Abstract
Temperate phages are common and prophages are abundant residents of sequenced bacterial genomes. Mycobacteriophages are viruses infecting mycobacterial hosts including Mycobacterium tuberculosis and Mycobacterium smegmatis, encompass substantial genetic diversity, and are commonly temperate. Characterization of ten Cluster N temperate mycobacteriophages reveals at least five distinct prophage-expressed viral defense systems that interfere with infection of lytic and temperate phages that are either closely-related (homotypic defense) or unrelated (heterotypic defense). Target specificity is unpredictable, ranging from a single target phage to one-third of those tested. The defense systems include a single-subunit restriction system, a heterotypic exclusion system, and a predicted (p)ppGpp synthetase, which blocks lytic phage growth, promotes bacterial survival, and enables efficient lysogeny. The predicted (p)ppGpp synthetase coded by the Phrann prophage defends against phage Tweety infection, but Tweety codes for a tetrapeptide repeat protein, gp54, that acts as a highly effective counter-defense system. Prophage-mediated viral defense offers an efficient mechanism for bacterial success in host-virus dynamics, and counter-defense promotes phage co-evolution.
Microbial warfare between prokaryotes and their viruses has been waging for perhaps over three billion years, with constant selection for bacterial survival in the face of viral infection and viral co-evolution to support ongoing viral replication1,2. Bacteriophages taken as a whole are abundant, dynamic and not unexpectedly highly diverse genetically, shaped by pervasive genomic mosaicism and rapid host range modulation3–5. Not surprisingly, prokaryotes rapidly acquire insensitivity to infection and numerous defense systems have been described including Clustered Regularly-Interspaced Short Palindromic Repeats (CRISPRs) that offer adaptive immunity6, restriction-modification systems7–9, Argonaute-based interference10, Bacteriophage Exclusion (BREX)11, and a collection of processes conferring abortive infection (abi), including Toxin-Antitoxin (TA) systems12,13. Phages have counter immunity systems including anti-restriction, anti-ToxIN and anti-CRISPR systems9,14,15.
Temperate phages are common and bacterial genomes are replete with integrated prophages16. Metagenomic studies show that at high microbial abundance temperate phages predominate and likely play key roles in microbial success17. Lysogeny offers numerous benefits through prophage-encoded genes that influence host physiology, provide virulence determinants18, or protect against phage infection through repressor-mediated immunity, superinfection exclusion, or restriction systems19. However, repressor-mediated immunity and exclusion typically target particles of the same or closely related phages (i.e. are homotypic)20,21. Given the long complex dynamic interaction of bacteria and their phages, it is likely that temperate phages provide a plethora of additional viral defense systems that have yet to be identified.
A large collection of >1000 completely sequenced mycobacteriophages provides a detailed genetic profile of phages infecting a single common host, Mycobacterium smegmatis mc215522. There are many groups (clusters) differing from each other in overall nucleotide sequence, and many clusters have high intra-cluster variation and can be divided into subclusters22,23. The genomes are pervasively mosaic as a consequence of horizontal genetic exchange mediated by illegitimate recombination events over their long period of evolution24,25. As a consequence, the cluster/subcluster groupings reflect uneven sampling of the viral communities, with a continuum of diversity22. The majority of mycobacteriophage genomes encode either integrase or partitioning functions and are either temperate or recent derivatives of temperate parents26. This phage collection provides an uncommon resource for exploring the phenomenon of prophage-mediated viral defense.
RESULTS
Isolation and characterization of Cluster N mycobacteriophages
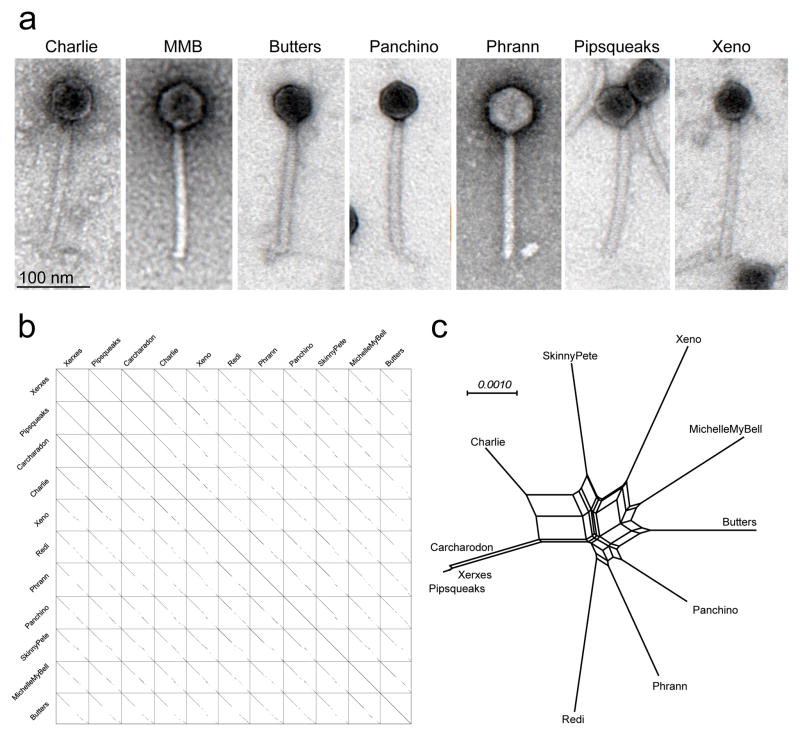
The phage discovery and genomics platform of the Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program27–29 has identified 11 closely-related phages grouped in Cluster N (Supplementary Table 1). All were isolated from environmental samples using M. smegmatis mc2155, have relatively small genomes (41.5 – 44.8 kbp), are morphologically similar (Fig. 1a), and share extensive but interrupted nucleotide sequence similarity (Fig. 1b). Nucleotide and gene content comparisons (Fig. 1b, c) show there is substantial variation among the genomes but not such as to warrant division into subclusters.
Figure 1. Cluster N phage genotypes and morphotypes.
(a) Electron micrographs of Cluster N mycobacteriophages. Images are representative of more than 30 particles examined from two preparations of each phage. (b) Dotplot comparison of Cluster N mycobacteriophages. A concatenated string of 10 cluster N phage genomes (as shown) was compared against itself using default parameters in Gepard51. (c) Network phylogenetic analysis of Cluster N mycobacteriophages based on gene content and displayed using the Network function in Splitstree52. Xerxes, Pipsqueaks, and Carcharadon have near-identical gene content, and differ by 30–50 single nucleotide polymorphisms (SNPs) and a few small insertions/deletions. Nucleotide sequence and gene content analyses do not support subclusters groupings23.
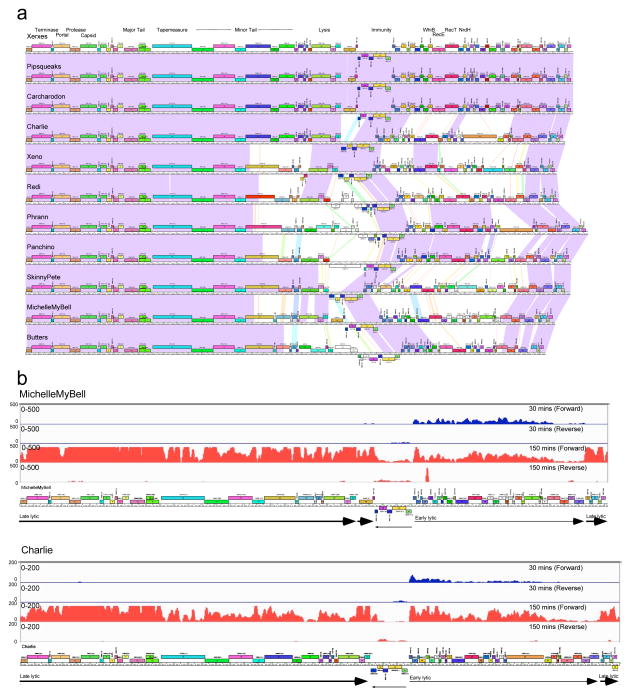
The Cluster N genomes are organized with the virion structure and assembly genes in their left arms, separated from the non-structural genes by lysis and immunity cassettes (Fig. 2a; see Supplementary Figures 1–11). The immunity cassettes include integrase and repressor genes, and a phage attachment site (attP) located within the repressor gene, a hallmark of integration-dependent immunity systems30. The repressors are divergently transcribed with the rightwards-facing genes traversing the genome right arms, and the first open reading frame in the operon is a Cro-like DNA binding protein implicated in lytic growth regulation30. All eleven phages have rightwards-facing early lytic PR promoters in a conserved region upstream of the rightwards operon. Note that genes are numbered and named in genomic order; similarly named genes are not necessarily functionally related.
Figure 2. Genomic organization of Cluster N mycobacteriophages.
(a) The genomes of 11 Cluster N mycobacteriophages are shown with pairwise nucleotide sequence similarity displayed by spectrum-coloring between the genomes; violet is the most similar, red is the least similar above threshold BLASTN E value of 10−5. The genes are shown as colored boxes above (transcribed-rightwards) or below (transcribed leftwards) each genome, and are color-coded according to the gene phamilies they are assigned to 44,53; the phamily number is shown above each gene with the number of phamily members in parentheses. Maps were generated using Phamerator and the database ‘Actinobacteriophage_554’ 44. (b) Lytic gene expression patterns in phages MichelleMyBell and Charlie. RNAseq profiles are shown for forward and reverse DNA strands (as indicated) at 30 mins (blue) or 150 minutes (red) after infection of M. smegmatis. The numbers of reads are shown on the y-axes and genome maps are shown below.
All of the Cluster N phages are temperate, forming turbid plaques from which stable lysogens can be recovered, and they are homoimmune with closely-related repressors (>80% amino acid identity) (Supplementary Figure 12a); a M. smegmatis strain expressing the repressor of phage Charlie reproduces the Cluster N immunity profile (Supplementary Figure 12). Lysogenic strains are immune to superinfection, release phage particles into culture supernatants, and contain integrated prophages (Supplementary Figure 12c). Superinfection immunity is tight, and escape from immunity is either not observed or at only very low levels (Supplementary Figure 12c).
In MichelleMyBell (MMB) lytic growth, early transcription (30 mins post-infection) starts at the repressor-regulated promoter PR and proceeds through genes 39 to 65 (although expression of 64 and 65 is quite low), with late gene expression (150 mins post-infection) starting between genes 65 and 66 and proceeding rightwards through the entire left arm and the lysis genes that follow (Fig. 2b). The transcripts at 150 mins stop at or near a predicted terminator between genes 28 and 29 (Supplementary Figure 11), and reinitiate for expression of genes 30 and 31 with transcription continuing at lower levels across the leftwards-transcribed downstream genes (31–35). The repressor, integrase, and downstream leftwards-transcribed genes are expressed at low levels at both time points, and the conditions used may support only low levels of lysogenic establishment. Charlie displays a closely-related pattern of lytic gene expression (Fig. 2b).
Lysogenic gene expression
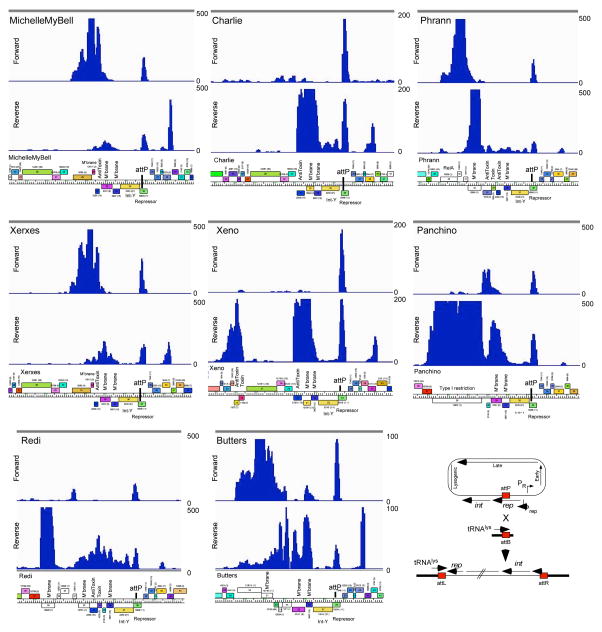
We examined gene expression profiles of eight Cluster N lysogens (Fig. 3; Supplementary Figures 13–15) that reflect the diversity in the lysis-immunity region (Fig. 4a). In each lysogen we see repressor expression as expected, as well as an RNA antisense to the repressor that arises from the chromosomal tRNA gene flanking the attachment site (Fig. 3). However, in all eight lysogens we also see expression of several genes adjacent to integrase, primarily between the lysis and integrase genes; a notable exception is the Xeno TA gene pair to the left of the lysis genes (Fig. 3). Expression levels vary greatly, but some are 10-fold higher than repressor transcription; pairs of closely related genomes (e.g. MMB/Xerxes, Charlie/Xeno) show similar patterns. There is considerable sequence variation in these genomic regions and many of the predicted genes are of unknown function. However, we note that Panchino 28 encodes a single-subunit restriction-modification (RM) system with sequence similarity to the HsdR, HsdM and HsdS domains of Type I RM systems; Xeno, Redi, and Phrann code for TA systems, and many have stand-alone antitoxin genes (e.g. MMB 31). Phrann 29 encodes a putative RelA-related (p)ppGpp synthetase that is lysogenically expressed (Fig. 3, 4a). Several of the functionally ill-defined genes are predicted to be membrane localized (e.g. MMB gp30; Figs. 3, 4a).
Figure 3. Transcription in Cluster N lysogens of M. smegmatis.
The central portions of eight Cluster N genomes are shown with their RNAseq profiles from lysogenic cultures. The numbers of reads are shown on the y-axes, and genome maps are shown below. Note that for simplicity the sequence reads are aligned to the viral rather than the prophage representation of the genomes, such that bacterial DNA sits adjacent to the left and right of attP. The cartoon at bottom right illustrates the integration system. RNAseq profiles of the entire genomes are shown in Supplementary Figures 13–15.
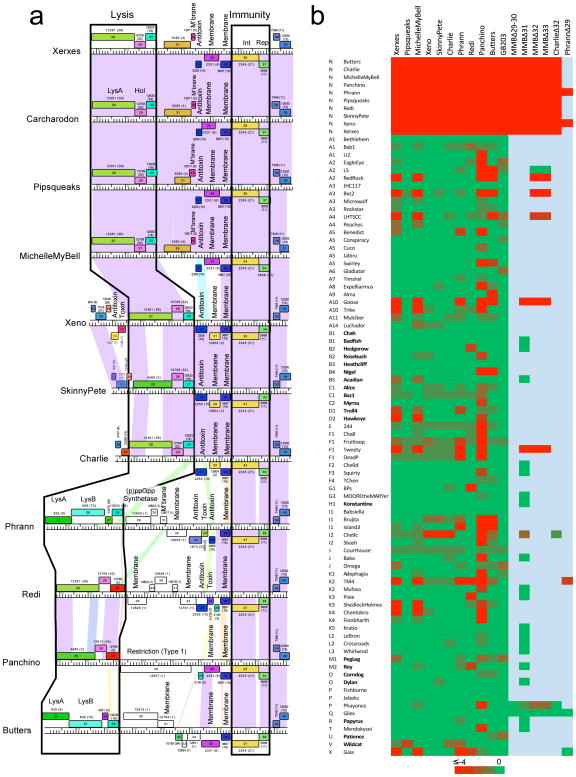
Figure 4. Cluster N prophage-mediated defense against phage infection.
(a) The central parts of 11 Cluster N mycobacteriophage genomes are aligned by their immunity cassettes and putative gene functions are indicated. Genomes are displayed as described for Figure 2, but ordered such that genomes with similarities in these regions are adjacent to each other, particularly Xerxes, Carcharodon, Pipsqueaks, and MMB as one group, and Xeno, SkinnyPete and Charlie as a second group. Note that in Pipsqueaks the open reading frame corresponding to MMB 29 is interrupted by a 25 bp deletion, leaving only the 3’ end of the gene intact and presumably inactivating it. (b) Heat map of prophage-mediated viral sensitivities, reporting efficiencies of plating of mycobacteriophages on Cluster N lysogenic strains or their deletion derivatives. Individual genome names and cluster designations are shown on the left. Phages that do not have an integrase gene and are presumably lytic are shown in bold type; all others are temperate or derivatives of temperate phages. Bright green and bright red corresponds to efficiencies of platings of 1 and less than 10−4, respectively. All efficiencies of plating were determined in at least two separate experiments. Efficiencies of platings are reported in Supplementary Table 2.
Cluster N prophage-mediated defense against viral attack
Prophage expression of the Panchino gp28 restriction system is anticipated to defend against viral attack19,31, raising the question whether other prophage-expressed genes are involved in heterotypic viral defense. Taking advantage of the large and diverse collection of sequenced mycobacteriophages, we determined efficiencies of plating (e.o.p.) of 80 phages on ten of the Cluster N lysogens; these include lytic and temperate phages and span the genomic diversity of the collection (Fig. 4b, Supplementary Table 2). We observed a remarkable pattern with over 70 instances where the e.o.p. is reduced by at least four orders of magnitude (Fig. 4b, Supplementary Table 2). This is not repressor-mediated as these phages plate efficiently on a repressor-expressing strain (GB203; Fig. 4b, Supplementary Table 2). The extent of viral defense differs among individual lysogens, with Pipsqueaks protecting against few if any phages (Wildcat is reduced by 10−3; Fig. 4b) and Panchino defending against over one-third of the phages (Fig. 4b). Strikingly, there is no clear correlation between a defense target and genome type; for example, U2 is subject to defense by Panchino, but the related Subcluster A1 phages Bethlehem and Bxb1 are not (Fig. 4b, Supplementary Table 2). Similarly, Tweety (F1) is targeted by Xerxes, MMB, Phrann and Panchino, but Che8 (and to a lesser extent Fruitloop), is targeted by Panchino alone (Fig. 4b). Defense can thus be highly specific, but not readily predicted from genomic information alone. It is likely though that these patterns are determined by a combination of prophage-expressed genes that confer defense against infection, and phage-encoded anti-defense systems.
Genes responsible for prophage-mediated viral defense
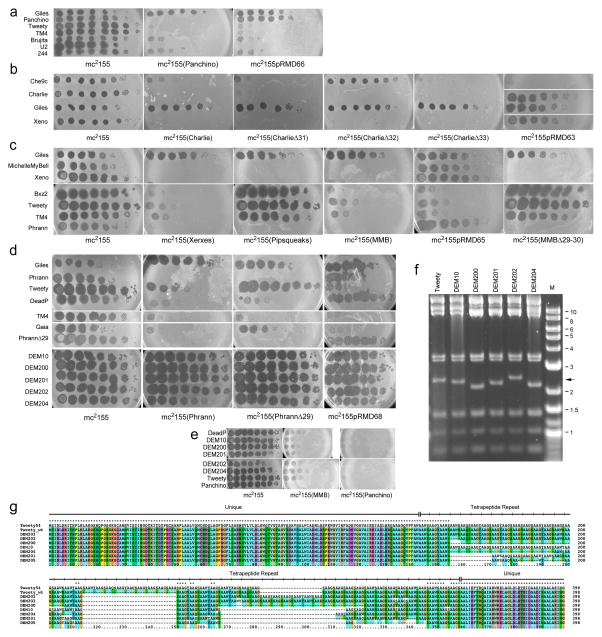
The Panchino prophage defends against 29 individual phages about one-half of which plate at normal efficiencies on other lysogens (Fig. 4b, 5a). Homologues of three Panchino genes (29, 30, and 31; Fig. 4a) are present in Butters and Redi, but Panchino defends against 21 phages that Butters and/or Redi do not. The Panchino restriction system (gp28) is a strong candidate for conferring this defense (Fig. 5a, Supplementary Table 2), and we confirmed this by showing that a M. smegmatis strain (mc2155pRMD66) expressing Panchino 28 alone has a defense profile similar to the Panchino lysogen, but does not prevent homotypic infection by itself or other Cluster N phages (Fig. 5a).
Figure 5. Genetics of Cluster N prophage-mediated defenses.
(a–e) Ten-fold serial dilutions of phage lysates were spotted onto lawns of M. smegmatis mc2155, lysogens and lysogens of mutant phage derivatives, as indicated beneath each panel. Phage names are shown to the left, including mutants (e.g. PhrannΔ29) and Defense Escape Mutant (DEM) derivatives of Tweety that overcome Phrann-mediated defense. All platings were performed in at least two separate experiments. (f) Bam HI restriction digestion of DEM DNAs shows changes in a 2.4 kbp fragment containing gene 54 (arrow). Marker (M) fragments are shown in kbp. (g) ClustalX alignment of gp54 of Tweety and DEM mutants. The wild type Tweety genome was re-sequenced and 54 contains 39 repeat units, rather than 48 in the original sequence 32; repeat consensus: AAXX, where XX is either GS, GY, WS, WY, QS, or QY) flanked by unique sequences.
Xeno, SkinnyPete and Charlie are closely-related in the lysis-immunity region and all three confer defense (e.o.p. <10−4) against a single phage, Che9c (Fig. 4b, 5b, Supplementary Table 2). The gene responsible is Charlie 32 (and presumably the analogous genes in Xeno and SkinnyPete), and a Charlie lysogen in which 32 is deleted loses defense against Che9c (Fig. 5b); deletion of Charlie 31 or 33 does not change the defense profile (Fig. 5b). Furthermore, a M. smegmatis strain expressing genes 31-33 (mc2155pRMD63) confers Che9c defense, presumably from the action of gp32, although a similar plasmid with 32 deleted fails to transform M. smegmatis and thus could not be tested. The defense is heterotypic, and Charlie gp32 does not prevent infection by Charlie and other Cluster N phages (Fig. 5b). Charlie gp32 is a 121-residue putative membrane protein containing a single transmembrane domain near the N-terminus; there are homologues in various actinobacterial strains but the functions are unknown. We propose that Charlie gp32 confers heterotypic exclusion, blocking Che9c DNA injection across the membrane. The narrow specificity of the defense against a single phage of those we tested is notable.
Phages MMB, Xerxes, and Pipsqueaks are closely-related in the lysis-immunity region (Fig. 4a) and MMB and Xerxes have almost identical patterns of defense against eleven individual phages (e.o.p <10−4; Fig. 4b, Supplementary Table 2); Pipsqueaks lacks this defense (Fig. 4). These patterns suggest that MMB gp29 (and Xerxes gp31) are involved, as their Pipsqueaks homologue is interrupted by a 25 bp deletion. To address this, we constructed a MMB mutant in which genes 29 and 30 are removed (deletions of individual genes appear to generate non-viable mutants; Supplementary Table 3) and showed that prophage-mediated defense is lost (Fig. 5c). In addition, a M. smegmatis strain (mc2155pRMD65) expressing MMB 29 and 30 confers the same pattern of defense as the MMB lysogen (Fig. 5c). A similar plasmid carrying only MMB 29 fails to transform M. smegmatis, and expression of MMB gp29 alone is toxic to M. smegmatis (Supplementary Figure 17); MMB gp30 – which is predicted to be membrane localized – alleviates this toxicity.
The Phrann prophage confers heterotypic defense against six phages from five clusters/subclusters (Fig. 4b, 5d, Supplementary Table 2). Phrann genes 29 and/or 30 are involved in defense against Tweety and Gaia, as a mutant lysogen lacking Phrann 29 loses defense against these phages (Fig. 5d); we have not yet identified the genes involved in defense against other phages (Fig. 5d). M. smegmatis strains (mc2155pRMD68) expressing Phrann genes 29 and 30 reproduce the Phrann defense against Tweety and Gaia (Fig. 5d). They also defend against TM4 indicating that Phrann encodes two defense systems targeting TM4, as both the Phrann 29-30 recombinant strains and the mc2155(PhrannΔ29) lysogen defend against TM4 infection (Fig. 5d). We were not able to test a similar M. smegmatis strain expressing only Phrann 29, as the plasmid fails to transform M. smegmatis, suggesting that – like MMB gp29 –Phrann gp29 expression alone is toxic to M. smegmatis, and that Phrann gp30 alleviates this toxicity. Phrann prophage-mediated defense is strictly heterotypic, and Phrann gp29/gp30 does not defend against Phrann or other Cluster N phages (Fig. 5d).
Outcomes of defense
The phage infection assays (Figs. 4b, 5) show that the prophage-expressed genes in the Cluster N lysogens reduce lytic growth of the infecting phages, but this could result from cellular survival as with CRISPR-cas or RM systems, or from suicidal-like abortive infection13. We thus tested the bacterial outcomes of infection by plating Cluster N lysogenic strains (or recombinant M. smegmatis strains expressing individual genes) onto phage-seeded solid media and recovering the bacterial survivors (Table 1). We anticipated that lytic and temperate attacking phages may behave differently, since temperate phages have the potential to establish lysogeny, whereas lytic phages do not.
Table 1.
Bacterial survival and lysogeny following phage infection
|
|
||||
|---|---|---|---|---|
| Tweety | Che9c | |||
|
| ||||
| Strains | Genes | Wild type1 | Δ451 | |
| mc2155 | 75% (15/15) | 0 | 50%(16/16) | |
| mc2155(Phrann) | 75% (67/82) | 100% (5/8) | n/a | |
| mc2155pRMD68 | Phrann 29-30 | 63% (8/8) | 33% (5/8) | n/a |
| mc2155(MMB) | 75% (71/72) | 50% (8/8) | n/a | |
| mc2155pRMD64 | MMB 29-30 | 100% (5/8) | 70%(8/8) | n/a |
| mc2155(Panchino) | 75% (5/65) | 75% (2/8) | n/a | |
| mc2155pRMD66 | Panchino 28 | 75% (0/5) | 75% (4/8) | n/a |
| mc2155(Charlie) | n/a | n/a | 100% (1/38) | |
| mc2155pRMD63 | Charlie 31-33 | n/a | n/a | 89%(1/8) |
Strains were plated onto solid media seeded with Tweety (wild type or a lytic derivative, Δ45) or Che9c, and the proportions of colonies recovered relative to non-seeded plates are shown.
The numbers of colonies showing phage release (an indicator of lysogeny) of the numbers tested are shown in parentheses.
M. smegmatis strains expressing Panchino gp28 efficiently survive infection by temperate phage Tweety, but the survivors are not lysogenic, consistent with the action of a RM system (Table 1). Likewise, Charlie gp32 confers survival to Che9c infection, which also is a temperate phage, but the survivors again are not lysogenic, consistent with the prediction that Charlie gp32 is a membrane protein that blocks Che9c DNA injection through heterotypic exclusion (Table 1). In contrast, MMB and Phrann lysogens and strains expressing either MMB 29-30 or Phrann 29-30 efficiently survive infection by temperate phage Tweety, and the majority of survivors are lysogenic for Tweety (Table 1). Because lysogeny can be efficiently established and the lytic-lysogenic decision is made post-infection, the MMB 29-30 and Phrann 29-30 defense systems do not block DNA injection, and presumably are triggered by lytic growth specifically. However, lysogeny is not required for survival of Tweety infection, and MMB 29-30 and Phrann 29-30 also confer survival to infection by a lytic Tweety derivative in which the immunity repressor has been deleted (TweetyΔ45; Table 1). Interestingly, the colonies recovered on TweetyΔ45-seeded plates show some phage-release suggesting that the defense system may not be fully active in all cells within the colony. However, phage release is not observed after subsequent rounds of colony purification. Defense by MMB 29-30 and Phrann 29-30 is thus not bactericidal like some abortive infection systems13, and survival is relatively efficient, as reported for type III TA systems12.
Target specificity and counter-defense measures
To explore target identity, we isolated and characterized Tweety Defense Escape Mutants (DEMs; ‘200” series) that are no longer subject to Phrann-mediated defense. The mutants plate efficiently on a Phrann lysogen (Fig. 5d) but remain subject to defense by Xerxes, MichelleMyBell and Panchino (Fig. 5d, 5e). All of the mutations map to Tweety gene 54 (see Supplementary Information) that encodes an unusual protein containing a large number of tetrapeptide repeats32. The mutants all have more (+5, +6) or fewer (−6, −11, −16, −21, −22; Figs. 5f, 5g, Supplementary Figure 21) copies of the repeat, relative to the wild type Tweety parent used in these experiments (which has 39 copies); these appear to follow a pattern in which variation in multiples of 5–6 repeats are associated with the DEM phenotype (Supplementary Figure 21). Tetrapeptide repeat proteins are uncommon, but include Plasmodium circumsporozoite protein33 and Human homeodomain protein HPRX134, which have 40–60 copies of tetrapeptide repeats (NAAG and PIPG, respectively); Gaia does not code for a related protein.
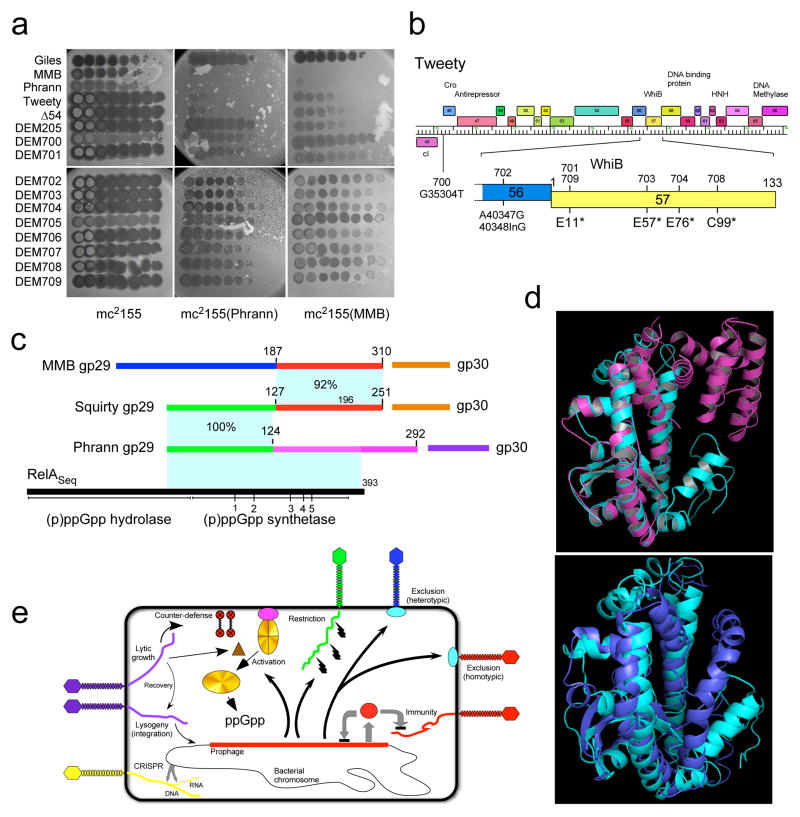
These Tweety 200-series DEM mutants presumably escape Phrann prophage-mediated defense either due to inactivation of gene 54 or are gain-of-function mutants that have acquired counter-defense ability. To test this, we constructed a 54 deletion derivative of Tweety (Tweety Δ54) which is viable (indicating that 54 is not essential for lytic growth) but is targeted by Phrann-mediated defense (Fig. 6a). This suggests that the DEM 200-series derivatives are gain-of-function mutants. We propose that Tweety 54 is a tunable counter-defense system in which the numbers of repeats can be altered – presumably by recombination or replication errors – followed by selection for specific variants that are active against a particular defense system. Thus wild-type Tweety gene 54 is not active in counter defense against Phrann (or MMB), and can be deleted without affecting targeting of Tweety by the Phrann-mediated defense system. But the 54 variants in the DEM derivatives gain the ability to specifically counter the Phrann system, and we predict that similar yet distinct variants can be isolated that have specificity for the MMB system.
Figure 6. Mechanisms of prophage-mediated defense against viral attack.
(a) Ten-fold serial dilutions of Defense Escape Mutant (DEM700-DEM709) derivatives of a TweetyΔ54 parent that overcome Phrann defense were spotted on wild type M. smegmatis (mc2155) or Phrann and MMB lysogens. All platings were performed in at least two separate experiments. (b) Tweety Δ54 DEM mutants map in early lytic genes. Genome sequencing shows DEM 700-series mutants map in Tweety early lytic genes. DEM706 and DEM707 are siblings of DEM701 and DEM702, respectively (see text and Supplementary Information for details; Supplementary Figure 22). (c–e) Relationships among phage-encoded (p)ppGpp synthetase-like proteins. (c) Phrann gp29 is a homologue of RelA/SpoT proteins with similarity to the (p)ppGpp synthetase domain of Streptococcus equimilis RelA including 5 conserved motifs (1–5)37. The Cluster F phage Squirty encodes a related protein (gp29) sharing the N-terminal 124 residues with Phrann gp29, but with divergent C-termini. MMB gp29 is not predicted by HHpred to be related to RelSeq, but shares its C-terminus with Squirty gp29. MMB gp30 and Squirty gp30 are closely related and are both predicted to be membrane localized. (d) I-TASSER38 alignment of Phrann gp29 (top, cyan) to Streptococcus equimilis RelA (PDB: 1vj7A, magenta; TM-score 0.539) and to a PHYRE239 structural prediction of Phrann gp29 (bottom, cyan). (e) Models for prophage-mediated viral defense. An integrated prophage (red line) confers defense against viral attack through numerous mechanisms, either homotypically (i.e. against the same or closely viruses) or heterotypically (against unrelated phages). Homotypic defense includes repressor-mediated immunity (repressor, red circle) and superinfection exclusion (blue circle) against itself (red phage). Heterotypic defense includes an exclusion-like system illustrated by Charlie gp32 defense against Che9c (blue phage), and restriction against many viruses (illustrated by the green phage) by Panchino gp28. Defense is also mediated by a predicted (p)ppGpp synthetase (e.g. Phrann gp29; gold circle), which we propose is kept in an inactive form (gold circle with crossed lines) by an inhibitor (purple circle), which for Squirty gp30 is membrane located. Lytic growth by specific phages activates the defense through early lytic protein, which is proposed to dissociate the (p)ppGpp synthetase from its inhibitor, enabling rapid accumulation of (p)ppGpp and growth arrest. Tweety encodes a counter-defense system (gp54) that may prevent activation of (p)ppGpp synthesis.
Removal of the counter-defense provides an opportunity to explore how Phrann and MMB target Tweety to prevent lytic growth. We thus isolated and sequenced a second series of DEM derivatives (“700” series) of a Tweety Δ54 parent phage that are able to efficiently infect a Phrann lysogen (Fig. 6a); e.o.p’s on the Phrann lysogen range from 1 to 10−1 relative to wild-type M. smegmatis (Fig. 6a). All of these DEMs also escape MMB defense (Fig. 6a), and MMB gp29 and Phrann gp29 presumably target the same Tweety locus. Sequencing of these mutants showed that five of them (DEM701, 703, 704, 708, 709) have single base substitutions in gene 57, which codes for a putative WhiB-like transcriptional regulator, all introducing translation stop codons (Fig. 6b). One mutant (DEM702) has both a base substitution and a single base insertion in gene 56, and DEM700 has a single base change in the putative ribosome binding site (RBS) of gene 46 encoding a putative Cro-like protein. DEM700 has a notably more turbid plaque phenotype than the parent phage, consistent with reduced expression of gp46 (Fig. 6a), but how this relates to defense escape is unclear. It is unlikely that Tweety gp56 or gp57 are directly required for targeting as preliminary observations show that deletions of genes 56 and 57 do not result in efficient escape from defense. Thus, although early gene expression in the 46–58 interval of the Tweety genome are implicated in the specificity of Tweety targeting, the particular gene(s) responsible is not yet clear.
(p)ppGpp synthetase-mediated viral defense
Bioinformatic analyses strongly suggest that Phrann gp29 is a (p)ppGpp synthetase (Fig. 6c, d). HHpred35 shows gp29 is related to RelA/SpoT homologues (RSH’s) including RelA of Streptococcus equimilis RelA (RelSeq), and contains key conserved residues36,37 (Fig. 6c, Supplementary Figure 18). Unlike the large (~750 aa) bacterial RelA and SpoT proteins that include (p)ppGpp hydrolysis and synthetase domains as well as a large C-terminal domain, Phrann gp29 (292 aa) contains only the (p)ppGpp synthetase domain (Fig. 6c). I-TASSER38 structure prediction shows compelling similarity with the (p)ppGpp synthetase domain of RelSeq (Fig. 6d), including the (p)ppGpp binding site, PHYRE239 predicts a similar structure (Fig. 6d).
The finding that Tweety DEM 700-series mutants escape both Phrann 29-30 and MMB 29-30 defense suggest that these systems may function similarly, even though there is no detectable amino acid similarity between the two sets of genes. A clue to the relationship is provided by comparisons with the Subcluster F1 phage Squirty, which is the only mycobacteriophage other than Phrann to encode a putative (p)ppGpp synthetase (gp29, 251 aa; Fig. 6c). Phrann gp29 and Squirty gp29 are identical over their N-terminal 124 residues, but share little or no sequence similarity at their C-termini (Fig. 6c); Squirty gp29 aligns to RelSeq by HHpred and I-TASSER (Supplementary Figures 19, 20). Although MMB gp29 has no similarity to Phrann gp29 or RSHs, the C-terminal 123 residues of MMB gp29 have 92% aa similarity with Squirty gp29 (Fig. 6c, Supplementary Figure 19), suggesting it may have a related although presumably distinct function.
As we noted above, both MMB gene 29 and Phrann gene 29 cannot be expressed individually in M. smegmatis without the genes immediately downstream (Phrann 30 and MMB 30, respectively) being also present. Although Phrann 30 and MMB 30 are not related, MMB 30 is a closely-related homologue of Squirty gene 30 (92% aa identity) and both encode putative membrane-localized proteins. The conservation of the C-termini of MMB gp29 and Squirty gp29, and of MMB gp30 and Squirty gp30, suggests the possibility that the proteins may interact directly.
Together, these observations suggest a model (Fig. 6e) in which Phrann 29 codes for a (p)ppGpp synthetase-like protein that when active promotes synthesis of the alarmone (p)ppGpp, leading to cessation of cell growth (as reported previously40), and concomitant interference with lytic phage growth. Because Phrann 29 is expressed in lysogenic cells (Fig. 3) when not under phage attack, Phrann gp29 activity must be strongly down-regulated. Because Phrann gp30 is required for M. smegmatis tolerance to Phrann gp29, we propose that Phrann gp29 and gp30 interact directly, maintaining gp29 in an inactive state. In this model, Tweety early lytic gene expression leads to dissociation of Phrann gp29 and gp30, activation of (p)ppGpp synthesis, and a halt to bacterial and phage growth (Fig. 6e). The Tweety gp54 counter-defense system could then act by binding to the gp29-gp30 complex and preventing dissociation. We propose that MMB gp29 and gp30 operate similarly, although the biochemical activity of MMB gp29 may be distinct from that of Phrann gp29, and the different configuration of the MMB gp29-gp30 complex may require different tuning of the gp54 counter-defense system than that required for Phrann counter-defense. The Squirty gp29-gp30 system has not been explored experimentally, but is likely to share similarities with these systems.
DISCUSSION
Viral attack of bacteria and bacterial responses to attack are dominant features of microbial evolution. It is therefore not surprising that a variety of mechanisms have evolved for bacterial defense against phage infection, as well as co-evolution of viral mechanisms for circumventing these defenses. However, the roles played by temperate phages and their prophages in conferring defense against other phages are poorly investigated. This is in part because heterotypic viral defense is not easy to distinguish from repressor-mediated superinfection immunity in the absence of genomic information. Furthermore, as illustrated here, heterotypic defense can be highly specific, and could easily escape detection without having a suitable collection of characterized phages to test for infection. Patterns of defense are also influenced by phage-encoded counter-defense systems, such as Tweety gp54. Nonetheless, it seems likely that prophage-mediated viral defense is a common and general phenomenon involving many different mechanisms. The Cluster N phages alone have multiple distinct defense systems, and genomic analysis of temperate mycobacteriophages in Clusters A, E, F, K, I, L, P, and Y suggests these may also be rich in viral defense systems. A recent report demonstrated prophage-mediated defense in Pseudomonas phages41.
The variety of distinct systems encoded by a small group of related Cluster N mycobacteriophages is remarkable. The restriction system of Panchino is not unexpected, although there are relatively few examples of prophage-encoded restriction systems where the defense profile has been explored31. The Charlie gp32 defense system is notable in that although it has the characteristics of superinfection exclusion systems – being membrane located and preventing DNA injection – it has remarkable specificity for a single phage among those tested, and does not show homotypic exclusion, unlike other phage exclusion systems20,21. Prophage-mediated defense by (p)ppGpp synthetases is a new and intriguing system for defense against viral attack. We propose that the putative (p)ppGpp synthetase associates with a regulator (e.g. Phrann gp30) holding it in an inactive form, and that lytic phage growth triggers dissociation, (p)ppGpp synthetase activation, and (p)ppGpp synthesis, rapidly and without requirement for de novo protein expression (Fig. 6e). Defense escape mutants show that Tweety early genes are involved in activation of both Phrann and MMB defenses, although the mechanism of action remains to be determined. Notably, defense does not lead to cell death as in some other abi systems13, and we propose that phage attack induces a persistence-like state in which (p)ppGpp accumulation shuts down cellular growth such that phage lytic development is arrested.
As with phage-encoded anti-restriction and anti-CRISPR systems, it should be no surprise that there are phage-encoded anti-prophage-defense systems. The Tweety gp54 counter-defense system appears tunable, and was likely only identified because the parental form does not counter Phrann prophage-mediated defense. Variants with differing repeat numbers have a gain-of-function phenotype highly active in counter-defense. We note that these variants are not active against MMB prophage-mediated defense, presumably because an alternatively tuned series of variants is required for MMB specificity.
Because prophages are common in bacterial genomes we predict that both prophage-mediated defense systems and phage-encoded counter-defense systems are common and play major roles in bacterial-viral dynamics.
METHODS
Phage isolation, propagation, and analysis
M. smegmatis mc2155 was used for phage isolation and growth as described previously3. Phage lysates typically contained greater than 5x109 pfu/ml and were used for plaque assays and DNA extraction. Electron microscopy42, genome sequencing, annotation, and analysis were as reported previously22. In brief, genomes were annotated using DNAMaster (http://cobamide2.bio.pitt.edu) beginning with an autoannotation with Glimmer and GenMark, followed by manual inspection and revision where necessary for each predicted open reading frame. Gene functions were predicted using BLAST43, HHPred35, and searches against the protein conserved domain database using Phamerator44. Phamerator44 was used for comparative genomic analyses and genome map representations, using the database ‘Actinobacteriophage_554’. Electron microscopy was performed on phage samples precipitated from lysates and resuspended in phage buffer (10 mM Tris pH 7.5, 68 mM NaCl, 10 mM MgSO4). Phage particles were spotted onto formvar and carbon coated 400 mesh copper grids, rinsed with distilled water and stained with 1% uranyl acetate. Images were taken using a FEI Morgagni transmission electron microscope. Phage dilutions for microbiological assays were made using phage buffer including 1 mM CaCl2.
Construction and characterization of lysogenic strains
Lysogens were isolated by spotting a dilution series of each high-titer Cluster N phage lysate on a lawn of mc2155, and incubating at 37° C for 24 hours. Cells from the cloudy centers of phage spots were streaked onto solid media, and incubated at 37° C until visible colonies appeared (approximately 3–4 days). Individual colonies were picked and re-purified by streaking onto solid media three times (to remove all exogenous phage from the initial infection) prior to inoculation into liquid Middlebrook 7H9 medium. After growing to saturation, cultures were verified for lysogeny by testing the supernatant for infectious phage particles indicating phage release, and by determining superinfection immunity by plating as a lawn and spotting with appropriate phage lysates. While propagating lysogens of the Cluster N phages we observed no evidence for loss of lysogeny, and all lysogens appear to be stably maintained. We tested a Xerxes lysogen by growing in liquid culture for 28 generations, plating for single colonies, and then testing these for phage release as a characteristic feature of lysogeny. The experiment was repeated in duplicate and all of ten colonies from each experiment were lysogenic.
The 39 bp attP common core is identical in all Cluster N phages except for Butters and Redi that have two and four base departures from them respectively, although only the Redi sequence is identical to the attB site overlapping a M. smegmatis tRNAlys gene (Msmeg_5758). All of the Cluster N prophages were shown by PCR to integrate into this attB site (see Supplementary Figure 12d), but we also examined RNAseq data from the lysogenic strains for evidence of secondary integration. Sufficient reads were available for the Butters, MMB, Phrann and Xerxes samples that mapped to the predicted attachment junctions (50, 115, 71, and 19, respectively) to show that integration occurred only at this site, and no integration events were identified that mapped to alternative attB loci.
Phage Genome Sequencing
Phage lysates were prepared and DNA extracted using the Wizard DNA kit (Promega). Libraries were constructed from phage DNA using the Illumina TruSeq Nano kit or the NEB Ultra II kit, then sequenced using an Illumina MiSeq – with either unpaired 140 bp reads or with paired-end 300 bp reads – to a minimum of 200-fold depth of coverage. Assembly was performed using Newbler version 2.9, and Consed version 29 was used for quality control of assembled genomes. In some cases, tandem repeats were manually assembled to ensure accurate complete sequences.
Isolation and characterization of Defense Escape Mutants
Tweety DEM mutants were isolated by plating four independent lysates onto lawns of a M. smegmatis mc2155(Phrann) lysogen, picking plaques from each plate and recovering on M. smegmatis mc2155. Individual plaques were then tested for the escape phenotype (i.e. efficiency of plating of one on the Phrann lysogen relative to M. smegmatis mc2155). A total of seven mutants (DEM10, DEM200, DEM201, DEM202, DEM203, DEM204, and DEM205) were isolated, of which two groups (DEM200 and DEM205; DEM202 and DEM203) were recovered from the same initial lysate, and could be siblings (see Supplementary Figure 22). We also prepared DNA from two wild-type Tweety lysates, both of which are targeted by the Phrann prophage; the wild type Tweety lysates used were derived from a stock prepared in March 2013. Complete genomes were sequenced using Illumina Mi-Seq with unpaired 140 bp read, or with paired-end 300 bp reads, and assembled using Newbler and Consed. (Supplementary Figures 21, 22). All of the phages (including both wild type Tweety lysates) contain a three base deletion corresponding to coordinates 34188–34190 in the reported Tweety sequence (GenBank accession number EF536069)32 at a sequence of five repeats of 5’-GAC; this same 3 bp deletion is present in more than 30 homologues including those in closely related phages such as Sisi, Mantra, Mumulus, and DotProduct. This therefore is unlikely to be involved in the escape phenotype, and is most likely accounted for by an error in the original Tweety sequence. Four of the mutants (DEM200, DEM201, DEM202, and DEM203) have a 1.3 kbp deletion that likely results from recombination within two directly repeated 115 bp motifs at 46219 – 46333 and 44879 – 44992; DEM10, DEM204, and DEM205 do not have this deletion and this also does not correlate with the escape phenotype (Supplementary Figures 21, 22).
The two newly sequenced wild type Tweety isolates are identical to each other, but differ from the originally reported sequence in having a 108 bp deletion within a repeated region of Tweety gene 54. The repeat is evident as a 12 bp nucleotide motif (or four amino acid repeat) that is present in the original sequence that spans ~576 bp, or 48 copies of the repeat. The newly sequenced wild-type Tweety isolates have nine fewer copies of the repeat (Supplementary Figure 21). However, this repeat presents challenges in assembly of 140 bp Illumina raw sequenced reads, and manual inspection of the minor differences between the repeats was needed to confirm correct alignment. Assembly of the two independent Tweety ‘wild type’ isolates into identical sequences adds confidence that these are correctly assembled. In addition, one of the wild type samples was sequenced using an Illumina long read (300 bp) paired-end library, which facilitated assembly through the repeated sequence and confirmed the consensus sequence derived from the 140 bp reads.
All of the DEM mutants have changes in the numbers of tetrapeptide repeats. Five of the mutants (DEM10, DEM200, DEM201, DEM204 and DEM205) have deletions that shorten gene 54 relative to the two newly sequenced wild-type isolates, corresponding to 72 bp, 252 bp, 132 bp, 192 bp, and 264 bp deletions respectively (Supplementary Figures 21, 22). Mutants DEM202 and DEM203 have an expansion of the tetrapeptide repeat and could not be confidently assembled using 140 bp Illumina reads and these were re-sequenced with 300 bp paired end reads, and shown to have insertions of 60 and 72 bp in gene 54 relative to the newly sequence wild type genomes (Fig. 5g, Supplementary Figure 21).
A second series of DEMs was isolated similarly using TweetyΔ54. Four independent cultures were used for mutant isolation, and a total of 10 isolates were purified and characterized (DEM700 – DEM709; Supplementary Figure 22). All of these DEMs infect mc2155(Phrann) more efficiently than wild type M. smegmatis, and similarly infect mc2155pRMD68 expressing Phrann 29-30; e.o.p.’s vary between 1 and 10−1. All ten isolates were fully sequenced and DEM701, 709, 703, 704 and 708 have single base changes within gene 57 that introduce translation termination codons. DEM701 and 709 were isolated independently but have the same mutation (Supplementary Figure 22). DEM706 is a sibling of DEM701 derived from the same phage lysate and are identical in sequence (Supplementary Figure 22). DEM702 and its sibling DEM707 have a base substitution in gene 56 (A40347G) and an adjacent single base insertion (40348InG) introducing a frameshift. Although we cannot rule out a role of gp56 in the defense system, it is likely that the frameshift mutation is polar and reduces expression of gp57. DEM700 and its sibling DEM705 have a single base change G35304T within the 5’ end of gene 46, which is predicted to act similarly to the Lambda Cro protein. However, the currently assigned translation start site at 35251 is probably incorrect and we predict that the start site used is more likely to be at 35314, based on comparative genomic analysis. The transcription start site for Tweety early rightwards transcription is not known, but the DEM700 base change could either alter the ribosome binding site of 46, or alternatively could affect the promoter of a leaderless transcript.
Construction of phage mutants and recombinant strains
Phage mutants were constructed with a modification of previously described phage recombineering45. A ~400 bp gBlock (Integrated DNA Technologies) was used as a DNA substrate containing the mutant allele (Supplementary Table 5). Approximately 179 bp upstream of the gene to be deleted plus 21 bp of the 5’ end of the gene was merged with 21 bp at the 3’ end of the gene plus 179 bp downstream to design the gBlock. The gBlock was then amplified by PCR with Q5 High-Fidelity DNA Polymerase (New England BioLabs) using primers that annealed to either end. The PCR reaction was verified on an agarose gel, cleaned using a PCR clean-up kit (Macherey-Nagel), and the concentration was determined using a Nanodrop. The pure substrates (~400 ng) were co-electroporated with 150 ng phage genomic DNA, prepared using a Wizard genomic DNA purification kit (Promega), into recombineering-proficient M. smegmatis cells. Individual plaques were screened by PCR for the presence of the mutant allele, mixed wild-type/mutant plaques re-plated, and secondary plaques tested by PCR. For several mutant constructions we were able to identify mixed primary plaques confirming the recombination occurred, but could not identify homogenous mutant secondary plaques, nor the mutant PCR product in a lysate of thousands of secondary plaques, indicating that the mutant is not viable. Coordinates of deletions and construction details are provided in Supplementary Table 3, and oligonucleotide and gBlock substrates are shown in Supplementary Table 5.
Recombinant plasmids (Supplementary Table 4) were constructed in the integration-proficient vector pMH9446 using Gibson assembly (New England Biolabs) and PCR products. Oligonucleotides used to generate PCR products are shown in Supplementary Table 5. All plasmids constructed were verified using restriction digest and sequencing (Genewiz).
Outcomes of infection and lysogeny
Outcomes of infection were determined by plating either lysogenic strains or recombinant plasmid containing strains onto solid media seeded with 107 pfu of phage, and plating of the same cultures on plates seeded with 108 pfu phage particles gave similar survival numbers. To prepare phage-seeded plates, phage lysates were diluted to 108 pfu/ml in phage buffer (65 mM NaCl, 1 mM Tris, pH 7.5, 1 mM MgCl2), and 100 μl was spread onto a Middlebrook 7H10 agar plate and allowed to dry. Bacterial cultures were diluted 104, 105, and 106 -fold into phage buffer, and 100 μl of each dilution was spread onto the phage-seeded plates, or onto control plates without phage. Three replicate plates were created for the 10−5 dilution. At least two separate platings with independent cultures were performed for each lysogenic strain. Plates were incubated at 37° C.
Colonies growing on phage-seeded plates were tested for phage release as follows. First, 250 μl of either mc2155, mc2155(Charlie), or mc2155(Phrann) saturated cultures were plated with top agar to generate lawns, and once set, individual colonies were speared with a pointed a toothpick such as to avoid touching the agar surface, and patched to the bacterial lawns. Plates were incubated at 37° C for 48 hours and then analyzed for clearings surrounding the patched colonies due to phage release. Lawns of mc2155(Charlie) and mc2155(Phrann) were used to differentiate between release of particles from the Cluster N lysogen being tested, and the attacking phage. At least five colonies of each strain were tested for phage release.
Plating of M. smegmatis mc2155 on Tweety or Che9c seeded plates resulted in 75% and 50% survival respectively, and all colonies tested released phage particles and are presumed to be lysogenic. However, on the Tweety-seeded plates the colonies of M. smegmatis were small compared to colonies plated and incubated for the same time on non-phage seeded plates. The lysogenic and recombinant cultures all exhibited normal sized colonies when plated on Tweety. However, on TweetyΔ45, approximately 50% of the colonies from the clones expressing MMB 29-30 or Phrann 29-30 were very small, while the rest were normal-sized. On the Che9c seeded plates, approximately 50% of the M. smegmatis colonies were small while the rest were normal-sized, and the lysogen and recombinant strain colonies were all normal sized. The basis for the difference in colony sizes is not known, although all colonies tested behaved the same in the phage release assay regardless of size. When M. smegmatis mc2155 was plated onto a derivative of Tweety in which the putative repressor is deleted (TweetyΔ45), no survivors were recovered after plating up to 105 colony forming units, as expected. Survivors of lysogenic or recombinant strains were recovered efficiently after plating on TweetyΔ45, and most released Tweety phage particles, presumably because at least a portion of the surviving cells in the colony were able to undergo productive phage infection. However, after three rounds of colony purification, no phage release was observed.
RNAseq
Total RNA was isolated from M. smegmatis lysogens in late logarithmic growth, or from phage infected cells (multiplicity of infection of three) 30 or 150 mins after infection. DNA removal and rRNA depletion was performed using a DNA-free kit (Ambion) and Ribo-Zero kit (Illumina) respectively. Libraries were prepared using a TruSeq Stranded RNAseq kit (Illumina), verified using a BioAnalyzer, and run on an Illumina MiSeq using multiplexed lanes., Quarter lanes were used for all lysogens, while half lanes were used for both MMB and Charlie 30 and 150 mins samples. The fastq reads were analyzed for overall quality using FastQC (Andrews, S. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), trimmed at the 5’ and 3’ ends with cutadapt (http://code.google.com/p/cutadapt/) using a quality score threshold of 30, and then simultaneously mapped to the genomes of M. smegmatis and the phage of interest with Bowtie247. SAMtools48 and BEDtools49 were used to process reads that aligned to exactly one locus (as computed by Bowtie2) and calculate strand-specific genome coverage. The Integrative Genomics Viewer50 was used to visualize and present the RNAseq coverage. RNAseq data sets, with additional method details, are deposited in the Gene Expression Omnibus (GEO) with accession number GSE82004.
All Cluster N phages have rightwards-facing early lytic PR promoters upstream of the rightwards operon, and this region is highly conserved in nine phages (all except Butters and Redi) with no more than a single base change in the 100 bp upstream of the rightwards-transcribed operon. An 18 bp region of dyad symmetry (5’-AATTTCTCtgtGAGAAATT) overlaps the putative -10 promoter motif and is present even in the more distantly related Butters and Redi genomes (with a single base difference in Redi) and is a candidate for the operator regulating PR activity.
I-TASSER and PHYRE2 structural predictions
Phrann gp29, Squirty gp29, and MMB gp29 were each submitted to the I-TASSER server at the University of Michigan (http://zhanglab.ccmb.med.umich.edu/I-TASSER/). Five models for Phrann gp29 were generated, with model 1 having a C-score of -2.60. Alignment with protein structures in the PDB identified the poorly characterized protein smu.1046c from Streptococcus mutans UA159 (PDB: 3l9da; TM-score = 0.539) and the well-characterized RelA (RelSeq) from S. equimilis (PDB: 1vj7a; TM-score = 0.535) as having the two highest scores. PHYRE239 also predicts a strong match with the same protein and scores as having 100% confidence. The I-TASSER- and PHYRE2-generated structures are closely superimposable across the region in which Phrann gp29 closely models with RelSeq as shown in Fig. 6 of the manuscript. Five models for Squirty gp29 were generated by I-TASSER with model 1 having a C-score of -3.29. Alignment with protein structures in the PDB identified closest similarity to RelSeq (PDB: 1vj7a; TM-score = 0.571). For Phrann gp29 and Squirty gp29 the residues 6, 78, 89, 133, 135, 140, 141,143, 147, 158, 160, 170, 173, 174, and 74, 76, 78, 79, 82, 86, 89, 100, 103, 104, 105, 156, 158, 211, respectively, are predicted to be associated with binding of a (p)ppGpp ligand. We note that Squirty residues E156 and E158 are in a beta sheet that is also present in RelSeq. MMB gp29 does show evidence structural similarity to known proteins using a combination of I-TASSER and PHYRE2.
Acknowledgments
We thank the many students in the SEA-PHAGES program that contributed to the isolation, annotation, and characterization of the phages described here, and specific contributions are noted at http://phagesdb.org. We thank Johnathon Schiebel, Audrey Jonas, Ty Stoner, Daryn Green, Rachael Rush, and Leon Lin for help with escape mutant isolation, Ching-Chung Ko for help with plasmid construction, and David Asai, Vic Sivanathan, Kevin Bradley, and Lucia Barker for support of the SEA-PHAGES program. This work was supported by grants from the National Institutes of Health (GM116884) and Howard Hughes Medical Institute (54308198) to GFH, and National Science Foundation pre-doctoral fellowship to TNM (1247842).
Footnotes
AUTHOR CONTRIBUTIONS
RMD, DJS, CAG, TNM, WHP, VCW, JW, GFH conceived and designed the experiments; RMD, DJS, CAG, RAG, WHP, JCCR, DAR, BRB, CFH, CMM, MTM JNT, performed the experiments; RMD, DJS, CAG, RAG, TNM, WHP, BRB, JCCR, DAR, TA, RA, JAB, JSB, DB, SGC, WBD, LAD, NPE, AMF, UG, JHG, CFH, LEH, KWH, SI, AAJ, MAK, KKK, CMM, SFM, SDM, MTM, JN, STP, MCP, MKP, CAR, CJR, MRR, JNT, EV, VCW, JW, GFH analyzed the data; SGC Contributed Material/analysis tools; RMD, DJS, TNM, WHP, DAR, TA, RA, JAB, JSB, DB, SGC, WBD, LAD, NPE, AMF, UG, JHG, LEH, KWH, SI, AAJ, MAK, KKK, CMM, SFM, SDM, JN, STP, MCP, MKP, CAR, CJR, MRR, EV, VCW, JW, GFH wrote the paper.
Data availability
Genbank accession numbers for the Cluster N mycobacteriophages used in this study are shown in Supplementary Table 1, and information of these and all other phages used in this study are available at http://phagesdb.org. The plating efficiencies of all phages tested and summarized in Fig. 4b are reported in Supplementary Table 1. A summary of the pedigrees of the Defense Escape Mutants (DEMs) is shown in Supplementary Figure 22. RNAseq data sets are deposited in the Gene Expression Omnibus (GEO) with accession number GSE82004. All data supporting the findings of this study are available from the corresponding author upon request.
References
- 1.Hambly E, Suttle CA. The viriosphere, diversity, and genetic exchange within phage communities. Curr Opin Microbiol. 2005;8:444–450. doi: 10.1016/j.mib.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Abedon ST. Phage evolution and ecology. Adv Appl Microbiol. 2009;67:1–45. doi: 10.1016/S0065-2164(08)01001-0. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs-Sera D, et al. On the nature of mycobacteriophage diversity and host preference. Virology. 2012;434:187–201. doi: 10.1016/j.virol.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendrix RW. Bacteriophages: evolution of the majority. Theor Popul Biol. 2002;61:471–480. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- 5.Suttle CA. Marine viruses--major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 6.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 7.Arber W, Dussoix D. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage lambda. J Mol Biol. 1962;5:18–36. doi: 10.1016/s0022-2836(62)80058-8. [DOI] [PubMed] [Google Scholar]
- 8.Dussoix D, Arber W. Host specificity of DNA produced by Escherichia coli. II. Control over acceptance of DNA from infecting phage lambda. J Mol Biol. 1962;5:37–49. doi: 10.1016/s0022-2836(62)80059-x. [DOI] [PubMed] [Google Scholar]
- 9.Tock MR, Dryden DT. The biology of restriction and anti-restriction. Curr Opin Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Swarts DC, et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldfarb T, et al. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 2015;34:169–183. doi: 10.15252/embj.201489455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fineran PC, et al. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci U S A. 2009;106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chopin MC, Chopin A, Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr Opin Microbiol. 2005;8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blower TR, Evans TJ, Przybilski R, Fineran PC, Salmond GP. Viral evasion of a bacterial suicide system by RNA-based molecular mimicry enables infectious altruism. PLoS Genet. 2012;8:e1003023. doi: 10.1371/journal.pgen.1003023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 17.Knowles B, et al. Lytic to temperate switching of viral communities. Nature. 2016;531:466–470. doi: 10.1038/nature17193. [DOI] [PubMed] [Google Scholar]
- 18.Fortier LC, Sekulovic O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence. 2013;4:354–365. doi: 10.4161/viru.24498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kita K, Kawakami H, Tanaka H. Evidence for horizontal transfer of the EcoT38I restriction-modification gene to chromosomal DNA by the P2 phage and diversity of defective P2 prophages in Escherichia coli TH38 strains. J Bacteriol. 2003;185:2296–2305. doi: 10.1128/JB.185.7.2296-2305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofer B, Ruge M, Dreiseikelmann B. The superinfection exclusion gene (sieA) of bacteriophage P22: identification and overexpression of the gene and localization of the gene product. J Bacteriol. 1995;177:3080–3086. doi: 10.1128/jb.177.11.3080-3086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cumby N, Edwards AM, Davidson AR, Maxwell KL. The bacteriophage HK97 gp15 moron element encodes a novel superinfection exclusion protein. J Bacteriol. 2012;194:5012–5019. doi: 10.1128/JB.00843-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pope WH, et al. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. Elife. 2015;4:e06416. doi: 10.7554/eLife.06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatfull GF, et al. Comparative Genomic Analysis of 60 Mycobacteriophage Genomes: Genome Clustering, Gene Acquisition, and Gene Size. J Mol Biol. 2010;397:119–143. doi: 10.1016/j.jmb.2010.01.011. S0022-2836(10)00026-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci U S A. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedulla ML, et al. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/s0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 26.Hatfull GF. The secret lives of mycobacteriophages. Adv Virus Res. 2012;82:179–288. doi: 10.1016/B978-0-12-394621-8.00015-7. B978-0-12-394621-8.00015-7 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Jordan TC, et al. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. MBio. 2014;5:e01051–01013. doi: 10.1128/mBio.01051-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatfull GF, et al. Complete genome sequences of 63 mycobacteriophages. Genome announcements. 2013;1 doi: 10.1128/genomeA.00847-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatfull GF, et al. Complete Genome Sequences of 61 Mycobacteriophages. Genome announcements. 2016;4 doi: 10.1128/genomeA.00389-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broussard GW, et al. Integration-dependent bacteriophage immunity provides insights into the evolution of genetic switches. Mol Cell. 2013;49:237–248. doi: 10.1016/j.molcel.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliveira PH, Touchon M, Rocha EP. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 2014;42:10618–10631. doi: 10.1093/nar/gku734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pham TT, Jacobs-Sera D, Pedulla ML, Hendrix RW, Hatfull GF. Comparative genomic analysis of mycobacteriophage Tweety: evolutionary insights and construction of compatible site-specific integration vectors for mycobacteria. Microbiology. 2007;153:2711–2723. doi: 10.1099/mic.0.2007/008904-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tahar R, Ringwald P, Basco LK. Heterogeneity in the circumsporozoite protein gene of Plasmodium malariae isolates from sub-Saharan Africa. Mol Biochem Parasitol. 1998;92:71–78. doi: 10.1016/s0166-6851(97)00226-0. [DOI] [PubMed] [Google Scholar]
- 34.Booth HA, Holland PW. Annotation, nomenclature and evolution of four novel homeobox genes expressed in the human germ line. Gene. 2007;387:7–14. doi: 10.1016/j.gene.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 35.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. 33/suppl_2/W244 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected] Cell. 2004;117:57–68. doi: 10.1016/s0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- 37.Steinchen W, Bange G. The magic dance of the alarmones (p)ppGpp. Mol Microbiol. 2016;101:531–544. doi: 10.1111/mmi.13412. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Zhang Y. Protein Structure and Function Prediction Using I-TASSER. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis ... [et al.] 2015;52:5 8 1–15. doi: 10.1002/0471250953.bi0508s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 41.Bondy-Denomy J, et al. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016 doi: 10.1038/ismej.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pope WH, et al. Cluster M Mycobacteriophages Bongo, PegLeg, and Rey with Unusually Large Repertoires of tRNA Isotypes. J Virol. 2014;88:2461–2480. doi: 10.1128/JVI.03363-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 44.Cresawn SG, et al. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics. 2011;12:395. doi: 10.1186/1471-2105-12-395. 1471-2105-12-395 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marinelli LJ, et al. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS ONE. 2008;3:e3957. doi: 10.1371/journal.pone.0003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee MH, Pascopella L, Jacobs WR, Jr, Hatfull GF. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci U S A. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krumsiek J, Arnold R, Rattei T. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23:1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- 52.Huson DH. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 53.Hatfull GF, et al. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet. 2006;2:e92. doi: 10.1371/journal.pgen.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]