021301. [DOI] [PubMed] [Google Scholar]
- 18.Souryal TO, Freeman TR. Intercondylar notch size and anterior cruciate ligament injuries in athletes. A prospective study. Am J Sports Med. 1993;21(4):535–539. doi: 10.1177/036354659302100410. [DOI] [PubMed] [Google Scholar]
- 19.Hashemi J, Chandrashekar N, Mansouri H, et al. Shallow medial tibial plateau and steep medial and lateral tibial slopes: new risk factors for anterior cruciate ligament injuries. Am J Sports Med. 2010;38(1):54–62. doi: 10.1177/0363546509349055. [DOI] [PubMed] [Google Scholar]
- 20.Slauterbeck JR, Fuzie SF, Smith MP, et al. The menstrual cycle, sex hormones, and anterior cruciate ligament injury. J Athl Train. 2002;37(3):275–278. [PMC free article] [PubMed] [Google Scholar]
- 21.Hewett TE, Myer GD, Ford KR. Anterior cruciate ligament injuries in female athletes: part 1, mechanisms and risk factors. Am J Sports Med. 2006;34(2):299–311. doi: 10.1177/0363546505284183. [DOI] [PubMed] [Google Scholar]
- 22.Flynn RK, Pedersen CL, Birmingham TB, et al. The familial predisposition toward tearing the anterior cruciate ligament: a case control study. Am J Sports Med. 2005;33(1):23–28. doi: 10.1177/0363546504265678. [DOI] [PubMed] [Google Scholar]
- 23.Hewett TE, Lynch TR, Myer GD, et al. Multiple risk factors related to familial predisposition to anterior cruciate ligament injury: fraternal twin sisters with anterior cruciate ligament ruptures. Br J Sports Med. 2010;44(12):848–855. doi: 10.1136/bjsm.2008.055798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.John R, Dhillon MS, Sharma S, et al. Is there a genetic predisposition to anterior cruciate ligament tear? A systematic review. Am J Sports Med. 2016 doi: 10.1177/0363546515624467. [DOI] [PubMed] [Google Scholar]
- 25.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Offringa M, Assendelft WJJ, Scholten RJPM. Inleiding in evidence-based medicine (Introduction to evidence-based medicine). Klinisch handelen gebaseerd op bewijsmateriaal (Clinical practice based on evidence). 3rd ed. Houten: Bohn, Stafleu, Van Loghum; 2008.
- 27.van Tulder M, Furlan A, Bombardier C, et al. Updated method guidelines for systematic reviews in the Cochrane Collaboration Back Review Group. Spine (Phila Pa 1976) 2003;28(12):1290–1299. doi: 10.1097/01.BRS.0000065484.95996.AF. [DOI] [PubMed] [Google Scholar]
- 28.Ficek K, Cieszczyk P, Kaczmarczyk M, et al. Gene variants within the COL1A1 gene are associated with reduced anterior cruciate ligament injury in professional soccer players. J Sci Med Sport. 2013;16(5):396–400. doi: 10.1016/j.jsams.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Ficek K, Stepien-Slodkowska M, Kaczmarczyk M, et al. Does the A9285G polymorphism in collagen type XII alpha1 gene associate with the risk of anterior cruciate ligament ruptures? Balk J Med Genet. 2014;17(1):41–46. doi: 10.2478/bjmg-2014-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khoschnau S, Melhus H, Jacobson A, et al. Type I collagen alpha1 Sp1 polymorphism and the risk of cruciate ligament ruptures or shoulder dislocations. Am J Sports Med. 2008;36(12):2432–2436. doi: 10.1177/0363546508320805. [DOI] [PubMed] [Google Scholar]
- 31.Khoury LE, Posthumus M, Collins M, et al. ELN and FBN2 gene variants as risk factors for two sports-related musculoskeletal injuries. Int J Sports Med. 2015;36(4):333–337. doi: 10.1055/s-0034-1390492. [DOI] [PubMed] [Google Scholar]
- 32.Malila S, Yuktanandana P, Saowaprut S, et al. Association between matrix metalloproteinase-3 polymorphism and anterior cruciate ligament ruptures. Genet Mol Res. 2011;10(4):4158–4165. doi: 10.4238/2011.October.31.1. [DOI] [PubMed] [Google Scholar]
- 33.Mannion S, Mtintsilana A, Posthumus M, et al. Genes encoding proteoglycans are associated with the risk of anterior cruciate ligament ruptures. Br J Sports Med. 2014;48(22):1640–1646. doi: 10.1136/bjsports-2013-093201. [DOI] [PubMed] [Google Scholar]
- 34.O’Connell K, Knight H, Ficek K, et al. Interactions between collagen gene variants and risk of anterior cruciate ligament rupture. Eur J Sport Sci. 2015;15(4):341–350. doi: 10.1080/17461391.2014.936324. [DOI] [PubMed] [Google Scholar]
- 35.Posthumus M, September AV, Keegan M, et al. Genetic risk factors for anterior cruciate ligament ruptures: COL1A1 gene variant. Br J Sports Med. 2009;43(5):352–356. doi: 10.1136/bjsm.2008.056150. [DOI] [PubMed] [Google Scholar]
- 36.Posthumus M, September AV, O’Cuinneagain D, et al. The COL5A1 gene is associated with increased risk of anterior cruciate ligament ruptures in female participants. Am J Sports Med. 2009;37(11):2234–2240. doi: 10.1177/0363546509338266. [DOI] [PubMed] [Google Scholar]
- 37.Posthumus M, September AV, O’Cuinneagain D, et al. The association between the COL12A1 gene and anterior cruciate ligament ruptures. Br J Sports Med. 2010;44(16):1160–1165. doi: 10.1136/bjsm.2009.060756. [DOI] [PubMed] [Google Scholar]
- 38.Posthumus M, Collins M, van der Merwe L, et al. Matrix metalloproteinase genes on chromosome 11q22 and the risk of anterior cruciate ligament (ACL) rupture. Scand J Med Sci Sports. 2012;22(4):523–533. doi: 10.1111/j.1600-0838.2010.01270.x. [DOI] [PubMed] [Google Scholar]
- 39.Rahim M, Gibbon A, Hobbs H, et al. The association of genes involved in the angiogenesis-associated signaling pathway with risk of anterior cruciate ligament rupture. J Orthop Res. 2014;32(12):1612–1618. doi: 10.1002/jor.22705. [DOI] [PubMed] [Google Scholar]
- 40.Raleigh SM, Posthumus M, O’Cuinneagain D, et al. The GDF5 gene and anterior cruciate ligament rupture. Int J Sports Med. 2013;34(4):364–367. doi: 10.1055/s-0032-1316361. [DOI] [PubMed] [Google Scholar]
- 41.Stępień-Słodkowska M, Ficek K, Eider J, et al. The +1245g/t polymorphisms in the collagen type I alpha 1 (col1a1) gene in Polish skiers with anterior cruciate ligament injury. Biol Sport. 2013;30(1):57–60. doi: 10.5604/20831862.1029823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stępień-Słodkowska M, Ficek K, Maciejewska-Karlowska A, et al. Overrepresentation of the COL3A1 AA genotype in Polish skiers with anterior cruciate ligament injury. Biol Sport. 2015;32(2):143–147. doi: 10.5604/20831862.1144416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stępień-Słodkowska M, Ficek K, Kaczmarczyk M, et al. The variants within the COL5A1 gene are associated with reduced risk of anterior cruciate ligament injury in skiers. J Hum Kinet. 2015;45:103–111. doi: 10.1515/hukin-2015-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossert J, Terraz C, Dupont S. Regulation of type I collagen genes expression. Nephrol Dial Transpl. 2000;15(Suppl 6):66–68. doi: 10.1093/ndt/15.suppl_6.66. [DOI] [PubMed] [Google Scholar]
- 45.Liu X, Wu H, Byrne M, et al. Type III collagen is crucial for collagen I fibrillogenesis and for normal cardiovascular development. Proc Natl Acad Sci USA. 1997;94(5):1852–1856. doi: 10.1073/pnas.94.5.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birk DE, Fitch JM, Babiarz JP, et al. Collagen type I and type V are present in the same fibril in the avian corneal stroma. J Cell Biol. 1988;106(3):999–1008. doi: 10.1083/jcb.106.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiquet M, Birk DE, Bonnemann CG, et al. Collagen XII: protecting bone and muscle integrity by organizing collagen fibrils. Int J Biochem Cell Biol. 2014;53:51–54. doi: 10.1016/j.biocel.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalamajski S, Oldberg A. The role of small leucine-rich proteoglycans in collagen fibrillogenesis. Matrix Biol. 2010;29(4):248–253. doi: 10.1016/j.matbio.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 49.Pufe T, Harde V, Petersen W, et al. Vascular endothelial growth factor (VEGF) induces matrix metalloproteinase expression in immortalized chondrocytes. J Pathol. 2004;202(3):367–374. doi: 10.1002/path.1527. [DOI] [PubMed] [Google Scholar]
- 50.Wacholder S, Rothman N, Caporaso N. Population stratification in epidemiologic studies of common genetic variants and cancer: quantification of bias. J Natl Cancer Inst. 2000;92(14):1151–1158. doi: 10.1093/jnci/92.14.1151. [DOI] [PubMed] [Google Scholar]
- 51.van Meurs JB, Uitterlinden AG. Osteoarthritis year 2012 in review: genetics and genomics. Osteoarthr Cartil. 2012;20(12):1470–1476. doi: 10.1016/j.joca.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 52.Zeggini E, Panoutsopoulou K, Southam L, et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet. 2012;380(9844):815–823. doi: 10.1016/S0140-6736(12)60681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evangelou E, Valdes AM, Kerkhof HJ, et al. Meta-analysis of genome-wide association studies confirms a susceptibility locus for knee osteoarthritis on chromosome 7q22. Ann Rheum Dis. 2011;70(2):349–355. doi: 10.1136/ard.2010.132787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castano Betancourt MC, Cailotto F, Kerkhof HJ, et al. Genome-wide association and functional studies identify the DOT1L gene to be involved in cartilage thickness and hip osteoarthritis. Proc Natl Acad Sci USA. 2012;109(21):8218–8223. doi: 10.1073/pnas.1119899109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Birkedal-Hansen H, Moore WG, Bodden MK, et al. Matrix metalloproteinases: a review. Crit Rev Oral Biol Med. 1993;4(2):197–250. doi: 10.1177/10454411930040020401. [DOI] [PubMed] [Google Scholar]
- 56.Frank CB. Ligament structure, physiology and function. J Musculoskelet Neuronal Interact. 2004;4(2):199–201. [PubMed] [Google Scholar]
- 57.Claessen FM, de Vos RJ, Reijman M, et al. Predictors of primary Achilles tendon ruptures. Sports Med. 2014;44(9):1241–1259. doi: 10.1007/s40279-014-0200-z. [DOI] [PubMed] [Google Scholar]
- 58.Ben-Zaken S, Meckel Y, Lidor R, et al. Genetic profiles and prediction of the success of young athletes’ transition from middle- to long-distance runs: an exploratory study. Pediatr Exerc Sci. 2013;25(3):435–447. doi: 10.1123/pes.25.3.435. [DOI] [PubMed] [Google Scholar]
- 59.Massidda M, Scorcu M, Calo CM. New genetic model for predicting phenotype traits in sports. Int J Sports Physiol Perform. 2014;9(3):554–560. doi: 10.1123/ijspp.2012-0339. [DOI] [PubMed] [Google Scholar]
- 60.Taylor JB, Waxman JP, Richter SJ, et al. Evaluation of the effectiveness of anterior cruciate ligament injury prevention programme training components: a systematic review and meta-analysis. Br J Sports Med. 2015;49(2):79–87. doi: 10.1136/bjsports-2013-092358. [DOI] [PubMed] [Google Scholar]
- 61.Swart E, Redler L, Fabricant PD, et al. Prevention and screening programs for anterior cruciate ligament injuries in young athletes: a cost-effectiveness analysis. J Bone Jt Surg Am. 2014;96(9):705–711. doi: 10.2106/JBJS.M.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bahr R, Krosshaug T. Understanding injury mechanisms: a key component of preventing injuries in sport. Br J Sports Med. 2005;39(6):324–329. doi: 10.1136/bjsm.2005.018341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.September AV, Posthumus M, Collins M. Application of genomics in the prevention, treatment and management of achilles tendinopathy and anterior cruciate ligament ruptures. Recent Pat DNA Gene Seq. 2012;6(3):216–223. doi: 10.2174/187221512802717358. [DOI] [PubMed] [Google Scholar]
- 64.McGowan ML, Glinka A, Highland J, et al. Genetics patients’ perspectives on clinical genomic testing. Per Med. 2013;10(4):339–347. doi: 10.2217/pme.13.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Callier S. Genetic privacy in sports: clearing the hurdles. Recent Pat DNA Gene Seq. 2012;6(3):224–228. doi: 10.2174/187221512802717303. [DOI] [PubMed] [Google Scholar]


