Abstract
The two major isoforms of the oncogenic Bcr–Abl tyrosine kinase, p210 and p190, are expressed upon the Philadelphia chromosome translocation. p210 is the hallmark of chronic myelogenous leukemia, whereas p190 occurs in the majority of B-cell acute lymphoblastic leukemia. Differences in protein interactions and activated signaling pathways that may be associated with the different diseases driven by p210 and p190 are unknown. We have performed a quantitative comparative proteomics study of p210 and p190. Strong differences in the interactome and tyrosine phosphoproteome were found and validated. Whereas the AP2 adaptor complex that regulates clathrin-mediated endocytosis interacts preferentially with p190, the phosphatase Sts1 is enriched with p210. Stronger activation of the Stat5 transcription factor and the Erk1/2 kinases is observed with p210, whereas Lyn kinase is activated by p190. Our findings provide a more coherent understanding of Bcr–Abl signaling, mechanisms of leukemic transformation, resulting disease pathobiology and responses to kinase inhibitors.
Introduction
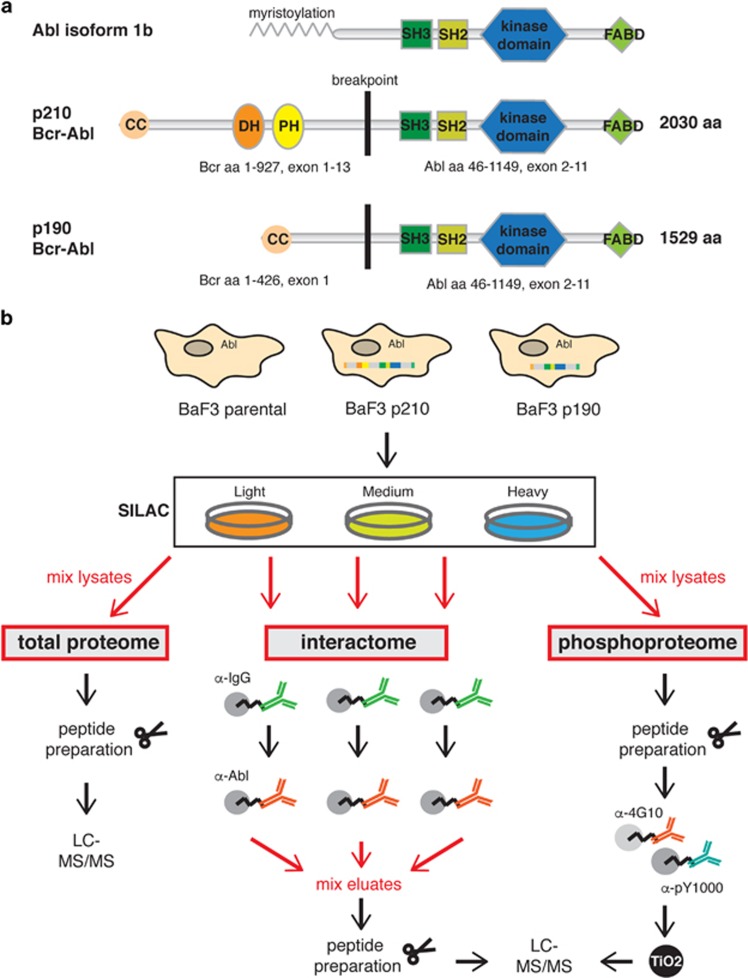
The Bcr–Abl kinase and its inhibitors (imatinib and successors) are a paradigm for targeted cancer therapy.1 Bcr–Abl is a constitutively active tyrosine kinase, expressed by the Philadelphia (Ph) chromosome. It is formed upon the t(9;22) reciprocal translocation that fuses the breakpoint cluster region (BCR) gene with the Abelson tyrosine kinase (ABL1).2 Depending on the translocation breakpoint in BCR, different Bcr–Abl protein isoforms are expressed, which all contain exons 2–11 of the ABL1 gene, but differ in the length of their BCR component.3 The most common Bcr–Abl isoforms are p210 and p190 (alternatively named: p185). p190 is 501 amino acids, that is, ~25%, shorter than p210 because it lacks a DH–PH domain unit; otherwise p210 and p190 have an identical sequence and domain organization (Figure 1a).4
Figure 1.
Bcr–Abl domain organization and workflow of the proteomics experiments. (a) The Abl tyrosine kinase and the two isoforms of the fusion protein Bcr–Abl, p210 and p190, are shown with their sizes and domain arrangement. The p210 isoform is 501 amino acids longer than p190 as it contains the DH–PH tandem domain. Domain abbreviations: CC, coiled-coil; DH, Dbl-homology; PH, Pleckstrin-homolgy; SH3/SH2, Src-homology 3/2; FABD, F-actin binding domain. (b) SILAC labeling was employed to allow quantitative comparison of three BaF3 cell lines (Supplementary Table S1). BaF3 parental cells express Abl endogenously. BaF3 p210 and BaF3 p190 cells express human Bcr–Abl p210 and p190. An immunoaffinity purification strategy was used to enrich for Bcr–Abl complexes for the interactome analysis and sample mixing was performed just prior to peptide preparation. For analysis of the tyrosine phosphoproteome cell lysates were mixed prior to enrichment of the pY peptides using the pY1000 and 4G10 antibodies and an additional TiO2 purification step. For both experiments, the analysis of the total proteome served for different normalization steps. LC-MS, liquid chromatography-mass spectrometry.
The expression of p210 is the molecular hallmark of chronic myelogenous leukemia (CML).3 The Ph-chromosome is also present in 20–30% of adult B-cell acute lymphoblastic leukemias (B-ALL), where approximately one-fourth of these patients express p210 and approximately three-fourth express p190 Bcr–Abl.3 Treating CML patients with the Bcr–Abl tyrosine kinase inhibitor (TKI) imatinib leads to durable remissions in most patients and the survival of those patients is not different from that of the general population.5 In contrast, in Ph-positive B-ALL, relapse and TKI resistance are frequent, and overall survival is still dramatically low, despite the increased remission rates and survival that can be achieved with Bcr–Abl TKIs.6, 7
p210 is the sole oncogenic driver that is sufficient to establish and maintain CML. In contrast, in Ph-positive B-ALL, additional mutations are frequently observed.8 Various mouse models that express Bcr–Abl in hematopoietic stem cells or progenitor cells were developed and recapitulate many features of human CML and B-ALL.9, 10 Only a few studies have compared the in vivo leukemogenic activity of p190 and p210 directly. Under specific experimental conditions, the expression of p190 lead to a disease with a shorter latency and more B-ALL, whereas p210 mice developed CML-like leukemias.9, 11, 12, 13 This may argue that the specific intrinsic differences in the p190 and p210 proteins contribute to the two different disease pathologies, in addition to the described different cell-of-origin of the observed p210 and p190-driven leukemias.12 Differences in activity and signaling between p210 and p190 have long been hypothesized but never studied in a comprehensive and quantitative manner. Early studies on selected signaling molecules indicated that qualitatively the same pathways are activated by p210 and p190,14 whereas kinase assays tended toward a mildly higher kinase activity for p190.11, 15, 16 The p210 interaction network has been mapped by affinity purification mass spectrometry experiments with p210 interactors as baits using non-quantitative proteomics.17, 18 To date, very little is known regarding specific protein interaction partners and substrates of p190, and most importantly, the two Bcr–Abl isoforms have not been compared directly in a uniform cellular background.
Being aware of the large amount, but heterogeneous data regarding Bcr–Abl interacting proteins and activated downstream pathways, we performed a first comparative, quantitative and systematic proteomics study to chart the common and differential interactome and tyrosine phosphoproteome of p210 and p190 Bcr–Abl. We show that the differences in interactome and phosphoproteome of p210 and p190 are surprisingly large despite similar kinase activation. Our study provides the first consolidated view on oncogene-intrinsic signaling differences of p210 and p190 in a defined cellular background.
We also refer to a parallel study by Cutler et al.19 (co-submitted with this manuscript) that used the same cell line models to map the p210 and p190 interactome and phosphoproteome. Our two studies represent a rare case of cross-validation of proteomics studies and emphasize the need to increase inter-laboratory reproducibility of large-scale proteomics datasets.20
Materials and methods
Interactome sample preparation
The optimized final protocol used 80–100 mg total cell lysate per cell line, corresponding to approx. 7–10 × 108 BaF3 cells. Cell lysis and purification were performed in TAP buffer (50 mm TRIS pH 7.5, 100 mm NaCl, 5% glycerol, 0.2% (w/v) NP-40 Alternative) containing protease and phosphatase inhibitors.21 Normal mouse IgG antibody (Thermo Scientific, Waltham, MA, USA; catalog no. 10400C) and the monoclonal Abl antibody (clone 24-21)22 were covalently coupled to NHS-activated sepharose (GE Healthcare, Pittsburgh, PA, USA; catalog no. 17-0906-01). The lysates were pre-cleared for 1 h at 4 °C with the IgG-NHS resin and the supernatant then transferred to the Abl-NHS resin for another 3 h at 4 °C. Elution fractions containing Bcr–Abl were pooled prior to mixing of the three individual immunoprecipitates of BaF3 parental, p210 and p190 with equal volumes and subsequently lyophilized.
Phosphoproteome sample preparation
Cell lysis was accomplished in urea buffer (8 m urea, 50 mm Hepes, pH 7.6) using 2 ml buffer for 1 × 108 cells. Ten milligrams of total cell lysate of each sample (light, medium and heavy labeled) were mixed to have a final amount of 30 mg SILAC sample for peptide preparation. Phosphotyrosine peptides were enriched using a mixture of two phosphotyrosine antibodies: The pY1000 antibody that is part of the PTMScan Phospho-Tyrosine Rabbit mAb (P-Tyr-1000) Kit (Cell Signaling Technologies, Danvers, MA, USA; catalog number 8803), in addition to 50 μl NHS-coupled 4G10 antibody. For the pY eluates, a second step of phosphopeptide enrichment was performed on TiO2 tips as described23 prior to mass spectrometry (MS) analysis.
Data analysis and MS raw data
A detailed description of the mass spectrometry data analysis to identify differential hits is given in Supplementary Information. Detailed lists of all interactors and phosphosites are provided in Supplementary Tables S4–S8 and an annotated Excel file with complete data. In addition, the mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD005149.24
A detailed description of the methods used in this study is given in the Supplementary Methods.
Results
Bcr–Abl interactome analysis
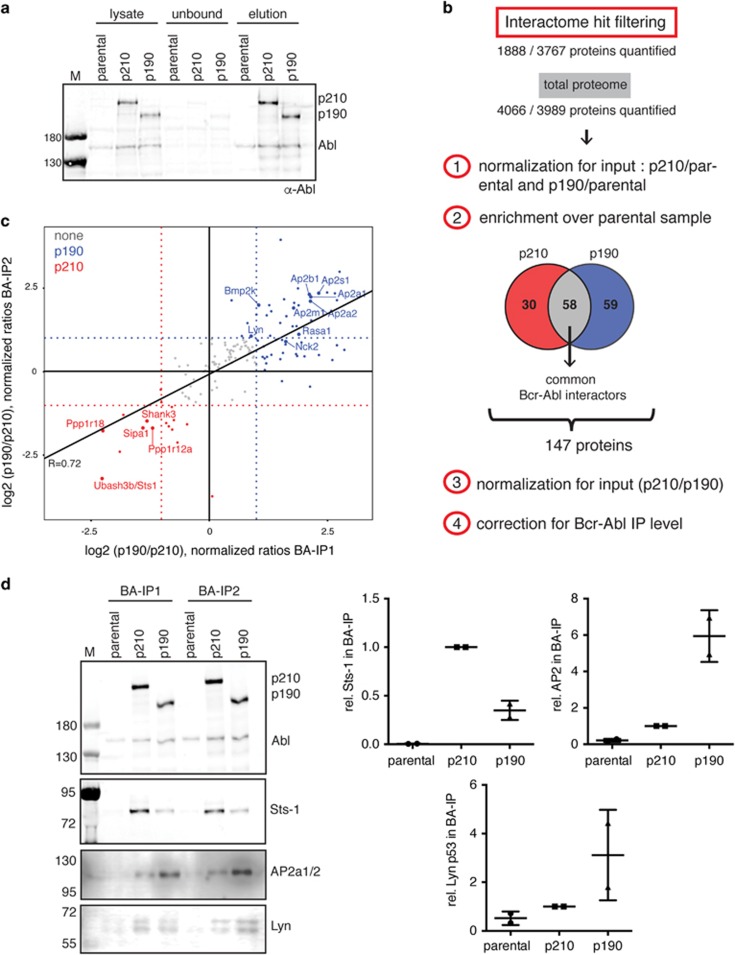
To identify and quantify the protein interaction network of p210 and p190 Bcr–Abl, we used a quantitative proteomics workflow based on SILAC (stable isotope labeling with amino acids in cell culture; Figure 1b). Murine BaF3 cells that express Abl endogenously were retrovirally transduced with the human Bcr–Abl p210 and p190 cDNAs25 and purification of Bcr–Abl complexes was achieved by immunoprecipitation experiments using an immobilized anti-Abl antibody with 80–100 mg of total protein lysate in biological duplicates. Parental (untransduced) BaF3 cells were used as a control to distinguish (endogenous) Abl from Bcr–Abl interactors (Figure 1b, Supplementary Tables S1 and S2). We achieved the quantification of >1800 proteins and a strong enrichment of Bcr–Abl p210 and p190 complexes over the parental control (Figure 2a and Supplementary Figures S1a and b). Furthermore, we identified most of the previously suggested p210 core interactors,17 which we found to be enriched with Bcr–Abl compared with the parental control (Supplementary Figures S1a and b).
Figure 2.
Analysis of the Bcr–Abl p210 and p190 interactome. (a) Quality control anti-Abl immunoblot after immunoaffinity purification of the Bcr–Abl complexes for a representative experiment. For the lysate samples 50 μg of total protein was loaded and an equivalent volume was used for the unbound fraction. The ratio of the two Bcr–Abl isoforms p210:p190 was 1.4:1 in the eluate, which was in accordance with the quantification by mass spectrometry (Supplementary Figure S1). For the elution fraction, 0.7% of the total SILAC eluate was loaded and quantification of bands enabled the calculation of the immunoprecipitation (IP) efficiency/Bcr–Abl complex recovery of 29% of the input amount. (b) Schematic representation of filtering criteria to select the differential Bcr–Abl interactors. Quantified proteins (1888 and 3767 proteins, respectively) were normalized to their input amounts (4066 and 3989 proteins quantified, respectively) and only those proteins with significant enrichment (according to Significance B) over the parental control were selected. This reduced number of proteins were further normalized for the Bcr–Abl IP amounts and finally selected if differentially enriched by at least twofold. A detailed description of the data analysis workflow is described in the Supplementary Methods. (c) Scatter plot representation depicting the final list of 147 Bcr–Abl interactors (see panel b) with the respective log2 ratios in both experiments (Bcr–Abl (BA) IP1 and IP2) showing an overall good correlation (R=0.72). Those proteins considered differential are colored in red (p210) and blue (p190). Selected proteins are highlighted with bigger dot size and gene name labels. (d) Validation of differential Bcr–Abl interactors by immunoblotting. Elution fractions of both replicates were analyzed corresponding to 0.7% of the total SILAC elution fraction. We chose to validate the interactome hits Ubash3b/Sts1 (enriched in p210) and AP2a1/2 and Lyn (both enriched in p190). The quantified signal after correction for the Bcr–Abl IP amounts is shown on the right side. Individual values are plotted together with the mean±s.d.
To subsequently identify common Bcr–Abl interactors and proteins that preferentially or exclusively interact with either p210 or p190, we first identified proteins that are enriched in either p210 or p190 Bcr–Abl samples compared with the parental control (Figure 2b). This strategy removed Abl interacting and contaminating proteins from further analysis. To compensate for the differential expression levels in the three different cell lines, we normalized the protein ratios for the input amounts, where ~4000 proteins of the expressed total proteome could be quantified (Figure 2b). The resulting 147 proteins included 56 proteins that were common to both Bcr–Abl isoforms (defined as common Bcr–Abl interactors; Supplementary Table S4), 30 proteins that were only found in the p210 samples and 59 proteins in the p190 samples (Figure 2b). The excellent sequence coverage from the Bcr–Abl region common to p210 and p190 allowed a precise quantification of and subsequent correction for the Bcr–Abl bait amount in the large-scale immunoprecipitation samples (Supplementary Figures S1c and d). For the comparison of the p210 and p190 interactors, this correction step is essential, because higher amounts of Bcr–Abl bait protein in one of the samples would lead to an overestimation of the amount of interactor. In both biological replicates p210 was ~1.4-fold enriched over p190 based on the mass spectrometry quantification (Figure 2b and Supplementary Figures S1c and d). After these normalization steps, we concentrated on those proteins that were at least twofold enriched, with 13 proteins for p210 and 34 proteins for p190 (Figure 2c and Supplementary Table S5).
Common Bcr–Abl interactors
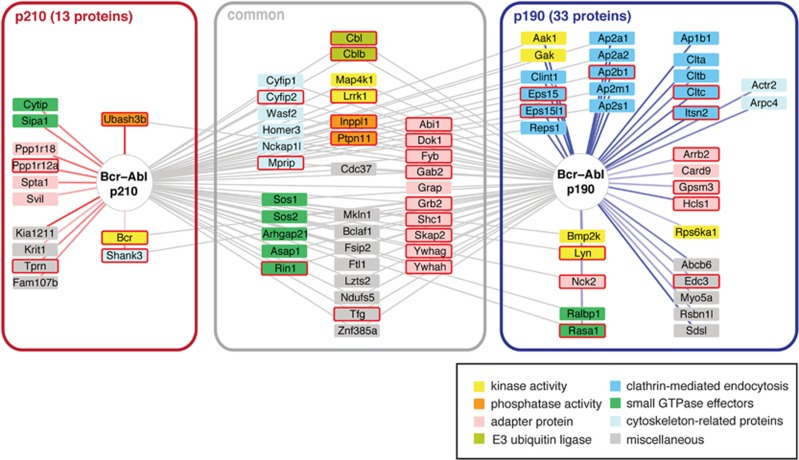
Among the 56 common Bcr–Abl interactors (Figure 3 and Supplementary Table S4), network analysis using the STRING database (http://string-db.org/) showed previously annotated experimentally determined interactions between the majority of these proteins. Only 15 molecules remain without previously mapped interactions (Supplementary Figure S2a). Two major clusters were identified with one subnetwork covering many cytosolic signaling components from tyrosine kinase and Ras–MAPK signaling pathways. These proteins include prototypic signaling adaptors (Grb2, Shc1, Dok1, Gab2), E3 ubiquitin ligases (Cbl, Cblb), phosphatases (Inppl1/Ship2 and Ptpn11/Shp2), kinases (Map4k1, Lrrk1), GTPase effector proteins (including the Ras GEFs Sos1/2) and cytoskeleton remodeling proteins (e.g., the Abi1–Wasf2–Cyfip2 complex), which is also reflected in the KEGG pathway and GO term enrichment (Supplementary Figure S2 and Figure 3). The three most significantly enriched domains using the SMART database were the SH2, SH3 and PH domains.
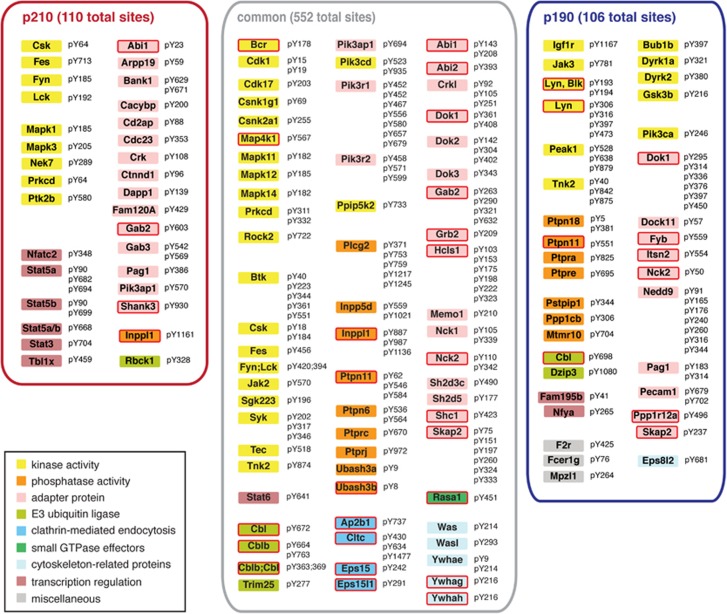
Figure 3.
Network of Bcr–Abl interacting proteins. Common and differential interactors of Bcr–Abl p210 and p190 are shown and summarized in Supplementary Tables S4 and S5. Connections for the differential interactors are color-coded according to their log2 ratios (from ‘light red’ for a weak p210 enrichment to ‘dark red’ for a strong p210 enrichment; ‘light blue’ for a weak p190 enrichment to ‘dark blue’ for a strong p190 enrichment). Boxes around the individual proteins are color-coded according to the function and a red frame indicates that the protein was also found phosphorylated in the phosphoproteome analysis. Note that certain proteins enriched with either of the two Bcr–Abl isoforms can also interact with the other isoform and thus have a connection to both proteins.
The very tightly interconnected second subnetwork contained the tetrameric adaptor protein complex 2 (AP2), of which all four subunits were identified, with various associated proteins that play a key role in regulating clathrin-mediated endocytosis of receptor proteins and other membrane trafficking processes (Figure 3). In addition to these two clusters, a small group of proteins with no previously experimentally mapped connections is included in the network of common Bcr–Abl interactors, such as the kinase co-chaperone Cdc37 and two members of the 14-3-3 proteins (Ywhag and Ywhah) (Figure 3).
Notably, there is a large overlap of the mapped common Bcr–Abl interactors with the previously identified p210 Bcr–Abl interactors using non-quantitative proteomics by affinity purification mass spectrometry of known Bcr–Abl interacting adaptor proteins in either the human K562 cell line17 or BaF3 cells.18 Owing to the use of SILAC, we achieve accurate quantitation and unbiased analyses.
Preferential and exclusive interactors of p210 and p190
The proteins that were differentially quantified between the two Bcr–Abl isoforms partly belong to the group of common Bcr–Abl interactors (preferential interactors), whereas another fraction of proteins is exclusive to one of the Bcr–Abl isoforms and not part of the common Bcr–Abl interactome. With regard to p190 Bcr–Abl, 17 of the 34 identified proteins are also part of the common interactors including most of the AP2 complex members and the Nck2 (SH3/SH2 adaptor)-Rasa1 (Ras GAP)-Lyn (Src family tyrosine kinase) complex. The remaining 17 proteins are only found in p190 and include additional members of the endocytosis machinery, such as the clathrin light and heavy chains (Cltca/b/c) (Figure 3, Supplementary Figure S2 and Supplementary Table S5).
Conversely, 3 of the 13 proteins that differentially bind to p210, are also part of the common Bcr–Abl interactors (Ubash3b/Sts1, Bcr and Shank3; Figure 3 and Supplementary Table S5). In contrast to p190, the differential proteins for p210 have no annotated experimental evidence for interaction with each other. Remarkably, the non-canonical tyrosine phosphatase Ubash3b/Sts1 shows a mean 6.5-fold enriched interaction with p210 Bcr–Abl (Figure 2c).
Interactome target validation
Validation of the interactome data was conducted by immunoblot analysis of the SILAC samples used for the MS analysis as an independent analytical method. Furthermore, anti-Abl immunoprecipitations were conducted for a set of human p210 and p190 expressing cell lines that were derived from CML and B-ALL patients (Supplementary Table S3 and Supplementary Figure S3). In addition, an independent set of BaF3 p210 and p190 cells was created and used as a further control.
We first focused on the tyrosine phosphatase Sts1 that was found as a common Bcr–Abl interactor and greatly enriched with p210 Bcr–Abl. The enrichment was confirmed in the immunoblot analysis of the SILAC samples (Figure 2d) and the independent BaF3 set (Supplementary Figure S3). For the human cell lines a less pronounced preference of Sts1 for p210 was observed (Supplementary Figure S3). The interpretation of such experiments is severely challenged by the different Bcr–Abl and interactor expression levels in the human cell lines (Supplementary Figure S3c), as well as the different cellular proteome background of the cells.
Validation of proteins that were enriched with p190 included Ap2a1/2, which is a subunit of the tetrameric AP2 complex, and the protein tyrosine kinase Lyn. Immunoblot analysis of the SILAC samples confirmed the findings by mass spectrometry (Figure 2d). The validation of Lyn in the human cell lines was complicated by limited antibody sensitivity and differential antibody binding to human and mouse Lyn, but with a possible trend towards p190 (Supplementary Figure S3). Nonetheless, these data are nicely in line with several Lyn tyrosine sites that are stronger phosphorylated in p190 cells.
Among the common interactors of p210 and p190, we validated the Inppl1/Ship2 interaction. Although the Ship2 antibody surprisingly did not recognize the mouse form of the protein, the human cells showed uniform interactions of Ship2 with p210 or p190 (Supplementary Figure S3).
In conclusion, we found and validated intriguingly strong differences in the protein interaction network of p210 and p190. Interestingly, our data set shows a number of proteins preferentially interacting with p190, even though the longer isoform p210 contains the additional DH–PH domains. Our data thus suggest that the DH–PH domain does not act as a protein-interaction scaffold. Alternatively, a different degree of kinase activation could result in rewiring of the cellular signaling network and thus change Bcr–Abl interaction partners.
Analysis of phosphorylation sites and activity of p210 and p190 Bcr–Abl
To test our hypothesis of possible quantitative differences in p210 and p190 activation, we studied the kinase activation state of Bcr–Abl by mapping phosphorylation sites by mass spectrometry, immunoblotting for major activating phosphorylation sites and in vitro kinase activity assays.
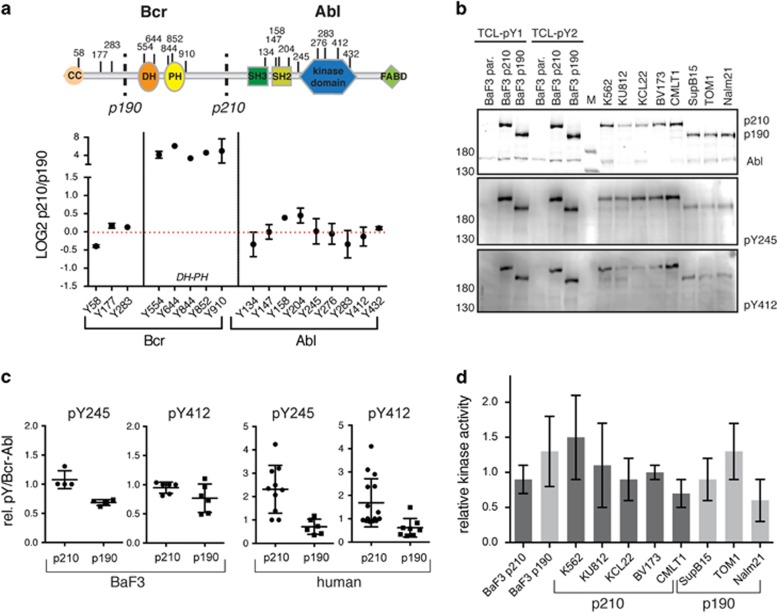
We could quantify 17 tyrosine phosphorylation sites of Bcr–Abl by mass spectrometry (Figure 4a). As expected, the five tyrosine phosphorylation sites (Y554, Y644, Y844, Y852 and Y910) that are unique to the p210 sequence were found strongly enriched in the p210 sample. In contrast, Y177 was phosphorylated to almost identical levels in p210 and p190, in line with our interactome data showing equal amounts of Grb2, which binds Y177 via its SH2 domain (Figure 4a). For the remaining Bcr–Abl phosphorylation sites, including Y412 in the activation loop and Y245, both commonly used Bcr–Abl activation markers,26 insignificant differences were observed (Figure 4a). In parallel, we monitored the phosphorylation state of Y412 and Y245 by immunoblotting using the total cell lysates of the SILAC samples and the human cell lines. Whereas the BaF3 cells show a slight increase of pY245 in the p210 cells, there is no major difference for pY412 between the isoforms (Figures 4b and c). The human cell lines, albeit quite heterogeneous, revealed a higher tendency of both phosphorylation sites towards p210 (Figures 4b and c). Similar results were obtained when equal amounts of Bcr–Abl after immunoprecipitation were loaded to facilitate normalization (Supplementary Figure S4).
Figure 4.
Bcr–Abl p210 and p190 phosphorylation sites and kinase activity. (a) Several Bcr–Abl pY sites were quantified in our phosphoproteome data set. The domain organization and location of the pY sites are shown and the sites are plotted together with their respective log2 ratio between p210/p190. Dots represent mean values ±s.d. Five out of the 17 sites could only be quantified in one experiment. Residues are numbered according to the Bcr and Abl 1b protein numbering. (b) Immunoblot analysis of the two autophosphorylation sites that are important for Abl catalytic activity: pY245 and pY412. Equal amounts of total cell lysates of the indicated BaF3 and human p210 and p190 expressing cell lines were loaded, immunoblotted with the indicated antibodies and quantified (see (c)). Note that only Bcr–Abl is phosphorylated on Y245 and Y412, while Abl is not or only very weakly phosphorylated on these sites. (c) Quantified pY245 and pY412 immunoblot signals (from panel (b)) after normalization to the total Bcr–Abl protein in BaF3 (left two graphs) or human cell lines (right two graphs). Individual values are plotted together with the mean±s.d. (d) Bcr–Abl was immunoprecipitated from the indicated BaF3 and human p210 and p190 cell lines and a radioactive in vitro kinase activity was performed measuring phosphotransfer to an optimal Abl substrate peptide. Each kinase assay was run in triplicate for each immunoprecipitate and the resulting averaged activity value was normalized to the Bcr–Abl amount determined by immunoblot analysis. The bar graph shows the mean±s.d. from three biological replicates.
We next compared the Bcr–Abl in vitro kinase activity after anti-Abl immunoprecipitation from BaF3 and our panel of eight human cell lines. We observed only minor differences without a consistent trend towards higher in vitro kinase activity of one of the two Bcr–Abl isoforms (Figure 4d). We additionally probed global p210 and p190 kinase activity using the PamChip PTK assay (PamGene). We observed only minor differences between the samples with only 7 of the 142 peptides phosphorylated stronger by the p210 lysate (Supplementary Figure S5) in contrast to a previous study in which we compared Nup214-Abl and p210 activity in BaF3 cells.27
Collectively, our data suggest that p210 and p190 do not differ significantly in kinase autophosphorylation or in vitro kinase activity. These results argue against quantitative differences in kinase activation that may account for the observed differences in the p210 vs p190 interactome and phosphoproteome.
Bcr–Abl phosphoproteome analysis
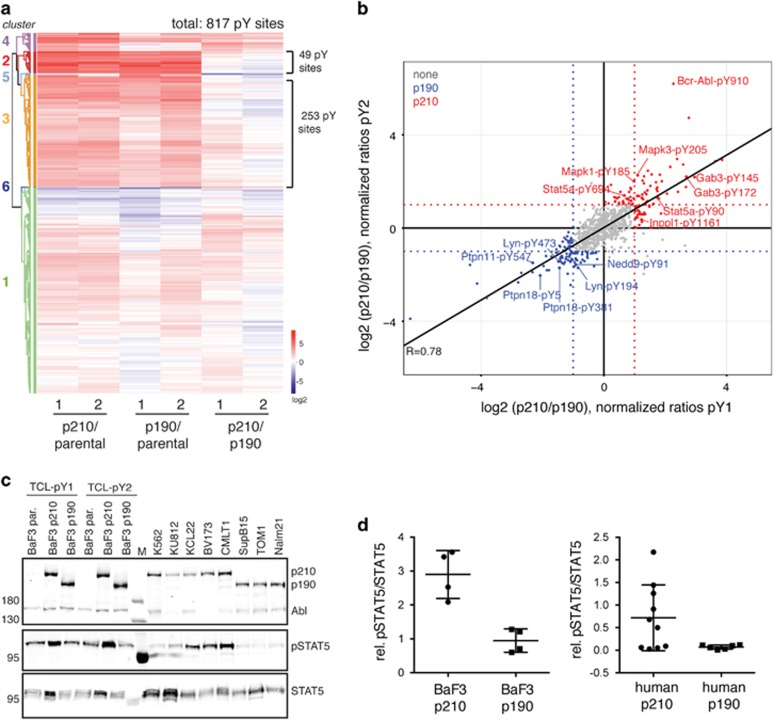
In addition to the phosphotyrosine sites of Bcr–Abl, we mapped the cellular tyrosine phosphoproteome and quantified 817 phosphotyrosine sites in 573 protein groups (Figure 5a and Supplementary Figure S6). Unsupervised hierarchical clustering of the quantified phosphopeptides revealed six clusters with cluster 1 comprising all the sites that remained unchanged among the three cell lines (Figure 5a). Clusters 2, 3 and 4 depict the phosphopeptides that are upregulated in the Bcr–Abl cell lines compared with the parental cell line. Here in particular, clusters 2 and 3 (with 302 phosphopeptides corresponding to 37% of all quantified pY peptides) stood out due to the high intensity of phosphorylation as compared with the BaF3 parental cells, thus representing the Bcr–Abl pY signature (Figure 5a). Among those peptides, 45 sites map to 27 of the Bcr–Abl interacting proteins identified in the interactome study, which makes it the cluster with the largest accumulation of Bcr–Abl interactors (Supplementary Figure S7). In-depth analysis of the phosphosites within this cluster revealed many known and amply characterized substrates of Bcr–Abl such as Abi1, Cbl, Gab2, Pik3r1, Pxn (paxillin) and Stat528 that is also reflected in the GO term enrichment analysis of these proteins containing CML and other tyrosine kinase signaling pathways (Supplementary Figure S7). In addition, phosphomotif analysis showed enrichment of the Abl phosphorylation site consensus in cluster 2 and 3 (67 out of 273 unique peptides) with a proline in position +3 following the phosphorylated tyrosine (Supplementary Figure S7). Altogether, our data set indicated that the mapped tyrosine phosphoproteome is dominated by Bcr–Abl kinase signaling and included many known substrates and phosphorylated interactors (Figure 3).
Figure 5.
Bcr–Abl pY phosphoproteome. (a) Heatmap representation of the 817 quantified phosphosites and their respective ratios in the comparison of the three samples: BaF3 parental, BaF3 p210 and BaF3 p190. The two biological replicates are plotted next to each other. The six clusters identified by unsupervised hierarchical clustering are highlighted. (b) Scatter plot for the normalized log2 ratios between the p210 and p190 Bcr–Abl samples (without correction for the total protein levels) in both biological replicates (pY1 and pY2). Each dot is representative of a phosphopeptide. Those pY sites considered as differential are colored in red (p210) and blue (p190). Selected phosphorylation sites are labeled. (c, d) Immunoblot validation of Stat5 pY694 using the BaF3 and human cell line panel. The quantified signals of two technical replicates were normalized to the total Stat5 protein and individual values are plotted together with the mean±s.d.
Differential phosphorylation sites of p210 and p190
The analysis of differential p210 and p190 phosphorylation events resulted in 106 pY sites in 78 proteins to be more highly phosphorylated in p190 and 110 pY sites in 92 proteins to be more highly phosphorylated in p210 (Figure 5b, Supplementary Tables S6 and S7).
Among the sites upregulated in p210, six phosphorylation sites of the transcription factor Stat5a (Y90, Y668, Y682, Y694) and Stat5b (Y90, Y668, Y699) were mapped, including the main site responsible for Stat5a activation (Y694 in Stat5a, Figure 6 and Supplementary Table S7).29 In addition, the activation loops of the tyrosine kinase Fes (Y713) and the Ser/Thr kinases Erk1 (Y205) and Erk2 (Y185) and sites of the Src family kinases Fyn (Y185) and Lck (Y192) were found to be more highly phosphorylated (Figure 6). This indicated a higher activation of these kinases in the p210-expressing cells. The high number of kinases that are phosphorylated in the p210 sample is also reflected in the GO term and KEGG pathway enrichment analysis (Supplementary Figure S7).
Figure 6.
Selected phosphorylation events of p210 and p190 Bcr–Abl. A selection of differential and common Bcr–Abl phosphorylation sites that were found in the pY data set is shown. The complete data set is listed in Supplementary Table S6. Boxes around the individual proteins are color-coded according to their function and a red frame indicates that a protein was also found in the interactome analysis.
Among the 106 pY sites upregulated in p190, 6 phosphosites of the Src family tyrosine kinase Lyn (two of which shared with Blk), including its activation loop (Y397), were found to be more strongly phosphorylated than in p210 (Figure 6). This is nicely in line with the stronger interaction of Lyn with p190 than p210 (Figure 2c). In addition, the non-receptor tyrosine phosphatases Shp2 (Ptpn11) and Flk1 (Ptpn18) showed higher phosphorylation in p190 cells, which may indicate a stronger activation of these enzymes in p190 cells. Furthermore, we found the adapter proteins Dok1 and Pag1 to be more highly phosphorylated in p190, and they, together with Ptpn11 and Ptpn18, were shown to play a central role in the negative-feedback regulation of Src family kinases (Figure 6).30
Noteworthy, among the 552 pY sites that remain unchanged between the two Bcr–Abl isoforms, we found many central signaling molecules mediating pY signaling (Supplementary Table S8). These included adaptor proteins (CrkL, Shc1, Gab2, Nck1/2, Abi1/2, Dok1/2/3, Sh2d3/5, Skap2, several 14-3-3 isoforms), E3 ubiquitin ligases (Cbl, Cblb), lipid and protein phosphatases (Inppl1/5d, Ptpn6, Ptprc, Ptprj), several PI3-kinase subunits and the transcription factor Stat6. In addition, a large number of activating pY sites on cytoplasmic tyrosine kinases (Btk, Tec, Jak2, Csk, Tnk2, Syk) as well as serine/threonine kinases (Cdk1/17, Mapk14/11/12=p38α/β/γ, Sgk233, Map4k1, Pkcδ) were found (Figure 6 and Supplementary Table S8). This shows the large deregulation of cellular signaling by Bcr–Abl and its link to core proliferative and antiapoptotic pathways that sustain leukemogenesis. Collectively, our analysis identified many differentially regulated phosphosites with annotated functional importance along with a large number of functionally uncharacterized sites in important signaling proteins.
Phosphoproteome site validation
Validation of the phosphoproteomic data set was conducted by immunoblot analysis of the SILAC samples in addition to the set of human p210 and p190 Bcr–Abl expressing cell lines (Supplementary Table S3). Among the six upregulated Stat5a/b phosphorylation sites in p210, we concentrated on Y694/Y699 in Stat5a/b, since it is the major activating site. We could confirm the MS results in BaF3 and human CML cell lines (Figures 5c and d). In line with the analysis of the total BaF3 Stat5 protein amount by MS, we also observed elevated Stat5 expression levels in p210-expressing cells. Nonetheless, pStat5 levels were increased also after correction for the elevated total protein levels showing increased Stat5 phosphorylation stoichiometry in p210 cells (Figures 5c and d). Another member of the Stat transcription factors, Stat3, also showed elevated phosphorylation on the main activating site (Y704) in p210 in line with the MS results (Supplementary Figure S8). In contrast, we found Stat6 to be equally phosphorylated on its main activating site (Y641; Figure 6).
Src family kinases were found to be phosphorylated in both Bcr–Abl cell lines with Lyn being activated in p190 cells, and Fyn and Lck phosphorylation being higher in p210 cells. Owing to the high sequence homology within this kinase family, antibodies for the activation loop of Src kinases cannot distinguish the different kinases of this family. Nonetheless, we attempted to monitor pSrc levels in our cell lines and found higher general pSrc levels in BaF3 p190 cells compared with the human cell lines, in which we observed a trend towards higher pSrc levels in the p210 Bcr–Abl cells (Supplementary Figure S8). A clear interpretation of these data is difficult, as the human cell lines have strongly different Src kinase expression patterns and levels. Therefore, reliable normalization of the pSrc signal is impossible. Interestingly, tyrosine kinase prediction based on the analysis of the PamChip PTK assay indicated overall stronger activation of Src kinases in p210 cells (Supplementary Figure S5). In contrast, we used an antibody that is specific for the C-terminal inhibitory phosphorylation site of Lyn (Y507) because we found several proteins of the Src negative-feedback circuit phosphorylated in p190. Immunoblot analysis showed higher levels of pY507 phosphorylation in Bcr–Abl-expressing BaF3 cells as compared with the parental cells but did not appear to differ between the two isoforms (Supplementary Figure S8). The same was found for the human cell lines, where high levels of Lyn pY507 correlate with a high expression level.
Kinase activity profiling on the PamChip STK assay (PamGene) revealed a much higher activity of Ser/Thr kinases in the lysate of p210 with more than 20% of the peptides being more phosphorylated as compared with the p190 lysate. The prediction of putative upstream kinases from these data suggested the involvement of Sgk2, Akt and Dclk2 (Supplementary Figure S5).
In summary, we found strongly activated Stat5 in p210, Dok1 in p190, and Src kinase members appear to be differentially activated by p210 and p190. In line with the observations of an isoform-dependent interactome, also the phosphoproteome varies significantly with the Bcr–Abl isoform and suggests important differential signaling nodes with possible therapeutic implications (Figure 7).
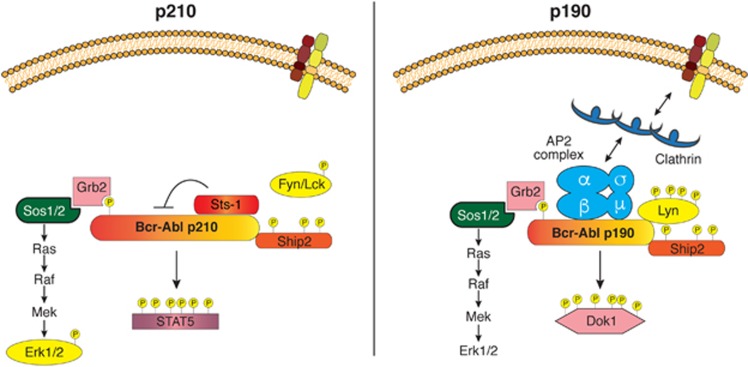
Figure 7.
Model of differential signaling networks of p210 and p190 Bcr–Abl. The main differences in Bcr–Abl interactors and phosphorylated proteins between p210 and p190 that we found, validated and discussed in this paper are summarized in this figure. As examples for common interactors/pY sites, the Ship2 phosphatase and the interaction of the Grb2/Sos complex with pY177 are shown. p210 shows stronger association with the Sts1 phosphatase and higher activation of the Stat5 transcription factor, as well as the Erk1/2 and Fyn/Lck kinases. p190 shows stronger association with the AP2 complex and clathrin, and higher activation of the Dok1 adaptor and the Lyn kinase.
Discussion
We observed many differences in both the interactome and the phosphoproteome between the two Bcr–Abl isoforms (Figures 3 and 6). Because p190 can be considered as an internal deletion mutant of p210, lacking 501 amino acids that encode for a DH–PH domain unit, significant gains in the p210 interactome were expected rather than the observed balanced number of interactors, excluding a scaffolding function of the DH–PH domain. This observation raises questions regarding the molecular mechanism. Our initial hypothesis on a different degree of kinase activity deregulation could be excluded. Even in BaF3 cells, that is, in the same proteomic 'background', differences in kinase activation were insignificant. In line with this, the human cell lines also showed no consistent differences of p210 and p190 Bcr–Abl kinase activation. It is also unlikely to assume that the different length of the Bcr portion of p190 and p210 may change substrate specificity because the substrate binding groove in the kinase domain primarily determines the kinase substrate specificity, which is identical in p210 and p190. A second attractive hypothesis includes a possible differential subcellular localization that is caused by the different overall structure and domain composition of the two isoforms. As a result, p210 would encounter another subset of the proteome than p190 and would consequently differentially activate signaling pathways due to different interacting proteins, including kinases and phosphatases, ultimately resulting in pronounced differences in the phosphoproteome. In line with this hypothesis, deletion of the F-actin-binding domain in p210 and p190 was found to have different effects on the leukemogenicity of Bcr–Abl.31, 32, 33 In addition, the presence of the largely uncharacterized PH domain in p210 may play an important role in differential subcellular localization. PH domains are classical phosphoinositide-binding domains and mediate targeting of PH domain containing proteins to membranes. If the p210 PH domain is important for Bcr–Abl localization, however, needs to be clarified by future studies. Similarly, we found that the AP2 adaptor protein complex and associated proteins strongly and preferentially interact with p190 (Figure 7). The tetrameric AP2 complex mediates the internalization of membrane receptors by regulating the assembly of clathrin-coated pits resulting in endocytosis. The functional consequences of the 'hijacking' of the AP2 complex on Bcr–Abl localization or receptor trafficking in p190 cells warrant in-depth future analysis. Among the strongest differential interactors of p210, we found the non-canonical tyrosine phosphatase Sts1, which was described as a negative regulator of the Zap-70 tyrosine kinases in T-cell receptor signaling.34 It is tempting to speculate that its strong interaction with p210 might be a mechanism to negatively regulate Bcr–Abl (Figure 7).
Among the strongest upregulated phosphorylation events in p210 was Stat5 on six tyrosine phosphorylation sites, including the major site that activates Stat5 dimerization, nuclear translocation and transcriptional activation (Figure 7). In addition, we found two phospho-tyrosines in the SH2 domain (pY668 and pY682) and one phosphotyrosine in the N-terminal domain of Stat5 (pY90) (Supplementary Figure S8). Contributions of these three phosphorylation sites to signaling of Stat5 have not been investigated and structural information remains limited35, 36 (Supplementary Figure S8). Both domains, SH2 and N-terminal domain, are protein–protein interaction domains and introduction of a phosphoryl group would change the electrostatic potential of the respective position which could in turn influence the interaction with binding partners.
Importantly, Stat5 is an essential Achilles heel in CML, as it is absolutely required for disease initiation as well as for disease maintenance, and Stat5 upregulation mediates TKI resistance.37, 38 Interestingly, the strong activation of Stat5 by p210 but not p190 is mirrored by our previous data on the requirement of the upstream JAK2 kinase for initial lymphoid transformation by p190 (or v-Abl) but not for myeloid transformation by p210.13 These findings have important obvious implications for the ongoing studies of Jak2 TKIs in Ph-positive leukemias.
Another critical class of downstream signaling mediators of Bcr–Abl is the Src kinases. We found that the Src family kinase Lyn and its interactor Hcls1 (hematopoietic cell-specific Lyn substrate 1) show stronger interactions with p190 than p210 and that Lyn further shows stronger phosphorylation in p190 cells at six sites, including the activation loop (Figure 7). Lyn, along with Hck and Fgr, was shown to be required for B-lymphoid, but not for myeloid transformation.39 Lyn upregulation was shown to mediate imatinib resistance40 and lymphoid, but not myeloid blast crisis. CML cells are strongly dependent on Lyn expression and activity.41 Similarly, the Fes tyrosine kinase that we found activated by p210 was previously shown to drive myeloid differentiation in K562 cells.42
Our data have revealed differential activation of prominent, druggable cell signaling pathways, such as the Jak–Stat pathway, Src family kinases as well as Erk as part of the Ras–Raf–Mek–Erk pathway. Based on our results, a rational drug treatment strategy could be envisaged targeting these pathways with for instance FDA-approved Jak2, Src or Mek inhibitors in combination with Abl TKIs for patients with p210 or p190 expression.
Importantly, the parallel study by Cutler et al.19 also found intriguing differences in the interactome and phosphoproteome of p210 and p190. Key molecules highlighted in our study such as the p210-interacting phosphatase Sts1/Ubash3b or the differential activation of Stat5 and Src family kinases are confirmed by their independent approach. Some findings are also unique to one of the two studies as, for example, the enrichment of the AP2 complex in our study and members of the cytoskeleton by Cutler et al.19 with p190. Nevertheless certain cytoskeletal components such as the Wiskott–Aldrich syndrome protein family member Wasf2, components of the WAVE regulatory complex, Cyfip1 and 2 as well as members of the Arp2/3 complex were identified in our study, but we did not observe a strong enrichment with p190.
We believe that the differences in the two studies are due to the individual experimental set-ups with different BaF3 cell lines and construct design as well as sample preparation, data acquisition and analysis workflows. In particular, the interactome analyses of the two studies are targeting different layers of interacting proteins. The purification of Bcr–Abl complexes used in our study is the more stringent approach and captures only those proteins that are tight interactors of Bcr–Abl with slow off rates.43 In contrast, the BioID experiment employed by Cutler et al.19 relies on the N-terminal fusion of the biotin ligase BirA that enzymatically modifies proteins in proximity to Bcr–Abl. This experiment captures also weak and transient interactions such as substrates of Bcr–Abl providing an additional layer of information.44 Altogether the two independent proteomics studies are highly similar presenting an important cross-validation.20
Collectively, our results show that the mapped differential activation of tyrosine kinase and phosphatase pathways may indeed be tightly associated or causal to drive either myeloid or B-lymphoid transformation by p210 or p190. Finally, these kinases are readily druggable targets that may help patients to better cope with TKI resistance and offer more effective treatment options, particularly for Ph-positive B-ALL patients.
Acknowledgments
This work was supported by the ISREC Foundation (to SR, SG and OH), Swiss National Science Foundation (grant 31003A_140913; to SR and OH) and National Center of Competence in Research (NCCR) in Chemical Biology (to OH). We thank B Correia for his critical input on the manuscript, G Müller-Newen for insightful discussions on Stat5 signaling, the PamGene team (D Pijnenburg, R de Wijn) for support on the PamGene array analysis, the EPFL Protein Expression Core Facility for help with antibody production and all members of the Hantschel lab for continuous support and discussions. OH wishes to dedicate this paper to the memory of Dr John Goldman.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
The authors declare no conflict of interest.
Supplementary Material
References
- O'Hare T, Zabriskie MS, Eiring AM, Deininger MW. Pushing the limits of targeted therapy in chronic myeloid leukaemia. Nat Rev Cancer 2012; 12: 513–526. [DOI] [PubMed] [Google Scholar]
- Wong S, Witte ON. The BCR-ABL story: bench to bedside and back. Annu Rev Immunol 2004; 22: 247–306. [DOI] [PubMed] [Google Scholar]
- Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood 2000; 96: 3343–3356. [PubMed] [Google Scholar]
- Hantschel O. Structure, regulation, signaling, and targeting of abl kinases in cancer. Genes Cancer 2012; 3: 436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambacorti-Passerini C, Antolini L, Mahon F-X, Guilhot F, Deininger M, Fava C et al. Multicenter independent assessment of outcomes in chronic myeloid leukemia patients treated with imatinib. J Natl Cancer Inst 2011; 103: 553–561. [DOI] [PubMed] [Google Scholar]
- Fielding AK. How I treat Philadelphia chromosome positive acute lymphoblastic leukaemia. Blood 2010; 116: 3409–3417. [DOI] [PubMed] [Google Scholar]
- Rousselot P, Coude MM, Gokbuget N, Gambacorti Passerini C, Hayette S, Cayuela JM et al. Dasatinib and low-intensity chemotherapy in elderly patients with Philadelphia chromosome-positive ALL. Blood 2016; 128: 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Downing JR. Genome-wide profiling of genetic alterations in acute lymphoblastic leukemia: recent insights and future directions. Leukemia 2009; 23: 1209–1218. [DOI] [PubMed] [Google Scholar]
- Wong S, Witte ON. Modeling Philadelphia chromosome positive leukemias. Oncogene 2001; 20: 5644–5659. [DOI] [PubMed] [Google Scholar]
- Van Etten RA. Studying the pathogenesis of BCR-ABL+ leukemia in mice. Oncogene 2002; 21: 8643–8651. [DOI] [PubMed] [Google Scholar]
- Li S, Ilaria RL Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med 1999; 189: 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacic B, Hoelbl A, Litos G, Alacakaptan M, Schuster C, Fischhuber KM et al. Diverging fates of cells of origin in acute and chronic leukaemia. EMBO Mol Med 2012; 4: 283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantschel O, Warsch W, Eckelhart E, Kaupe I, Grebien F, Wagner K-U et al. BCR-ABL uncouples canonical JAK2-STAT5 signaling in chronic myeloid leukemia. Nat Chem Biol 2012; 8: 285–293. [DOI] [PubMed] [Google Scholar]
- Okuda K, Golub TR, Gilliland DG, Griffin JD. p210BCR/ABL, p190BCR/ABL, and TEL/ABL activate similar signal transduction pathways in hematopoietic cell lines. Oncogene 1996; 13: 1147–1152. [PubMed] [Google Scholar]
- Lugo TG, Pendergast AM, Muller AJ, Witte ON. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science 1990; 247: 1079–1082. [DOI] [PubMed] [Google Scholar]
- Ilaria RL Jr, Van Etten RA. The SH2 domain of P210BCR/ABL is not required for the transformation of hematopoietic factor-dependent cells. Blood 1995; 86: 3897–3904. [PubMed] [Google Scholar]
- Brehme M, Hantschel O, Colinge J, Kaupe I, Planyavsky M, Kocher T et al. Charting the molecular network of the drug target Bcr-Abl. Proc Natl Acad Sci USA 2009; 106: 7414–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titz B, Low T, Komisopoulou E, Chen SS, Rubbi L, Graeber TG. The proximal signaling network of the BCR-ABL1 oncogene shows a modular organization. Oncogene 2010; 29: 5895–5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler J, Tahir R, Sreenivasamurthy SK, Mitchell C, Renuse S, Nirujogi RS. Differential signaling through p190 and p210 BCR-ABL fusion proteins revealed by interactome and phosphoproteome analysis. Leukemia 2017. [DOI] [PubMed]
- Varjosalo M, Sacco R, Stukalov A, van Drogen A, Planyavsky M, Hauri S et al. Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat Methods 2013; 10: 307–314. [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G et al. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods 2006; 3: 1013–1019. [DOI] [PubMed] [Google Scholar]
- Schiff-Maker L, Burns MC, Konopka JB, Clark S, Witte ON, Rosenberg N. Monoclonal antibodies specific for v-abl- and c-abl-encoded molecules. J Virol 1986; 57: 1182–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach AN, Gerber SA. Rapid and reproducible single-stage phosphopeptide enrichment of complex peptide mixtures: application to general and phosphotyrosine-specific phosphoproteomics experiments. Anal Chem 2011; 83: 7635–7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I et al. update of the PRIDE database and its related tools. Nucleic Acids Res 2016; 44: D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmuth M, Kim S, Gu XJ, Xia G, Adrian F. Ba/F3 cells and their use in kinase drug discovery. Curr Opin Oncol 2007; 19: 55–60. [DOI] [PubMed] [Google Scholar]
- Hantschel O, Superti-Furga G. Regulation of the c-Abl and Bcr-Abl tyrosine kinases. Nat Rev Mol Cell Biol 2004; 5: 33–44. [DOI] [PubMed] [Google Scholar]
- De Keersmaecker K, Versele M, Cools J, Superti-Furga G, Hantschel O. Intrinsic differences between the catalytic properties of the oncogenic NUP214-ABL1 and BCR-ABL1 fusion protein kinases. Leukemia 2008; 22: 2208–2216. [DOI] [PubMed] [Google Scholar]
- Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer 2005; 5: 172–183. [DOI] [PubMed] [Google Scholar]
- Shuai K, Horvath CM, Huang LH, Qureshi SA, Cowburn D, Darnell JE Jr. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell 1994; 76: 821–828. [DOI] [PubMed] [Google Scholar]
- Rubbi L, Titz B, Brown L, Galvan E, Komisopoulou E, Chen SS et al. Global phosphoproteomics reveals crosstalk between bcr-abl and negative feedback mechanisms controlling SRC signaling. Sci Signal 2011; 4: ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisterkamp N, Voncken JW, Senadheera D, Gonzalez-Gomez I, Reichert A, Haataja L et al. Reduced oncogenicity of p190 Bcr/Abl F-actin-binding domain mutants. Blood 2000; 96: 2226–2232. [PubMed] [Google Scholar]
- Wertheim JA, Perera SA, Hammer DA, Ren R, Boettiger D, Pear WS. Localization of BCR-ABL to F-actin regulates cell adhesion but does not attenuate CML development. Blood 2003; 102: 2220–2228. [DOI] [PubMed] [Google Scholar]
- Hantschel O, Wiesner S, Guttler T, Mackereth CD, Rix LL, Mikes Z et al. Structural basis for the cytoskeletal association of Bcr-Abl/c-Abl. Mol Cell 2005; 19: 461–473. [DOI] [PubMed] [Google Scholar]
- Mikhailik A, Ford B, Keller J, Chen Y, Nassar N, Carpino N. A phosphatase activity of Sts-1 contributes to the suppression of TCR signaling. Mol Cell 2007; 27: 486–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neculai D, Neculai AM, Verrier S, Straub K, Klumpp K, Pfitzner E et al. Structure of the unphosphorylated STAT5a dimer. J Biol Chem 2005; 280: 40782–40787. [DOI] [PubMed] [Google Scholar]
- Fahrenkamp D, Li J, Ernst S, Schmitz-Van de Leur H, Chatain N, Kuster A et al. Intramolecular hydrophobic interactions are critical mediators of STAT5 dimerization. Sci Rep 2016; 6: 35454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelbl A, Schuster C, Kovacic B, Zhu B, Wickre M, Hoelzl MA et al. Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia. EMBO Mol Med 2010; 2: 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warsch W, Kollmann K, Eckelhart E, Fajmann S, Cerny-Reiterer S, Holbl A et al. High STAT5 levels mediate imatinib resistance and indicate disease progression in chronic myeloid leukemia. Blood 2011; 117: 3409–3420. [DOI] [PubMed] [Google Scholar]
- Hu Y, Liu Y, Pelletier S, Buchdunger E, Warmuth M, Fabbro D et al. Requirement of Src kinases Lyn, Hck and Fgr for BCR-ABL1-induced B-lymphoblastic leukemia but not chronic myeloid leukemia. Nat Genet 2004; 36: 453–461. [DOI] [PubMed] [Google Scholar]
- Wu J, Meng F, Kong LY, Peng Z, Ying Y, Bornmann WG et al. Association between imatinib-resistant BCR-ABL mutation-negative leukemia and persistent activation of LYN kinase. J Natl Cancer Inst 2008; 100: 926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptasznik A, Nakata Y, Kalota A, Emerson SG, Gewirtz AM. Short interfering RNA (siRNA) targeting the Lyn kinase induces apoptosis in primary, and drug-resistant, BCR-ABL1(+) leukemia cells. Nat Med 2004; 10: 1187–1189. [DOI] [PubMed] [Google Scholar]
- Yu G, Smithgall TE, Glazer RI. K562 leukemia cells transfected with the human c-fes gene acquire the ability to undergo myeloid differentiation. J Biol Chem 1989; 264: 10276–10281. [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Burke B. BioID: a screen for protein-protein interactions. Curr Protoc Protein Sci 2013; 74, Unit 19.23.1–19.34.14. [DOI] [PubMed] [Google Scholar]
- Varnaite R, MacNeill SA. Meet the neighbors: mapping local protein interactomes by proximity-dependent labeling with BioID. Proteomics 2016; 16: 2503–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.