Summary
Inflammasome signalling induces the processing and secretion of interleukin (IL)‐1β and IL‐18 which, coupled with pyroptosis, activate further the inflammatory response. In the present study we evaluated the expression of genes involved in inflammasome signalling pathways in septic patients, their interaction networks and the predicted functions modulated in survivors and non‐survivors. Twenty‐seven patients with sepsis secondary to community‐acquired pneumonia admitted to intensive care units from three general hospitals in São Paulo were included into the study. We performed a polymerase chain reaction (PCR) array encompassing 35 genes related to the nucleotide‐binding oligomerization domain and leucine‐rich repeat‐containing (NLR)‐inflammasome in peripheral blood mononuclear cells obtained at admission and after 7 days of follow‐up. Eleven healthy volunteers were used as the reference group. Increased NLRC4 and NLRP3 and decreased nucleotide‐binding oligomerization domain (NOD1), and NLRP1 expression was observed in septic patients compared to healthy individuals; the IL‐1β and IL‐18 expression levels were also high in the patients. The gene expression changes followed the same patterns in surviving and non‐surviving patients, with higher magnitudes observed in non‐survivors. Functional analyses revealed, however, that activation and inhibition intensity for representing functions were different in survivors and non‐survivors, as for production of reactive oxygen species, synthesis of nitric oxide and for the control of bacterial infections. Our results showed that the genes involved in the activation of the NLR‐inflammasome cascades were altered substantially in septic patients, with a higher number of altered genes and a higher intensity in the disturbance of gene expression found among patients dying of sepsis.
Keywords: interleukin‐18, interleukin‐1beta, NLRP3, pyroptosis, Toll‐like receptor
Introduction
Sepsis has been defined as a systemic inflammatory response syndrome (SIRS) triggered by an infection 1. This concept has been reviewed recently, and sepsis is now defined as a life‐threatening organ dysfunction caused by a dysregulated host response to infection 2.
Bacterial sensing and induced cell signalling are modulated during sepsis, with up‐ and down‐regulated functions observed during the ongoing infection process 3.
The nucleotide‐binding oligomerization domain and leucine‐rich repeat‐containing (NLR) proteins are intracellular receptors that signal for pathogen‐associated molecular patterns (PAMPs) and damage or danger signals (DAMPs) 4. The inflammasome is a multi‐protein complex composed of NLR or interferon (IFN)‐inducible protein or ALR member absent in melanoma 2 (AIM2), an adaptor protein [an apoptosis‐associated speck‐like protein containing a CARD domain (ASC)] and proinflammatory caspases 5. Following stimulation, NLRs recruit and activate caspase 1, leading to the conversion of pro‐interleukin (IL)‐1β and pro‐IL‐18 into their active forms 6, 7. Inflammasome signalling also induces pyroptosis that further activates the inflammatory response 8.
Interactions exist between Toll‐like receptors (TLR) and NLR signalling. NLRP3 activation requires two signals; the first is dependent upon nuclear factor kappa B (NF‐κB) activation, which is induced through the TLR or nucleotide‐binding oligomerization domain (NOD) receptors, and the second is triggered by one of the NLRP3‐activating stimuli, such as extracellular adenosine triphosphate (ATP) 8. Bacterial flagellin is recognized at the cell surface by TLR‐5, leading to NF‐κB activation, whereas the NLRC4 inflammasome senses flagellin in the cytosol, resulting in caspase‐1 activation 9. The TLR signalling pathways have been studied extensively in experimental models and in clinical sepsis 3, 10, 11, and there is increasing interest in evaluating inflammasome activation in these settings.
Disturbances in inflammasome activation have been implicated in the pathogenesis of several diseases, including diabetes and atherosclerosis 12. Conflicting results have been reported in septic patients 13, 14.
In this study, we evaluated the expression of genes involved in the NLR‐inflammasome signalling pathways in septic patients. To avoid patient heterogeneity, we selected patients with community‐acquired pneumonia as a source of sepsis. Gene expression was evaluated at admission and after 7 days of follow‐up in the surviving and non‐surviving patients.
Material and methods
Septic patients and healthy volunteers
Patients with a clinical diagnosis of severe sepsis and septic shock admitted to the intensive care units of three large hospitals in São Paulo, Brazil, were enrolled prospectively into the cohort 15. This study was approved by the ethics committees of the participating hospitals, São Paulo Hospital (study number 1477/06), Albert Einstein Hospital (study number 07/549) and Sirio Libanes Hospital (study number 2006/27). The diagnosis was made according to the American College of Chest Physicians/Society of Critical Care Medicine consensus 1, 16, and corresponded approximately to the revised concepts of sepsis and septic shock 2. Samples were obtained within 48 h of the first organ dysfunction or shock and after 7 days, after informed consent was obtained from the donor or relative. Patients less than 18 years old, on immunosuppressive therapy, with AIDS or end‐stage chronic disease or on experimental therapy were excluded. After approval by the ethics committee, we selected patients with sepsis secondary to community pneumonia and healthy volunteers for the present study.
Isolation of peripheral blood mononuclear cells (PBMCs)
PBMCs were isolated using the Ficoll gradient method 17, 18 (Ficoll‐Paque PLUS; GE Healthcare, Uppsala, Sweden) and stored in liquid nitrogen prior to use.
Reverse transcription–polymerase chain reaction (RT–PCR) array
Gene expression was evaluated using the RT–PCR array (Qiagen, Valencia, CA, USA) in customized plates consisting of 35 genes involved in the NLR‐inflammasome pathways. The selected genes and their functions are described in Supporting information, Table S1. Amplification, data acquisition and melting curve analysis were performed using the Applied Biosystems 7500 RT–PCR System (Applied Biosystems, Carlsbad, CA, USA). The SUGT1 gene was used as an internal control. The fold change ratio for each gene (2(–ΔΔCt)) was compared between the septic groups and healthy controls, and was considered relevant when it was ≥ 1·5. The P‐values are calculated based on a Student's t‐test of the 2(–ΔCt) values for pairwise comparison between the control group and septic group (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php). Analysis of variance (anova) followed by Dunnett's post‐hoc multiple comparisons test was used to compare the results between septic groups (survivors and non‐survivors, D0 and D7) against the control group. Gene expression was considered modulated differentially compared to the healthy volunteers when the fold change was ≥ 1·5 and the P‐value was ≤ 0·05.
Hierarchical cluster analysis was performed using Genesis (http://genome.tugraz.at). Ingenuity pathway analysis (IPA) (Ingenuity Pathway Knowledge Database, Redwood City, CA, USA) was used for the functional analysis.
Validation of IL‐18 and IL‐1β
IL‐18 and IL‐1β were measured in plasma samples using ELISA (MBL‐Immunotech; Nagoya, Japan) and cytometric bead array (CBA) (BD Biosciences, San Jose, CA, USA), respectively, according to the manufacturer's instructions.
Results
Patients and healthy volunteers
Twenty‐seven patients with sepsis secondary to community‐acquired pneumonia were enrolled into the study. The hospital mortality rate was 29·6%. Eleven healthy volunteers, matched for age (56·4 years ± 22) and gender (72·7% male), were included as the control group (Table 1).
Table 1.
Demographic data and outcomes from septic patients included into the study
| Septic patients (n = 27) | |
|---|---|
| Age [mean (s.d.)] | 62·7 (20·7) |
| Gender [n (%)] | |
| Male | 19 (70·4) |
| Female | 8 (29·6) |
| Stages of sepsis [n (%)] | |
| Sepsis | 9 (33·3) |
| Septic shock | 18 (66·7) |
| SOFA D0 score [mean] | 7·5 ± 3·55 |
| APACHE II score [mean] | 19 ± 6·46 |
| In‐hospital mortality [n (%)] | |
| Survivors | 19 (70·4) |
| Non‐survivors | 8 (29·6) |
| Outcome accordingly to stage at enrolment [n (%)] | |
| Sepsis | |
| Survivors | 8 (42·2) |
| Non‐survivors | 1 (12·5) |
| Septic shock | |
| Survivors | 11 (57·8) |
| Non‐survivors | 7 (87·5) |
SOFA = sepsis‐related organ failure assessment; APACHE II = acute physiology and chronic health evaluation; s..d. = standard deviation.
Gene expression in septic patients
Gene expression was analysed in the septic patients at D0 and at D7, and according to the patients' outcomes (survivors and non‐survivors).
Among the 35 genes evaluated, 10 presented with increased fold change (FC ≥ 1·5) in patients at D0 compared with the healthy volunteers. When gene expression was analysed according to the outcomes, a similar pattern was found in the surviving patients, who presented 10 up‐regulated genes. In contrast, 19 genes showed increased fold changes in the non‐surviving patients: 12 were up‐regulated, six with P ≤ 0·05, and seven were down‐regulated, five with P ≤ 0·05. At D7, seven genes were expressed differentially in the septic patients, six of which were up‐regulated. Similar results (eight genes) were observed in the surviving patients on D7, whereas in the non‐surviving patients 13 genes presented altered fold changes, with 10 up‐regulated (four with P ≤ 0·05) (Table 2).
Table 2.
Fold change ratios of gene expression in septic patients compared to the healthy volunteers at admission and after 7 days of follow‐up and according to the outcomes
| Total patients n = 27 | Survival n = 19 | Non‐survival n = 8 | ||||
|---|---|---|---|---|---|---|
| Genes | D0 | D7 | D0 S | D7 S | D0 NS | D7 NS |
| BCL10 | 1·2852 a | 1·1625 | 1·2866 | 1·0937 | 1·2823 | 1·3623 |
| CARD6 | 1·7259 | 1·7322 | 1·699 | 1·6294 | 1·7847 | 2·031 |
| CARD8 | −1·0865 | −1·014 | −1·119 | −1·0508 | −1·0207 | 1·0818 |
| CARD9 | 1·1883 | 1·1182 | 1·1291 | 1·0502 | 1·3248 | 1·3164 |
| CASP1 | 1·1385 | 1·1853 | 1·0595 | 1·1655 | 1·3265 | 1·2383 |
| CASP5 | 1·6884 | 1·2059 | 1·6287 | 1·0388 | 1·8226 | 1·7768 |
| CASP8 | −1·1404 | −1·2408 | −1·0067 | −1·216 | −1·4862 | −1·3078 |
| CASP9 | 1·2352 | 1·0748 | 1·3021 | 1·1089 | 1·1041 | −1·009 |
| CD40 | −1·3963 | −1·3183 | −1·2715 | −1·2805 | –1·7037 | −1·4217 |
| CHUK | 1·3304 a | 1·3166 a | 1·33 | 1·2731 | 1·3313 a | 1·437 a |
| CIAPIN1 | −1·1318 | −1·1084 | −1·0605 | −1·0289 | −1·2996 | −1·3452 a |
| ERBB2IP | −1·2545 | −1·1564 | −1·1756 | −1·0748 | −1·4403 | −1·399 |
| NDUFA13 | −1·2554 | −1·446 | −1·091 | −1·2525 | –1·6916 | –2·1008 |
| HSP90AA1 | 1·0541 | −1·0323 | 1·0337 | −1·0007 | 1·099 | −1·1191 |
| IKBKB | −1·3712 a | −1·2313 a | −1·2572 | −1·231 | –1·649 a | −1·2321 |
| IKBKG | −1·0041 | −1·034 | 1·0893 | 1·1095 | −1·2146 | −1·4778 |
| IL‐10 | 2·451 | 1·282 | 2·7971 | 1·3173 | 1·851 | 1·1946 |
| IL‐18 | 1·8261 a | 1·7549 a | 1·5988 | 1·6802 | 2·4223 a | 1·9648 a |
| IL‐1β | 4·1022 a | 4·2624 | 3·0019 | 2·9438 | 7·9654 a | 11·1579 a |
| MALT1 | −1·4651 | −1·2681 | −1·3045 | −1·2153 | –1·8751 a | −1·4164 |
| NAIP | 1·4891 | 1·1998 | 1·5794 | 1·2226 | 1·3138 | 1·1423 |
| NFKB1 | 1·1827 | 1·0914 | 1·1759 | 1·2177 | 1·1973 | −1·2182 |
| NFKB2 | 1·0346 | −1·1139 | 1·1035 | −1·059 | −1·1084 | −1·2704 |
| NLRC4 | 1·976 a | 1·7127 | 1·9406 | 1·5586 | 2·0535 | 2·1883 |
| NLRP1 | –1·6496 | –1·5266 | −1·373 | −1·364 | –2·4362 a | –2·0461 |
| NLRP3 | 2·375 | 1·943 | 1·9546 | 1·6138 | 3·5931 a | 3·1486 |
| NOD1 | −1·481 a | −1·373 | −1·3405 | −1·2786 | –1·8303 a | –1·6524 |
| NOD2 | 1·2297 | 1·0907 | 1·1524 | 1·1157 | 1·4119 | 1·0284 |
| PSTPIP1 | 1·0881 | 1·0785 | 1·1441 | 1·1392 | −1·0224 | −1·0689 |
| PYCARD | 1·4954 a | 1·2964 | 1·4645 a | 1·2265 | 1·5632 a | 1·4975 |
| RIPK2 | 1·3811 | 1·3609 | 1·2448 | 1·2229 | 1·7223 a | 1·7973 |
| TLR‐5 | 1·4752 a | 1·2241 | 1·3831 | −1·0024 | 1·6918 a | 2·0839 a |
| TNF | 2·5498 | 2·8503 | 1·9068 | 2·2815 | 4·7281 a | 5·0841 a |
| TNFAIP3 | 1·6389 | 1·7467 | 1·5695 | 1·6243 | 1·7967 | 2·1098 |
| TRAF6 | −1·2829 | −1·1356 | −1·1577 | −1·0951 | –1·5958 a | −1·2482 |
D0 represent total septic patients after admission and D7 represent after 7 days. We further separated the gene fold changes on the basis of survival (D0S and D7S) and non‐survival (D0NS and D7NS). The bold letters show fold change ≥ 1·5; a P‐value ≤ 0·05.
NOD1 was expressed at a lower level in the septic patients and was down‐regulated significantly in the non‐surviving patients at D0. NOD2 did not show differences between the patients and healthy volunteers. Among the genes related to the NOD signalling pathway, TRAF6, IKBKB and MALT1 were down‐regulated in the non‐surviving patients. NLRP1 showed lower expression in the septic patients; this down‐regulation was more pronounced and significant in the non‐surviving patients. In contrast, the flagellin sensors NLRC4 and TLR5 were up‐regulated in the septic patients. NLRP3 was up‐regulated in the septic patients at D0 and D7, with a higher FC and significance (D0) in the non‐surviving patients. The IL‐1β and IL‐18 cytokines were up‐regulated, again with more robust regulation in the non‐surviving patients (Table 2).
Overall, the gene expression changes followed the same pattern in the surviving and non‐surviving patients, with higher changes observed in non‐survivors. This result is illustrated in the clustergram (Fig. 1), which segregated the surviving and non‐surviving patients and demonstrated that few genes showed divergent regulation.
Figure 1.
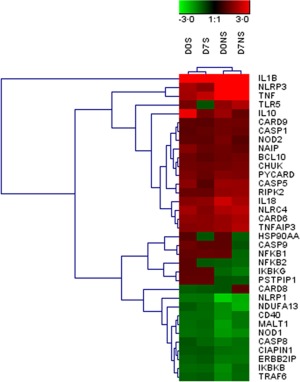
Hierarchical clustering of differentially expressed genes when compared with healthy volunteers. D0S represents survival day 0, D0NS represents non‐survival day 0, D7S represents survival after day 7 and D7NS represents the day 7 non‐survival group. The red colour indicates up‐regulation and green indicates down‐regulation. The colour intensity depends upon the change ratio; the higher the ratio, the higher the intensity.
Network and biological functions altered in septic patients
The network analysis showed that tumour necrosis factor (TNF), IL‐18, T helper type 1 (Th1) cytokines and NLRP3 occupied central NODS in the septic patients. Several genes related to NLRP signalling and NF‐κB signalling were identified in the network analysis. NLRC4, NLRP3, CARD6, Casp5 and IL‐18 were up‐regulated in the septic patients, whereas NLRP1 was down‐regulated. When the analyses were performed according to the outcomes, a wider gene alteration was illustrated clearly in non‐survivors than in survivors at day 0 (Supporting information, Fig. S1). The changes in gene expression in the admission samples in septic patients resulted in the activation of various functions, such as macrophage cell death, apoptosis, proinflammatory response, nitric oxide (NO) synthesis, macrophage activation, and a cellular immune response that inhibited bacterial infections (Fig. 2a). When we differentiated the expression profiles based on the final outcome, most of the altered functions were common in both the surviving and non‐surviving patients. However, the activation and inhibition intensities (Z‐scores) representing the functions were different (e.g. for reactive oxygen species (ROS) production, NO synthesis, pyroptosis and control of bacterial infections) (Fig. 2b,c). Most of the functional pattern alterations were similar in the follow‐up samples, but had different activation or inhibition Z‐scores (data not shown).
Figure 2.
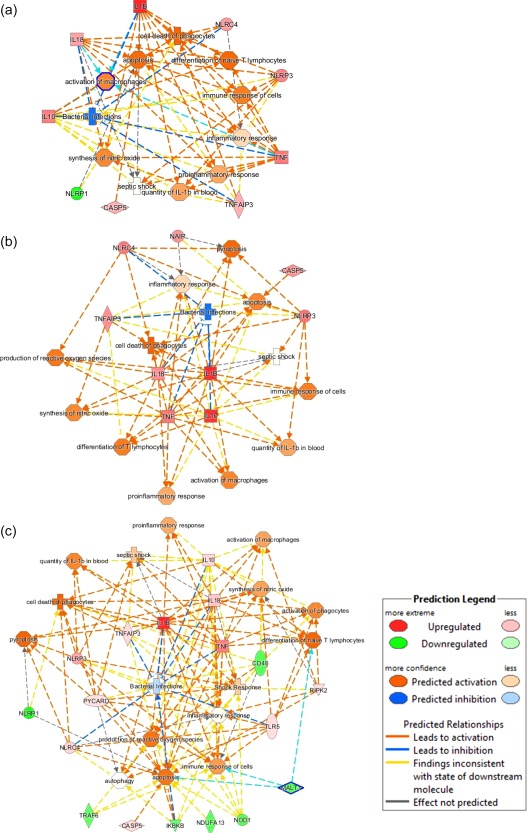
Identification of altered functions in septic patients (a), survival (b) and non‐survival (c) septic patients by ingenuity pathway analysis (IPA). Samples were obtained at D0. The node colour intensity represents activation (orange with positive Z‐score) and inhibition (blue with negative Z‐score) of functions, while genes up‐regulation is shown in red and down‐regulation in green.
IL‐18 and IL‐1β protein expression levels
We observed higher IL‐18 and IL‐1β expression at the gene level in the septic patients, and measured their plasma levels. IL‐18 plasma levels were significantly higher in septic patients at D0 and D7 than in the healthy volunteers (Fig. 3). No significant differences were detected between the surviving (981·4 pg/ml; range: 363·6–3464·2 pg/ml) and non‐surviving patients (936·0 pg/ml; range: 362·6–3097·6 pg/ml). IL‐1β was not detected in plasma of septic patients.
Figure 3.
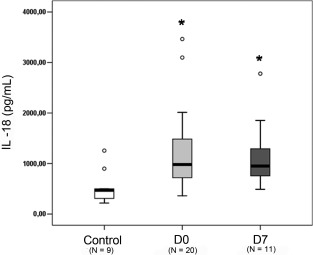
Detection of interleukin (IL)‐18 in septic patients. Plasma levels were measured by enzyme‐linked immunosorbent assay (ELISA) and values are expressed in pg/ml. The data are represented as box‐plot, where D0 and D7 represent the days of sample collection. *P ≤ 0·05 compared to healthy volunteers.
Discussion
Our results show that the genes involved in the activation of the NLR‐inflammasome cascades are altered substantially in septic patients. Increased expression of the NLRC4 and NLRP3 receptors and decreased expression of NOD1 and NLRP1 were observed in the septic patients compared to the healthy individuals. Importantly, IL‐1β and IL‐18, as well as TNF‐α, showed high expression levels in the patients. This scenario summarized the general disturbance in inflammasome‐related genes that we found in clinical sepsis; the relevance of these results for recovery or disease progression was supported by the remarkable differences between the surviving and non‐surviving patients, with a higher number of altered genes and a higher magnitude of gene regulation found among the patients dying of sepsis.
The inflammasome has emerged as a key modulator of the immune response and has been related to the pathogenesis of inflammatory and infectious diseases through the release of proinflammatory cytokines and pyroptic cell death 8. Bacterial recognition by the inflammasome is fundamental for the restriction of bacterial growth and host survival 9, 19. Nevertheless, few studies have addressed NOD and NLR‐inflammasome signalling in clinical sepsis.
We found that NLRP3 gene expression was up‐regulated in septic patients, with higher expression in non‐surviving patients than in survivors compared with healthy volunteers. ROS and NO are mediators that can induce the NLRP3‐inflammasome 8, 9. Accordingly, we have shown previously that ROS and NO generation was increased in monocytes and neutrophils from septic patients 20, including patients belonging to the same cohort in this study 21, even in the presence of suppressed TNF‐α and IL‐6 production 22. ROS‐driven NLRP‐3 activation may be derived from the nicotinamide adenine dinucleotide phosphate (NADPH)‐oxidase, which we found to be assembled in PBMCs from septic patients 22 or from dysfunctional mitochondria 23, which are reported commonly to be present in septic patients 24, 25. Accordingly, in a previous study of patients with sepsis secondary to community‐acquired pneumonia, we found differences in the expression profiles of genes related to the mitochondrial electron transport chain (ETC) I–V between survivors and non‐survivors 26.
There are consistent data showing the induction of the NLRP3‐inflammasome in experimental sepsis. Garcia and co‐workers demonstrated the interaction of NF‐κB and NLRP3, which led to a proinflammatory and pro‐oxidant status in the heart tissues of septic mice 27. Kebaier et al. 28 showed that NLRP3 inflammasome activation was deleterious rather than protective to the lung tissues during pneumonia.
NLRC4 and TLR5 were up‐regulated in septic patients, again with more robust regulation in the non‐surviving patients. Thus, these important membrane and cytosolic sensors of flagellin are up‐regulated similar to NLRP3 and may contribute to the conversion of IL‐1β and IL‐18 into mature proteins and to pyroptosis. The increased TLR5 gene expression was supported by a previous work in which we found that TLR‐5 detection on the cell surface was higher among septic patients than healthy volunteers 29. Interestingly, pretreatment of monocytes from human volunteers with lipopolysaccharide (LPS)‐induced tolerance to LPS and macrophage‐activating lipopeptide‐2 (MALP‐2) (a TLR‐2/6 agonist), but did not change the intracellular detection of IL‐6 after challenge with flagellin. The recognition of flagellin by NLRC4 (in addition to TLR‐5) was proposed as one possible explanation for this finding 30. TLR‐5 and NLRC4 in combination are required for maximal protective lung innate mucosal immunity against Pseudomonas aeruginosa. A significant increase in mortality was shown in TLR‐5/NLRC4−/− mice, which was associated with an increase in P. aeruginosa colony‐forming units (CFUs) in the lung and systemic bacterial dissemination 31.
IL‐1β and IL‐18 gene expression was up‐regulated in septic patients, again with more robust modulation in the non‐surviving patients. We were unable to detect circulating IL‐1β in the plasma samples, due possibly to the kinetics of the cytokine and the preservation of the samples. Increased levels of IL‐1β have been reported previously in septic patients 32, 33. IL‐18 was detected in higher levels in the plasma from septic patients than in the plasma from the healthy volunteers, as reported previously 34, 35. Both cytokines play major roles in the differentiation of T lymphocytes into the Th1 and Th17 subpopulations 36. Accordingly, the network analysis predicted differentiation of T lymphocytes as one of the functions modulated in septic patients. In our previous work, we found an increased proportion of Th17 cells and a decreased proportion of Th1 cells in septic patients in the context of decreased T lymphocyte cell numbers 37. We were unable to evaluate the expression of IL‐33, a member of the IL‐1 family which shows similarity to IL‐18 38. Madouri et al. demonstrated the reduced production of IL‐33 through caspase‐1 and NLRP3 activation in allergic lung inflammation 39. Furthermore, IL‐33 activates neutrophils by inhibition of G protein‐coupled receptor kinase (GRK2) through TLR signalling and maintains the level of CXCR2 for pathogen clearance 40.
In contrast to NLRC4, NOD1 was down‐regulated in the septic patients, again with a more robust disturbance in the non‐surviving patients. The same was true for NLRP1. The NOD1 and NOD2 receptors recognize peptides derived from PGN, which is present in the bacterial cell wall. This complex recruits the inhibitor of NF‐κB (IKK), leading to NF‐κB activation 41. NF‐κB gene expression was unaltered in our present study, whereas IKBKB was down‐regulated in the non‐survivor admission samples. NLRP1 is a component of the originally described inflammasome and is characterized by its activation by the anthrax lethal toxin 8.
Thus, our results show up (NLRP3 and NLRC4)‐ and down (NOD1 and NLRP1)‐regulation of NLRs in patients with sepsis. Previous studies have led to conflicting inflammasome activation results in critically ill and septic patients. In the study by Fahy and co‐workers, decreased NALP1 and CASP1 gene expression was found in septic shock patients compared with critically ill patients and healthy volunteers, whereas NOD1, NOD2 and NALP3 expression was similar between the groups. The authors concluded that the changes in the inflammasome were part of the monocyte deactivation process that occurred in septic patients 13. In contrast, Dolinay and co‐workers reported increased CASP1, IL‐1β and IL‐18 expression in septic patients with ARDS compared to SIRS 14. Similarly, increased IL‐18 plasma levels were found recently in septic patients, with higher levels in non‐surviving patients 42.
Pattern recognition receptors (PRRs) co‐operate to recognize a variety of microbial infections by sensing microbial or danger molecules 4. The same is true for the diverse NLR‐inflammasomes that play redundant and complementary roles, as shown for NLRC4 and TLR‐5 31 and NLRP3 and NLRC4 43. Thus, one of the most relevant contributions of the present work derives from the network analysis showing gene interactions and the predicted activation and inhibition of cellular functions. This analysis is particularly important because we found up‐ and down‐regulation of diverse NLR genes. The functional analyses indicated macrophage activation, NO synthesis, apoptosis, pyroptosis and control of bacterial infections in septic patients. The individual gene expression analyses showed that the differences between survivors and non‐survivors were related mainly to the magnitude and duration of the disturbance.
Limitations
Sepsis is a complex syndrome with several primary sources of infection. To avoid patient heterogeneity, we selected patients with sepsis secondary to community‐acquired pneumonia from a cohort of septic patients 15. Thus, our data might not be representative of the spectrum of septic patients or even of patients with less severe pneumonia. To illustrate this issue, we found differential proteomic responses in septic patients secondary to community and hospital‐acquired pneumonia in a preliminary analysis of plasma proteomics in septic patients 44. Our primary focus was to study genes which are participating actively in inflammasome signalling. Thus, our data are based on the gene expression profile of 35 selected genes. Despite the limited number of genes, we tried to identify affecting signalling and function using IPA from significant differentially expressed genes. Nevertheless, our networking analysis and predicted cell functions are consistent with previous functional studies in sepsis. Finally, the sample size was small, even though a single source of sepsis was investigated.
Conclusion
Previous studies evaluating inflammasome gene expression in septic patients have led to conflicting results. In this study, evaluating a broad array of genes involved in inflammasome signalling, we show NLR up‐regulation (NLRP3 and NLRC4) and down‐regulation (NOD1 and NLRP1) in patients with sepsis, with more intense disturbances in non‐survivors than in survivors. The functional analysis showing gene interactions and the predicted activation and inhibition of cellular functions showed that the activation of ROS production, NO synthesis and inhibition of bacterial infections was higher in survivors than in non‐survivors.
Ethical approval and consent to participate
This study was approved by the ethics committees of the participating hospitals, São Paulo Hospital (study number 1477/06), Albert Einstein Hospital (study number 07/549) and Sírio Libanes Hospital (study number 2006/27). Samples were obtained prospectively after informed consent was obtained from the donor or relative. After approval by the ethics committee, we selected patients with sepsis secondary to community pneumonia and healthy volunteers for the present study.
Disclosure
Authors declare no competing interests.
Author contributions
K. F. E., N. K. S., M. K. C. B., G. L. B. Z. and R. S. contributed to the design of the study as well as the acquisition and analysis of the data. A. T. B., L. C. P. A. and M. A. contributed to the design of the study, selection, enrolment and monitoring of patients and the revision of the manuscript. K. F. E., N. K. S., M. K. C. B. and R. S. wrote the manuscript.
Supporting information
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. Identification of functional networks for sepsis patients after admission in the intensive care unit (ICU) (a), classified further in survival patients (b) and non‐survival patients (c) septic patients by ingenuity pathway analysis (IPA). The cellular movement, hematological system development and function, immune cell trafficking functions in septic patients after admission are represented. Furthermore, cell death and survival, hematological disease, immunological disease in survivals and cellular function and maintenance, hematological system development and function, cell death and survival is also represented. The intensity of node colour represents up‐regulation (red), down‐regulation (green) or no regulation (no colour).
Table S1. List of selected genes and their functions.
Acknowledgements
This work was supported by Fundacao de Amparo a Pesquisa do Estado de São Paulo (FAPESP), grant number 2011/20401‐4 and Conselho Nacional de Desenvolvimento Cientifico e Tecnologico – CNPq, grant number 305685/2011‐2. K. F. E. has a fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – CAPES. N. K. S. has a fellowship from FAPESP.
References
- 1. Bone RC, Sibbald WJ, Sprung CL. The ACCP–SCCM consensus conference on sepsis and organ failure. Chest 1992; 101:1481–3. [DOI] [PubMed] [Google Scholar]
- 2. Singer M, Deutschman CS, Seymour CW et al The third international consensus definitions for sepsis and septic shock (sepsis‐3). JAMA 2016; 315:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Salomao R, Brunialti MK, Rapozo MM, Baggio‐Zappia GL, Galanos C, Freudenberg M. Bacterial sensing, cell signaling, and modulation of the immune response during sepsis. Shock 2012; 38:227–42. [DOI] [PubMed] [Google Scholar]
- 4. Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe 2008; 3:352–63. [DOI] [PubMed] [Google Scholar]
- 5. Takeda K, Akira S. Toll‐like receptors in innate immunity. Int Immunol 2005; 17:1–14. [DOI] [PubMed] [Google Scholar]
- 6. dos Santos G, Kutuzov MA, Ridge KM. The inflammasome in lung diseases. Am J Physiol Lung Cell Mol Physiol 2012; 303:L627–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL‐beta. Mol Cell 2002; 10:417–26. [DOI] [PubMed] [Google Scholar]
- 8. Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell 2014; 157:1013–22. [DOI] [PubMed] [Google Scholar]
- 9. von Moltke J, Ayres JS, Kofoed EM, Chavarria‐Smith J, Vance RE. Recognition of bacteria by inflammasomes. Annu Rev Immunol 2013; 31:73–106. [DOI] [PubMed] [Google Scholar]
- 10. Brunialti MK, Martins PS, Barbosa de Carvalho H, Machado FR, Barbosa LM, Salomao R. TLR2, TLR4, CD14, CD11B, and CD11C expressions on monocytes surface and cytokine production in patients with sepsis, severe sepsis, and septic shock. Shock 2006; 25:351–7. [DOI] [PubMed] [Google Scholar]
- 11. Salomao R. Sepsis: the challenges in developing and translating knowledge. Shock 2008; 30:1–2. [DOI] [PubMed] [Google Scholar]
- 12. Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol 2011; 29:707–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fahy RJ, Exline MC, Gavrilin MA et al Inflammasome mRNA expression in human monocytes during early septic shock. Am J Respir Crit Care Med 2008; 177:983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dolinay T, Kim YS, Howrylak J et al Inflammasome‐regulated cytokines are critical mediators of acute lung injury. Am J Respir Crit Care Med 2012; 185:1225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Machado FR, Salomao R, Rigato O et al Late recognition and illness severity are determinants of early death in severe septic patients. Clinics 2013; 68:586–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Levy MM, Fink MP, Marshall JC et al 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Intensive Care Med 2003; 29:530–8. [DOI] [PubMed] [Google Scholar]
- 17. Boyum A. Separation of white blood cells. Nature 1964; 204:793–4. [DOI] [PubMed] [Google Scholar]
- 18. Boyum A. Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand J Clin Lab Invest Suppl 1968; 97:77–89. [PubMed] [Google Scholar]
- 19. Lamkanfi M, Dixit VM. Modulation of inflammasome pathways by bacterial and viral pathogens. J Immunol 2011; 187:597–602. [DOI] [PubMed] [Google Scholar]
- 20. Martins PS, Brunialti MK, Martos LS et al Expression of cell surface receptors and oxidative metabolism modulation in the clinical continuum of sepsis. Crit Care 2008; 12:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Santos SS, Brunialti MK, Rigato O, Machado FR, Silva E, Salomao R. Generation of nitric oxide and reactive oxygen species by neutrophils and monocytes from septic patients and association with outcomes. Shock 2012; 38:18–23. [DOI] [PubMed] [Google Scholar]
- 22. Santos SS, Carmo AM, Brunialti MK et al Modulation of monocytes in septic patients: preserved phagocytic activity, increased ROS and NO generation, and decreased production of inflammatory cytokines. Intensive Care Med Exp 2016; 4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011; 469:221–5. [DOI] [PubMed] [Google Scholar]
- 24. Bozza FA, D'Avila JC, Ritter C, Sonneville R, Sharshar T, Dal‐Pizzol F. Bioenergetics, mitochondrial dysfunction, and oxidative stress in the pathophysiology of septic encephalopathy. Shock 2013; 39:10–6. [DOI] [PubMed] [Google Scholar]
- 25. Singer M. The role of mitochondrial dysfunction in sepsis‐induced multi‐organ failure. Virulence 2014; 5:66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Severino P, Silva E, Baggio‐Zappia GL et al Gene expression profiling of mononuclear cells from patients with sepsis secondary to community‐acquired pneumonia. Genom Data 2014; 2:332–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garcia JA, Volt H, Venegas C et al Disruption of the NF‐kappaB/NLRP3 connection by melatonin requires retinoid‐related orphan receptor‐alpha and blocks the septic response in mice. FASEB J 2015; 29:3863–75. [DOI] [PubMed] [Google Scholar]
- 28. Kebaier C, Chamberland RR, Allen IC et al Staphylococcus aureus alpha‐hemolysin mediates virulence in a murine model of severe pneumonia through activation of the NLRP3 inflammasome. J Infect Dis 2012; 205:807–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Silva SC, Baggio‐Zappia GL, Brunialti MK et al Evaluation of Toll‐like, chemokine, and integrin receptors on monocytes and neutrophils from peripheral blood of septic patients and their correlation with clinical outcomes. Braz J Med Biol Res 2014; 47:384–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fernandes ML, Mendes ME, Brunialti MK, Salomao R. Human monocytes tolerant to LPS retain the ability to phagocytose bacteria and generate reactive oxygen species. Braz J Med Biol Res 2010; 43:860–8. [DOI] [PubMed] [Google Scholar]
- 31. Tolle L, Yu FS, Kovach MA et al Redundant and cooperative interactions between TLR5 and NLRC4 in protective lung mucosal immunity against Pseudomonas aeruginosa . J Innate Immun 2015; 7:177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Casey LC, Balk RA, Bone RC. Plasma cytokine and endotoxin levels correlate with survival in patients with the sepsis syndrome. Ann Intern Med 1993; 119:771–8. [DOI] [PubMed] [Google Scholar]
- 33. Cannon JG, Tompkins RG, Gelfand JA et al Circulating interleukin‐1 and tumor necrosis factor in septic shock and experimental endotoxin fever. J Infect Dis 1990; 161:79–84. [DOI] [PubMed] [Google Scholar]
- 34. Oberholzer A, Steckholzer U, Kurimoto M, Trentz O, Ertel W. Interleukin‐18 plasma levels are increased in patients with sepsis compared to severely injured patients. Shock 2001; 16:411–4. [DOI] [PubMed] [Google Scholar]
- 35. Grobmyer SR, Lin E, Lowry SF et al Elevation of IL‐18 in human sepsis. J Clin Immunol 2000; 20:212–5. [DOI] [PubMed] [Google Scholar]
- 36. Dinarello CA. Immunological and inflammatory functions of the interleukin‐1 family. Annu Rev Immunol 2009; 27:519–50. [DOI] [PubMed] [Google Scholar]
- 37. Brunialti MK, Santos MC, Rigato O, Machado FR, Silva E, Salomao R. Increased percentages of T helper cells producing IL‐17 and monocytes expressing markers of alternative activation in patients with sepsis. PLOS ONE 2012; 7:e37393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schmitz J, Owyang A, Oldham E et al IL‐33, an interleukin‐1‐like cytokine that signals via the IL‐1 receptor‐related protein ST2 and induces T helper type 2‐associated cytokines. Immunity 2005; 23:479–90. [DOI] [PubMed] [Google Scholar]
- 39. Madouri F, Guillou N, Fauconnier L et al Caspase‐1 activation by NLRP3 inflammasome dampens IL‐33‐dependent house dust mite‐induced allergic lung inflammation. J Mol Cell Biol 2015; 7:351–65. [DOI] [PubMed] [Google Scholar]
- 40. Alves‐Filho JC, Sonego F, Souto FO et al Interleukin‐33 attenuates sepsis by enhancing neutrophil influx to the site of infection. Nat Med 2010; 16:708–12. [DOI] [PubMed] [Google Scholar]
- 41. Meylan E, Tschopp J. The RIP kinases: crucial integrators of cellular stress. Trends Biochem Sci 2005; 30:151–9. [DOI] [PubMed] [Google Scholar]
- 42. Eidt MV, Nunes FB, Pedrazza L et al Biochemical and inflammatory aspects in patients with severe sepsis and septic shock: the predictive role of IL‐18 in mortality. Clin Chim Acta 2016; 453:100–6. [DOI] [PubMed] [Google Scholar]
- 43. Qu Y, Misaghi S, Newton K et al NLRP3 recruitment by NLRC4 during Salmonella infection. J Exp Med 2016; 213:877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sharma NK, Tashima AK, Brunialti MKC et al Identification of differential proteomic response in septic patients secondary to community and hospital acquired pneumonia [abstract]. Crit Care 2016; 20:22. 26847450 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. Identification of functional networks for sepsis patients after admission in the intensive care unit (ICU) (a), classified further in survival patients (b) and non‐survival patients (c) septic patients by ingenuity pathway analysis (IPA). The cellular movement, hematological system development and function, immune cell trafficking functions in septic patients after admission are represented. Furthermore, cell death and survival, hematological disease, immunological disease in survivals and cellular function and maintenance, hematological system development and function, cell death and survival is also represented. The intensity of node colour represents up‐regulation (red), down‐regulation (green) or no regulation (no colour).
Table S1. List of selected genes and their functions.


