Abstract
CYP3A4 is one of the major drug-metabolizing enzymes in human and is responsible for the metabolism of 60% of clinically used drugs. Many drugs are able to induce the expression of CYP3A4, which usually causes drug-drug interactions and adverse drug reactions. This study aims to explore the role of histone modifications in rifampicin-induced expression of CYP3A4 in LS174T cells. We found that the induction of CYP3A4 mRNA (4- to 15-fold) by rifampicin in LS174T cells was associated with increased levels of histone H3 lysine 4 trimethylation (H3K4me3, above 1.8-fold) and H3 acetylation (above 2-fold) and a decreased level of histone H3 lysine 27 trimethylation (H3K27me3, about 50%) in the CYP3A4 promoter. Rifampicin enhanced recruitment to the CYP3A4 promoter of nuclear receptor coactivator 6 (NCOA6, above 3-fold) and histone acetyltransferase p300 (p300, above 1.6-fold). Silencing NCOA6 or p300 by short-hairpin RNAs resulted in inhibition of the CYP3A4 induction as well as altered levels of H3K4me3, H3K27me3, or H3 acetylation in the CYP3A4 promoter. Knockdown of pregnane X receptor (PXR) expression not only suppressed the recruitment of NCOA6 and p300 but also abolished the changes caused by rifampicin in H3K4me3, H3K27me3, and H3 acetylation levels in the CYP3A4 promoter. Moreover, rifampicin treatment enhanced the nuclear accumulation and interactions between PXR and NCOA6/p300. In conclusion, we show that the alterations of histone modifications contribute to the PXR-mediated induction of CYP3A4 by rifampicin.
Introduction
Human CYP3A4 plays an important role in the metabolism of 60% of clinical drugs and is responsible for many drug-drug interactions owing to inducibility by numerous inducers (Liu et al., 2007; Raucy, 2003; Novotna and Dvorak, 2014). Drug-drug interactions caused by the induction of CYP3A4 have posed many challenges to drug development and clinical safety (Niemi et al., 2003; Cheng et al., 2009; Baciewicz et al., 2013). Over the past few years, the molecular mechanisms accountable for the regulation of CYP3A4 have been well studied. The basis of induction of CYP3A4 is thought to be activated pregnane X receptor (PXR) (Goodwin et al., 1999), constitutive androstane receptor (Moore et al., 2000), or vitamin D receptor (Thummel et al., 2001) binding to the CYP3A4 promoter (Schuetz, 2004). However, individual variability in CYP3A4 induction has been observed (Tang et al., 2005). Previously, genetic variations in CYP3A4, its related transporters, and transcriptional factor genes were thought to contribute to the variability (Tang et al., 2005; Lamba et al., 2006; Thirumaran et al., 2012). However, the genetic polymorphisms cannot sufficiently explain the large interindividual variability in the CYP3A4 induction owing to their limited frequencies in the general population (Rahmioglu et al., 2011).
Epigenetic modifications, such as histone methylation and acetylation, are important factors in chromatin remodeling and play essential roles in the regulation of gene transcription (Schübeler et al., 2004). The induction of genes by different stimulations, including xenobiotics and endogenous compounds, were found to be associated with histone modifications, such as histone methylation and acetylation (Xydous et al., 2012; Zhang et al., 2013; Xie et al., 2015). Likewise, drug metabolism enzymes and transporters are also regulated by different epigenetic modifications (Ivanov et al., 2014; Peng and Zhong, 2015). In HepG2 cells, histone H3 acetylation was showed to be associated with trichostatin A-mediated regulation of CYP2E1 (Yang et al., 2010). Transcriptional activation of CYP2C9 was found to be regulated by the epigenetic modification of histone H3 lysine 27 (H3K27) (Englert et al., 2015). Dynamic alterations of histone methylation are associated with ontogenic expression of the Cyp3a genes during mouse liver maturation (Li et al., 2009). In addition, MDR1 gene was proven to be regulated by histone methyltransferase mixed lineage leukemia 1 (MLL1) in chemoresistance (Huo et al., 2010). More interestingly, histone deacetylase 1 (HDAC1) was shown to be required in carbamazepine-induced CYP3A4 expression in HepG2 cells (Wu et al., 2012). These findings suggest that histone modifications may also be involved in the regulation of CYP3A4 and contribute to its individual variability as well as inducibility.
Epigenetic modifications are often regulated by nuclear receptors via recruitment of cofactors that are necessary for catalyzing distinct types of histone modifications (Green and Han, 2011). Nuclear receptor coactivator 6 (NCOA6, also named ASC-2, PRIP, AIB3, RAP250, NCR, and TRBP) is a transcriptional coactivator of multiple nuclear receptors and belongs to a novel, steady-state complex called ASCOM (for ASC-2 complex), which contains two different histone-modifying enzymes linked to transcriptional activation, histone H3 lysine 4 (H3K4), methyltransferase mixed lineage leukemia 3 (MLL3)/mixed lineage leukemia 4 (MLL4), and H3K27-demethylase lysine demethylase 6A (UTX) (Lee et al., 2009). Trimethylation of histone 3 lysine 4 (H3K4me3) is an active epigenetic mark for gene transcription, but trimethylation of histone 3 lysine 27 (H3K27me3) is a gene-silencing mark. It has been demonstrated that NCOA6 plays an essential role in ligand-dependent regulation of target gene expression by transcriptional factors through recruitment of MLL3/MLL4 to promote H3K4 methylation (Lee et al., 2008; Ananthanarayanan et al., 2011). The demethylation of H3K27me3 regulated by UTX has also been reported as a requirement for transcriptional activation (Zha et al., 2015). Moreover, PXR and hepatic nuclear factor 4α (HNF4α) were found to interact with NCOA6 in vitro (Surapureddi et al., 2011). Apart from that, histone acetylation catalyzed by p300, an essential coactivator recruited by transcriptional factors, is reported to be involved in gene induction (Zhang et al., 2013).
In the present study, we investigated whether NCOA6 and p300 as well as histone methylation and acetylation were involved in PXR-mediated induction of CYP3A4 by rifampicin in LS174T cells. LS174T is a human intestinal cell line derived from a 58-year-old female with adenocarcinoma of the colon and has been used in studies of the PXR-mediated induction of CYP3A4 by rifampicin (Liu et al., 2012; Tan et al., 2016). We found that NCOA6 and p300 were recruited to the CYP3A4 promoter by PXR after rifampicin treatment and were responsible for the changes of H3K4me3, H3K27me3, and H3 acetylation in the CYP3A4 promoter, further contributing to the induction of CYP3A4.
Materials and Methods
Reagents.
Rifampicin was purchased from Sigma-Aldrich (St. Louis, MO). Fetal bovine serum (FBS) was purchased from Biologic Industries (BI, Kibbutz Beit Haemek, Israel). Life Technologies Protein A and G magnetic beads were purchased from Thermo Fisher Scientific (Carlsbad, CA). Proteinase inhibitor cocktail was purchased from Roche (Basel, Switzerland). Penicillin, streptomycin, puromycin, ampicillin, dimethylsulfoxide (DMSO), and all other chemicals were purchased from Solarbio Company (Beijing, China), unless otherwise noted.
Plasmids.
PSG5-hPXR, a human PXR expression vector, was provided generously by Dr. Wen Xie (Center of Pharmacogenomics, University of Pittsburgh, Pittsburgh, PA). Human NCOA6 expression vector, pCMV2-hNCOA6:V5, was a gift from Kenneth Irvine (Addgene plasmid no. 63585) (Oh et al., 2014). Human p300 expression vector, pcDNA3.1-p300, was a gift from Warner Greene (Addgene plasmid no. 23252) (Chen et al., 2002). Short-hairpin RNA (shRNA) plasmids targeting NCOA6 (HSH005707-LVRU6P) or p300 (HSH004897-LVRU6P) were provided by GeneCopoeia (Rockville, MD), and shRNA plasmid targeting PXR (sc-44057-SH) was provided by Santa Cruz Biotechnology (Dallas, TX).
Cell Culture and Treatment.
Human colon cancer LS174T cells (obtained from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China) were maintained in Dulbecco’s modified Eagle’s medium (DMEM, GE Healthcare, Chicago, IL) supplemented with 10% FBS, 100 IU penicillin, and 100 IU streptomycin under 5% CO2 at 37°C. For induction study, LS174T cells were treated with rifampicin (10 μM) or solvent (DMSO, 0.1% v/v) for 48, 72, or 96 hours before mRNA and protein determination.
Transfection and RNA Interference.
All transfections were performed using Life Technologies Lipofectamine 3000 (Thermo Fisher Scientific), following the manufacturer’s instructions. To generate stable cell lines with continuous suppression of specific gene expression, LS174T cells were transiently transfected with a shRNA plasmid that targets NCOA6 (shNCOA6) or p300 (shp300), respectively. After 48 hours of transfection, a puromycin selection with a concentration of 2 μg/ml was performed for 4–6 weeks until small colonies occurred. Then, the colonies were tested for mRNA determination and positive colonies were selected to generate stable cell lines using puromycin selection. Knockdown of PXR expression was performed by transient transfection with a shRNA plasmid (shPXR). For overexpression studies, PSG5-PXR, pCMV2-hNCOA6:V5, and pcDNA3.1-p300 plasmids were transfected into LS174T cells, individually or together.
RNA Isolation and Gene Expression Analysis.
RNA was extracted using TriPure isolation reagent (Roche, Basel, Switzerland) and 1 μg of total RNA was reversely transcribed to cDNA using a PrimeScript RT Reagent Kit (Takara Bio, Dalian, China). Relative mRNA expression of genes was determined by real-time quantitative PCR (qPCR) and normalized to GAPDH mRNA using a Life Technologies SYBR green method (Thermo Fisher Scientific). Primers are shown in Supplemental Table 1.
Western Blot Analysis.
Total proteins of treated LS174T cells were prepared using RIPA buffer and protein concentrations were determined by a bicinchoninic acid method, according to the manufacturer’s instructions (Beyotime Institute of Biotechnology, Hangzhou, China). Total proteins were separated on SDS-PAGE gels (∼6 to 12%) and transferred to polyvinylidene fluoride membranes (Millipore Sigma, Billerica, MA). Membranes were incubated with a primary antibody: anti-CYP3A4 (Abcam, Cambridge, UK), anti-PXR (Abcam), anti-NCOA6 (Proteintech, Wuhan, China), anti-p300 (Abcam), or anti-GAPDH after blocked with 5% nonfat milk and were visualized with enhanced chemiluminescence followed by densitometry analysis of blot bands using FluorChem E system (Bio-Techne, Ltd., Abingdon, UK). The relative fold changes in CYP3A4, PXR, NCOA6, and p300 proteins were normalized to GAPDH.
Chromatin Immunoprecipitation Analysis.
Chromatin immunoprecipitation analysis (ChIP) was performed on treated cells to determine enriched levels of H3K4me3 (cat. no. 17-678; Millipore Sigma), H3K27me3 (cat. no. 17-622; Millipore Sigma), and H3 acetylation (cat. no. 06-599; Millipore Sigma) as well as binding levels of PXR, NCOA6, and p300 in the promoter region of CYP3A4 gene. After treatment, cells were crosslinked with 1% (v/v) formaldehyde for 10 minutes, then stopped by glycine (125 mM) for 5 minutes at room temperature. Cells were washed twice with cold phosphate-buffered saline, harvested, and lysed in SDS lysis buffer (0.1% SDS, 50 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 1% Triton-X) with a proteinase inhibitor cocktail on ice for 15 minutes. Cell lysates were sonicated with a Bioruptor Pico sonication system [Diagenode, Seraing (Ougrée), Belgium] to achieve DNA fragments between ∼200 and 1000 bp and centrifuged at 12,000g for 10 minutes at 4°C. Ten percent of the supernatants were taken as chromatin input, and aliquots of supernatants were incubated with antibodies overnight at 4°C after dilution with ChIP dilution buffer (1 mM EDTA, 50 mM Tris-HCl, pH 8.0). A mouse immunoglobulin G (IgG)(Millipore Sigma) was used as a negative control. Prewashed Dynabeads Protein A and G were then added to each sample and incubated for 2 hours and washed three times with a high-salt wash buffer (0.1% SDS, 1% Triton X-100, 1 mM EDTA, 20 mM Tris-HCl, pH 8.0, and 500 mM NaCl), three times with a low-salt wash buffer (0.1% SDS, 1% Triton X-100, 1 mM EDTA, 20 mM Tris-HCl, pH 8.0, and 150 mM NaCl), once with a LiCl buffer (0.25 M LiCl, 1% NP40, and 1% sodium deoxycholate), twice with a TE buffer (1 mM EDTA and 20 mM Tris-HCl, pH 8.0). After all washes, samples were eluted with an elution buffer (1% SDS, 0.5 M EDTA, and 1 M Tris-HCl, pH 8.0) and incubated with 200 mM NaCl at 65°C for overnight to reverse crosslinking. Then samples were incubated with Proteinase K for 1.5 hours at 55°C, and DNA was purified using a PCR purification kit (QIAGEN GmbH, Hilden, Germany) followed by qPCR analysis with the primers (Supplemental Table 1) using a SYBR green method. Enrichment was calculated as percentage of chromatin input.
Immunofluorescence.
LS174T cells that were grown on coverslip-coated 24-well plates and treated with rifampicin or solvent for 48 hours were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100, then blocked with 1% bovine serum albumin for 1.5 hours followed by overnight incubation of a primary antibody at 4°C and 1.5-hour incubation of a secondary antibody at room temperature: For PXR localization, an anti-PXR (1:50; Santa Cruz Biotechnology) and a Alexa Fluor 647-conjugated rabbit secondary antibody (1:200; Abcam) were used; for NCOA6 or p300 localization, an anti-NCOA6 (1:50; Proteintech, Wuhan, China) or anti-p300 (1:100; Abcam) with a Alexa Fluor 488-conjugated mouse secondary antibody (1:200; Abcam) were used. DAPI (Beyotime Institute of Biotechnology) was used to stain nuclei. Fluorescence was observed using a laser confocal microscope FluoView FV1000 (OLYMPUS, Tokyo, Japan).
Coimmunoprecipitation.
LS174T cells were transiently cotransfected with PSG5-hPXR and pCMV2-hNCOA6:V5 or pcDNA3.1-p300 plasmids, 24 hours after transfection cells were treated with rifampicin or solvent for an additional 48 hours and were harvested to perform coimmunoprecipitation (Co-IP) using a Pierce Classic IP Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions with a few modifications. Briefly, whole cell lysates were diluted with 1× Tris-buffered saline and immunoprecipitated with an anti-NCOA6 or anti-p300 antibody, with a rabbit anti-IgG as negative control. The immunoprecipitates were captured with Dynabeads Protein A/G followed by washes and then eluted for Western blot analysis.
Statistical Analysis.
Data are expressed as mean ± S.D. Statistical significance between groups was analyzed by Student t test, or one-way analysis of variance with Bonferroni’s post-hoc test or Dunnett’s test using SPSS for Windows, version 17.0 (IBM, Armonk, NY). A value of P < 0.05 was considered statistically significant.
Results
Rifampicin-Induced CYP3A4 Expression Was Accompanied by Altered Levels of H3K4me3, H3K27me3, and H3 Acetylation in the CYP3A4 Promoter.
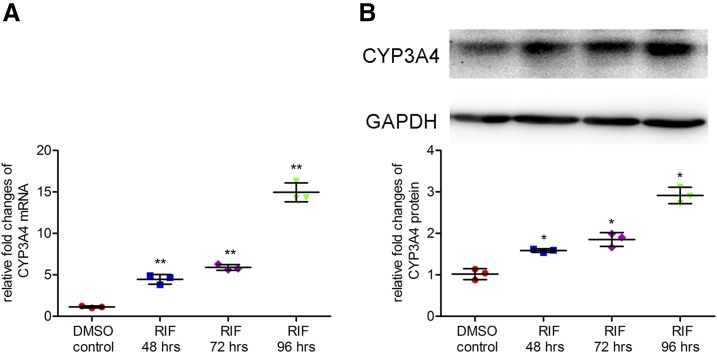
The induction of CYP3A4 by rifampicin (10 μM) in LS174T cells was assessed by qPCR and Western blot. Rifampicin treatment of 48, 72, or 96 hours induced the expression of CYP3A4 mRNA by 4.5-fold (95% CI: 1.76 to 4.91), 5.9-fold (95% CI: 3.20 to 6.34), and 15.0-fold (95% CI: 12.25 to 15.40), respectively (Fig. 1A), and protein by 1.6-fold (95% CI: 0.22 to 0.91), 1.8-fold (95% CI: 0.49 to 1.79), and 2.9-fold (95% CI: 1.56 to 2.24), respectively (Fig. 1B), compared with a solvent control.
Fig. 1.
Effect of rifampicin on the expression of CYP3A4 mRNA and protein in LS174T cells. LS174T cells were seeded onto 24-well plates and treated with a solvent DMSO control (0.1%, v/v) or rifampicin (10 μM) for 48, 72, and 96 hours, respectively. Then cells were harvested for determination of mRNA by qPCR (A) and protein by Western blot (B). Data are shown as mean ± S.D. of three independent experiments. *p < 0.05, **p < 0.01 versus control (one-way analysis of variance followed by Dunnett’s test). RIF, rifampicin.
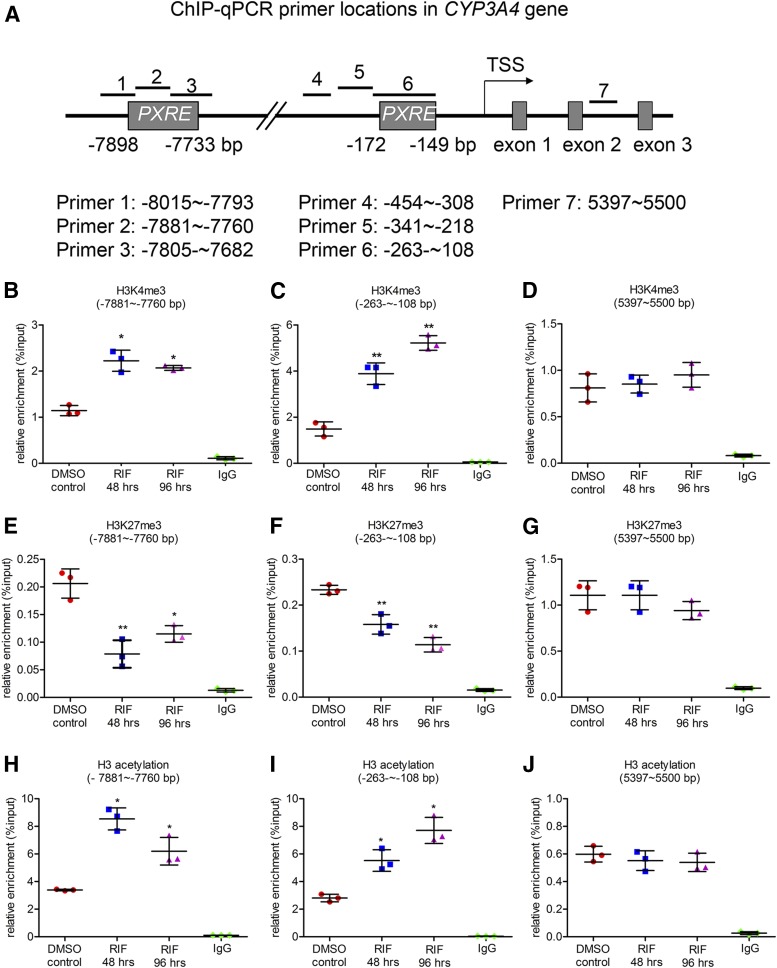
To reveal the influence of rifampicin on histone modifications, the enriched levels of H3K4me3, H3 acetylation, and H3K27me3 around the PXR response elements (PXRE) in the promoter of CYP3A4 were determined by ChIP analysis in rifampicin-treated LS174T cells. Since the basis of induction of CYP3A4 by rifampicin is activation of PXR, a set of ChIP-qPCR primers were designed to cover the distal (∼–7,898 to –7,733 bp) and proximal (∼–172 to –149 bp) PXRE in the promoter of CYP3A4 gene, accompanied with a pair of primers (∼5,397 to 5,500 bp) located in intron 2 of CYP3A4 gene as a negative control (Fig. 2A). After rifampicin treatment of 48 or 96 hours, the levels of H3K4me3 were increased by 1.8-fold in distal PXRE region (95% CI: 0.30 to 1.16 and 95% CI: 0.16 to 1.02, respectively), and higher increase (2.6-fold, 95% CI: 0.12 to 3.44 and 3.5-fold, 95% CI: 1.47 to 4.80, respectively) was observed in the proximal PXRE region (Fig. 2, B–D, and Supplemental Fig. 1A). A high level of H3K27me3 in promoter region of a gene is a mark associated with gene silencing. In this study, rifampicin treatment (48 and 96 hours) was found to decrease the levels of H3K27me3 in both distal (60%, 95% CI: –0.91 to –0.34 and 45%, 95% CI: –0.71 to –0.13, respectively) and proximal (30%, 95% CI: –0.49 to –0.17 and 53%, 95% CI: –0.68 to –0.36, respectively) PXRE regions (Fig. 2, E–G, and Supplemental Fig. 1B). Moreover, H3 acetylation, which is often observed in promotion of an active gene, was also increased by rifampicin in the same regions (Fig. 2, H–J, and Supplemental Fig. 1C). These results suggest that alterations of histone methylation and acetylation are involved in rifampicin-induced CYP3A4 expression in LS174T cells.
Fig. 2.
Effect of rifampicin on the levels of histone modifications around the PXR response elements (PXRE) in the CYP3A4 promoter. LS174T cells were treated with a solvent DMSO control (0.1%, v/v) or rifampicin (10 μM) for 48 or 96 hours, followed by ChIP analysis. Enrichment was calculated as percentage of chromatin input. (A) qPCR primers that cover different sites near the PXRE in the CYP3A4 promoter, a pair of primers located in intron 2 was performed as a negative control. The relative levels of H3K4me3 (B–D), H3K27me3 (E–G), and H3 acetylation (H–J) in the CYP3A4 promoter region and were determined. Data are shown as mean ± S.D. of three independent experiments. *p < 0.05, **p < 0.01 versus control (one-way analysis of variance followed by Dunnett’s test).
Rifampicin Increased NCOA6 and p300 Binding to PXRE in the CYP3A4 Promoter.
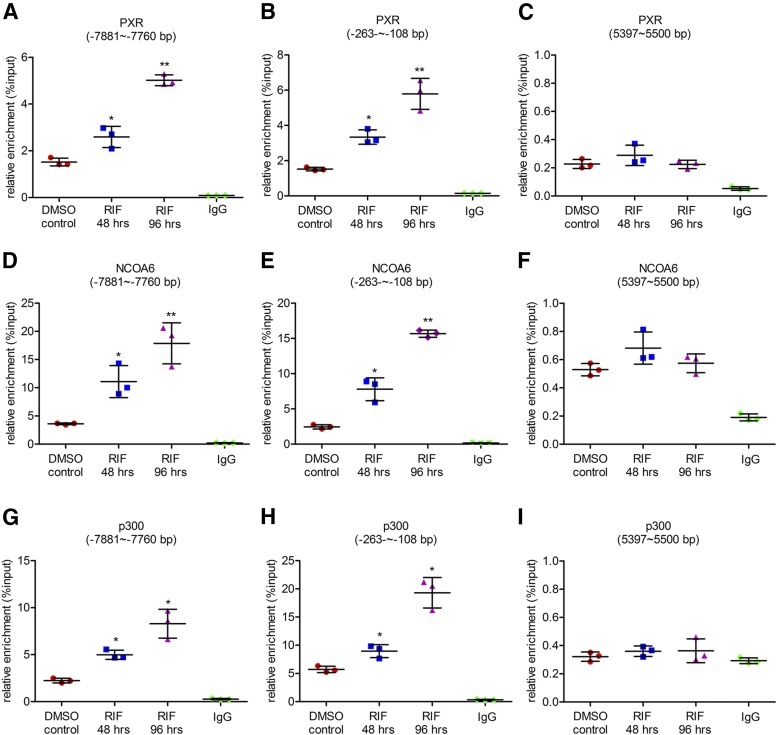
Since NCOA6 and p300 have been reported to be associated with H3K4me3, H3K27me3, and H3 acetylation levels, respectively, the binding of NCOA6 and p300 to the PXRE regions in the CYP3A4 promoter was analyzed by ChIP analysis. PXR binding was also analyzed as the induction of CYP3A4 by rifampicin is on the basis of its occupancy. The enrichments of PXR at both distal and proximal PXRE regions were increased by 1.7-fold (95% CI: 0.29 to 1.10) and 2.1-fold (95% CI: 0.50 to 2.33) after rifampicin treatment of 48 hours, and increased by 3.3-fold (95% CI: 1.90 to 2.72) and 3.8-fold (95% CI: 1.68 to 3.97) after treatment of 96 hours, respectively (Fig. 3, A–C, and Supplemental Fig. 1D). The enrichments of NCOA6 at both regions were increased by 3.1-fold (95% CI: 0.10 to 4.09) and 3.2-fold (95% CI: 1.01 to 3.98) after rifampicin treatment of 48 hours, and were increased by 4.9-fold (95% CI: 1.98 to 5.97) and 6.4-fold (95% CI: 4.31 to 6.58) after 96 hours of the treatment (Fig. 3, D–F, and Supplemental Fig. 1E). The enrichments of p300 were increased by 2.2-fold (95% CI: 0.24 to 2.23) and 1.6-fold (95% CI: 0.12 to 0.98) after rifampicin treatment of 48 hours and increased by 3.7-fold (95% CI: 1.72 to 3.70) and 3.4-fold (95% CI: 1.93 to 2.79) after 96 hours of the treatment (Fig. 3, G–I, and Supplemental Fig. 1F). These findings indicate that recruitment of NCOA6 and p300 is involved in PXR-mediated induction of CYP3A4 by rifampicin and can be associated with the changes of H3K4me3, H3K27me3, and H3 acetylation levels in the CYP3A4 promoter.
Fig. 3.
Effect of rifampicin on the enrichment of NCOA6 and p300 around the PXRE in the CYP3A4 promoter. LS174T cells were treated with a solvent control (0.1% DMSO, v/v) or rifampicin (10 μM) for 48 or 96 hours, followed by ChIP analysis. (A–C) Enrichment of nuclear receptor PXR, (D–F) cofactor NCOA6, and (G–I) histone acetyltransferase p300 in the CYP3A4 promoter after LS174T cells were treated with the solvent control or rifampicin. Data are shown as mean ± S.D. of three independent experiments. *p < 0.05, **p < 0.01 versus control (one-way analysis of variance followed by Dunnett’s test). RIF, rifampicin.
NCOA6 and p300 Are the Essential Factors in PXR-Meditated Induction of CYP3A4 by Rifampicin.
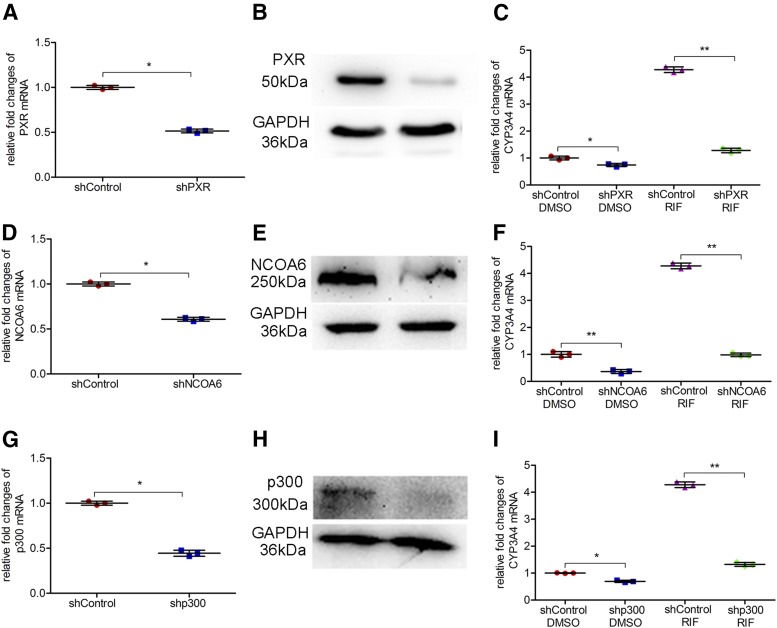
To explore the specific roles of NCOA6 and p300 in the CYP3A4 induction, stable cell lines with decreased expression of NCOA6 or p300 were generated as mentioned in Materials and Methods. Transient transfection was performed to knock down the expression of PXR in LS174T cells to confirm its function in the CYP3A4 induction. The levels of mRNA and protein expression of PXR were decreased after shRNA transfection compared with a negative control group (Fig. 4, A and B); the basal expression of CYP3A4 mRNA was decreased by 31% (95% CI: 0.08 to 0.53) (Fig. 4C). To study the effect of silencing PXR on rifampicin-induced CYP3A4 expression, shRNA-transfected cells were treated with rifampicin and CYP3A4 expression was determined. As a result, silencing the expression of PXR abolished the induction of CYP3A4 (Fig. 4C). Moreover, similar results were observed in stably transfected LS174T cells that possessed a lower expression level of NCOA6 or p300. Silencing the expression of NCOA6 not only reduced 64% (95% CI: 0.33 to 0.94) of the basal expression of CYP3A4 but also totally abolished the induced expression of CYP3A4 (Fig. 4, D–F). In addition, silencing of p300 also reduced the basal and rifampicin-induced expression of CYP3A4 [decreased by 30% (95% CI: 0.07 to 0.54) and 70% (95% CI: 2.72 to 3.18), respectively] (Fig. 4, G–I). All together, these results show that NCOA6 and p300 are indispensable in PXR-mediated induction of CYP3A4 by rifampicin.
Fig. 4.
Functions of PXR, NCOA6, and p300 in the induction of CYP3A4 by rifampicin. Knockdown of target genes by RNA interference (RNAi) in LS174T cells was performed by transfection of shRNA plasmids. Transiently or stably transfected cells were treated with a solvent DMSO control (0.1%, v/v) or rifampicin (10 μM) for 48 hours before RNA isolation. Knockdown of PXR by RNAi on mRNA (A) and protein levels (B). Effect of silencing PXR expression on CYP3A4 mRNA and the induction by rifampicin (C). Knockdown of NCOA6 by RNAi on mRNA (D) and protein levels (E). Effect of silencing NCOA6 expression on CYP3A4 mRNA and induction by rifampicin (F). Knockdown of p300 by RNAi on mRNA (G) and protein levels (H). Effect of silencing p300 expression on CYP3A4 mRNA and induction by rifampicin (I). Data are shown as mean ± S.D. of three independent experiments. *p < 0.05, **p < 0.01versus control (Student’s t test or one-way analysis of variance followed by Bonferroni’s post-hoc test). RIF, rifampicin.
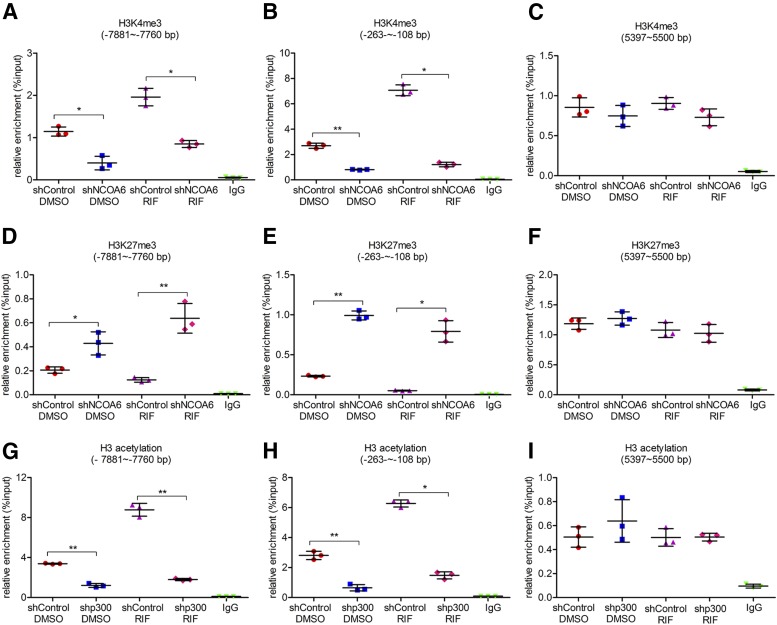
NCOA6 and p300 Are Required in Rifampicin-Induced Changes of Histone Modifications in the CYP3A4 Induction.
To further confirm the effects of NCOA6 and p300 on histone modifications in the induction of CYP3A4 by rifampicin, ChIP analysis was performed after silencing the expression of NCOA6 and p300, respectively (Fig. 5 and Supplemental Fig. 2). First, knockdown of NCOA6 expression was performed by stable transfection to study the effect of silencing NCOA6 expression on rifampicin-induced alterations of H3K4me3 and H3k27me3 levels. Silencing of NCOA6 decreased the basal level of H3K4me3 and increased the basal level of H3K27me3 in the CYP3A4 promoter compared with a transfection control group (Fig. 5, A–F). The changes of H3K4me3 and H3K27me3 in the CYP3A4 promoter by rifampicin were also abolished after silencing of NCOA6: The levels of H3K4me3 in rifampicin-treated shNCOA6 group were decreased by 57% (95% CI: 0.78 to 1.44) in the distal PXRE region and 83% (95% CI: 5.16 to 6.58) in the proximal PXRE region compared with the rifampicin-treated control group (Fig. 5, A–C, and Supplemental Fig. 2A); the levels of H3K27me3 were increased by 5.3-fold (95% CI: –0.69 to –0.34) and 14.0-fold (95% CI: –0.94 to –0.54), respectively (Fig. 5, D–F, and Supplemental Fig. 2B). Besides, a decrease of H3 acetylation in the CYP3A4 promoter was observed after knockdown of p300 expression, the increase of H3 acetylation by rifampicin was reduced by 80% (95% CI: 5.94 to 8.01) and 76% (95% CI: 4.18 to 5.41) in the distal and proximal PXRE regions, respectively, compared with the control transfection group treated with rifampicin (Fig. 5, G–I, and Supplemental Fig. 2C). These findings demonstrate that NCOA6 and p300 are responsible for the alterations of H3K4me3, H3K27me3, and H3 acetylation in the CYP3A4 promoter with or without rifampicin.
Fig. 5.
Effects of silencing NCOA6 or p300 expression on alteration of histone methylation or acetylation levels of the CYP3A4 promoter. Knockdown of NCOA6 or p300 was performed by transfecting shRNA plasmids into LS174T cells followed by treatment with a solvent DMSO control (0.1%, v/v) or rifampicin (10 μM) for 48 hours, and cells were harvested for ChIP analysis. The relative levels of H3K4me3 (A–C) or H3K27me3 (D–F) in the CYP3A4 promoter in the control and treated groups. The relative levels of H3 acetylation (G–I) in the CYP3A4 promoter in control and treated groups. Data are shown as mean ± S.D. of three independent experiments. *p < 0.05, **p < 0.01 (one-way analysis of variance followed by Bonferroni’s post-hoc test). RIF, rifampicin.
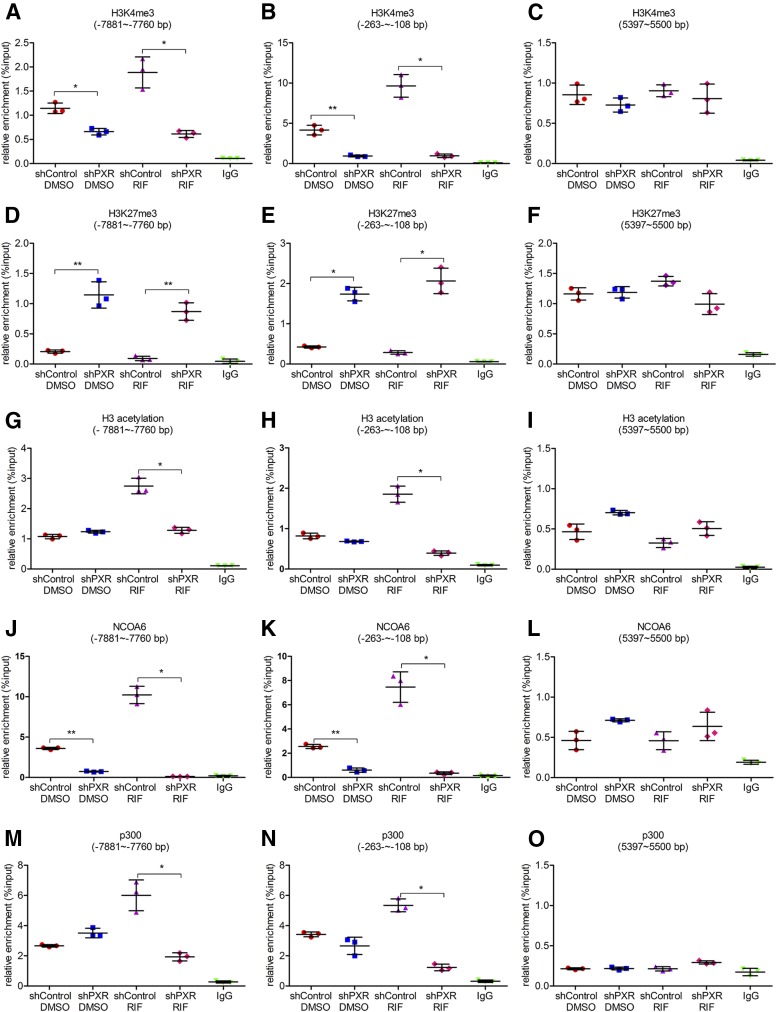
PXR Recruits NCOA6 and p300 to the CYP3A4 Promoter and Modulates the Epigenetic Changes in H3K4me3, H3K27me3, and H3 Acetylation.
The effects of silencing PXR expression on levels of H3K4me3, H3K27me3, and H3 acetylation, as well as the enrichment of NCOA6 and p300 in the CYP3A4 promoter were determined next (Fig. 6 and Supplemental Fig. 2). Knockdown of PXR expression reduced the levels of H3K4me3 and H3 acetylation and increased the level of H3K27me3 in the CYP3A4 promoter regardless of rifampicin treatment. The levels of H3K4me3 in the distal and proximal PXRE regions were reduced by 68% (95% CI: 0.81 to 1.74) and 90% (95% CI: 6.48 to 10.90) in rifampicin-treated PXR silencing group compared with rifampicin-treated control group (Fig. 6, A–C, and Supplemental Fig. 2D), whereas the levels of H3K27me3 were increased by 8.9-fold (95% CI: –1.11 to –0.44) and 6.6-fold (95% CI: –2.25 to –1.30) (Fig. 6, D–F, and Supplemental Fig. 2E); the H3 acetylation levels were reduced by 52% (95% CI: 1.07 to 1.86) and 80% (95% CI: 1.15 to 1.77) in rifampicin-treated PXR-silencing group (Fig. 6, G–I, and Supplemental Fig. 2F). Moreover, silencing the expression of PXR also decreased the recruitment of NCOA6 and p300 to the CYP3A4 promoter in both rifampicin-treated and untreated cells, the enrichment of NCOA6 was reduced by at least 80%, and p300 was reduced by 70% in rifampicin-treated PXR-silencing group (Fig. 6, J–O, and Supplemental Fig. 2G and 2H). These results suggest that PXR is essential for the enrichment of NCOA6 and p300 and the epigenetic changes of histone modifications in the CYP3A4 promoter.
Fig. 6.
Effects of silencing PXR expression on alterations of histone methylation and acetylation levels as well as enrichment of NCOA6 and p300 in the CYP3A4 promoter. Knockdown of PXR was performed by transiently transfecting shRNA plasmids into LS174T cells followed by treatment with a solvent DMSO control (0.1%, v/v) or rifampicin (10 μM) for 48 hours, and cells were harvested for ChIP analysis. The relative levels of H3K4me3 (A–C), H3K27me3 (D–F), H3 acetylation (G–I) in the CYP3A4 promoter in control and treated groups. The relative enrichment of NCOA6 (J–L) and p300 (M–O) in the CYP3A4 promoter in control and treated groups. Data are shown as mean ± SD of three independent experiments. *p < 0.05, **p < 0.01 (one-way analysis of variance followed by Bonferroni’s post-hoc test). RIF, rifampicin.
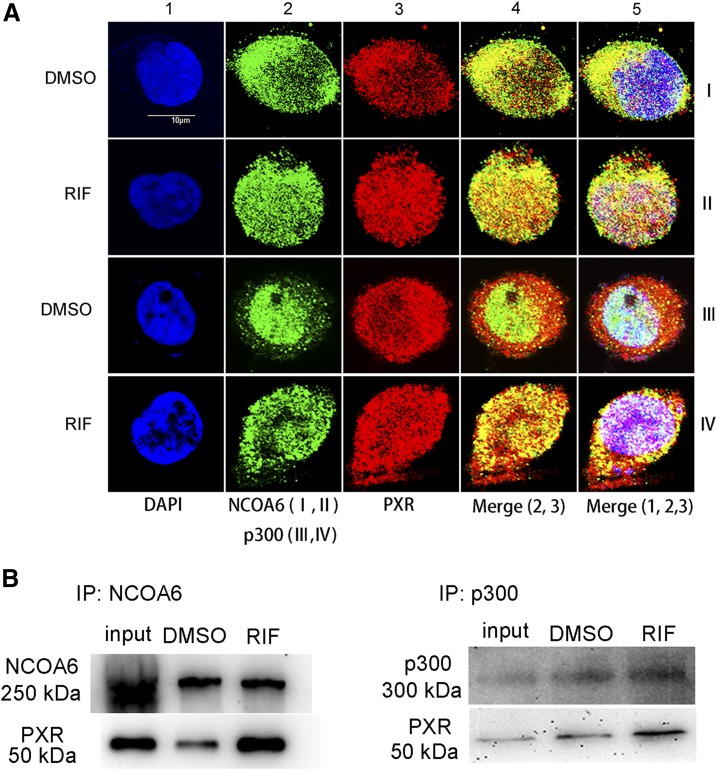
We next performed an immunofluorescence staining experiment to determine the effect of rifampicin on nuclear accumulation of PXR, NCOA6, and p300. Rifampicin increased the nuclear accumulation of PXR, NCOA6, and p300 (Fig. 7A, column 5). In addition, the colocalization of PXR and NCOA6/p300 was increased in rifampicin-treated cells (Fig. 7A, column 5, rows II and IV). Moreover, the Co-IP results showed that NCOA6 and p300 directly interacted with PXR, and the interaction was enhanced in rifampicin-treated LS174T cells (Fig. 7, B and C). These results suggest that rifampicin increases the nuclear accumulation of PXR, NCOA6, and p300, and enhances the interactions between PXR and NCOA6/p300.
Fig. 7.
Effects of rifampicin on the nuclear accumulation of PXR, NCOA6, and p300, as well as interactions between PXR and NCOA6/p300. (A) Representative immunofluorescence images of PXR, NCOA6, and p300 in the treated LS174T cells. Bar, 10 μm. All cells were stained with DAPI (blue, column 1, rows I, II, III, and IV) to visualize the nuclei. Cells were stained with an antibody of anti-NCOA6 (green, column 2, rows I and II), anti-p300 (green, column 2, rows III and IV), and anti-PXR (red, column 3, rows I, II, III, and IV) to visualize the nuclear and cytoplasmic distribution of NCOA6, p300, and PXR. (B) LS174T cells were cotransfected with PXR and NCOA6 or p300 expression plasmids, and further incubated with rifampicin (10 μM) or a solvent DMSO control (0.1%, v/v) for 48 hours after the transfection. Cells were harvested for coimmunoprecipitation and detected by Western blot. All experiments were performed in three independent replicates. RIF, rifampicin.
Discussion
Induction of drug-metabolizing enzymes and transporters is an important cause of drug-drug interactions, especially for the induction of CYP3A4, the most abundant isoform of cytochrome P450. PXR is the most important nuclear receptor that is responsible for the induction of CYP3A4 by a broad range of xenobiotics. Increasing evidence indicates that histone modifications are important to nuclear receptor-mediated transcription of target genes (Xie et al., 2009; Huo et al., 2010). In this study, we found that levels of H3K4me3 and H3 acetylation in the CYP3A4 promoter, two marks for activation of gene transcription, were elevated by rifampicin treatment in LS174T cells, whereas the level of H3K27me3 (a mark for gene silencing) was decreased. We then found that NCOA6 and p300 were required in the PXR-mediated induction of CYP3A4 and were associated with histone methylation and acetylation in the CYP3A4 promoter.
Epigenetic modifications like histone methylation, acetylation, phosphorylation, and ubiquitination have been shown to play important roles in the regulation of gene expression. Drug-metabolizing enzymes have also been reported to be regulated by histone modifications. For instance, CYP2C9 was found to be regulated by mediator complex subunit 25 (MED25) through the epigenetic modification of H3K27. MED25 was recruited to the CYP2C9 promoter by HNF4α and promoted H3K27 acetylation and repressed its methylation (Zha et al., 2015). In the current study, we determined the status of H3K4me3, H3K27me3, and H3 acetylation in the CYP3A4 promoter upon rifampicin exposure, as these histone modifications are commonly seen in the regulation of gene expression. We found that rifampicin elevated the levels of H3K4me3 and H3 acetylation but attenuated the levels of H3K27me3. These results indicate that histone methylation and acetylation are involved in the induction of CYP3A4 by rifampicin.
However, epigenetic modifications occur as executors rather than initiators in the regulation of gene expression. Alterations of environment, physiology, and pathology are considered the causes of epigenetic changes (Peng and Zhong, 2015). Nuclear receptors have been suggested as the sensors that respond to extracellular changes, and they have been identified as interacting with epigenetic-modification enzymes after their activation by ligands to control gene expression (Green and Han, 2011). We investigated the mechanism underlying the changes of histone methylation and acetylation in the CYP3A4 promoter caused by rifampicin. The enrichment of NCOA6 and p300 at the PXRE regions in the CYP3A4 promoter was significantly increased after rifampicin treatment compared with a control group, and the enrichment was consistent with changes of H3K4me3, H3 acetylation, and H3K27me3. Among the cofactors that related with nuclear receptors, NCOA6 has been demonstrated to be critical for the trimethylation of H3K4 in gene transcription, as it belongs to a part of ASCOM that contains H3K4 methyltransferase MLL3/4 (Ananthanarayanan et al., 2011). We found that silencing NCOA6 resulted in a remarkable decrease of H3K4me3 but a considerable increase of H3K27me3 in the CYP3A4 promoter. Apart from H3K4 methyltransferase MLL3/4, the ASCOM also contains UTX, the specific H3K27 demethylase. Thus, the involvement of ASCOM in transcriptional activation was carried out by altering the trimethylation of H3K4 and H3K27 (He et al., 2015). In addition, histone acetyltransferase p300 plays an important role in nuclear receptor-mediated gene expression through elevation of levels of histone H3 and H4 acetylation near nuclear receptor binding sites in a promoter of a target gene (Tsai and Fondell, 2004; Green and Han, 2011). Indeed, increase of p300 enrichment near the PXR binding sites in the CYP3A4 promoter was observed after LS174T cells were incubated with rifampicin. The increase was correlated with the induction of CYP3A4 and increase of H3 acetylation by rifampicin, as they were both diminished after the silencing of p300 expression. More importantly, we found that the recruitment of NCOA6 and p300 to the CYP3A4 promoter was mediated by PXR. Knockdown of the expression of PXR resulted in significant inversion of alteration of histone modifications and enrichment of NCOA6 and p300 in the CYP3A4 promoter, which was consistent with the decrease of CYP3A4 induction. It has been reported that NCOA6 interacts with PXR through the first LXXLL motif (Surapureddi et al., 2011). In the immunofluorescence and Co-IP experiments, we confirmed the interaction between NCOA6/p300 and PXR and found that rifampicin enhanced the interaction. Thus, these results indicate that PXR recruits NCOA6 and p300 to the CYP3A4 promoter region and modulates histone methylation and acetylation to facilitate the CYP3A4 induction.
In addition to NCOA6 and p300, some of the coactivators that interact with PXR could affect chromatin remodeling or histone modification patterns, such as steroid receptor coactivator 1 (SRC-1). It has been reported that SRC-1 together with p300 play crucial roles in dioxin-induced expression of CYP1A1 and CYP1B1 in MCF-7 cells (Taylor et al., 2009). SRC-1 has been reported to interact with PXR and regulate CYP3A4 induction (Li and Chiang, 2006). However, its effect on histone modifications, like H3 acetylation in the CYP3A4 promoter, remains unclear. Further studies focusing on SRC-1 and its role in histone modification patterns in the CYP3A4 promoter induced by PXR ligands will provide valuable findings in understanding the epigenetic regulation of CYP3A4 induction.
The importance of epigenetic regulation of drug metabolism-related genes has been reviewed recently (Ivanov et al., 2014; Peng and Zhong, 2015; Tang and Chen, 2015). Epigenetic regulation of gene expression is modulated by such environmental factors as drugs, diets, and environmental pollutants (Choudhuri et al., 2010). For example, it has been demonstrated that dynamic H3K27 acetylation is associated with vascular endothelial growth factor A–stimulated changes in chromatin conformation and transcriptional responses (Zhang et al., 2013). Apparently, epigenetic changes in drug-metabolizing enzymes caused by environmental factors could be a reason for the interindividual differences in drug responses and drug-drug interactions. In addition to CYP3A4, PXR regulates a variety of other P450 enzymes, phase II drug-metabolizing enzymes, and drug transporters in response to xenobiotics (Chai et al., 2013). Further research using primary hepatocytes and transgenic mice in the determination of epigenetic regulation of drug-metabolizing enzymes and transporters mediated by PXR will be performed to provide new insights into the variations of drug responses and will be beneficial to precision medicine in future.
In conclusion, this study reveals that alterations of histone methylation and acetylation are involved in the CYP3A4 induction by rifampicin through the activation of PXR and recruitment of histone methyltransferase–related factor NCOA6 and histone acetyltransferase p300.
Acknowledgments
The authors thank Dr. Wen Xie at Center of Pharmacogenomics, University of Pittsburgh for providing the PSG5-hPXR plasmid.
Abbreviations
- ASCOM
ASC-2 complex
- ChIP
chromatin immunoprecipitation analysis
- Co-IP
coimmunoprecipitation
- DMSO
dimethylsulfoxide
- H3K27me3
histone 3 lysine 27 trimethylation
- H3K4me3
histone 3 lysine 4 trimethylation
- HNF4α
hepatic nuclear factor 4 α
- IgG
immunoglobulin G
- MLL3
mixed lineage leukemia 3
- MLL4
mixed lineage leukemia 4
- NCOA6
nuclear receptor coactivator 6
- PXR
pregnane X receptor
- PXRE
PXR response element
- qPCR
real-time quantitative polymerase chain reaction
- sh
short-hairpin
- SRC-1
steroid receptor coactivator 1
- UTX
lysine demethylase 6A.
Authorship Contributions
Participated in research design: Yan, Zhong, Kan, Zhang.
Conducted experiments: Yan, Wang, Liu, Nie.
Performed data analysis: Yan, Wang, Zhang.
Wrote or contributed to the writing of the manuscript: Yan, Zhong, Zhang.
Footnotes
This work was supported by the National Natural Science Foundation of China [Grants 81173127 and U1604163] and the National Institutes of Health National Institute of General Medical Sciences [Grants R01GM-087376 and R01GM-118367].
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Ananthanarayanan M, Li Y, Surapureddi S, Balasubramaniyan N, Ahn J, Goldstein JA, Suchy FJ. (2011) Histone H3K4 trimethylation by MLL3 as part of ASCOM complex is critical for NR activation of bile acid transporter genes and is downregulated in cholestasis. Am J Physiol Gastrointest Liver Physiol 300:G771–G781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baciewicz AM, Chrisman CR, Finch CK, Self TH. (2013) Update on rifampin, rifabutin, and rifapentine drug interactions. Curr Med Res Opin 29:1–12. [DOI] [PubMed] [Google Scholar]
- Chai X, Zeng S, Xie W. (2013) Nuclear receptors PXR and CAR: implications for drug metabolism regulation, pharmacogenomics and beyond. Expert Opin Drug Metab Toxicol 9:253–266. [DOI] [PubMed] [Google Scholar]
- Chen LF, Mu Y, Greene WC. (2002) Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J 21:6539–6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Ma X, Krausz KW, Idle JR, Gonzalez FJ. (2009) Rifampicin-activated human pregnane X receptor and CYP3A4 induction enhance acetaminophen-induced toxicity. Drug Metab Dispos 37:1611–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri S, Cui Y, Klaassen CD. (2010) Molecular targets of epigenetic regulation and effectors of environmental influences. Toxicol Appl Pharmacol 245:378–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert NA, Luo G, Goldstein JA, Surapureddi S. (2015) Epigenetic modification of histone 3 lysine 27: mediator subunit MED25 is required for the dissociation of polycomb repressive complex 2 from the promoter of cytochrome P450 2C9. J Biol Chem 290:2264–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin B, Hodgson E, Liddle C. (1999) The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol 56:1329–1339. [DOI] [PubMed] [Google Scholar]
- Green CD, Han JD. (2011) Epigenetic regulation by nuclear receptors. Epigenomics 3:59–72. [DOI] [PubMed] [Google Scholar]
- He X, Chen X, Zhang X, Duan X, Pan T, Hu Q, Zhang Y, Zhong F, Liu J, Zhang H, et al. (2015) An Lnc RNA (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res 43:3712–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo H, Magro PG, Pietsch EC, Patel BB, Scotto KW. (2010) Histone methyltransferase MLL1 regulates MDR1 transcription and chemoresistance. Cancer Res 70:8726–8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov M, Barragan I, Ingelman-Sundberg M. (2014) Epigenetic mechanisms of importance for drug treatment. Trends Pharmacol Sci 35:384–396. [DOI] [PubMed] [Google Scholar]
- Lamba J, Strom S, Venkataramanan R, Thummel KE, Lin YS, Liu W, Cheng C, Lamba V, Watkins PB, Schuetz E. (2006) MDR1 genotype is associated with hepatic cytochrome P450 3A4 basal and induction phenotype. Clin Pharmacol Ther 79:325–338. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee J, Lee SK, Lee JW. (2008) Activating signal cointegrator-2 is an essential adaptor to recruit histone H3 lysine 4 methyltransferases MLL3 and MLL4 to the liver X receptors. Mol Endocrinol 22:1312–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Roeder RG, Lee JW. (2009) Roles of histone H3-lysine 4 methyltransferase complexes in NR-mediated gene transcription. Prog Mol Biol Transl Sci 87:343–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chiang JY. (2006) Rifampicin induction of CYP3A4 requires pregnane X receptor cross talk with hepatocyte nuclear factor 4alpha and coactivators, and suppression of small heterodimer partner gene expression. Drug Metab Dispos 34:756–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Cui Y, Hart SN, Klaassen CD, Zhong XB. (2009) Dynamic patterns of histone methylation are associated with ontogenic expression of the Cyp3a genes during mouse liver maturation. Mol Pharmacol 75:1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Lim YP, Hu ML. (2012) Fucoxanthin attenuates rifampin-induced cytochrome P450 3A4 (CYP3A4) and multiple drug resistance 1 (MDR1) gene expression through pregnane X receptor (PXR)-mediated pathways in human hepatoma HepG2 and colon adenocarcinoma LS174T cells. Mar Drugs 10:242–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YT, Hao HP, Liu CX, Wang GJ, Xie HG. (2007) Drugs as CYP3A probes, inducers, and inhibitors. Drug Metab Rev 39:699–721. [DOI] [PubMed] [Google Scholar]
- Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, et al. (2000) Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J Biol Chem 275:15122–15127. [DOI] [PubMed] [Google Scholar]
- Niemi M, Backman JT, Fromm MF, Neuvonen PJ, Kivistö KT. (2003) Pharmacokinetic interactions with rifampicin : clinical relevance. Clin Pharmacokinet 42:819–850. [DOI] [PubMed] [Google Scholar]
- Novotna A, Dvorak Z. (2014) Omeprazole and lansoprazole enantiomers induce CYP3A4 in human hepatocytes and cell lines via glucocorticoid receptor and pregnane X receptor axis. PLoS One 9:e105580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Slattery M, Ma L, White KP, Mann RS, Irvine KD. (2014) Yorkie promotes transcription by recruiting a histone methyltransferase complex. Cell Reports 8:449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Zhong X. (2015) Epigenetic regulation of drug metabolism and transport. Acta Pharm Sin B 5:106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmioglu N, Heaton J, Clement G, Gill R, Surdulescu G, Zlobecka K, Hodgkiss D, Ma Y, Hider RC, Smith NW, et al. (2011) Genetic epidemiology of induced CYP3A4 activity. Pharmacogenet Genomics 21:642–651. [DOI] [PubMed] [Google Scholar]
- Raucy JL. (2003) Regulation of CYP3A4 expression in human hepatocytes by pharmaceuticals and natural products. Drug Metab Dispos 31:533–539. [DOI] [PubMed] [Google Scholar]
- Schübeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, et al. (2004) The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18:1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz EG. (2004) Lessons from the CYP3A4 promoter. Mol Pharmacol 65:279–281. [DOI] [PubMed] [Google Scholar]
- Surapureddi S, Rana R, Goldstein JA. (2011) NCOA6 differentially regulates the expression of the CYP2C9 and CYP3A4 genes. Pharmacol Res 63:405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Xu C, Zeng H, Wang Y, Li Y, Fan X, Chen P, Jiang Y, Chen X, Huang M, et al. (2016) SUMOylation of pregnane X receptor suppresses rifampicin-induced CYP3A4 and P-gp expression and activity in LS174T cells. J Pharmacol Sci 130:66–71. [DOI] [PubMed] [Google Scholar]
- Tang C, Lin JH, Lu AY. (2005) Metabolism-based drug-drug interactions: what determines individual variability in cytochrome P450 induction? Drug Metab Dispos 33:603–613. [DOI] [PubMed] [Google Scholar]
- Tang X, Chen S. (2015) Epigenetic regulation of cytochrome P450 enzymes and clinical implication. Curr Drug Metab 16:86–96. [DOI] [PubMed] [Google Scholar]
- Taylor RT, Wang F, Hsu EL and Hankinson O (2009) Roles of coactivator proteins in dioxin induction of CYP1A1 and CYP1B1 in human breast cancer cells. Toxicol Sci 107(1): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumaran RK, Lamba JK, Kim RB, Urquhart BL, Gregor JC, Chande N, Fan Y, Qi A, Cheng C, Thummel KE, et al. (2012) Intestinal CYP3A4 and midazolam disposition in vivo associate with VDR polymorphisms and show seasonal variation. Biochem Pharmacol 84:104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel KE, Brimer C, Yasuda K, Thottassery J, Senn T, Lin Y, Ishizuka H, Kharasch E, Schuetz J, Schuetz E. (2001) Transcriptional control of intestinal cytochrome P-4503A by 1alpha,25-dihydroxy vitamin D3. Mol Pharmacol 60:1399–1406. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Fondell JD. (2004) Nuclear receptor recruitment of histone-modifying enzymes to target gene promoters. Vitam Horm 68:93–122. [DOI] [PubMed] [Google Scholar]
- Wu Y, Shi X, Liu Y, Zhang X, Wang J, Luo X, Wen A. (2012) Histone deacetylase 1 is required for Carbamazepine-induced CYP3A4 expression. J Pharm Biomed Anal 58:78–82. [DOI] [PubMed] [Google Scholar]
- Xie X, Song X, Yuan S, Cai H, Chen Y, Chang X, Liang B, Huang D. (2015) Histone acetylation regulates orphan nuclear receptor NR4A1 expression in hypercholesterolaemia. Clin Sci (Lond) 129:1151–1161. [DOI] [PubMed] [Google Scholar]
- Xie Y, Ke S, Ouyang N, He J, Xie W, Bedford MT, Tian Y. (2009) Epigenetic regulation of transcriptional activity of pregnane X receptor by protein arginine methyltransferase 1. J Biol Chem 284:9199–9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xydous M, Sekeri-Pataryas KE, Prombona A, Sourlingas TG. (2012) Nicotinamide treatment reduces the levels of histone H3K4 trimethylation in the promoter of the mper1 circadian clock gene and blocks the ability of dexamethasone to induce the acute response. Biochim Biophys Acta 1819:877–884. [DOI] [PubMed] [Google Scholar]
- Yang H, Nie Y, Li Y, Wan YJ. (2010) Histone modification-mediated CYP2E1 gene expression and apoptosis of HepG2 cells. Exp Biol Med (Maywood) 235:32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha L, Li F, Wu R, Artinian L, Rehder V, Yu L, Liang H, Xue B, Shi H. (2015) The histone demethylase UTX promotes brown adipocyte thermogenic program via coordinated regulation of H3K27 demethylation and acetylation. J Biol Chem 290:25151–25163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Day DS, Ho JW, Song L, Cao J, Christodoulou D, Seidman JG, Crawford GE, Park PJ, Pu WT. (2013) A dynamic H3K27ac signature identifies VEGFA-stimulated endothelial enhancers and requires EP300 activity. Genome Res 23:917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]