Abstract
In a previous genome-wide association study (GWAS) for musculoskeletal adverse events during aromatase inhibitor therapy for breast cancer, we reported that single nucleotide polymorphisms (SNPs) near the TCL1A gene were associated with this adverse drug reaction. Functional genomic studies showed that TCL1A expression was induced by estradiol, but only in cells with the variant sequence for the top GWAS SNP (rs11849538), a SNP that created a functional estrogen response element. In addition, TCL1A genotype influenced the downstream expression of a series of cytokines and chemokines and had a striking effect on nuclear factor κB (NF-κB) transcriptional activity. Furthermore, this SNP-dependent regulation could be reversed by selective ER modulators (SERMs). The present study was designed to pursue mechanisms underlying TCL1A SNP-mediated, estrogen-dependent NF-κB activation. Functional genomic studies were performed using a panel of 300 lymphoblastoid cell lines for which we had generated genome-wide SNP and gene expression data. It is known that toll-like receptors (TLRs) can regulate NF-κB signaling by a process that requires the adaptor protein MYD88. We found that TLR2, TLR7, TLR9, and TLR10 expression, as well as that of MYD88, could be modulated by TCL1A in a SNP and estrogen-dependent fashion and that these effects were reversed in the presence of SERMs. Furthermore, MYD88 inhibition blocked the TCL1A SNP and estrogen-dependent NF-κB activation, as well as protein-protein interaction between TCL1A and MYD88. These observations greatly expand the range of pathways influenced by TCL1A genotype and raise the possibility that this estrogen- and SNP-dependent regulation might be altered pharmacologically by SERMs.
Introduction
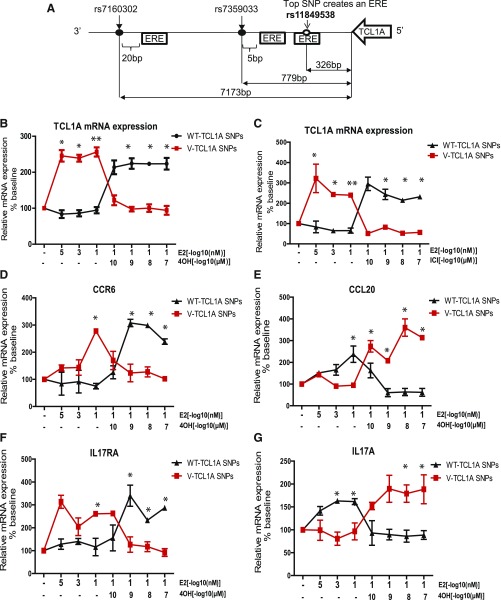
In a previous genome-wide association study (GWAS) addressing musculoskeletal adverse events in breast cancer patients treated with aromatase inhibitors, we identified single nucleotide polymorphisms (SNPs) 3′ of the T-cell leukemia/lymphoma 1A gene (TCL1A) that were associated with this adverse drug reaction (Ingle et al., 2010). The minor allele frequency for the TCL1A SNP (rs11849538) is ∼19% in European, African, Asian, and Caucasian Americans according to the 1000 Genome Project (Ingle et al., 2010). We subsequently performed a series of functional genomic studies using as a model system lymphoblastoid cell lines (LCLs) for which we had generated dense genome-wide genomic data. Those studies showed that TCL1A expression was upregulated by estradiol (E2), but only in cell lines homozygous for the variant TCL1A genotype for the top GWAS SNP (rs11849538) (Fig. 1A), a SNP that created a functional estrogen response element (ERE) (Ingle et al., 2010). We subsequently found that there were at least three SNPs 3′ of TCLIA, rs11849538, rs7359033, and rs7160302 (Fig. 1A)—all in tight linkage disequilibrium (LD)—that appeared to act in concert to influence the E2-dependent induction of TCL1A expression, in part because SNPs near, but not in, an ERE can affect estrogen receptor (ER) binding to the ERE and subsequent transcription (Ho et al., 2016). In addition, induction of TCL1A by E2 was associated downstream with variation in the expression of a series of proinflammatory cytokines (Liu et al., 2012) and chemokines (Ho et al., 2016), as well as their receptors (Fig. 1, D–G), and with striking variation in nuclear factor κB (NF-κB) transcriptional activity (Liu et al., 2012). For example, we found that the expression of immune mediators, such as the C-C motif chemokine receptor 6 (CCR6) and its only known ligand, CCL20, as well as the cytokine receptor IL17RA and its ligand, IL17A, could be modulated by TCL1A in a SNP-estrogen–dependent fashion (Ho et al., 2016). In addition, we found that the SNP and estrogen-dependent induction of TCL1A expression could be reversed after ER blockade, that is, when cells were exposed to 4-hydroxytamoxifen (4-OH-TAM), an active metabolite of tamoxifen, or to fulvestrant (formerly ICI 182,780) (Fig. 1, B and C), an ER antagonist (Liu et al., 2012; Ho et al., 2016). Specifically, cells homozygous for the wild-type (WT) genotype for the TCL1A SNPs displayed increased TCL1A expression, whereas cells homozygous for the variant genotype showed decreased expression in the presence of these drugs—the opposite of the situation seen after estrogen exposure alone (Fig. 1, B and C). Even more striking, downstream receptors for inflammatory mediators (CCR6 and IL17RA) responded in a parallel fashion, indicating that TCL1A was upstream of the cytokine and chemokine receptors in terms of these pathways (Ho et al., 2016). It should be emphasized that the SNPs involved were those 3′ of TCLIA, not SNPs that mapped to the IL17RA or CCR6 genes (Ho et al., 2016). Finally, SNP and estrogen-dependent TCL1A induction resulted in significantly increased NF-κB transcriptional activity after ER blockade, but only for the variant genotype (Liu et al., 2012). Taken as a whole, these observations suggest that TCL1A genotype is capable of influencing the expression of a series of genes that play important roles in inflammation and immunity, and they also raise the possibility of pharmacologic approaches (i.e., exposure to SERMs) that could be used to modify these effects.
Fig. 1.
The SNP- and estrogen-dependent effects on mRNA expression of TCL1A, cytokines, and chemokines in LCLs. (A) Schematic diagrams of the two TCL1A SNPs, rs7359033 and rs7160302, in tight LD with rs11849538, the top hit signal from the MA.27 GWAS. Locations of EREs are shown as boxes for these three SNPs that map between the 3′-termini of TCL1A and TCL1B. ER blockade by 4-hydroxytamoxifen (4OH) or fulvestrant (ICI) treatment resulted in the reversal of TCL1A SNP and estrogen-dependent TCL1A expression patterns (B and C) and downstream effects on the expression of CCR6, CCL20, IL17RA, and IL17A (D–G) Values are mean ± S.E.M. of three assays. *P < 0.0001. Adapted from Fig. 2 and Fig. 3 in Ho et al. (2016).
Although we have studied the transcriptional regulation of cytokines and chemokines and their receptors in relation to TCL1A SNPs and estrogens (Liu et al., 2012; Ho et al., 2016), significant gaps remain in our knowledge, particularly with regard to mechanisms that might be involved in the effect of TCL1A on NF-κB signaling. NF-κB activation is a multifactorial, complex process involving many biologic pathways, such as DNA damage (McCool and Miyamoto, 2012), proteasome-mediated degradation (Hayden et al., 2006), signal transduction via stimulation of T cells, or exposure to inflammatory molecules (Takeda et al., 2008). The expression of NF-κB varies among cell types and can be event-specific, depending on the stimulus (Tak and Firestein, 2001). Toll-like receptors (TLRs) are crucial immune mediators and can have a profound effect on inflammation as a result, in part, of NF-κB activation (Tak and Firestein, 2001; Kawasaki and Kawai, 2014). In the present experiments, we set out to study mechanisms that might be involved in the TCL1A SNP-mediated, estrogen-dependent activation of NF-κB (Liu et al., 2012). Specifically, we aimed to determine whether the expression of TLRs and their adaptor molecules might be altered in a TCL1A SNP-dependent fashion and, if so, their possible contribution to the TCL1A SNP-mediated, estrogen-dependent activation of NF-κB. In addition, we tested the hypothesis that SERMs might be able to modulate these effects. In summary, the experiments described subsequently involved the application of functional genomic studies to identify mechanisms underlying the effect of TCL1A SNPs on the regulation of inflammatory mediators.
Materials and Methods
Ethics Statement.
The Mayo Clinic Institutional Review Board determined that this work did not require their review or approval.
Human Variation Panel Lymphoblastoid Cell Lines.
A Human Variation Panel lymphoblastoid cell line (LCL) model system consisting of 300 LCLs from healthy subjects of three ethnicities (100 European American, 100 African American, and 100 Han Chinese American) was used to perform these studies. This cell line model system provides genome-wide mRNA expression as determined by Affymetrix U133 2.0 Plus GeneChip expression array as well as genome-wide SNP data generated with Illumina 550K and 510S SNP BeadChip SNP arrays (Illumina, San Diego, CA). The genotype data were used to impute approximately seven million SNPs per cell line.
RNA Interference and Transfection.
We purchased siRNA (TCL1A and MYD88) and negative controls from Dharmacon (Chicago, IL). LCLs were transfected with siRNA by electroporation using the nucleofector kit (Lonza, Allendale, NJ). Briefly, the electroporation reaction contained 2 × 106 cells, 100 µl of nucleofector solution, and 1 µM siRNA. After electroporation, cells were transferred into 12-well plates containing pre-equilibrated RPMI medium.
Drug Treatment.
LCLs with known genotypes were cultured in RPMI 1640 media (Cellgro, Manassas, VA) supplemented with 15% fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA). Before estrogen treatment, cells were cultured in RPMI media containing 5% (v/v) charcoal stripped FBS for 24 hours. The cells were then cultured in FBS-free RPMI media for another 24 hours and were treated with 0.1 nM E2 (Sigma, St. Louis, MO) for 24 hours. In some experiments, cells were treated with 10−7 µM 4-hydroxytamxifen (4-OH-TAM) (Sigma) or fulvestrant (formerly ICI 182,780) (Sigma), followed by E2 treatment. MYD88 inhibitor peptide was purchased from Novus Biologicals (Littleton, CO).
Real-Time Polymerase Chain Reaction.
RNA was isolated from the cells (Zymo, Irvine, CA). The polymerase chain reaction (PCR) mixture contained 50 ng of total RNA, 5µl of 2× SYBR Green real-time PCR nix (Affymetrix, Santa Clara, CA), 0.1 µl of DNA polymerase enzyme, 1 µl of gene-specific primer (Qiagen, Valencia, CA), and distilled water to achieve 10 µl per reaction. Real-time PCR reactions were performed in duplicate using Applied Biosystems ViiA 7 real-time PCR System (Life Technologies, Carlsbad, CA). The 2-ΔΔCt method was used for statistical data analysis.
Western Blot Analysis.
Protein samples were used to perform electrophoresis, followed by transfer to a polyvinylidene fluoride membrane. The membranes were incubated overnight with primary antibodies: TCL1A, TLR2, TLR7, TLR9, TLR10, MYD88, UNC93B1 (Novus Biologicals, Littleton, CO), and ACTB at a 1:500 dilution at 4°C. The washed membranes were then incubated with goat anti-rabbit or anti-mouse secondary antibody (Santa Cruz Biotechnology, Dallas, TX) at a 1:20,000 dilution. The washed membranes were subsequently incubated in Pierce ECL Western blotting substrate (Thermo Scientific, Madison, WI) and were visualized using Geldoc (Bio-Rad Laboratories, Hercules, CA).
NF-κB Reporter Assay.
Lymphoblastoid cell lines with known TCL1A SNP genotypes were transfected by electroporation with 2 µg of the pGL4.32[luc2P/NF-κB-RE.Hydro] vector (Promega, Madison, WI). A Renilla construct, pRL-TK (Promega), was used to determine transfection efficiency. The cells were plated in RPMI medium for 24 hours to allow them to recover from electroporation. Cells were then treated with 0.1 nM E2 for 24 hours, followed by 10−7 µM 4-OH-TAM or ICI for an additional 24 hours. In some experiments, cells were treated with MYD88 (100 µM) for 24 hours before E2 treatment. Luciferase assays were performed with the Dual-Luciferase Reporter assay system (Promega). Relative NF-kB/luciferase activities were normalized to Renilla luciferase activities.
Coimmunoprecipitation of TCL1A and MYD88.
LCLs (1 × 107) were resuspended in 650 µl of immunoprecipitation (IP) lysis buffer containing 2.2 µl of protease inhibitor cocktail (Qiagen, Valencia, CA) and incubated on ice for 30 minutes. Cells were then centrifuged at 12,000g at 4°C for 15 minutes. Supernatant was collected. Protein A agarose (Thermo Scientific) was prepared and washed three times with IP lysis buffer. A precleaning step was performed to clean the background. Cell lysates containing protein A agarose beads were rotated at 4°C for an hour. After centrifugation, supernatant was collected. At this point, input (50 µl) was collected and stored at −80°C. Anti-TCL1A (1:50) antibodies (Cell Signaling Technology, Danvers, MA) were used to perform IP. IgG (Cell Signaling Technology), used as negative control. Specifically, IP samples containing protein A agarose beads were rotated at 4°C overnight. Immunoprecipitates were washed three times with ice cold lysis buffer, and proteins were eluted with 50 µl of 1× Laemmli loading buffer. Proteins were separated on 4%–12% SDS-PAGE gels and transferred onto polyvinylidene fluoride membranes. After blocking, membranes were incubated with primary antibodies against TCL1A at 4°C overnight. The washed membrane was then incubated with secondary antibody (1:15,000 dilution) for an hour at room temperature. The membrane was visualized using super-signal ECL substrate (Thermo Scientific).
Statistics.
Data were analyzed using GraphPad Prism Software (San Diego, CA) and an R package. Data are presented as mean ± S.E.M. unless otherwise stated. Gene expression and NF-κB activities were analyzed using multivariate analysis of variance, followed by planed comparisons with appropriate post hoc tests. We also conducted t tests for all meaningful combinations (effect of treatments on WT and variant, effect of genotype on treatments vs. vehicle). To correct for the inflated type 1 error that results from multiple comparisons, we have obtained a false discovery rate for each comparison using the R package (the default “p.adjust” function with “BH” method).
Results
Correlation between the Expression of TCL1A and that of Toll-Like Receptors.
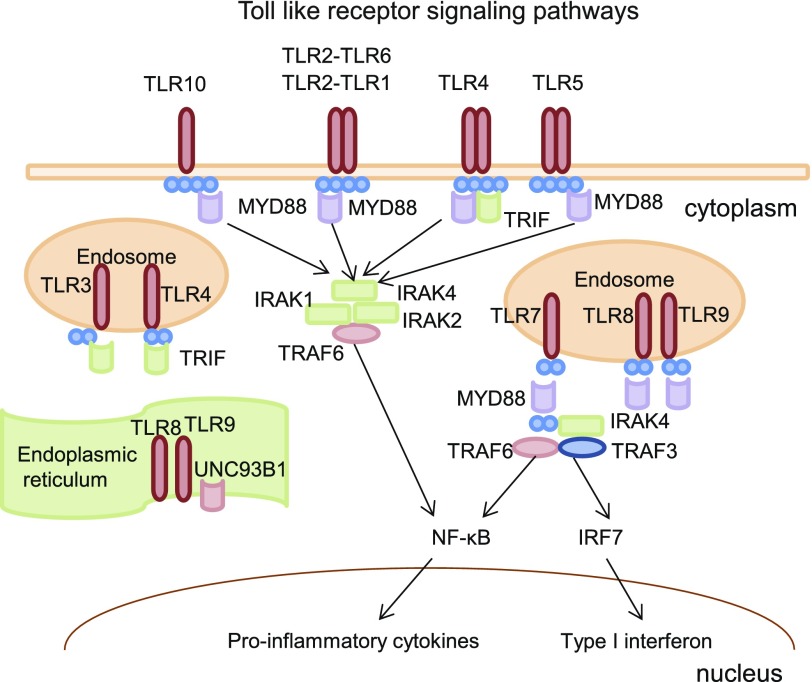
Excessive production of cytokines and chemokines mediated by toll-like receptor (TLR) pathways is often associated with an inflammatory response mediated though NF-κB signaling (Zeytun et al., 2010). The human TLR family consists of 10 isoforms (TLR1–TLR10). TLRs are synthesized in the endoplasmic reticulum (Kawasaki and Kawai, 2014). Most TLRs are located on the cell membrane, but TLR3, TLR7, TLR8, and TLR9 are located on the membranes of intracellular compartments, such as the endoplasmic reticulum, endosomes, and lysosomes (Fig. 2) (Joosten et al., 2016). The myeloid differentiation primary response gene 88 (MYD88) encodes one of the functional adapter molecules that has not been shown to interact with all TLRs except TLR3. MYD88 recruits interleukin-1R-activating kinase (IRAK) 1, IRAK2, IRAK4, and tumor necrosis factor receptor–associated factor 6, leading ultimately to NF-κB activation, proinflammatory cytokine secretion, and an inflammatory response (Fig. 2). In the present study, in an attempt to extend our observations with regard to TCL1A SNPs and estrogen-dependent expression of immune mediators, we set out to determine whether TLR expression might also be regulated by the TCL1A SNPs observed in our GWAS in an estrogen-dependent fashion and, if so, whether these observations might cast light on the role of TCL1A SNPs in NF-κB activation.
Fig. 2.
TLR intracellular localization and signaling. TLR3, TLR7, TLR8, and TLR9 are located on the membranes of intracellular compartments, such as the endoplasmic reticulum and endosomes. The myeloid differentiation primary response gene 88 (MYD88) is the one of the functional adapter molecules that have been reported to interact with all TLRs except TLR3. MYD88 recruits IL-IRAK1, IRAK2, IRAK4, and tumor necrosis factor–receptor associated factor 6 (TRAF6), leading ultimately to NF-κB activation and proinflammatory cytokine secretion and an inflammatory response. UNC93B1 is a multitransmembrane-domain-containing protein and plays a critical role in trafficking TLR7 and TLR9 from the endoplasmic reticulum to endosomes, where TLR7 and TLR9 transmit signals via MYD88/TRIF-dependent pathways (Kim et al., 2008).
We once again turned to the Human Variation Panel of LCLs, a cell line model system that has repeatedly demonstrated its power to generate pharmacogenomic hypotheses and to test hypotheses arising from clinical GWA studies (Li et al., 2008; Ingle et al., 2010, 2013; 2016; Niu et al., 2010, 2016; Liu et al., 2013, 2014; Ho et al., 2016). This model system consists of LCLs from 300 healthy subjects for whom we have generated seven million SNPs after imputation, as well as microarray data for basal levels of gene expression for each cell line. We began by asking whether TLR expression might be associated with TCL1A gene expression. Basal levels of expression, in the absence of E2, showed a significant correlation in these 300 cell lines of TCL1A expression with that of both TLR7 and TLR9, with P values of 6.43E–15 and 1.05E–09, respectively (Table 1). We also examined the correlation between the expression of MYD88, a critical adapter protein for inflammation signaling pathways downstream of TLRs (Takeuchi and Akira, 2010) and that of Unc93 Homolog B1 (UNC93B1), a molecule that plays a critical role in trafficking TLR7 and TLR9 from the endoplasmic reticulum to endosomes (Fig. 2) (Sasai and Iwasaki, 2011; Lee and Barton, 2014). Very significant correlations were also observed between basal expression levels of TLR9 and TLR7 with the expression of MYD88, with P values 1.71E–18 and 7.55E–09, respectively (Table 2). We also found that expression levels of MYD88 and UNC93B1 were themselves significantly correlated (r = 0.41, P = 7.66E–13) (Table 2); however, in the absence of E2, there was no significant SNP-dependent difference in the expression of the genes listed in Table 1 in these 300 LCLs (data not shown). Therefore, we also asked whether the expression of these genes might be influenced by the TCL1A SNPs in the presence of E2, as we had observed for proinflammatory cytokines, chemokines, and their receptors (Fig. 1, D–G) (Ho et al., 2016).
TABLE 1.
Correlations of TCL1A mRNA expression and those of toll-like receptors and MYDBB in the Human Variation Panel of 300 LCLs
| Gene | Gene | R | P Value |
|---|---|---|---|
| TCL1A | TLR1 | −0.089 | 1.36E-01 |
| TCL1A | TLR2 | 0.276 | 2.54E-06 |
| TCL1A | TLR3 | −0.051 | 3.98E-01 |
| TCL1A | TLR4 | 0.158 | 8.02E-03 |
| TCL1A | TLR5 | 0.027 | 6.47E-01 |
| TCL1A | TLR6 | −0.073 | 2.21E-01 |
| TCL1A | TLR7 | 0.443 | 6.43E-15 |
| TCL1A | TLR8 | 0.052 | 3.89E-01 |
| TCL1A | TLR9 | 0.354 | 1.05E-09 |
| TCL1A | TLR10 | −0.200 | 7.29E-04 |
| TCL1A | MYD88 | 0.273 | 3.50E-06 |
Statistical significance was considered P < 1.8E-07.
TABLE 2.
Correlations of MYD88 mRNA expression with those of toll-like receptors and UNC93B1 in the Human Variation Panel of 300 LCLs
| Gene | Gene | R | P Value |
|---|---|---|---|
| MYD88 | TLR1 | 0.19 | 1.36E-03 |
| MYD88 | TLR2 | 0.227 | 1.26E-04 |
| MYD88 | TLR3 | 0.061 | 3.10E-01 |
| MYD88 | TLR4 | 0.244 | 3.50E-05 |
| MYD88 | TLR5 | 0.057 | 3.42E-01 |
| MYD88 | TLR6 | 0.02 | 7.34E-01 |
| MYD88 | TLR7 | 0.491 | 1.71E-18 |
| MYD88 | TLR8 | -0.097 | 1.03E-01 |
| MYD88 | TLR9 | 0.336 | 7.55E-09 |
| MYD88 | TLR10 | 0.145 | 1.50E-02 |
| MYD88 | UNC93B1 | 0.41 | 7.66E-13 |
Statistical significance was considered P < 1.8E-07.
TLR Expression Can Be Regulated by TCL1A SNPs in an Estrogen-Dependent Fashion.
The first set of experiments used eight LCLs that were homozygous for WT and eight LCLs homozygous for variant genotypes for the TCL1A SNPs that we had shown to be associated with AI-induced musculoskeletal adverse events (Ingle et al., 2010) and that we had also shown to be associated with alterations in the expression of a series of cytokines and chemokines in a SNP- and estrogen-dependent fashion (Fig. 1, D–G) (Ho et al.). All the cell lines used were homozygous for either WT or variant genotype for all three TCL1A SNPs, as shown graphically in Fig. 1A. TCL1A and all the immune mediators listed in Table 1 are highly expressed in LCLs—which are Epstein-Barr virus–transformed B cells—which made the functional genomic studies described subsequently possible.
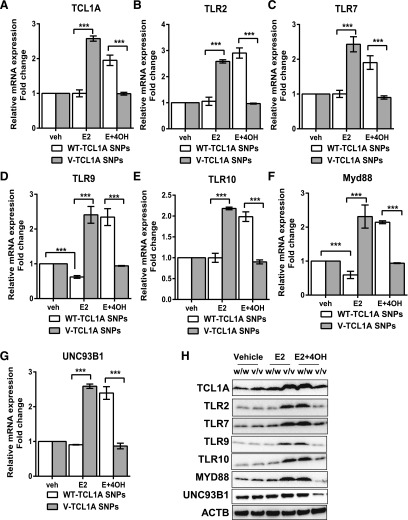
The initial experiment represented an attempt to determine whether the expression of TLRs and MYD88 could be modulated by TCL1A in a SNP-dependent fashion in response to E2 treatment, and to determine whether the direction of that change in expression could be reversed in response to 4-OH-TAM treatment—implying that the expression might be regulated by TCLIA genotype. Concentrations of E2 and 4-OH-TAM used to perform these experiments were those that were found to be optimal in previous studies of TCL1A induction by E2 and the reversal of that effect by 4-OH-TAM (Liu et al., 2012; Ho et al., 2016). TCL1A expression was induced by 0.1 nM E2 treatment, but only in cells homozygous for variant genotypes for the TCL1A SNPs (Fig. 3A)—confirming our previous results (Liu et al., 2012). This genotype-dependent gene expression pattern was reversed, however, when 4-OH-TAM (10−7µM) was added to E2—also confirming our previous results (Fig. 3A) (Ho et al., 2016). In parallel, the expression of TLR2, TLR7, TLR9, and TLR10 were upregulated (∼2- to 3-fold) in the presence of E2, but only in cells homozygous for variant genotypes for the TCL1A SNPs. In addition, as anticipated, in the presence of 4-OH-TAM, expression levels for these four TLRs increased significantly in cells homozygous for WT genotypes for the TCL1A SNPs (Fig. 3, B–E). Furthermore, other TLRs (TLR1, TLR3, TLR4, TLR5, TLR6, and TLR8) did not display alternation in their expression in a TCL1A SNP-dependent fashion in the presence of E2 (data not shown). In a similar fashion, we also observed that the expression of MYD88 and UNC93B1 were significantly upregulated in parallel with TCL1A in a SNP-estrogen-dependent fashion (Fig. 3, F–G). In addition, expression of those immune genes was significantly changed at the protein level as determined by Western blot analysis (Fig. 3H). It should be pointed out once again that these SNPs were in the TCL1A gene, as shown in Fig. 1A, not in genes encoding the immune mediators, that is, not in the TLR2, TLR7, TLR9, TLR10, MYD88, and UNC93B1 genes (Fig. 3).
Fig. 3.
SNP- and estrogen-dependent mRNA expression of TCL1A (A), TLR2 (B), TLR7 (C), TLR9 (D), TLR10 (E), MYD88 (F), and UNC93B1 (G) in LCLs (H). Western blot analysis was performed for TCL1A, TLR2, TLR7, TLR9 TLR10, MYD88, UNC93B1, and ACTB in LCLs with known TCL1A SNP genotypes. The cells were treated with 0.1 nM E2 or with 0.1 nM E2 plus 10−7µM 4-hydroxytamoxifen (4OH-TAM) for an additional 24 hours. Eight cell lines homozygous for the variant (V) genotypes for all three of the TCL1A SNPs and eight cell lines homozygous for WT genotypes were used in these experiments. ***P < 0.0001.
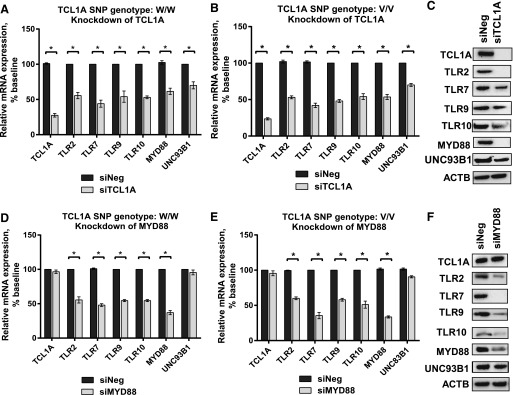
Knockdown of TCL1A Resulted in Decreased Expression of TLRs.
We next determined whether TCL1A itself was involved in the differences in expression for TLR2, TLR7, TLR9, TLR10, MYD88, and UNC93B1 shown in Fig. 3 using cell lines with differing genotypes for the TCL1A SNPs. Specifically, siRNA knockdown studies were performed using four independent siRNAs, as well as one pooled siRNA (Dharmacon Chicago, IL), and the results for all of these approaches were consistent. Knockdown of TCL1A to 25% of its baseline level resulted in significant downregulation of the expression of TLR2, TLR7, TLR9, TLR10, MYD88, and UNC93B1. We observed no SNP-dependent effect when TCL1A was knocked down (Fig. 4, A and B). In a similar fashion, knockdown of MYD88 resulted in significantly decreased TLR2, TLR7, TLR9, and TLR10 expression in LCLs, but TCL1A expression did not change (Fig. 4, D and E). Finally, protein expression was altered in parallel with the changes seen for mRNA after TCLIA or MYD88 knockdown (Fig. 4, C and F). These results indicated that the expression of TLR2, TLR7, TLR9, and TLR10 could be modulated by TCL1A and MYD88. It should be pointed out that knockdown of MYD88 did not alter the expression of TCL1A, suggesting that TCL1A is upstream of MYD88 in this signaling pathway. At this point, we had determined that TCL1A can regulate the expression of TLRs as well as that of MYD88, an important adaptor molecule that can trigger downstream signaling, including signaling through the NF-κB pathway.
Fig. 4.
TCL1A could modulate TLR2, TLR7, TLR9, TLR10, MYD88, and UNC93B1 expression in LCLs. Relative mRNA expression (A and B) of TCL1A, TLR2, TLR7, TLR9, TLR10, MYD88, and UNC93B1 after knockdown of TCL1A in LCLs with known TCL1A SNP genotypes using pooled siRNA. Eight cell lines of each genotype were used in these experiments. *P < 0.05. Student’s t test was performed to compare gene expression in LCLs with differing TCL1A SNP genotypes before and after gene knockdown, *P value ≤ 0.05 was considered statistically significant. All values are mean ±S.E.M for three separate independent assays. Protein expression was determined by Western blot analysis (C). Relative mRNA expression (D and E) of TCL1A, TLR2, TLR7, TLR9, TLR10, MYD88, and UNC93B1 after knockdown of MYD88 in LCLs with known TCL1A SNP genotypes using pooled siRNA. (F) Western blot analysis was performed for TCL1A, TLR2, TLR7, TLR9, TLR10, MYD88, and UNC93B1 after knockdown of MYD88.
TCL1A SNP and SERM-Mediated MYD88-Dependent NF-κB Activation.
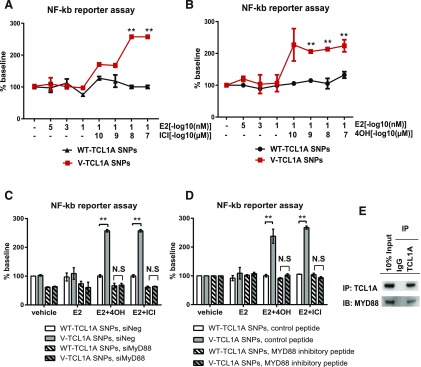
We had previously reported that changes in TCL1A expression after ER blockade using fulvestrant (ICI) could influence NF-κB transcriptional activity in a striking SNP genotype-dependent fashion (Liu et al., 2012). Specifically, we treated the same cells that were used to generate the results shown in Fig. 3, LCLs with differing genotypes for the TCL1A genotypes, with ICI or 4-OH-TAM. Cells homozygous for the variant genotype for the TCL1A SNPs increased NF-κB transcriptional activity approximately 3-fold in the presence of E2 plus either ICI or 4-OH-TAM (Fig. 5, A and B). MYD88 is the adaptor protein for all TLRs except TLR3 and is capable of activating the NF-κB signaling pathway and of inducing the production of inflammatory cytokines (Broad et al., 2007; Kawai and Akira, 2007). We had observed that MYD88 expression could be regulated by TCL1A in a SNP- and estrogen-dependent fashion (Fig. 3F) and that it is downstream of TCL1A (Fig. 4, A and B). The next series of experiments was designed to study the possible contribution of MYD88, a known adaptor molecule for TLR-mediated NF-κB signaling, to TCL1A SNP and SERM-dependent NF-κB activation. We found that, after knockdown of MYD88 to ∼30% of its baseline level, TCL1A SNP-dependent NF-κB transcriptional activation in the presence of either 4-OH-TAM or ICI was lost (Fig. 5C). In addition, when MYD88 was silenced by the application of MYD88 inhibitory peptide, the effects of the TCL1A SNP-dependent and ER blocker-dependent NF-κB activation were also lost, as anticipated (Fig. 5D). We also determined by coimmunoprecipitation that TCL1A could interact directly with MYD88 (Fig. 5E). These results further confirmed that TCL1A SNP and estrogen-dependent NF-κB activation may occur, in part, through a TLR-MYD88–dependent pathway. They also greatly extended our original observations and served to highlight a novel pharmacogenomic mechanism by which TCL1A can influence and modulate the immune response and inflammation.
Fig. 5.
TCL1A SNP- and estrogen-dependent NF-κB activation as determined by NF-κB reporter assays could be altered by the knockdown or inhibition of MYD88. ER blockade by fulvestrant (ICI) or 4-hydroxytamoxifen (4OH) treatment resulted in TCL1A SNP -dependent NF-κB activation (A and B). TCL1A SNP and estrogen-dependent NF-κB activation could be blocked by MYD88 siRNA knockdown (C) or by exposure to MYD88 inhibitory peptide (100 µM) (D). Specifically, LCLs were cotransfected with an NF-κB reporter construct and siRNA (MYD88 or control siRNA); 24 hours after transfection, cells were treated with either vehicle or 0.1 nM E2 for 24 hours, followed by 10−7µM 4OH or ICI for an additional 24 hours. In some experiments, cells were exposed to MYD88 inhibitory peptide for 24 hours before E2 treatment. Luciferase activity was measured 72 hours after transfection. The firefly luciferase activity derived from the NF-κB responsive reporter was normalized by the use of Renilla luciferase activity as a control to correct for possible variation in transfection efficiency. All experiments were repeated three times in triplicate. **P < 0.001. (E) Coimmunoprecipitation was used to determine whether TCL1A protein could interact with MYD88 in LCLs. Whole-cell lysates from 1 × 107 LCLs were immunoprecipitated with anti-TCL1A (1:50) antibodies or anti-IgG antibodies. Whole-cell lysate (input, left panel) and immunoprecipitated samples (middle and right panels) were immunoblotted and probed with antibodies against TCL1A and MYD88.
Discussion
We previously reported that SNPs 3′ of the TCL1A gene that we identified during a GWAS for musculoskeletal adverse events that occur during aromatase inhibitor therapy of breast cancer could have significant effects on the expression of a series of genes encoding inflammatory mediators in an SNP- and estrogen-dependent fashion. Even more striking, those effects could be reversed in a TCL1A SNP-dependent fashion by exposure to 4-OH-TAM or ICI (Ho et al., 2016). We also reported that variant TCL1A SNP genotypes were associated with increased NF-κB transcriptional activity after ER blockade, but that was not true for the WT genotype (Liu et al., 2012). NF-κB activation is known to play an important role in chronic inflammation (Hayden et al., 2006). As a result, there is great interest in developing specific molecular inhibitors of NF-κB activation. Therefore, the present study represents a step toward a better understanding of molecular and genetic mechanisms responsible for variation in NF-κB activation—understanding that could potentially have implications for the management of NF-kB–related inflammation.
In the present study, we pursued mechanisms associated with the TCL1A SNP-mediated, SERM-dependent activation of NF-κB. It has been reported that TCL1A can function as a transcriptional regulator, which interacts with many transcription factors, including AP1 and cAMP response element binding/p300 and, as a result, that it can influence NF-κB activity in B-cell chronic lymphocytic leukemia (Pekarsky et al., 2008). Since TLR signaling, signaling that occurs—in part—through MYD88, can influence NF-kB activity (Broad et al., 2007), MYD88 has been suggested as a potential therapeutic target for regulating the immune response (Olson et al., 2015). When TCL1A was knocked down in our cell line model system, MYD88 was significantly downregulated and, in parallel, expression levels of TLR2, TLR7, TLR9, and TLR10 were also decreased significantly (Fig. 4, C and D). Several TLRs can interact with MYD88 and NF-kB, as determined by STRING protein interaction network analysis (Szklarczyk et al., 2015). We also observed by IP studies that TCL1A protein could interact with MYD88 (Fig. 5E). We have reported previously that ER blockade by fulvestrant (ICI) can result in the activation of NF-κB transcriptional activity, but only in cells with variant genotypes for the TCL1A SNPs (3) (Fig. 5A). In the present study, we extended that finding by observing a similar response after exposure to 4-OH-TAM (Fig. 5B). We also demonstrated that the TCL1A SNP- and SERM-dependent activation of NF-κB through TLRs could be blocked by MYD88 siRNA or MYD88 inhibitory peptide (Fig. 5, C and D). Therefore, the present study has raised the possibility that TLR-MYD88–dependent NF-κB signaling could be involved in TCL1A SNP-dependent NF-kB activation and, as a result, in the regulation of inflammation and immune response. The fact that expression of a single gene, TCL1A, can influence signaling through this pathway represents an important advance in our understanding of the regulation of the pathway and suggests novel ways to influence signaling through this pathway.
Obviously, further studies will be required to determine the possible therapeutic implications of this series of observations. Specifically, we found that TCL1A expression was positively correlated with the basal expression of toll-like receptors TLR7 and TLR9 in 300 LCLs (r = 0.443, P = 6.43E–15, and r = 0.354, P = 1.05E–09, respectively), as listed in Table 1. Both TLR7 and TLR9 are therapeutic targets for the treatment of lupus erythematosus, an autoimmune disorder for which more than 90% of patients are women. We also found that TCL1A knockdown could influence the expression of TLR2 and TLR10, both of which can play a role in cytokine production, inflammation, and NF-κB activation via MYD88 (Joosten et al., 2016). Crosstalk between estrogens and the immune response is well documented (Seillet et al., 2012; Hughes and Choubey, 2014; Kovats, 2015). In the present study, we have demonstrated the differential expression of a series of genes encoding immune mediators in response to estrogen treatment that was dependent on TCL1A SNP genotypes. Even more striking, we showed that 4-OH-TAM, an active metabolite of tamoxifen, a Food and Drug Administration–approved treatment of breast cancer and for breast cancer chemopreventive that has been suggested for the treatment of millions of women at high risk for breast cancer (Maximov et al., 2013), can reverse the TCL1A SNP-dependent expression of those immune mediators—all of which play a critical role in inflammatory disease (Iwakura et al., 2011; Bartlett and Million, 2015).
We should emphasize that the LCL model system used in our studies has both strengths and weaknesses, but it has repeatedly demonstrated that it can be a powerful tool for both generating and testing genomic and pharmacogenomics hypotheses (Li et al., 2008, 2012, 2013, 2014; Ingle et al., 2010, 2013; Niu et al., 2010; Wheeler and Dolan, 2012). The availability of comprehensive genotype and gene expression data for this model system makes it possible to study the functional implications of genetic variants found to be associated with clinical phenotypes, as demonstrated by the present studies. We have previously used this model system successfully to demonstrate novel SNP- and estrogen-dependent mechanisms underlying variation in the regulation of the expression of cytokines and chemokines by TCL1A (Liu et al., 2012). In the present study, we have extended those observations with regard to the transcriptional regulation of immune mediators (cytokines and chemokines) to include TLRs and NF-κB transcriptional activity. Obviously, these results will have to be verified using both additional cell-based systems and clinical samples, but the present results represent an important step in the process of providing functional and mechanistic explanations for the association of SNPs in the TCL1A gene with inflammation and the immune response.
In summary, the series of functional genomic studies described here confirm that TCL1A expression is E2 inducible in an SNP-dependent manner and have demonstrated that TCL1A can influence the downstream expression of a series of immune mediators, including TLR2, TLR7, TLR9, TLR10, and MYD88. Furthermore, inhibition of MYD88 resulted in the blockade of TCL1A SNP-dependent NF-κB activation, indicating that the TLR-MYD88–dependent NF-κB signaling pathway might contribute, at least in part, to the TCL1A SNP- and estrogen-dependent effects that we had observed. TCL1A SNP- and estrogen-dependent variation in NF-κB transcriptional activity and the transcriptional regulation of other immune mediators suggests that this pathway may play a role in the complex interactions that are known to exist between the endocrine and immune systems. Of equal importance is the fact that SERMs are able to modulate this pathway in a striking fashion—opening the way for the pharmacologic regulation of this pathway in clinical setting.
Abbreviations
- 4-OH-TAM
4-hydroxytamxifen
- E2
estradiol
- ER
estrogen receptor
- ERE
estrogen response element
- FBS
fetal bovine serum
- GWAS
genome-wide association study
- ICI
fulvestrant
- IP
immunoprecipitation
- IRAK
interleukin-1R-activating kinase
- LCL
lymphoblastoid cell lines
- NFκB
nuclear factor κB
- PCR
polymerase chain reaction
- SERM
selective estrogen receptor modulator
- SNP
single nucleotide polymorphisms
- TCL1A
T-cell leukemia/lymphoma 1A
- TLR
toll-like receptor
- WT
wild-type
Authorship Contributions
Participated in research design: Ho, Ingle, Bongartz, Goss, Shepherd, Mushiroda, Kubo, Wang, Weinshilboum.
Conducted experiments: Ho.
Performed data analysis: Ho, Ingle, Bongartz, Wang, Weinshilboum.
Wrote or contributed to the writing of the manuscript: Ho, Ingle, Bongartz, Kalari, Goss, Shepherd, Mushiroda, Kubo, Wang, Weinshilboum.
Footnotes
References
- Bartlett HS, Million RP. (2015) Targeting the IL-17-T(H)17 pathway. Nat Rev Drug Discov 14:11–12. [DOI] [PubMed] [Google Scholar]
- Broad A, Kirby JA, Jones DEJ, Applied Immunology and Transplantation Research Group (2007) Toll-like receptor interactions: tolerance of MyD88-dependent cytokines but enhancement of MyD88-independent interferon-β production. Immunology 120:103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, West AP, Ghosh S. (2006) NF-kappaB and the immune response. Oncogene 25:6758–6780. [DOI] [PubMed] [Google Scholar]
- Ho M-F, Bongartz T, Liu M, Kalari KR, Goss PE, Shepherd LE, Goetz MP, Kubo M, Ingle JN, Wang L, et al. (2016) Estrogen, SNP-dependent chemokine expression and selective estrogen receptor modulator regulation. Mol Endocrinol 30:382–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GC, Choubey D. (2014) Modulation of autoimmune rheumatic diseases by oestrogen and progesterone. Nat Rev Rheumatol 10:740–751. [DOI] [PubMed] [Google Scholar]
- Ingle JN, Liu M, Wickerham DL, Schaid DJ, Wang L, Mushiroda T, Kubo M, Costantino JP, Vogel VG, Paik S, et al. (2013) Selective estrogen receptor modulators and pharmacogenomic variation in ZNF423 regulation of BRCA1 expression: individualized breast cancer prevention. Cancer Discov 3:812–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle JN, Schaid DJ, Goss PE, Liu M, Mushiroda T, Chapman J-AW, Kubo M, Jenkins GD, Batzler A, Shepherd L, et al. (2010) Genome-wide associations and functional genomic studies of musculoskeletal adverse events in women receiving aromatase inhibitors. J Clin Oncol 28:4674–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle JN, Xie F, Ellis MJ, Goss PE, Shepherd LE, Chapman JW, Chen BE, Kubo M, Furukawa Y, Momozawa Y, et al. (2016) Genetic polymorphisms in the long noncoding RNA MIR2052HG offer a pharmacogenomic basis for the response of breast cancer patients to aromatase inhibitor therapy. Cancer Res 76:7012–7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakura Y, Ishigame H, Saijo S, Nakae S. (2011) Functional specialization of interleukin-17 family members. Immunity 34:149–162. [DOI] [PubMed] [Google Scholar]
- Joosten LAB, Abdollahi-Roodsaz S, Dinarello CA, O’Neill L, Netea MG. (2016) Toll-like receptors and chronic inflammation in rheumatic diseases: new developments. Nat Rev Rheumatol 12:344–357. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. (2007) Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med 13:460–469. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Kawai T. (2014) Toll-like receptor signaling pathways. Front Immunol 5:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-M, Brinkmann MM, Paquet M-E, Ploegh HL. (2008) UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature 452:234–238. [DOI] [PubMed] [Google Scholar]
- Kovats S. (2015) Estrogen receptors regulate innate immune cells and signaling pathways. Cell Immunol 294:63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BL, Barton GM. (2014) Trafficking of endosomal Toll-like receptors. Trends Cell Biol 24:360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Fridley B, Kalari K, Jenkins G, Batzler A, Safgren S, Hildebrandt M, Ames M, Schaid D, Wang L. (2008) Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res 68:7050–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Goss PE, Ingle JN, Kubo M, Furukawa Y, Batzler A, Jenkins GD, Carlson EE, Nakamura Y, Schaid DJ, et al. (2014) Aromatase inhibitor-associated bone fractures: a case-cohort GWAS and functional genomics. Mol Endocrinol 28:1740–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Ingle JN, Fridley BL, Buzdar AU, Robson ME, Kubo M, Wang L, Batzler A, Jenkins GD, Pietrzak TL, et al. (2013) TSPYL5 SNPs: association with plasma estradiol concentrations and aromatase expression. Mol Endocrinol 27:657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wang L, Bongartz T, Hawse JR, Markovic SN, Schaid DJ, Mushiroda T, Kubo M, Nakamura Y, Kamatani N, et al. (2012) Aromatase inhibitors, estrogens and musculoskeletal pain: estrogen-dependent T-cell leukemia 1A (TCL1A) gene-mediated regulation of cytokine expression. Breast Cancer Res 14:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximov PY, Lee TM, Jordan VC. (2013) The discovery and development of selective estrogen receptor modulators (SERMs) for clinical practice. Curr Clin Pharmacol 8:135–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool KW, Miyamoto S. (2012) DNA damage-dependent NF-κB activation: NEMO turns nuclear signaling inside out. Immunol Rev 246:311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu N, Liu T, Cairns J, Ly RC, Tan X, Deng M, Fridley BL, Kalari KR, Abo RP, Jenkins G, et al. (2016) Metformin pharmacogenomics: a genome-wide association study to identify genetic and epigenetic biomarkers involved in metformin anticancer response using human lymphoblastoid cell lines. Hum Mol Genet 25:4819–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu N, Qin Y, Fridley BL, Hou J, Kalari KR, Zhu M, Wu T-Y, Jenkins GD, Batzler A, Wang L. (2010) Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res 20:1482–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MA, Lee MS, Kissner TL, Alam S, Waugh DS, Saikh KU. (2015) Discovery of small molecule inhibitors of MyD88-dependent signaling pathways using a computational screen. Sci Rep 5:14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y, Palamarchuk A, Maximov V, Efanov A, Nazaryan N, Santanam U, Rassenti L, Kipps T, Croce CM. (2008) Tcl1 functions as a transcriptional regulator and is directly involved in the pathogenesis of CLL. Proc Natl Acad Sci USA 105:19643–19648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai M, Iwasaki A. (2011) Love triangle between Unc93B1, TLR7, and TLR9 prevents fatal attraction. Immunity 35:3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C, Laffont S, Trémollières F, Rouquié N, Ribot C, Arnal J-F, Douin-Echinard V, Gourdy P, Guéry J-C. (2012) The TLR-mediated response of plasmacytoid dendritic cells is positively regulated by estradiol in vivo through cell-intrinsic estrogen receptor α signaling. Blood 119:454–464. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak PP, Firestein GS. (2001) NF-kappaB: a key role in inflammatory diseases. J Clin Invest 107:7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Harada Y, Watanabe R, Inutake Y, Ogawa S, Onuki K, Kagaya S, Tanabe K, Kishimoto H, Abe R. (2008) CD28 stimulation triggers NF-kappaB activation through the CARMA1-PKCtheta-Grb2/Gads axis. Int Immunol 20:1507–1515. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140:805–820. [DOI] [PubMed] [Google Scholar]
- Wheeler HE, Dolan ME. (2012) Lymphoblastoid cell lines in pharmacogenomic discovery and clinical translation. Pharmacogenomics 13:55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeytun A, Chaudhary A, Pardington P, Cary R, Gupta G. (2010) Induction of cytokines and chemokines by toll-like receptor signaling: strategies for control of inflammation. Crit Rev Immunol 30:53–67. [DOI] [PubMed] [Google Scholar]