Abstract
Objective
This work investigates the problem of clinical coreference resolution in a model that explicitly tracks entities, and aims to measure the performance of that model in both traditional in-domain train/test splits and cross-domain experiments that measure the generalizability of learned models.
Methods
The two methods we compare are a baseline mention-pair coreference system that operates over pairs of mentions with best-first conflict resolution and a mention-synchronous system that incrementally builds coreference chains. We develop new features that incorporate distributional semantics, discourse features, and entity attributes. We use two new coreference datasets with similar annotation guidelines – the THYME colon cancer dataset and the DeepPhe breast cancer dataset.
Results
The mention-synchronous system performs similarly on in-domain data but performs much better on new data. Part of speech tag features prove superior in feature generalizability experiments over other word representations. Our methods show generalization improvement but there is still a performance gap when testing in new domains.
Discussion
Generalizability of clinical NLP systems is important and under-studied, so future work should attempt to perform cross-domain and cross-institution evaluations and explicitly develop features and training regimens that favor generalizability. A performance-optimized version of the mention-synchronous system will be included in the open source Apache cTAKES software.
Keywords: Coreference, clinical NLP, portability, generalizability, machine learning
1 Introduction and Background
Coreference resolution is the task of grouping entity and event mentions in a discourse into the set of “chains” of real world entities or events they describe.1 Human beings perform this task naturally in the course of everyday linguistic processing, to track events, entities, and actors across time in conversation or writing. In the general domain, a newspaper article may refer to multiple people, organizations, events, dates, and locations, using a variety of names (United States, USA, ISIS, ISIL), pronouns (they), and terms (explosion, blast). Similarly, in the context of the clinical narrative, a disease (e.g., “colon cancer”) may be referred to dozens of times over the course of its progression, using different terms (cancer, adenocarcinoma) and pronouns (it), within and between different encounters and notes. Each of these mentions may introduce novel information about the cancer (e.g., primary site, tumor size, metastasis location) to the total picture of the disease, making it essential to reconcile the multiple mentions in order for an extracted representation to be accurate and complete.
Deep phenotyping of cancer patients in particular requires coreference for accurate phenotype representations; how many primary tumors, how many metastases, and which tumor is which size. Downstream research applications making use of deep phenotypes may depend on this information. For example, consider a clinical researcher who wants to conduct a retrospective study of patients with breast cancer that metastasized to the lungs, and study the importance of initial tumor size. Coreference resolution will need to correctly link the mention of the initial tumor's location to the mention of that tumor's size, and also detect a tumor mention in the lung and correctly infer that it is a different tumor altogether.
There are a variety of method types that have been applied to the coreference resolution problem. The earliest approaches focused on pronouns only and were rule-based,[1] navigating syntax trees using a hard-coded search algorithm to find the most likely antecedent. One of the most successful early machine learning approaches was the mention-pair paradigm that used supervised classifiers.[2] In this paradigm, a coreference chain would be converted into a set of pairwise decisions between mentions, where the label of a mention pair was TRUE if the two mentions belonged to the same chain. A statistical classifier can be trained on these labels, and used to label new documents. This approach requires a reconciliation mechanism for resolving globally inconsistent local classifier decisions, since it is possible to make inconsistent pairwise decisions (e.g., f(A,B) = TRUE, f(B,C) = TRUE, f(A,C) = FALSE). More recent rule-based approaches have built on upstream statistical NLP components and have had some success. These so-called “sieve”-based approaches[3] apply a series of rules ranked in order of how reliable (precise) they are. For example, a rule linking both parts of appositive constructions such as The German Chancellor, Angela Merkel is very reliable and so is applied early in the process.
Coreference resolution has seen progress in the clinical domain recently, largely due to a shared task, the 2012 i2b2 challenge on coreference resolution[4], and shared datasets for the task.[5]–[7] Approaches used in the clinical domain include some of those mentioned above, including rule-based sieve approaches,[8], [9] traditional pairwise mention classification approaches,[10] and hypergraph factorization approaches.[11], [12] Before the recent release of shared datasets for training, machine learning approaches were uncommon.[13]
Since the shared task, much work has been done in the general domain that has not yet filtered into the clinical version of this research topic. Some of the work has focused on improving the search through the possibility space. Instead of attempting to find links between a new mention and all previous mentions, some of these systems instead incrementally build chains, [14]–[16] and then attempt to find links between a new mention and partial chains created by previous decisions. These “entity-centric” approaches allow more global features to be considered by the classifier. In pairwise approaches, by contrast, global information can only be considered in the reconciliation post-process mentioned above. Some entity-centric approaches use agglomerative clustering, initializing the set of mentions as singleton clusters, and iteratively merging clusters until some stopping criterion is met. “Mention synchronous” approaches, including the work described in this manuscript, are entity-centric, but more specifically build chains by attempting to merge newly encountered mentions with already-built partial chains, processing the document in the same way a human reader does.
Coreference resolution is still an unsolved problem, with new features, approaches, and datasets being developed across domains. But even at the modest levels of published performance that currently represent the state of the art, systems trained in one domain do not always generalize to other domains. In fact, this work was motivated by preliminary experiments that found the existing cTAKES coreference resolution system [17] performed poorly on a new gold standard coreference dataset that our lab and collaborators created as part of the THYME (Temporal History of Your Medical Events) project.[7] It is worth considering whether clinical coreference resolution is a meaningful task that warrants domain-specific research, or whether clinical text should just be considered a domain to adapt generally-trained coreference systems to. There are reasons to consider the problems separate. In the general domain, there is a distinction drawn between entity and event coreference, where entity coreference performance is well-studied and has seen some improvements over time,[18] though it is still far from perfect. General domain event coreference, however, is still in its early stages of development and is considered a very difficult unsolved problem. In clinical texts, this distinction is not as clear, and the task is essentially mixed event and entity coreference. While anatomical sites and people behave like traditional entities, diseases, symptoms, and procedures are more like events, in that they have possibly finite time spans, can change state over time, and can have different attributes for different instances. They are potentially more tractable than general domain events, however, because most events in a given patient record will belong to that patient. This is compared to, for example, the news domain where different mentions of the same explosion event might occur across articles, days, newspapers, etc.
If clinical coreference resolution task is indeed unique enough, then it will be even more crucial to continue developing and growing coreference corpora specific to the clinical domain. If not, it may be preferable to build ever-larger corpora in the general domain. In either case, however, questions of generalizability and domain adaptation loom large, as it is simply not feasible to develop labeled corpora for every clinical sub-domain on which one might want to extract coreference information.
In this paper, we begin to explicitly examine issues of generalizability in the development of a new coreference resolution system. We consider three research questions: 1) Can approaches that center on coreference chains rather than mentions improve performance? 2) How well do systems optimized for one domain generalize to new domains? And 3) How well do certain feature types perform when applied to new domains? We develop a new mention-synchronous coreference resolution system for the clinical domain, several new features focused on generalizability, and evaluate the system against existing baselines.
2 Materials and Methods
2.1 Methods
Mention-synchronous Coreference Resolution
The first step in any coreference resolution system is identifying the set of markables, or candidate phrases for membership in coreference chains. Many existing systems build chains using a mention-pair approach, computing pairwise scores for all markable pairs and adding a second pass to create chains using these scores and reconcile inconsistencies. In contrast, we use the mention-synchronous approach, which was first described by Luo et al. [19] In the mention-synchronous approach, coreference chains are incrementally built up as a document is processed from left-to-right. Coreference decisions involve comparing a new markable to the set of partial chains, and either adding the markable to an existing chain or starting a new chain. Many subsequent approaches following this approach have attempted to model this process as a search procedure, tracking multiple hypotheses through time. Here, rather than performing a search, we keep track of the one-best hypothesis for efficiency reasons. Figure 1 formalizes our description of the mention-synchronous algorithm, while Figure 2 shows a schematic comparison of the mention-pair vs. mention synchronous approach. The two subsections below describe in detail how candidate markables are selected, and how candidate antecedent chains are selected, which determines the search order and thus impacts the greedy search.
Figure 1.
Algorithm describing the greedy mention-synchronous coreference system for a single document D. c is the running set of coreference chains for the document that is built with the algorithm. The outer loop is over markables m, and the inner loop is over candidate chains c in c. The function extractFeatures(m, c) results in a feature vector f. At classification time, if the classifier returns True, the markable m is added to the current chain under consideration c. If the inner loop terminates without adding m to any chain, the variable singleton will be True and a new singleton chain will be created and appended to c.
Figure 2.
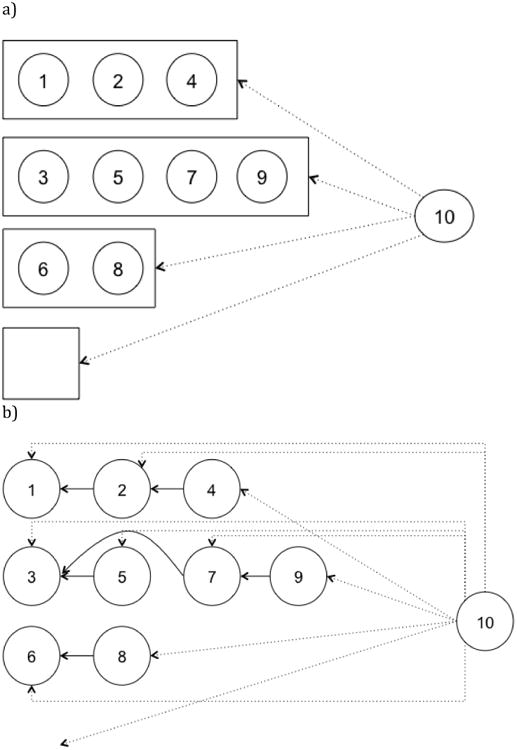
a) Mention-synchronous representation. Circles represent markables. Boxes represent explicit coreference chains being built. Dotted arrows indicate decision that mention-synchronous model must make when evaluating markable 10.
b) Mention-pair representation. Solid arrows represent links between markables, implicitly defining chains. Dotted arrows indicate decisions a mention-pair system must make when evaluating markable 10.
Rule-based markable detection
The first stage in coreference resolution is finding markables, or candidate phrases for membership in coreference chains. Here we follow recent work[20] by focusing on recall at the markable detection stage. We start by running a dependency parser on the entire corpus (the clinically trained parser in Apache cTAKES). This builds a syntactic graph of the sentence in which every word is tagged with a part of speech and has an outgoing arc pointing to its syntactic head. We create a markable for every word tagged as a noun, expanding it to include all words with an arc into that word. For example, in the phrase ascending colon cancer, both colon and cancer will be tagged as nouns, so we will create two markables. The first will include all terms syntactically dominated by the disease name (ascending colon cancer), and the second will include all terms dominated by the anatomical site name (ascending colon).
Time expressions may also be coreferent in THYME (see the Datasets section for a description of the THYME corpus), though they do not always include nouns (e.g., today, January 15). Thus, we also run the THYME-trained time expression recognizer[21] to tag time expressions and we deterministically add these as markables. While this may bias our training process because the THYME-trained system will be better at finding time expressions in our training data than we might expect on new data, there is no information leakage from the test set, because the system included in Apache cTAKES was not trained on any THYME test data.
Pair selection heuristics
One of the challenges in coreference resolution is the sheer number of potential coreference links that need to be evaluated. Besides the issue of system speed, the vast majority of proposed pairs are not coreferent, so the training data will be extremely unbalanced towards negative examples. In such cases a classifier can achieve high training accuracy by always predicting the majority class, but will be useless for the actual task.
These concerns led us to the conclusion that instead of exhaustively comparing markables and chains, we should use heuristic rules to create a smaller, but high-recall (sensitivity), set of comparisons to make for every markable. First, we consider links with any chains already consisting of two or more elements. This is motivated by the intuition that entities mentioned more than once are important and more likely to be mentioned again. Remaining singleton chains are considered for pairing first based on distance. We consider all singleton chains mentioned in the five sentences prior to the markable under consideration. We next consider singleton chains in which their markable element appears in a section header. We define section headers to be paragraphs that consist of single sentences. Paragraphs are defined as blocks of text starting and ending with two or more newline characters. Finally, we consider all singleton markables whose headwords match the headword of the candidate anaphor markable. In the pairwise baseline, we use the same sentence distance heuristic, section header heuristic, and headword matching heuristic. These heuristics are encapsulated in the getCandidateAntecedentChains() function in Figure 1.
System Features
We use a variety of manually engineered features to attempt to represent important aspects of the relationship between an anaphor and its antecedent. Many of these features are derived from earlier work, in particular, the seminal works of Soon et al.[2] and Ng and Cardie,[22] but also our previous work,[17] and the work of other recent systems. These features are summarized in Supplementary Table 1.
The strongest of the features taken from existing coreference literature are string matching features. These features take different configurations of each markable, including the whole string, the first word, last word, and whole string without determiners, and perform all these permutations against markables in candidate chains. String-matching features are valuable, but the difficulty in coreference resolution is learning how to link markables without strong string matches. Additional standard features for the clinical domain use the Unified Medical Language System (UMLS). One strong feature indicates whether two markables map to the same UMLS Concept Unique Identifier (CUI). Another, weaker feature, indicates whether they map to any of the same Type Unique Identifiers (TUIs).
We first implement three new feature sets, attempting to model discourse, semantic, and entity attributes aspects of coreference. Our goal in defining these higher-level linguistic features is to move away from surface level features that are believed to be more domain-specific. These new feature sets are summarized in Table 1, and Supplementary Table 1 includes these as well.
Table 1.
The Type column is brief description of the feature class. The Name column is the specific feature description. Match type refers to how the feature is distributed over chains in the mention-synchronous model. Any means that any element of the chain can match the mention. Max means that the maximum match between a chain element and the mention is the feature value. Description gives a brief explanation or example of the feature where space allows.
| Type | Name | Match type | Description |
|---|---|---|---|
| Discourse Features | Stack position (all) Stack position (no singletons) Antecedent salience Anaphor salience |
Max | Number of intervening chains since last mention Number of non-singleton intervening chains See text See text |
| Entity Attribute Features | Negation Uncertainty Generic Subject History |
Any |
No tumor Possible tumor Discussed chemotherapy Breast cancer mother Hx headaches |
| Semantic Features | Cosine similarity Selectional preferences UMLS Ancestry |
Max Max |
Similarity of headwords in word2vec (see text) Probability of governing semantic type (see text) Markables are in an ancestral relation (see text) |
| Surrounding Token Context | Word POS (Part of speech) Vector |
N/A | Token identities within markable and context window POS tags of markable and context Word embeddings of markable head and context words |
Discourse features take into account the fact that our antecedents are chains instead of markables. While pairwise systems have features that count the number of markables between the two candidate pairs under consideration, we count the number of partial chains between the candidate anaphor and candidate chain. This converts an entity mention distance to an entity distance. One version of this feature counts all intervening chains, while a second version only counts intervening non-singleton chains. We also create a new feature representing the salience of entities in the discourse. For this, we borrow from existing work by Recasens et al. on detecting singleton mentions.[23] Detecting singletons has been used in previous work to filter mentions from the pair-matching stage. Here we invert the sign and call it a salience classifier. Our intuition is that salient discourse entities will be ones that are mentioned multiple times and singletons are not very salient. We incorporate the features from Recasens et al. with a few new features, including UMLS Semantic Group and a feature that encodes which third of the sentence the mention occurs in (beginning, middle, end).
To train the salience classifier we use gold standard coreference chains and the deterministic markable detector described above. Training data consists of positive examples that are members of chains (salient markables) and negative examples that are not in chains (non-salient markables). We train a logistic regression classifier that outputs probabilities, and we treat this probability as the salience. We then extract two features, one for the salience of the candidate anaphor and one for the maximum salience of any markable in the candidate chain.
Entity attribute features attempt to enforce compatibility between entity attributes in anaphors and antecedents. We run the cTAKES assertion module[24] on each markable to classify four binary attributes (negation, uncertain, generic, historical) of each markable and one multi-class attribute (subject). There is one feature for each attribute category, whose value is TRUE only if the value of the anaphor for that category matches the value of some member of the candidate antecedent chain. For example, if the anaphor is marked as Negated by cTAKES, the feature will only fire if some member of the candidate antecedent chain is also marked Negated.
The first new semantic feature uses word embeddings to compute semantic similarity between the headwords of the candidate anaphor and the candidate chain. Word embeddings[25] are continuous vector representations of words that are typically created by training a neural network to predict neighboring contexts, usually other words. The resulting vector representation of a word can then be used for a variety of purposes, including predicting similarity between two words. This can be done by simply computing the cosine similarity between the vector representations of two words. We create 200-dimensional vectors for each word type, training on all 1.2 million notes in the MIMIC II dataset (for details on MIMIC II and other datasets see the Datasets section below) using word2vec.[25] We use cosine similarity as a feature, computing the maximum cosine distance between the vector for the anaphor headword and any antecedent chain mention headword. Another semantic feature uses the IsA relation from SnomedCT that is present in the UMLS Metathesaurus. This feature encodes whether the candidate anaphor is connected via IsA relations to any mention in the candidate antecedent chain and the direction of that relation.
The final semantic feature attempts to model semantic group selectional preferences of pronouns. [26] Pronouns cannot be mapped to UMLS semantic groups, so they can be difficult to resolve when there are multiple candidate antecedents in previous sentences. One way to resolve these difficult cases is to look at what the pronoun is doing. For example, for it reoccurred the mention it is more likely to corefer to a disease or sign/symptom, while in tolerated it well, the pronoun it is more likely to be a drug or procedure. To model these selectional preferences we processed the entire MIMIC-II corpus (for details on MIMIC and other datasets see the Datasets section) with the cTAKES UMLS dictionary lookup and dependency parser and extracted tuples of (Governing verb, Dependency relation, Semantic group) – for example: (reoccurred, subject, sign/symptom) is extracted from the phrase the rash reoccurred, indicating an instance of a sign/symptom being the subject of the verb reoccurred. After compiling these counts for the whole corpus we estimated the distribution P(Semantic group | Verb, depRel). With this distribution, if we see a new pronoun as the subject to the verb reoccurred, we can estimate that the pronoun is most likely to refer to a procedure. This feature fires only for pronoun anaphor candidates, and its value is the maximum of the estimated probability of any semantic group found in the current candidate chain.
Finally, we introduce features to represent the surrounding context of a markable and experiment with different kinds of abstraction to represent this context. These feature types have been successful for us on other relation extraction tasks[27] and we found they were surprisingly effective on the development set. This is a potentially valuable representation for learning whether a candidate is coreferent at all, but surface features like tokens may overfit to the training domain, so we designed experiments attempting to measure this. This feature is also summarized in Table 1.
For this markable context feature, we only extract features for the candidate markable anaphor and not the candidate antecedent chain. We experiment with token identity, part of speech (POS) and vector features. For the token identity features we extract a feature for every token identity in the markable as a bag, as well as features for three words of context on either side of the markable. For the POS feature we use the system-tagged POS tags for the tokens instead of their token identities.
We also experiment with word embeddings (described above) for the token context feature. For this feature, since it only attaches to the anaphor and not any potential antecedent, we simply use the dimensions of the word vector as continuous features. We represent words with 200-dimensional vectors, and we use one word of context on either side of the markable, as well as the headword of the markable, for a total of 600 continuous features.
Resolving Person Mentions
In the general domain, person mentions are frequent and important to resolve, as there may be ambiguity about who committed which actions. In the clinical domain, there is significantly less ambiguity – nearly all person mentions are of the patient, physician mentions can be handled with a few rules, and the bulk of the few remaining person mentions are in the family history section, which can be detected with other means. However, in our datasets (THYME and DeepPhe, described in more detail in the Datasets section), person coreference chains are annotated, so without some extraction mechanism, scores will appear artificially low. We built a simple pattern-based extractor for detecting person mentions without a name dictionary, and used this for resolving names in the THYME corpus. But while both corpora are de-identified, only the THYME corpus has realistic-looking fake names. The DeepPhe corpus does not always have realistic depictions of names due to the way de-identification was implemented. For example, a patient's name might be substituted with tags such as PERSON1. Because of this de-identification method, overall performance on DeepPhe may be lower due to poor performance on person mentions.
2.2 Datasets
In our work we used two datasets. The Clinical TempEval sets of the THYME corpus (both 2015[28] and 2016 tasks[29]) annotated for coreference have already been made available to the research community. Of note, the THYME corpus is not exhaustively annotated for coreference -- the annotated set used here consists of 98 training documents, 32 development documents and 55 test documents. The documents are pathology and clinical notes from colorectal cancer patients at the Mayo Clinic. The training set contains 2216 coreference chains, the development set contains 1272 chains, and the test set contains 1343 chains. The corpus was double-annotated by a linguistics student and a domain expert, followed by an adjudication phase of the disagreements through a discussion between the two annotators. The inter-annotator agreement (IAA) for coreference is reported as a CoNLL F1 score (see a description of the CoNLL metric in the Evaluation Section below). The IAA between two annotators is 62.2, while average annotator agreement with the gold standard is 71.9. For final testing, the training and development sets were combined, so that the final tested system was trained using 3488 chains.
The second set consists of the documents (pathology, radiology, oncology, clinical notes) of breast cancer patients from the UPMC and is part of a bigger project on the topic of deep cancer phenotyping (DeepPhe2). The subset we used includes 48 documents for 4 patients (the DeepPhe training split), containing 191 chains. The DeepPhe corpus is single-annotated therefore no IAA is available. DeepPhe annotation used the THYME annotation guidelines as a starting point, so the annotation guidelines are quite similar. Some divergence was introduced to improve annotation speed and reliability. In particular, non-clinical entity chains were not annotated in DeepPhe, reducing the total number of chains. These were more difficult annotations to perform while also being less useful for downstream use cases than clinical events and entities.
The documents from both datasets are completely de-identified except for temporal expressions, which were left unchanged according to the THYME project limited data use agreement. The coreference annotation guidelines are posted on the THYME project page.3
We consider these datasets to be different domains since they represent different disease populations (colon vs. breast cancer) and are sourced from different institutions (Mayo Clinic vs. UPMC). While these two diseases are distinct, and there will undoubtedly be lexical variation, we also expect there to be some lexical overlap, as terms like tumor should be shared. Therefore, the generalizability experiments we perform probably represent a conservative estimate of the difficulty of adapting a coreference system to a new disease domain.
We use one large external data source, the MIMIC II corpus,[30] for training word embeddings as described above, to use as features in our classifiers. Learning these representations requires large unlabeled text datasets, typically several orders of magnitude larger than gold standard annotated coreference datasets. Therefore, the relatively large MIMIC corpus is more appropriate for this purpose than the THYME or DeepPhe corpora.
2.3 Research Questions
In our evaluation, we are concerned with several variables around coreference system development. Our first research question is whether consideration of chains (mention-synchronous system) can improve performance over pairwise systems (mention-pair system). For the fairest possible comparison, we strive for feature parity. Since the two systems use different architectures they are not exactly the same, but this is part of what the experiment is meant to measure – whether the different form of features used by a cluster-based system has value. For both systems we use Liblinear[31] for our machine learning, a fast, high-performing linear support vector machine implementation, with L2 regularization. Early experiments with non-linear kernels did not show any improvement and took substantially longer to train. We performed a grid search to find the best value for the regularization parameter C on the development set, then retrained the system on the training and development data with the optimal parameter, finally testing on the held out test data.
Our second research question is how systems optimized for one corpus perform on a new corpus, and whether relative rankings hold up. For this experiment we took the best performing THYME-trained systems from experiment one above and tested them on the unseen DeepPhe data. Since the DeepPhe corpus annotation guidelines focus on chains containing UMLS entities, if we run the THYME-trained system in end-to-end mode, precision on DeepPhe will appear artificially low. To eliminate this confusion, at test time we give the system only gold markables that are not singletons (i.e., those in chains). While this decision removes the ability to analyze the importance of discovering markables on DeepPhe, it allows us to do more meaningful cross-corpus comparisons and isolate the performance on the core coreference resolution task. Since DeepPhe corpus annotation is not complete, we do not yet have enough data to train a good system only on DeepPhe data. Therefore we cannot perform the reverse experiment where we train on DeepPhe and test on THYME, so we focus on how the trained system performs on brand new data compared to a baseline.
Our third research question is the effect that different features have on generalization performance. We hypothesize that token-level features are a cause of overfitting and removing them may improve generalization at the expense of some in-domain optimization. To test this we look at four separate token feature conditions: no token-level features, token identity features, POS tag token features, and word vector features. In each case we optimize a system on the THYME development set and evaluate it on the THYME test set and the DeepPhe set. For this experiment we give both THYME and DeepPhe systems non-singleton gold standard markables at test time so that we can compare across corpora.
2.4: Evaluation Methodology
Evaluating coreference resolution systems is difficult and there are a number of proposed metrics for doing so. The most prominent are MUC,[32] B3,[33] and CEAF.[34] While the original papers describing these metrics focused on gold standard mentions, they have recently been standardized[35] in the way that they handle end-to-end coreference for a shared task in the general domain sponsored by CoNLL (Computational Natural Language Learning – an NLP/machine learning conference)[36]. The standard scoring tool that was built for that shared task has been made a community standard, and computes these three metrics (in addition to BLANC[37]), as well as an average of the three metrics that was used as the official metric for the task. Here we report the MUC, B3, and CEAF score, as well as the “CoNLL score” that averages the F1 of those three metrics.
MUC computes recall by counting the percentage of inter-cluster links in the gold standard that are found by the system. Precision is computed by reversing the gold and system outputs. B3 computes precision and recall scores for each mention, as the number of correct elements in the chain containing that mention, divided by the number of elements in the system chain or gold chain, respectively. Document precision and recall are computed as weighted averages, with the weights determined by the size of the chains. CEAF first computes an optimal alignment between the reference and gold mention sets, and compares that alignment to an alignment of gold outputs to itself (for recall) and system output to itself (for precision). For details on these algorithms, especially how they apply to end-to-end coreference, see Pradhan et al.[35]
We test for statistical significance using a Wilcoxon signed-rank test. Testing for significance in coreference resolution is difficult because it is a structured prediction problem. We can't compare performance at each classification decision because two different systems will eventually diverge and encounter different classification decisions. The compromise test setting we consider is over document F1 scores. The standard CoNLL scoring tool computes coreference metrics at the document level, so we can compare two systems for each document. The limitation of this approach is that it removes information about document length. For example, a 5% performance gain on a long document represents more decisions affected than a 5% loss on a short document, but this difference cannot be accounted for by a document-by-document significance test. This is not optimal, since coreference performance is probably correlated with document length (longer documents are harder). Nevertheless, given the difficulty of significance testing for coreference resolution, we find this to be the best method.
3 Results
The results to answer the first research question — how the mention-synchronous system compares to a mention-pair system – are in Table 2. They show that the both systems obtain similar F1 scores on the THYME test set (mention synchronous system CoNLL score of 55.3 vs. mention-pair system CoNLL score of 54.7; p=0.9). The mention-synchronous system improves recall but precision decreases. This result makes clear that representing chains is not the panacea that will solve coreference resolution.
Table 2.
Results of experiment 1 – end to end coreference on THYME test set. Comparison between Mention-Synchronous and Mention-Pair Systems. Comparison between mention-synchronous system CoNLL score of 55.3 and mention-pair system CoNLL score of 54.7 was not significant (p=0.9).
| Mention-Pair System | Mention-Synchronous System | |||||
|---|---|---|---|---|---|---|
| Metric | Recall | Precision | F1 | Recall | Precision | F1 |
| MUC | 53.1 | 75.2 | 62.3 | 55.7 | 71.0 | 62.4 |
| Bˆ3 | 44.5 | 69.0 | 54.1 | 48.0 | 63.4 | 54.7 |
| CEAF | 41.1 | 56.9 | 47.7 | 43.6 | 55.3 | 48.8 |
| CoNLL | 54.7 | 55.3 | ||||
The results for the second research question — how well mention-synchronous and mention-pair systems generalize — are in Table 3. Both systems see performance degradation on the DeepPhe data, despite being given gold standard markables, highlighting the need for domain adaptation. The mention-synchronous system performs much better than the mention-pair system (mention synchronous system CoNLL score of 52.5 vs. mention-pair system CoNLL score of 30.4; p < 0.0001), largely by having very high precision. This suggests that, given a domain-adapted markable detection strategy, the mention-synchronous system has a better chance of performing well in new domains than the mention-pair system.
Table 3.
Results of experiment 2 – coreference generalizability with gold markables on DeepPhe set. Comparison between mention-synchronous system CoNLL score of 52.5 and mention-pair system CoNLL score of 30.4 is significant (p < 0.0001)
| Mention-Pair System | Mention-Synchronous System | |||||
|---|---|---|---|---|---|---|
| Metric | Recall | Precision | F1 | Recall | Precision | F1 |
| MUC | 37.8 | 45.0 | 41.1 | 36.5 | 94.0 | 52.5 |
| Bˆ3 | 28.6 | 35.4 | 31.6 | 31.3 | 95.1 | 47.1 |
| CEAF | 17.7 | 19.3 | 18.5 | 39.1 | 84.8 | 53.5 |
| CoNLL | 30.4 | 52.5 | ||||
The results supporting the third research question – what features are most generalizable – are in Table 4. The baseline system (no token features) is worse than the token identity feature condition on THYME (CoNLL score of 59.7 vs. 62.9; p < 0.001). POS features on the DeepPhe corpus were 3.6 points higher than token identity features at the corpus level, but this result was not found significant in the document-aligned test, likely due to high variation in performance between documents (CoNLL score of 52.6 vs. 56.2; p=0.42). Surprisingly, the vector features, which were included because they had a positive effect on development set performance in the end-to-end setting, did not have a positive impact in the setting where gold markables are provided (CoNLL score dropped from 54.2 to 51.9; p=0.16). This suggests that vector features play the role of detecting anaphoricity more than choosing the correct antecedent.
Table 4.
Results of different token feature configurations – coreference generalizability given gold markables. Significance tests were performed within each corpus, comparing each feature configuration against the baseline. The symbol * indicates significance at p < 0.001, while ˆ indicates significance at p=0.005.
| CoNLL Scores | THYME | DeepPhe |
|---|---|---|
| Baseline (No token features) | 59.7 | 54.2 |
| Token identity only | 62.9* | 52.6 |
| Token POS only | 61.8ˆ | 56.2 |
| Token vector only | 51.8* | 51.9 |
4 Discussion
Our results show that while system gains for within-document coreference resolution are not significant (mention synchronous system CoNLL score of 55.3 vs. mention-pair system CoNLL score of 54.7; p=0.9), they generalize better than existing approaches (mention synchronous system CoNLL score of 52.5 vs. mention-pair system CoNLL score of 30.4; p < 0.0001).
4.1 Error Analysis
To further gain insights into our mention synchronous method performance, we conducted an error analysis on all mentions in all documents for one randomly chosen patient in the DeepPhe corpus, 14 documents in total. This is about the same number of documents as other patients, with slightly longer documents on average. We categorized errors into four mutually exclusive categories – false new (FN), wrong link (WL), false anaphor (FA), and annotator error (AE). [38] FN errors occur when the system creates a new chain for a markable when it should be added to an existing chain, WL errors are when the system adds an anaphoric markable to the wrong chain, and FA errors occur when the system adds a markable to an existing chain when it should start its own chain. We observed 54% of the errors were FN, indicating that our main problem was a failure to link (roughly, recall errors). These errors occur when two mentions have different headwords (e.g., tumor vs. mass), have very different modifiers (left breast cancer vs. cancer), or they are very far away from each other in the note. Recall is a common problem with coreference systems, with string matching features often being the only strong-enough features to create new links. One reason for the recall problem is that coreference has many more negative instances than positive, so coming up with features that are strong enough to overcome the number of negative instances to favor linking is a major challenge in a feature-engineering approach. Our features attempted to remedy this using UMLS CUI similarity and word-embedding-based lexical similarity to link mentions with semantically similar words. Since this is still the main error source we clearly have a long way to go. One interesting possible next step is to make use of methods that merge word embeddings with ontologies [39], “retrofitting” learned word embeddings so that they align to a known hierarchy. This would presumably improve the quality of the embeddings for the task and may combine the strengths of UMLS and embedding features.
Over a third of the errors (35%) were FA links that should not have been created. In the patient whose notes we examined, there were multiple tumors, and the system falsely conflated them as the same tumor due to similarity of terms used to describe them, as well as their being located closely together in the document. Similarly, the multiple tumors necessitated multiple imaging studies, which were also sometimes conflated. While this is an important case for a coreference system to handle, its relative rarity means that this analysis perhaps overestimates the FA error type. This kind of error may be addressed by taking into account entity relations as well as attributes. While we currently use agreement between negation and other entity attributes as features, we find that this feature does not fire very frequently. A related feature type that may be useful is one that enforces agreement between, for example, location relations. In the example described above, each tumor has a location (right breast, lymph node, etc.) and the cTAKES relation extraction system can find relations between tumors and anatomical sites. The main difficulty is finding enough examples where this occurs for the system to learn this constraint properly (e.g., a relation to a right breast is incompatible with a left breast, but a relation between a right breast and breast is compatible). It may be necessary to collapse all constraints (attributes, relations) into a single feature for the feature to be strong enough to make a decisive difference.
Annotator errors (AE), where the gold standard was judged by the authors to be erroneous, were next most common (8%), and these errors will be used to perform another pass of error correction on the gold standard. Finally, WL errors were rare (3% of errors).
4.2 Directions for Future Work
Future work will examine the degree to which using search procedures during learning can be made efficient and whether they improve performance. The core issue is that coreference resolution is a structured problem, where the inputs to a coreference decision later in the document are determined by the sequence of earlier decisions. By tracking only a single best hypothesis, a single incorrect early decision can lead to a “dead end” where there are no good decisions. If search is used during inference, the system can backtrack out of dead ends to return to an earlier decision and go down another path. The search space for the coreference resolution problem is quite large, so constraining this search is necessary for the system to run in an acceptable amount of time. The first step in this work will be a deeper error analysis looking at the impact of errors in the input to decisions.
This work also laid the groundwork for reasoning about entities rather than pairs of mentions. Future work will extract patient-level representations of entities and events, by performing the even more difficult task of cross-document coreference. Cross-document coreference is an elaboration of the coreference task that allows for tracking entities across multiple notes in the patient record. The cross-document setting is even more challenging because there are fewer linguistic cues that relate mentions in different documents. There are as of right now no publicly available clinical datasets annotated for cross-document coreference, so this work focused on within-document coreference. However, phase two of the THYME project is undertaking cross-document coreference annotations. The approach we described here is entity-centric, allowing the system to carry over entity representation from one document to another.
5 Conclusion
This paper has described a system for clinical coreference resolution that represents resolved entities as chains as it proceeds through the note. The mention-synchronous system developed to use chains as antecedents rather than individual mentions does not result in significant gains for within-document coreference resolution, but it generalizes better than existing approaches. A performance-optimized version of the mention-synchronous system will be included in the open source Apache cTAKES software.
Supplementary Material
Highlights.
Coreference resolution is important for extracting information from clinical documents
We implement and evaluate a model that tracks entities across a document
We explore generalizability of some important coreference features
Acknowledgments
We thank Profs. Rebecca Jacobson and Harry Hochheiser at University of Pittsburgh Department of Biomedical Informatics for expert review of the manuscript. This research was supported by grants from several NIH institutes: NLM award R01LM010090 (THYME), NIGMS award R01GM103859 (iPGx), and NCI award U24CA184407 (DeepPhe). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
While our system tracks clinically relevant entities and events, rather than referring to “entities and events” in every instance, we hereafter just say “entities” in the interest of brevity.
Disclosure statement: TM has worked as a paid consultant, and GS is on the Advisory Board for Wired Informatics, which provides services and products for clinical NLP applications.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hobbs JR. Resolving pronoun references. Readings Nat Lang Process. 1986;3(1):339–352. [Google Scholar]
- 2.Soon WM, Ng HT, Lim DCY. A machine learning approach to coreference resolution of noun phrases. Comput Linguist. 2001;27(4):521–544. [Google Scholar]
- 3.Raghunathan K, Lee H, Rangarajan S, Chambers N, Surdeanu M, Jurafsky D, Manning CD. A Multi-Pass Sieve for Coreference Resolution. Proc 2010 Conf Empir Methods Nat Lang Process (EMNLP '10) 2010:492–501. [Google Scholar]
- 4.Uzuner O, Bodnari a, Shen S, Forbush T, Pestian J, South BR. Evaluating the state of the art in coreference resolution for electronic medical records. J Am Med Informatics Assoc. 2012;19(5):786–791. doi: 10.1136/amiajnl-2011-000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savova GK, Chapman WW, Zheng J, Crowley RS. Anaphoric relations in the clinical narrative: corpus creation. J Am Med Inform Assoc. 2011;18(4):459–465. doi: 10.1136/amiajnl-2011-000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albright D, Lanfranchi A, Fredriksen A, Styler WF, Warner C, Hwang JD, Choi JD, Dligach D, Nielsen RD, Martin JH, Ward W, Palmer M, Savova GK. Towards comprehensive syntactic and semantic annotations of the clinical narrative. J Am Med Informatics Assoc JAMIA. 2013;20(5):922–930. doi: 10.1136/amiajnl-2012-001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Styler WF, IV, Bethard S, Finan S, Palmer M, Pradhan SS, de Groen PC, Erickson B, Miller TA, Lin C, Savova GK, Pustejovsky J. Temporal annotation in the clinical domain. Trans Assoc Comput Linguist. 2014;2:143–154. [PMC free article] [PubMed] [Google Scholar]
- 8.Jonnalagadda S, Li D, Sohn S, Wu S. Coreference analysis in clinical notes: a multi-pass sieve with alternate anaphora resolution modules. J Am Med Informatics Assoc. 2012;19(5):867–874. doi: 10.1136/amiajnl-2011-000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rink B, Roberts K, Harabagiu SM. A supervised framework for resolving coreference in clinical records. J Am Med Informatics Assoc. 2012;19(5):875–882. doi: 10.1136/amiajnl-2012-000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Liu J, Wu J, Wang Y, Tu Z, Sun JT, Tsujii J, Chang EIC. A classification approach to coreference in discharge summaries: 2011 i2b2 challenge. J Am Med Informatics Assoc. 2012;19(5):897–905. doi: 10.1136/amiajnl-2011-000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai J, Mújdricza-Maydt E, Strube M. Unrestricted coreference resolution via global hypergraph partitioning. Proc Fifteenth …. 2011 Jun;:56–60. [Google Scholar]
- 12.Cai J, Mujdricza E, Hou Y. Weakly supervised graph-based coreference resolution for clinical texts. Proc 2011 i2b2/VA/Cincinnati Work …. 2011 [Google Scholar]
- 13.Zheng J, Chapman WW, Crowley RS, Savova GK. Coreference resolution: A review of general methodologies and applications in the clinical domain. J Biomed Inform. 2011;44(6):1113–1122. doi: 10.1016/j.jbi.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma C, Doppa J, Orr J, Mannem P. Prune-and-Score: Learning for Greedy Coreference Resolution. EMNLP. 2014 [Google Scholar]
- 15.Stoyanov V, Eisner J. Easy-first Coreference Resolution. COLING. 2012 [Google Scholar]
- 16.Clark K, Manning C. Entity-centric coreference resolution with model stacking. Assoc Comput Linguist (…. 2015 [Google Scholar]
- 17.Zheng J, Chapman WW, Miller TA, Lin C, Crowley RS, Savova GK. A system for coreference resolution for the clinical narrative. J Am Med Informatics Assoc. 2012;19(4):660–667. doi: 10.1136/amiajnl-2011-000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng V. Supervised Noun Phrase Coreference Research: The First Fifteen Years. Acl. 2010 Jul;:1396–1411. [Google Scholar]
- 19.Luo X, Ittycheriah A, Jing H. A mention-synchronous coreference resolution algorithm based on the bell tree. Proceedings of the 42nd …. 2004:135. [Google Scholar]
- 20.Lee H, Peirsman Y, Chang A, Chambers N, Surdeanu M, Jurafsky D. Stanford's Multi-Pass Sieve Coreference Resolution System at the CoNLL-2011 Shared Task. Proc Fifteenth Conf Comput Nat Lang Learn Shar Task Assoc Comput Linguist. 2011 Jun;:28–34. [Google Scholar]
- 21.Miller T, Bethard S, Dligach D, Lin C, Savova G. Extracting Time Expressions from Clinical Text. Proceedings of the 2015 Workshop on Biomedical Natural Language Processing (BioNLP 2015) Workshop on Biomedical Natural Language Processing. 2015 [Google Scholar]
- 22.Ng V, Cardie C. Improving Machine Learning Approaches to Coreference Resolution. Proc 40th Annu Meet Assoc Comput Linguist (ACL '02) 2002 Jul;:104–111. [Google Scholar]
- 23.Recasens M, de Marneffe M, Potts C. The Life and Death of Discourse Entities: Identifying Singleton Mentions. Proc NAACL-HLT. 2013 [Google Scholar]
- 24.Wu S, Miller T, Masanz J, Coarr M, Halgrim S, Carrell D, Clark C. Negation's Not Solved: Generalizability Versus Optimizability in Clinical Natural Language Processing. PLoS One. 2014;9(11):e112774. doi: 10.1371/journal.pone.0112774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikolov T, Corrado G, Chen K, Dean J. Efficient Estimation of Word Representations in Vector Space. Proc Int Conf Learn Represent (ICLR 2013) 2013:1–12. [Google Scholar]
- 26.Resnik P. Semantic classes and syntactic ambiguity. Proceedings of the workshop on Human Language Technology. 1993:278–283. [Google Scholar]
- 27.Dligach D, Bethard S. Discovering body site and severity modifiers in clinical texts. J …. 2014 doi: 10.1136/amiajnl-2013-001766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bethard S, Derczynski L, Savova G. Semeval-2015 task 6: Clinical tempeval. Proc SemEval. 2015 [Google Scholar]
- 29.Bethard S, Savova G, Chen WT, Derczynski L, Pustejovsky J, Verhagen M. SemEval-2016 Task 12: Clinical TempEval. Proceedings of the 10th International Conference on Semantic Evaluation (SemEval 2016) 2016 [Google Scholar]
- 30.Saeed M, Lieu C, Raber G, Mark RG. MIMIC II: a massive temporal ICU patient database to support research in intelligent patient monitoring. Comput Cardiol …. 2002 [PubMed] [Google Scholar]
- 31.Fan RE, Chang KW, Hsieh CJ, Wang XR, Lin CJ. LIBLINEAR: A Library for Large Linear Classification. J Mach Learn Res. 2008;9:1871–1874. [Google Scholar]
- 32.Vilain M, Burger J, Aberdeen J, Connolly D, Hirschman L. A Model-Theoretic Coreference Scoring Scheme. Proc 6th Conf Messag Underst (MUC6 '95) 1995:45–52. [Google Scholar]
- 33.Bagga A, Baldwin B. Algorithms for scoring coreference chains. first Int Conf Lang …. 1998 [Google Scholar]
- 34.Luo X. On coreference resolution performance metrics. Proc EMNLP. 2005 Oct;:25–32. [Google Scholar]
- 35.Pradhan S, Luo X, Recasens M, Hovy E. Scoring Coreference Partitions of Predicted Mentions: A Reference Implementation. ACL. 2014 doi: 10.3115/v1/P14-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pradhan S, Moschitti A, Uryupina O. CoNLL-2012 Shared Task: Modeling Multilingual Unrestricted Coreference in OntoNotes. Conll. 2012 Jun;:1–40. [Google Scholar]
- 37.Recasens M, Hovy E. BLANC: Implementing the Rand index for coreference evaluation. Nat Lang Eng. 2011;17(4):485–510. [Google Scholar]
- 38.Durrett G, Klein D. Easy victories and uphill battles in coreference resolution. Proc Conf Empir …. 2013 Oct;:1971–1982. [Google Scholar]
- 39.Faruqui M, Dodge J, Jauhar SK, Dyer C, Hovy E, Smith NA. Retrofitting Word Vectors to Semantic Lexicons. Proceedings of the 2015 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. 2015:1606–1615. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



