Abstract
Mycobacterium tuberculosis remains a leading cause of death worldwide, especially among individuals infected with HIV1. While phylogenetic analysis has revealed M. tuberculosis spread throughout history2–5 and in local outbreaks6–8, much less is understood about its dissemination within the body. Here, we report genomic analysis of 2693 samples collected postmortem from lung and extrapulmonary biopsies of 44 subjects in KwaZulu-Natal, South Africa who received minimal antitubercular treatment and most of whom were HIV seropositive. We found that purifying selection acted within individual patients, without the need for patient-to-patient transmission. Despite negative selection, mycobacteria diversified within individuals to form sub-lineages that co-existed for years. These sub-lineages, as well as distinct strains from mixed infections, were differentially distributed throughout the lung, suggesting temporary barriers to pathogen migration. As a consequence, samples taken from the upper airway often captured only a fraction of the population diversity, challenging current methods of outbreak tracing and resistance diagnostics. Phylogenetic analysis indicated that dissemination from the lungs to extrapulmonary sites was as frequent as between lung sites—supporting similar migration routes within and between organs, at least in subjects with HIV. Genomic diversity therefore provides a record of pathogen diversification and repeated dissemination across the body.
Mutations that pathogens accumulate over time can be used to reconstruct their transmission history. In theory, the same principles used to track spread across the globe and between individuals can inform our understanding of how M. tuberculosis spreads within the lungs and throughout the body9,10. While M. tuberculosis primarily causes lung infections, it can disseminate to extrapulmonary organs, particularly in subjects co-infected with HIV. Understanding how M. tuberculosis diversifies and spreads within individuals is crucial to outbreak tracing11, clinical diagnosis of resistance12–14, and designing strategies to minimize de novo emergence of antibiotic resistance15–17.
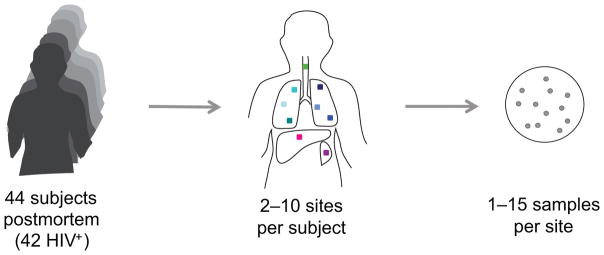
Here, we conducted a postmortem study in KwaZulu-Natal, South Africa to capture the spatial diversity of M. tuberculosis within each person (Fig. 1). Inclusion criteria included tuberculosis as cause of death, suspected disseminated infection, and fewer than 4 days of anti-tubercular treatment (Online Methods). Ninety-six of the 100 subjects enrolled were HIV positive, reflecting the high rate of HIV positivity among those dying from tuberculosis in the province. For each subject, a minimally invasive autopsy was performed, collecting specimens from each of six lung sites (3 regions within each lung, Supplementary Table 1), as well as from the liver and spleen. Each lung biopsy site was processed individually, while multiple biopsies from the other organs were pooled to form a single site per organ. Endotracheal aspirate, a proxy for what might have been detectable from a sputum specimen, was also collected. Specimens were cultured on solid media to select for M. tuberculosis. We focused on 44 subjects that had positive cultures for at least one lung site and at least one extrapulmonary site. Of these, all but two were HIV positive (Supplementary Table 2).
Figure 1. Postmortem analysis of 2693 M. tuberculosis samples from 329 sites across the body of 44 subjects.
Subjects who died in the emergency, admitting, or inpatient wards of Edendale Hospital in Kwazulu-Natal, South Africa were eligible to be enrolled in our study if they had been on anti-tubercular therapy for fewer than 4 days before death. For each subject, postmortem, biopsies were taken from each of six locations within the lung, each treated as a separate site and from multiple biopsies within the liver and spleen, pooled to form one site per organ. Aspirates of endotracheal secretions were taken, as were aspirates from clinically apparent ascitic fluid, pleural fluid, or pus collections when detected. Lung sites 1–3 were taken from the right lung and lung sites 4–6 were taken from the left lung (top to bottom, Supplementary Table 1). For some subjects, samples from ascitic fluid, pleural fluid, or pus collections were obtained. Each site was cultured separately for M. tuberculosis. DNA from one or more samples (up to 15 per site) was sequenced at the whole genome level.
A total of 2693 M. tuberculosis samples from 329 sites were analyzed for both fixed and polymorphic single-nucleotide polymorphisms (SNPs) at the whole-genome level (Supplementary Table 3). For the first 12 subjects, the diversity on solid media was scraped into a single tube, resulting in one DNA sample per site. For the remaining subjects, up to 15 individual colonies from each site were processed as separate samples (in many cases colonies were found to be formed by multiple cells). Genomic libraries were sequenced on the HiSeq platform to an average of 178x for scrape samples and 59x for colony samples. Reads were aligned to an approximate ancestral genome of the M. tuberculosis complex3, allowing inference of derived and ancestral alleles. Co-variation of mutation frequencies across samples from each subject distinguished pre-existing polymorphisms due to mixed infection from de novo mutations (Online Methods, Supplementary Fig. 1). We then used a constraint-based method to infer individual genotypes in non-clonal samples (Supplementary Fig. 2).
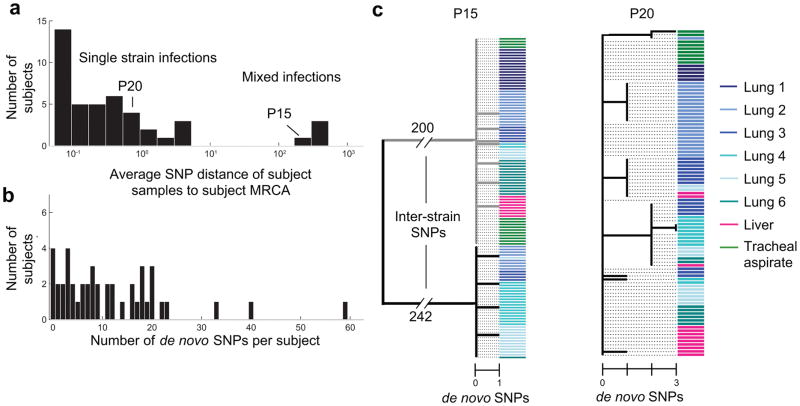
Four subjects had mixed infections, providing a prevalence estimate (9%) consistent with high endemic areas15,18,19. These subjects were identified by the average mutational distance of their M. tuberculosis population to its most recent common ancestor (〈dMRCA〉, Online Methods, Fig. 2a). Given the M. tb molecular clock rate of about 0.3–0.5 SNPs/year5,8,20, these subjects had values of 〈dMRCA〉 consistent with mixed infection (201–439 SNPs/cell), while all others had values of fewer than 5 SNPs/cell. In 2 of these 4 cases, the minority strain was detected at low frequency across samples (5% and 11%), highlighting the importance of deep sampling for detection of mixed infections.
Figure 2. Genomic sequencing reveals variation due to mixed infection and de novo mutation.
(a) A histogram of the average value of 〈dMRCA〉 across subjects, the average number of single nucleotide mutations per M. tuberculosis cell in that subject—relative to each subject’s population’s most recent common ancestor (MRCA). (b) A histogram of the number of de novo mutations within each subject’s population, after removal of polymorphisms arising due to mixed infection (Supplementary Fig. 1). (c) Two examples of within-subject phylogenies created from inferred genotypes. Genotypes were called if they were found at least 20% frequency within a sample (each sample can contain multiple genotypes); colored bars indicate site.
Mutations appearing de novo showed evidence of purifying selection. The number of de novo mutations detected varied considerably across subjects (median 8.5 SNPs, range 0–59; Fig. 2b) and was somewhat dependent on the number of specimens (Supplementary Fig. 3a), but was generally too low for selection to be analyzed on a subject-by-subject basis. Considering all de novo mutations together, the relative proportions of different nucleotide-to-nucleotide mutations closely matched that of mutations between subjects(Supplementary Fig. 4). De novo mutations had fewer amino-acid changing mutations than expected under a model of neutral evolution (dN/dS = 0.78, p<.01, Online Methods), similar to mutations observed between subjects (dN/dS = 0.80, p<.001). These data suggest that purifying selection known from between patient comparisons21,22 does not require patient-to-patient transmission, but can act within each subject.
De novo mutations led to sub-lineages that co-existed for many years in some patients. Focusing on single-strain infections, 〈dMRCA〉 estimates varied considerably across subjects (range 0–5 SNPs/cell, 0–16 years; Fig 2a) and were not sensitive to the number of samples (Supplementary Fig. 3c–d). Some subjects had values of 〈dMRCA〉 consistent with many years of within-patient diversification, yet most subjects had much lower 〈dMRCA〉 estimates (median 0.24 SNPs/cell, < 1 year), consistent with the fact that immunocompromised subjects are likely to have fulminant courses of disease. Subjects who were HIV seronegative or diagnosed with HIV infection only postmortem had higher values 〈dMRCA〉 on average (6 subjects, median 1.0 SNPs/cell, 2.2–3.4 years; p<.02, Wilcoxon rank sum test), consistent with a longer duration of infection in patients who are not immunocomprised23,24. For all subjects with de novo mutations, even when 〈dMRCA〉 was small, diversification led to coexisting short-branched sub-lineages (Fig. 2c).
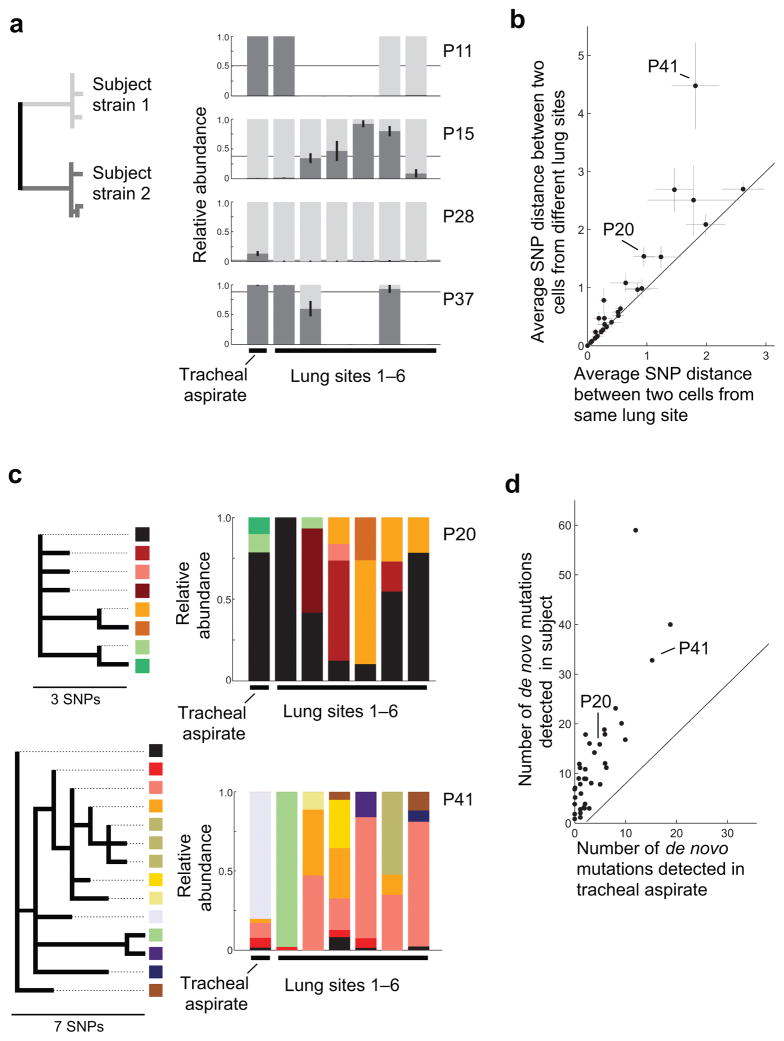
The diversity arising from de novo mutation and mixed infections was heterogeneously distributed across lung sites. In subjects with mixed infections, strain distribution varied considerably across sites, often with only one strain detected per lung site (Fig. 3a). Considering only de novo mutations, cells from the same site were significantly more similar to one another than cells from different sites (one-sided, two-sample t-test, p<0.05 for 17/30 subjects with multiple sites and at least one de novo mutation; Fig. 3b). Inspection of intrapersonal phylogenies further revealed that minority genotypes can be localized to a single site and that some sites were dominated by minority sub-lineages not seen elsewhere within the lungs (e.g. Fig. 3c). This spatial heterogeneity was neither universal nor long-lived; many new mutations were shared across lung sites and no long branches were unique to a single lung or site. These results are consistent with reports of spatial heterogeneity in other lung infections9 and in M. tuberculosis infections of HIV negative individuals25–27. Whatever the mechanism, this heterogeneity meant that many de novo mutations and strains infecting subjects’ lungs were undetected in tracheal aspirate samples (Fig. 3d; Supplementary Fig. 5). While sputum samples were not available, this suggests that spatial heterogeneity can compromise the reliability of antibiotic resistance profiling and other assays performed even on the entire diversity within respiratory samples28.
Figure 3. M. tuberculosis diversity within the lungs is spatially structured.
For each subject, we compared the distribution of strains, mutations, and genotypes across the lungs and tracheal aspirate, averaging across samples from each site. (a) The relative abundance of both strains in each respiratory site (Tracheal aspirate and lung sites 1–6, left to right) is indicated for subjects with mixed infection. Different shades of grey represent the two strains. Error bars indicate the standard error of the mean (s.e.m.) for subjects with multiple samples per site. Thin black lines indicate the average across sites from each subject. (b) For each subject with a single strain infection and multiple samples per site, the average distance between two cells from different lung sites is plotted against the average distance between two cells from the same lung site (averaging across sites). Error bars indicate the s.e.m. across sites or pairs of sites. (c) For two example subjects, the relative abundance of each genotype within each respiratory site is indicated (Tracheal aspirate and lung sites 1–6, left to right). Color represents genotype; the phylogenetic relationships of these genotypes are shown at left. (d) For each subject, the number of de novo mutations detected within the subject is plotted against the number of de novo mutations detected only within the tracheal aspirate. Low abundance mutations were the most likely to be missing from sputum (Supplementary Fig. 5a–b). Jitter added for visibility (Up to 0.5 mutations; same value added in x and y).
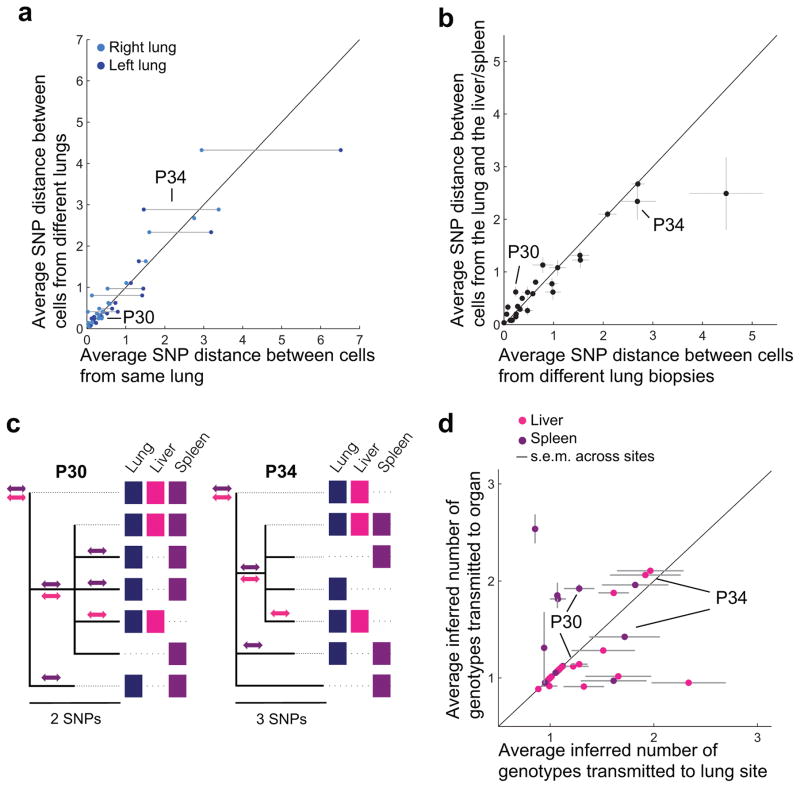
Surprisingly, the genetic distance between cells from different sites, both within the lungs and between organs, did not depend on spatial proximity. Cells from sites in different lungs (right vs. left) were, on average, no more different than cells taken from sites in the same lung (Fig. 4a). Further, two cells from different organs were no more different than two cells taken from different lung sites (Fig. 4b; Supplementary Fig. 6). Moreover, genotypes from the liver or spleen sites did not cluster phylogenetically, suggesting dissemination of multiple genotypes to each of these organs (Fig. 4c). To compare rates of transmission between organs and within the lungs, we estimated the number of transmitted genotypes between the lungs and extrapulmonary organs inferred by their joint phylogeny (Fig. 4c, Online Methods). We inferred multiple transmitted genotypes between the lungs and extrapulmonary organs for many subjects (Fig. 4c–d). These multiple genotypes might result from a single transmission event involving multiple genotypes or repeated transmission events29; we note that the detection of different genotypes in the liver and spleen of the same individual suggests independent multiple transmission events. Interestingly, the number of genotypes transmitted from the lungs to extrapulmonary sites was as high as the number of transmissions between different lung sites (Fig. 4d).
Figure 4. M. tuberculosis dissemination within lungs, between lungs, and between organs follows similar dynamics.
For each subject with a single strain infection: (a) the average distance between cells from the right lung and left lung is plotted against the average distance between cells from the same lung (right lung, dark blue; left lung, light blue); and (b) the average distance between a cell from a lung site and a cell from an extrapulmonary organ is plotted against the average distance between cells from the different lung sites. Error bars indicate s.e.m. across sites or pairs of sites. (c) Examples of intrasubject phylogenies showing shared genotypes. Genotypes are indicated as within an organ with a colored box in the corresponding column if they were found above 20% frequency in at least two samples from that organ. Potential transmission events between the lungs and extrapulmonary organs are identified by mutations shared across organs and indicated with a double-sided arrow of corresponding color. (d) For each subject, the minimum number of transmitted genotypes to the liver and/or spleen from the lungs was inferred (y-axis) and compared to mean of the minimum number of transmitted genotypes to each lung site from the rest of the lungs (averaging across sites, x-axis). Simulations were used to normalize for sampling efforts (Online Methods). Error bars indicate the s.e.m. across lung sites. Jitter added for visibility (Up to 10%; Same value added in x and y). Magenta indicates liver and purple indicates spleen.
The similar patterns of migration within and between organs suggest that dissemination of M. tuberculosis across the lungs of these subjects is no easier than dissemination across organs. These results support the hypothesis that both intralung and interorgan spread are mediated by similar dissemination mechanisms, such as by macrophage trafficking through the bloodstream or other mechanisms of hematogenous spread30–32. It would be interesting to investigate the generality of these results in subjects with less advanced disease or without HIV.
The diversity of genotypes within individuals and their stratification across the body have implications for outbreak tracing. A major impetus for whole-genome sequencing of M. tuberculosis, as well as other pathogens, has been to identify transmission links between individuals. While future approaches will incorporate the pathogen diversity within each person33–35, current approaches establish an epidemiological linkage between people if the mutational distance between single samples from each patient is below a set threshold7,8,22,36. Considering all genotypes within a subject, 20% of subjects harbored genotypes separated by more than a common threshold of 5 SNPs8 (Supplementary Table 2; 9% emerging from multiple-strain infection) and 11% harbored genotypes separated by more than an alternative threshold of 12 SNPs37. These estimates may be even greater when considering longer duration infections, such as those of HIV negative individuals. While the thresholds chosen will depend on the mutation detection sensitivity unique to each study, the frequency of substantial within-patient distances adds weight to the mounting evidence11,33,38,39 against ruling out epidemiological links on the basis of mutational thresholds between single samples.
This study represents the first large-scale genomic investigation of Mycobacterium tuberculosis diversity across the human body. Extensive sampling from multiple body sites and detection of minor alleles within each sample shows that even the notoriously slowly evolving M. tuberculosis presents substantial within-patient diversity. The distribution of this diversity across the body suggests that, at least in people with HIV, dissemination within the lungs follows a similar mechanism as dissemination from the lungs to extra-pulmonary organs. In total, these findings highlight the potential of whole-genome sequencing to make inferences about the history of individual bacterial infections from the diversity surveyed at a single time point. We anticipate that future studies will extend these approaches to address open questions about M. tuberculosis and other pathogens, including the contribution of spatial heterogeneity to evolution of antibiotic resistance and the dynamics of dormancy and reactivation.
Methods
Study cohort
Subjects were eligible for inclusion in our study if they died while at Edendale Hospital, were age 20 or over, were on anti-tubercular treatment for fewer than 4 days before death, and were either smear positive for acid fast bacilli or had at least one focal process compatible with tuberculosis without an alternative diagnosis. The number of subjects studied was determined based on feasibility. Subjects were de-identified prior to DNA analysis and mutation calling was performed while blind to subject information.
Sample collection
Following informed consent by family members under a protocol approved by the institutional review board at University of KwaZulu-Natal, a minimally invasive autopsy was performed for each subject. Briefly, three lung sites were collected from each side, at the second, fourth and sixth intercostal spaces. A new sterile needle was used at each site (see Supplementary Note for more detail). Specimens were stored on ice, and transported to the University of KwaZulu-Natal in Durban. Participants not documented in the case notes to be HIV seropositive were tested post-mortem using a lateral flow ELISA assay. Subject information is listed in Supplementary Table 2. Cause of death for all subjects was culture confirmed disseminated tuberculosis.
Biopsy specimens were cut into small pieces and decontaminated using Petroff’s method40 (to enrich for M. tuberculosis). Endotracheal aspirate specimens were decontaminated with a 4% NALC/NaOH solution. 500μl of the decontaminated specimen was inoculated into a MGIT tube and 100ul on 3 separate 7H11 plates41 to select for M. tuberculosis. For the first 12 subjects, the diversity on these 3 plates was scraped into a single tube for DNA analysis. For the remaining cultures, up to 5 single-colonies, demonstrating morphological variation, were selected from each plate (for a maximum of 15 colonies per specimen). Each colony was then sub-cultured onto dedicated 7H11 plates and incubated for three weeks at 37°C, under aerobic conditions.
Antibiotic resistance testing
Phenotypic antibiotic susceptibility testing was performed on all decontaminated specimens. The modified proportion method was used for antibiotic susceptibility testing. Positive MGIT tubes from each specimen were used to set up the antibiotic susceptibility tests. Tubes were confirmed to be uncontaminated and incubated for a further 24–48 hours after becoming instrument positive before the test was performed. Three week 7H11 cultures were used in the event the MGITs were found to be contaminated or to confirm resistance.
Quadrant 7H10 agar plates that comprised a control (antibiotic free) quadrant and antibiotic containing quadrants were used. The following concentrations were used (μg/mL): Isoniazid: 1, Rifampicin: 1, Ethambutol: 7.5, Streptomycin: 2, Ofloxacin: 2, Kanamycin: 5, Ethionamide: 5, Capreomycin: 10. Cultures were diluted so as to obtain approximately 100 colony forming units (CFU) on each quadrant and seeded onto each quadrant. The plates were sealed in CO2 permeable bags and incubated at 37°C, 5% CO2 for one week and under aerobic conditions for a further two weeks. Control strains (H37Rv and A169) were set up in parallel. Antibiotic containing quadrants with CFU > 1% of the CFU on the control quadrant was interpreted as resistant. No growth or <1% was interpreted as susceptible. Cultures were read in a double blind manner.
No significant variation was found across specimens from a given subject. Specimens from subjects P4, P9, and P12 were multi-drug-resistant, specimens from P16 were rifampicin resistant, and all others were pan-sensitive (Supplementary Table 2).
DNA extraction
M. tuberculosis colonies were scraped from agar and inoculated into 500μl of water in an eppendorf tube and heat killed for 30 minutes at 80°C. DNA was extracted using the CTAB method42. 70μl 10% sodium dodecyl sulfate and 50μl proteinase K (10 mg/ml stock solution) were added and the solution incubated for 1 hour at 60°C using a thermomixer on low mode shaking. Samples were removed from the thermomixer, and 100μl 5M NaCl (preheated to 60°C) was added and mixed thoroughly by hand inversion.. 100μl of 10% CTAB (N-acetyl-N,N,N-trimethyl ammonium bromide, preheated to 60°C) was added and the solution mixed by hand inversion. Tubes were then incubated at 60°C for 15 minutes (thermomixer on low-mode shaking). 700μl chloroform/isoamyl alcohol (24:1) was added and the solution mixed by hand inversion. Tubes were then centrifuged for 10 min at 13,000rpm. The upper (aqueous) phase was transferred to tube with 700μl of a cold isopropanol and mixed by hand inversion. DNA was precipitated by freezing –20°C for at least 30 minutes and centrifuged for 10 minutes at 13,000rpm. Tubes were drained and the pellet washed with 80% ethanol followed by centrifugation for 5–10min. These were then drained and the pellet was allowed to dry. DNA was suspended in 55μl of TE.
Illumina sequencing and mutation identification
Genomic libraries were constructed and barcoded using a previously described modified version of the Illumina Nextera protocol43. Genomic libraries were sequenced on the Illumina HiSeq 2000 platform by Macrogen using paired-end 100 bp reads. Reads were aligned to a modified H37rv reference genome in which each nucleotide reflects this ancestor of the M. tuberculosis complex3 (provided by Iñaki Comas). The limited mutation distance of all extant TB to this ancestor (< .05% of the genome) and limited recombination in M. tuberculosis3 gives us confidence that each difference from this reference that passed our conservative filters (Supplementary Note) represents a newly derived mutation. Standard approaches were used for read filtering and alignment44. Average coverage for each sample is listed Supplementary Table 4.
Potential cross-contamination or mislabeling events were identified by the presence of multiple mutations not found in any other sample from that subject and/or multiple mutations found in samples from different subjects processed on the same plate. We identified 68 such potentially contaminated samples. New repeated genomic libraries were prepared for 40 of these 68 samples. In 31 cases, the duplicate matched more closely with other samples from that subject and was used for analysis. In 9 cases, the duplicate also showed evidence of contamination and the two replicates of the sample were discarded (for a total of 37 discarded samples out of 2730, 1.4%). In our final data, subjects had distinct strains (Supplementary Fig. 7). As some of these samples may have been discarded in error, some true mixed infections may have been called as simple.
Many single-colony samples from subjects with mixed infections had hundreds of mutations at intermediate frequencies—suggesting that cells from different strains were comingled in a single colony (Supplementary Fig. 1c; consistent with the cording phenotype of M. tb45). We therefore searched for both fixed and polymorphic mutations within each sample, using a series of filters and statistical tests to distinguish false positives (e.g. alignment and sequencing errors) from real polymorphisms44. Additional filters regarding the proportion of samples in each subject with a polymorphism were also used to remove notoriously problematic sites, such as those in the proline-glutamate and proline-proline-glutamate genes46 (see Supplementary Note for details). Among these filters, polymorphic mutations were required to be at least 10% frequency and supported by at least 4 reads on each strand. As a consequence, rare variants may have not been detected, particularly in cases where few samples were available per site.
For each polymorphic position that met the filtering criteria in at least one sample, raw reads were used to determine the allele frequency across samples from that subject. The 518 de novo mutations found and their distribution across samples are listed in Supplementary Table 4. Mutations and genes were annotated according to H37rv (NC_000962.3). None of these mutations are known to be associated with antibiotic resistance47.
dN/dS
Calculations for dN/dS, the ratio of nonsynonymous mutations to synonymous mutations divided by the ratio under a neutrality, were performed normalizing for the spectrum of mutations observed as previously described44. P-values for depletion of non-synonymous mutations were calculated according to the binomial cumulative distribution function.
Identification of polymorphisms arising from multiple-strain infection
Mutations carried by each of the infecting strains covary across samples with one another and anti-covary with mutations from the other strain48. Mutations arising from multiple strain infection are defined by their contribution to the primary principal component (PC1) in a Principal Component Analysis of the mutation frequency matrix. De novo mutations are identified as those without a significant contribution to PC1 (absolute value < 0.015, cutoff determined empirically; Supplementary Fig. 1). Previously described SNP markers were used to assign each strain to a global lineage49.
Estimation of 〈dMRCA〉
The average mutational distance across cells in a subject’s M. tuberculosis population to its most recent common ancestor, 〈dMRCA〉, was calculated as the sum of the mutation frequencies at each polymorphic position called within the subject44. To normalize for sampling efforts, 〈dMRCA〉 was first calculated across samples from each specimen, and these values were averaged for each subject. For subjects previously diagnosed with HIV, the mean value of 〈dMRCA〉 was 0.42 SNPs/cell and for subjects who were HIV negative or diagnosed post-mortem, this mean was 1.8 SNPs/cell (Fig. 2a, Supplementary Table 2).
Interpretation of 〈dMRCA〉 as time of infection assumes that these subjects were infected with a single genotype; if subjects were infected by multiple closely related genotypes, these estimates are inflated. Alternatively, these estimates may be underestimations if mutations have swept the subject’s diversity since infection due to adaptation or bottlenecks (drift). Estimates of 〈dMRCA〉 for subjects with multiple-strain infection are lower bounds because the strict mutation caller was optimized for detecting de novo mutations. Other potential sources of error include Poisson error in the number of mutations accumulated in each lineage since each subject’s MRCA, underestimation due to limited sensitivity in detecting mutations, overestimation due to false positives, incorrect designation of ancestral versus derived alleles, and underestimation due to incomplete sampling of the diversity within each subject.
Genotype identification and phylogenetic inference
We used a conservative algorithm to infer a minimal set of distinct genotypes within each subject based the assumptions that genotypes are shared across samples from each subject and that each SNP occurred only once within a subject (strict parsimony). A direct consequence of the strict parsimony assumption is that two mutations at high frequency (majority) in the same sample must coexist on the same genotype50,51. For each sample within a subject, starting with the most homogenous and covered samples, high frequency polymorphisms were matched with previously identified genotypes, or if no such match was made, the polymorphisms found at high frequency were used to define a new genotype (Supplementary Fig. 2). Some de novo mutations could not be confidently assigned to a genotype and were omitted; combined with our stringent mutation caller, this approach produced conservative phylogenies in which some genotypes were not identified or called with fewer mutations than they really have. After genotypes were identified, each sample was then assigned to one or more genotypes. We note that our algorithm will not work well in cases where parallel nucleotide evolution is expected or where pure samples are not available and that the cutoffs in our implementation may need to be adjusted for different use cases (Supplementary Note).
Pairwise genetic distances
We calculated the average pairwise distance between cells in different samples, considering the frequency of each mutation f at each confident polymorphic position x on the genome, in samples i and j as:
Only lung, liver, and spleen sites with more than 2 samples per site were included in the analysis. To normalize for different sampling efforts across sites, we first calculate the mean at each site (x-axis, Fig. 3b) or pairs of sites and then average across site or pairs. Error bars indicate the s.e.m. of the means across sites or pairs of sites.
Transmission analysis
We developed an algorithm based on the number of shared mutations between sites (Fig 4c). Only genotypes found in two separate samples within an organ or lung site were considered to reduce the effects of potential cross-contamination. We iterated through mutations shared between the destination site and the rest of the lung. On each iteration, the genotype closest to the subject’s MRCA containing a shared mutation was chosen and counted as a transmission event. All mutations in that genotype were removed from the list of unaccounted for shared mutations, and the next iteration was performed. In this way, groups of mutations appearing on the same branch of the phylogeny were only counted as a single transmitted genotype. Lastly, the presence of the subject’s MRCA in both locations suggests a transmission of this ancestral genotype (Fig 4c.). Simulations were performed to normalize for different sampling efforts across sites and organs, omitting one site from the lung each trial (for estimations of intraorgan transfer, this omitted site was then treated as destination). Only sites with more than 6 samples were included and each site was randomly subsampled, taking the minimum number of samples across sites with >6 samples. For each omitted lung site, 1,000 trials were performed and averaged.
Code availability
Custom MATLAB code used for calling mutations and analyzing the data can be found at: https://github.com/kishonylab/TB-diversity-across-organs.
Supplementary Material
Acknowledgments
We are especially thankful to the subjects and their families, to L. Maziya, Z. Magcaba, N. Xengxe and M.J. Khumalo for their assistance with enrollment and postmortems, to the Edendale Hospital management, and to the KwaZulu-Natal Department of Health. We are grateful to M. Baym and members of the Kishony lab for helpful discussions, to C. Duvallet for comments on the manuscript, and to H. Chung for discussions of spatial diversity and comments on the manuscript. We thank the team at Macrogen Clinical Laboratories for their help with Illumina sequencing and I. Comas for providing the reference sequence. This work was funded in part by the US National Institutes of Health grants DP2 OD006663 (to T.C.) and R01-GM081617, Hoffman-LaRoche, and European Research Council FP7 ERC Grant 281891 (to R.K.).
Footnotes
Accession codes
Sequencing reads have been deposited in the NCBI Sequence Read Archive (SRA), under BioProject PRJNA323744.
Author Contributions
T.C. and D.W. conceived and coordinated the study. T.D.L., D.W., T.C., and R.K. designed the study. D.W. identified eligible decedents and conducted postmortem biopsies. R.M. and P.M. processed and cultured samples, performed antibiotic sensitivity testing, and extracted DNA. T.D.L. and L.L.X. prepared genomic libraries. T.D.L. and R.K. analyzed genomic data. T.D.L., D.W, T.C. and R.K. interpreted the results and wrote the manuscript.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Global tuberculosis report 2015. World Health Organization; 2015. pp. 1–204. [Google Scholar]
- 2.Cohen KA, et al. Evolution of Extensively Drug-Resistant Tuberculosis over Four Decades: Whole Genome Sequencing and Dating Analysis of Mycobacterium tuberculosis Isolates from KwaZulu-Natal. PLoS Med. 2015;12:e1001880. doi: 10.1371/journal.pmed.1001880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comas I, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45:1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kay GL, et al. Eighteenth-century genomes show that mixed infections were common at time of peak tuberculosis in Europe. Nat Commun. 2015;6:6717. doi: 10.1038/ncomms7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bos KI, et al. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature. 2014;514:494–497. doi: 10.1038/nature13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardy JL, et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med. 2011;364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 7.Walker TM, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant JM, et al. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect Dis. 2013;13:110. doi: 10.1186/1471-2334-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jorth P, et al. Regional Isolation Drives Bacterial Diversification within Cystic Fibrosis Lungs. Cell Host Microbe. 2015;18:307–319. doi: 10.1016/j.chom.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieberman TD, et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet. 2011;43:1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pérez-Lago L, et al. Whole genome sequencing analysis of intrapatient microevolution in Mycobacterium tuberculosis: potential impact on the inference of tuberculosis transmission. J Infect Dis. 2014;209:98–108. doi: 10.1093/infdis/jit439. [DOI] [PubMed] [Google Scholar]
- 12.Zetola NM, et al. Clinical outcomes among persons with pulmonary tuberculosis caused by Mycobacterium tuberculosis isolates with phenotypic heterogeneity in results of drug-susceptibility tests. J Infect Dis. 2014;209:1754–1763. doi: 10.1093/infdis/jiu040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Black PA, et al. Whole genome sequencing reveals genomic heterogeneity and antibiotic purification in Mycobacterium tuberculosis isolates. BMC Genomics. 2015;16:857. doi: 10.1186/s12864-015-2067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun G, et al. Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J Infect Dis. 2012;206:1724–1733. doi: 10.1093/infdis/jis601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen T, et al. Mixed-strain mycobacterium tuberculosis infections and the implications for tuberculosis treatment and control. Clin Microbiol Rev. 2012;25:708–719. doi: 10.1128/CMR.00021-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colijn C, Cohen T, Ganesh A, Murray M. Spontaneous emergence of multiple drug resistance in tuberculosis before and during therapy. PLoS ONE. 2011;6:e18327. doi: 10.1371/journal.pone.0018327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prideaux B, et al. The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat Med. 2015;21:1223–1227. doi: 10.1038/nm.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mankiewicz E, Liivak M. Phage types of mycobacterium tuberculosis in cultures isolated from Eskimo patients. Am Rev Respir Dis. 1975;111:307–312. doi: 10.1164/arrd.1975.111.3.307. [DOI] [PubMed] [Google Scholar]
- 19.Warren RM, et al. Patients with active tuberculosis often have different strains in the same sputum specimen. Am J Respir Crit Care Med. 2004;169:610–614. doi: 10.1164/rccm.200305-714OC. [DOI] [PubMed] [Google Scholar]
- 20.Ford CB, et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet. 2013;45:784–790. doi: 10.1038/ng.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pepperell CS, et al. The role of selection in shaping diversity of natural M. tuberculosis populations. PLoS Pathog. 2013;9:e1003543. doi: 10.1371/journal.ppat.1003543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee RS, et al. Population genomics of Mycobacterium tuberculosis in the Inuit. Proc Natl Acad Sci USA. 2015;112:13609–13614. doi: 10.1073/pnas.1507071112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiemersma EW, van der Werf MJ, Borgdorff MW, Williams BG, Nagelkerke NJD. Natural history of tuberculosis: duration and fatality of untreated pulmonary tuberculosis in HIV negative patients: a systematic review. PLoS ONE. 2011;6:e17601. doi: 10.1371/journal.pone.0017601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eldholm V, et al. Impact of HIV co-infection on the evolution and transmission of multidrug-resistant tuberculosis. Elife. 2016;5:306. doi: 10.7554/eLife.16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin PL, et al. Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing. Nat Med. 2014;20:75–79. doi: 10.1038/nm.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.García-de-Viedma D, Marín M, Ruiz Serrano MJ, Alcalá L, Bouza E. Polyclonal and compartmentalized infection by Mycobacterium tuberculosis in patients with both respiratory and extrarespiratory involvement. J Infect Dis. 2003;187:695–699. doi: 10.1086/368368. [DOI] [PubMed] [Google Scholar]
- 27.Liu Q, et al. Within patient microevolution of Mycobacterium tuberculosis correlates with heterogeneous responses to treatment. Sci Rep. 2015;5:17507. doi: 10.1038/srep17507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ford C, et al. Mycobacterium tuberculosis--heterogeneity revealed through whole genome sequencing. Tuberculosis (Edinb) 2012;92:194–201. doi: 10.1016/j.tube.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turajlic S, Swanton C. Metastasis as an evolutionary process. Science. 2016;352:169–175. doi: 10.1126/science.aaf2784. [DOI] [PubMed] [Google Scholar]
- 30.Krishnan N, Robertson BD, Thwaites G. The mechanisms and consequences of the extra-pulmonary dissemination of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2010;90:361–366. doi: 10.1016/j.tube.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 31.McMurray DN. Hematogenous reseeding of the lung in low-dose, aerosol-infected guinea pigs: unique features of the host-pathogen interface in secondary tubercles. Tuberculosis (Edinb) 2003;83:131–134. doi: 10.1016/s1472-9792(02)00079-3. [DOI] [PubMed] [Google Scholar]
- 32.Ssengooba W, de Jong BC, Joloba ML, Cobelens FG, Meehan CJ. Whole genome sequencing reveals mycobacterial microevolution among concurrent isolates from sputum and blood in HIV infected TB patients. BMC Infect Dis. 2016;16:371. doi: 10.1186/s12879-016-1737-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Worby CJ, Lipsitch M, Hanage WP. Within-host bacterial diversity hinders accurate reconstruction of transmission networks from genomic distance data. PLoS Comput Biol. 2014;10:e1003549. doi: 10.1371/journal.pcbi.1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Didelot X, Gardy J, Colijn C. Bayesian inference of infectious disease transmission from whole-genome sequence data. Mol Biol Evol. 2014;31:1869–1879. doi: 10.1093/molbev/msu121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paterson GK, et al. Capturing the cloud of diversity reveals complexity and heterogeneity of MRSA carriage, infection and transmission. Nat Commun. 2015;6:6560. doi: 10.1038/ncomms7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guerra-Assunção JA, et al. Large-scale whole genome sequencing of M. tuberculosis provides insights into transmission in a high prevalence area. Elife. 2015;4 doi: 10.7554/eLife.05166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker TM, et al. Assessment of Mycobacterium tuberculosis transmission in Oxfordshire, UK, 2007–12, with whole pathogen genome sequences: an observational study. Lancet Respir Med. 2014;2:285–292. doi: 10.1016/S2213-2600(14)70027-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatherell HA, et al. Interpreting whole genome sequencing for investigating tuberculosis transmission: a systematic review. BMC Med. 2016;14:21. doi: 10.1186/s12916-016-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eldholm V, et al. Evolution of extensively drug-resistant Mycobacterium tuberculosis from a susceptible ancestor in a single patient. Genome Biol. 2014;15:490. doi: 10.1186/s13059-014-0490-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petroff SA. A NEW AND RAPID METHOD FOR THE ISOLATION AND CULTIVATION OF TUBERCLE BACILLI DIRECTLY FROM THE SPUTUM AND FECES. J Exp Med. 1915;21:38–42. doi: 10.1084/jem.21.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohn ML, Waggoner RF, McClatchy JK. The 7H11 medium for the cultivation of mycobacteria. Am Rev Respir Dis. 1968;98:295–296. doi: 10.1164/arrd.1968.98.2.295. [DOI] [PubMed] [Google Scholar]
- 42.Somerville W, Thibert L, Schwartzman K, Behr MA. Extraction of Mycobacterium tuberculosis DNA: a question of containment. J Clin Microbiol. 2005;43:2996–2997. doi: 10.1128/JCM.43.6.2996-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baym M, et al. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS ONE. 2015;10:e0128036. doi: 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lieberman TD, et al. Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat Genet. 2014;46:82–87. doi: 10.1038/ng.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Middlebrook G, Dubos RJ, Pierce C. VIRULENCE AND MORPHOLOGICAL CHARACTERISTICS OF MAMMALIAN TUBERCLE BACILLI. J Exp Med. 1947;86:175–184. doi: 10.1084/jem.86.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casali N, et al. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 2012;22:735–745. doi: 10.1101/gr.128678.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coll F, et al. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med. 2015;7:51. doi: 10.1186/s13073-015-0164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cleary B, et al. Detection of low-abundance bacterial strains in metagenomic datasets by eigengenome partitioning. Nat Biotechnol. 2015;33:1053–1060. doi: 10.1038/nbt.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coll F, et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun. 2014;5:4812. doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiao W, Vembu S, Deshwar AG, Stein L, Morris Q. Inferring clonal evolution of tumors from single nucleotide somatic mutations. BMC Bioinformatics. 2014;15:35. doi: 10.1186/1471-2105-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischer A, Vázquez-García I, Illingworth CJR, Mustonen V. High-definition reconstruction of clonal composition in cancer. Cell Rep. 2014;7:1740–1752. doi: 10.1016/j.celrep.2014.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.