ABSTRACT
Background: The presence of Aggregatibacter actinomycetemcomitans in patients with periodontitis has been extensively studied for decades.
Objective: To study the prevalence of A. actinomycetemcomitans in younger and older periodontitis patients and to genetically characterize isolates of this bacterium.
Design: Data from microbiological analyses of 3459 subgingival plaque samples collected from 1445 patients, 337 ‘younger’ patients (≤35 yrs) and 1108 ‘older’ patients (>35 yrs) during 15 years (2000–2014), has been summerized. Isolates of A. actinomycetemcomitans were serotyped, leukotoxin promoter typed (JP2 and non JP2) and arbitrarily primed PCR (AP-PCR) genotyped. The origin of the JP2 genotype detected in the study population was determined.
Results: The prevalence of A. actinomycetemcomitans was higher among younger than older patients and samples from the younger patients contained higher proportions of the bacterium. Serotype b was more prevalent among younger patients and the majorty of these isolates was from the same AP-PCR genotype. The JP2 genotype was detected in 1.2% of the patients, and the majority of these carriers were of non-African origin.
Conslusions: For presence and charcteristics of A. actinomycetemcomitans in clinical samples the age of the carriers were a discriminating factor. Additional, apparently non-African carriers of the JP2 genotype of A. actinomycetemcomitans were identified.
KEYWORDS: Aggregatibacter actinomycetemcomitans, leukotoxin, JP2, serotypes, AP-PCR genotypes, microbiological diagnostics
Introduction
Periodontitis is an infectious disease with a concomitant host response associated with the destruction of the periodontium [1]. In contrast to many other infections, periodontitis is rarely associated with one or a few bacterial species; it is rather a polyinfection, with a collection of different oral microorganisms involved [2–5].
The bacterium Aggregatibacter actinomycetemcomitans is strongly associated with aggressive forms of the disease [6]. Based on longitudinal studies, the presence of this bacterium is considered a marker for progression of periodontal attachment loss (AL), that is, degradation of periodontal tissues around the teeth [7].
Seven serotypes (a–g) of A. actinomycetemcomitans have been described [8]. The prevalence of these serotypes is associated with a number of factors, such as geographic localization, ethnicity, and periodontal status of the carriers [8,9]. Among the serotypes, a, b, and c are the most common. Serotype b is more pathogenic due to its higher capacity to produce leukotoxin, the main virulence factor of A. actinomycetemcomitans [10–12]. The leukotoxin kills neutrophils and macrophages in cellular processes associated with the activation and release of proteases and interleukin-1β (IL-1β), respectively [13]. These properties of the leukotoxin are believed to attenuate the host response and predispose to periodontal breakdown [13].
Very high levels of leukotoxin production have been detected in serotype b isolates that lack a 530 base-pair (bp) DNA fragment within the leukotoxin promoter region [14,15]. Isolates with this characteristic are described as the JP2 genotype of A. actinomycetemcomitans [14], whereas the non-JP2 genotype has a full-length leukotoxin promoter region. Carriers of the JP2 genotype are at increased risk of developing periodontal disease compared to carriers of the non-JP2 genotype [16,17].
The JP2 genotype of A. actinomycetemcomitans emerged on the African continent >2,000 years ago, and since then, it has spread worldwide through human migration of African populations [18]. Although the JP2 genotype today can be detected in areas far from Africa, the carriers have been almost exclusively of African descent. However, a few exceptions are reported [19,20], which suggests that the bacterium has colonized hosts with other geographic origin in some cases. Despite being genetically conserved, the JP2 genotype can be divided into different sequence types based on the presence of a number of point mutations in housekeeping genes [18]. One of these mutations, detected in hbpA-2, distinguishes microevolutionary subtypes of the JP2 genotype, found in individuals living in the Mediterranean area from those populations from West Africa [18].
Another A. actinomycetemcomitans genotype that lacks 640 bp in the leukotoxin promoter region has been isolated from a teenager of Ethiopian origin [21]. It is considered to be ‘JP2 genotype-like’, since it shares the 530 bp deletion with the JP2 genotype, but lacks additional 110 bp [21]. Within serotype b, there are other genotypes reported than those with the 530 and the 640 bp deletion, respectively [12,22]. Recently, a subgroup of highly virulent serotype b that consists of both JP2 and non-JP2 genotypes was identified [23]. A common feature for this genotype is high leukotoxicity, an identical arbitrarly primed (AP) polymerase chain reaction (PCR) pattern, and the presence of an intact cagE gene.
For the elucidation of the association of specific virulent bacterial species to periodontal disease, ‘the presence’ in the site of infection is a weak parameter. To strengthen this association, the so-called ecological plaque hypothesis should be followed [24]. This means that the proportion of specific pathogenic bacterial species at diseased sites has to be quantified.
Aiming to investigate the role of A. actinomycetemcomitans and its different virulent genotypes in periodontal disease, this study used the local collection of clinical isolates. In the Clinical Laboratory of the Dental School in Umeå, Sweden, various periodontitis-associated bacterial species have been isolated from clinical samples and have been studied for >30 years. The present study shows the colonization pattern of A. actinomycetemcomitans in two age groups of periodontitis patients. In addition, it focuses on the genetic characterization of isolates of A. actinomycetemcomitans and hypothesizes that the genetic diversity of the bacterium is of major importance for the colonization pattern among groups of younger and older periodontitis patients.
Material and methods
Study population and collection
The study collection comprises data from microbiological analyzes of 3,459 subgingival plaque samples collected from 1,445 patients, 337 ‘younger’ patients (YP; ≤35 years of age) and 1,108 ‘older’ patients (OP; >35 years of age) during 15 years (2000–2014). The clinical diagnosis of the patients was not homogeneously reported in the patient information attached to the referral to the laboratory for microbiological diagnostics. Thus, the classification of the patients was digitomized only and was based on the old definition of early onset periodontitis, which distinguished patients ≤35 years versus those >35 years of age [25]. Samples were sent from the student’s clinic and from the Specialist Clinic of Periodontology at the Dental School in Umeå, as well as from external specialist dental clinics throughout Sweden. The students collected samples from periodontal pockets >5 mm, measured from the top of the gingival margin to the bottom of the periodontal pocket. Samples from the specialist clinics of periodontology were collected from individuals suffering from periodontitis, although clinical and other parameters were sometimes unsystematically reported due to the many different clinics referring to the laboratories and to the retrospective nature of this study. The patients were between 9 and 92 years old and were sampled one to six times within the study period, and one to eight samples were taken each time. The ancestry of the patients, infected by the JP2 genotype of A. actinomycetemcomitans, was reported by the clinicians, except for two patients (patient 3 and 4; Table 1) who were ancestry tested [21].
Table 1.
Proportion of the JP2 genotype of Aggregatibacter actinomycetemcomitans in samples collected from periodontis patients and characteristics of the carriers of this genotype
| Isolate | Aa1 | Origin | hbpA-22 | M/W3 | Age(years) | |
|---|---|---|---|---|---|---|
| 1 | 520A-01 | 57-90 | Sweden | A | W | 30 |
| 2 | 246A-04 | 0.1-25 | Algeria | G | M | 43 |
| 3 | 133A-08 | 78 | Sweden | A | W | 33 |
| 4 | BL1-08 | 4 | Sweden | G | M | 62 |
| 5 | 090A-10 | 0.9-90 | Sweden | G | M | 27 |
| 6 | 196A-10 | 3.2-4.1 | Croatia | G | M | 23 |
| 7 | 115A-11 | 73-97 | Iraq | G | M | 16 |
| 8 | 352B-11 | 92 | Iraq | G | M | 23 |
| 9 | 245A-12 | 90 | Sweden | G | M | 18 |
| 10 | 557A-12 | 0.1-62 | Cape Verde | A | W | 19 |
| 11 | 338A-13 | 28-39 | Sweden | G | M | 31 |
| 12 | 342A-13 | 20 | Morocco | G | M | 15 |
| 13 | 408A-13 | 0.6-0.7 | Gambia | A | W | 15 |
| 14 | 456A-135 | 42-68 | Ethiopia | G | M | 16 |
| 15 | 304A-14 | 0.3-2.7 | Sweden | G | M | 15 |
| 16 | 361A-14 | 76-98 | Morocco | G | M | 23 |
| 17 | 698A-145 | 2.5-18 | Morocco | G | M | 16 |
% of total viable counts.
G and A are nucleotides within the hemoglobin-binding gene (hbpA-2) at position 525285 of the genome (HK1651) [18].
Mediterranean (M) or West African (W) origin of the JP2 genotype carriers
The proportion of A. actinomycetemcomitans could not be calculated due to contamination of the sample [20].
This isolate of A. actinomycetemcomitans has a 640 bp deletion within the leukotoxin promoter region [21].
Isolates of A. actinomycetemcomitans were collected from the patients and stored in a freezer. However, A. actinomycetemcomitans was not isolated or could not be recovered from the freezer from all positive patients, especially those examined during the first years of the study period. Thus, the study collection comprises isolates from 347 (78%) of the 435 A. actinomycetemcomitans–positive patients. Since 88 isolates are missing, the 347 isolates belong to a subpopulation of 1,357 patients.
Quantification of A. actinomycetemcomitans
The samples were transported in an anaerobic medium (VMGAIII; Viable Medium of the Department of Bacteriology, University of Gothenburg) [26] to the Clinical Laboratory at the Dental School in Umeå, Sweden, serially diluted in a salt buffer [27], and spread on blood agar plates for the determination of the total number of bacteria. The dilutions were spread on Trypticase-Bacitracin plates for the determination of A. actinomycetemcomitans [28]. A. actinomycetemcomitans was identified according to established methods [28]. If the patients were sampled more than once, the sample containing the highest proportion of A. actinomycetemcomitans of the total viable count (TVC) were consequently selected for subsequent calculations.
Characterization of A. actinomycetemcomitans isolates
All isolates were serotyped. In addition, all serotype b isolates were leukotoxin promoter and AP-PCR genotyped. For the preparation of the PCR mixtures, PureTaq Ready-To-Go PCR (GE Healthcare; Little Chalfont, UK) was used. For serotyping and leukotoxin promoter typing, DNA was isolated by heating suspensions of the isolates in water for 8 min, while for the AP-PCR, the DNA was purified with GenElute Bacterial DNA kit (Sigma–Aldrich, St. Louis, MO).
The primers used and the PCR program for the serotyping and leukotoxin promoter typing are described elsewhere [29–31]. For the AP-PCR genotyping, the random sequence oligonucleotide OPB-3 (AGTCAGCCAC; Invitrogen, Carlsbad, CA; 0.4 µM) was used. The concentration of MgCl2 in the PCR mixture was increased to 2.5 µM. The amplification was carried out as previously described [32].
The sequencing of the house-keeping gene, hbpA-2, was carried out as described elsewhere [18].
Statistical analyses
Chi-square tests were used to identify significant differences in the distribution of the individuals and samples positive for A. actinomycetemcomitans in relation to the age group. The same test was used to examine differences in the distribution of A. actinomycetemcomitans AP-PCR types among YP and OP. The differences in the proportions of the bacterium in patients from the two age groups were calculated with the Mann–Whitney test. The rank test was used to calculate significant differences in the distribution of A. actinomycetemcomitans serotypes in samples from the two age groups.
Results
Presence of A. actinomycetemcomitans
A. actinomycetemcomitans could be isolated from around 30% of the sampled patients (Table 2). The detection frequencies of the bacterium were similar independently if the calculation was based on (a) the total number of subgingival plaque samples, or (b) the total number of patients (Table 2). Both ways for presentation showed that the detection frequency of the bacterium was significantly higher among YP than among OP (A; B: p < 0.001).
Table 2.
Detection frequencies of A. actinomycetemcomitans (Aa) in subgingival plaque samples collected from younger (YP; ≤35 years) and older patients (OP; >35 years) with periodontitis
| A |
B |
|||
|---|---|---|---|---|
| Age groups | n | Aa, n(%) | n | Aa, n(%) |
| YP | 934 | 379 (40.6) | 337 | 162 (48.1) |
| OP | 2,535 | 529 (20.9) | 1,108 | 273 (24.6) |
| YP + OP | 3,459 | 908 (26.3) | 1,445 | 435 (30.1) |
Aa was significantly more prevalent among YP than among OP (A, B; p < 0.001).
A, distribution of Aa-positive samples; B, distribution of Aa-positive patients.
Proportions of A. actinomycetemcomitans
When all patients carrying A. actinomycetemcomitans were distributed into different groups based on the proportion of the bacterium in the samples, approximately 14% of the patients were observed in the group of samples containing >50% of the bacterium (Table 3). However, when the patients were distributed in the two age groups, samples containing > 50% A. actinomycetemcomitans were five times more common among YP than among OP.
Table 3.
Distribution of patients, n (%), in groups with regard to age (YP and OP) and proportions (%) of A. actinomycetemcomitans of total viable count (TVC) in the samples from the patients (n = 435)
| Age groups | >0-1% | >1-5% | >5-25% | >25-50% | >50% | n |
|---|---|---|---|---|---|---|
| YP | 38 (23.5) | 30 (18.5) | 31 (19.1) | 17 (10.5) | 46 (28.3) | 162 |
| OP | 108 (39.6) | 51 (18.7) | 79 (28.9) | 20 (7.3) | 15 (5.5) | 273 |
| YP + OP | 146 (33.6) | 81 (18.6) | 110 (25.3) | 37 (8.5) | 61 (14.0) | 435 |
Samples containing >50% Aa were significantly more common among YP than they were among OP (p < 0.001).
Distribution of serotypes of A. actinomycetemcomitans
Serotyping of the collection of A. actinomycetemcomitans isolates showed that serotype b was significantly more prevalent among YP, while serotype c was more prevalent among OP (p < 0.001). For the remaining serotypes, the distribution was similar in the two age groups (Figure 1).
Figure 1.
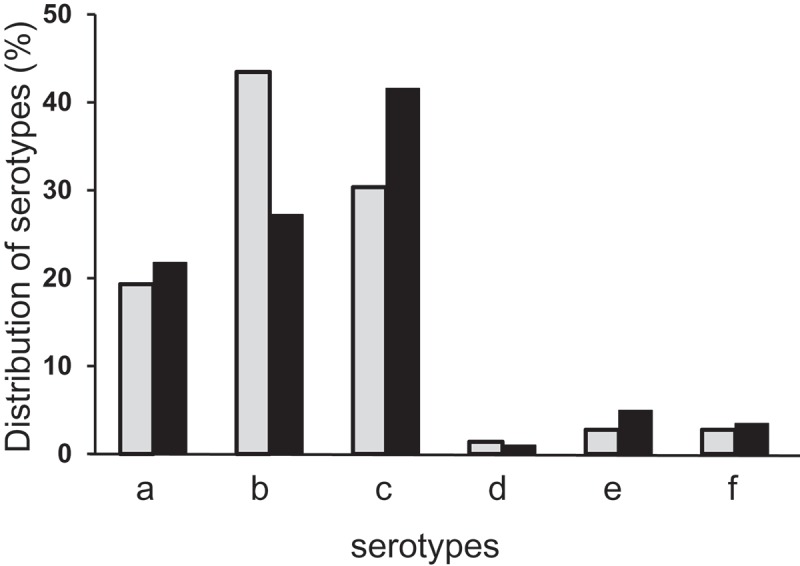
Percentage distribution of serotypes a–f of Aggregatibacter actinomycetemcomitans (Aa) among the isolates from younger patients (YP; gray columns; n = 145) and older patients (OP; black columns; n = 202). Serotyping of the collection of Aa isolates showed that serotype b was significantly more prevalent among YP, while serotype c was more prevalent among OP (p < 0.001).
When the three most prevalent serotypes of A. actinomycetemcomitans (a, b, and c) were distributed into proportions of the bacterium in the samples, serotype b was the most common in the category with the highest proportions (>50%) in both age groups. However, the prevalence of serotype b was almost five times higher among YP than in the corresponding category among OP (Table 4).
Table 4.
Distribution of A. actinomycetemcomitans-isolates, n (%), in groups with regard to age (YP and OP), serotype (a, b, and c) and proportions (%) of Aa of TVC in the samples (n = 317)
| Age groups | >0–1% | >1–5% | >5–25% | >25–50% | >50% | n |
|---|---|---|---|---|---|---|
| YP | ||||||
| a | 7 (25.0) | 3 (10.7) | 7 (25.0) | 2 (7.1) | 9 (32.1) | 28 |
| b | 8 (12.7) | 5 (7.9) | 14 (22.2) | 6 (9.5) | 30 (47.6) | 63 |
| c | 18 (40.9) | 11 (25.0) | 8 (18.2) | 4 (9.1) | 3 (6.8) | 44 |
| OP | ||||||
| a | 11 (25.0) | 10 (22.7) | 14 (31.8) | 6 (13.6) | 3 (6.8) | 44 |
| b | 18 (32.7) | 9 (16.4) | 20 (36.4) | 2 (3.6) | 6 (10.9) | 55 |
| c | 35 (42.2) | 20 (24.1) | 22 (26.5) | 5 (6.0) | 1 (1.2) | 83 |
Prevalence and proportions of the JP2 genotype of A. actinomycetemcomitans
Of 1,357 patients, 17 (1.2%) carried the JP2 genotype of A. actinomycetemcomitans (Table 1). The proportion (% of TVC) of the JP2 genotype in at least one sample per patients was >50% in 56% of these 17 patients. Based on information from clinicians, 10 (59%) of the JP2 genotype carriers were considered to be of non-African origin, although ancestry testing had not been performed for all. Only two (12%) of the 17 JP2 carriers belonged to the OP group (Table 1). Furthermore, 13/17 (76.5%) JP2 genotype carriers were colonized by the Mediterranean subtype of the JP2 genotype.
Distribution of AP-PCR genotypes of A. actinomycetemcomitans
Eleven different AP-PCR patterns were identified among serotype b isolates (n = 119; Figure 2). Among those, 72 A. actinomycetemcomitans isolates belonged to AP-PCR genotypes 1 or 2 (Figure 2). The AP-PCR banding pattern of all 17 JP2 and JP2-like genotype isolates was identical (not shown), as compared to the JP2-reference strain HK1651, and belonged to AP-PCR genotype 1 (Figure 2). In Figure 2, AP-PCR genotypes 1–7 are each represented by two isolates, whereas for genotypes 8–11, only one isolate was available from each genotype.
Figure 2.
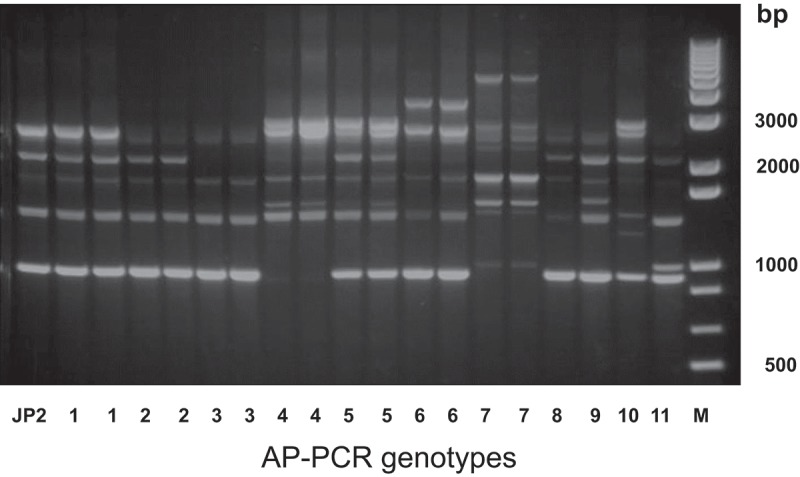
Different arbitrarily primed polymerase chain reaction (AP-PCR) genotypes of A. actinomycetemcomitans among serotype b isolates.
When the serotype b isolates were distributed among the AP-PCR genotypes with regard to the age of the carriers, >60% of the isolates from the YP group were identified as AP-PCR genotype 1, while only 15% of the isolates from the OP group were identified as AP-PCR genotype 1. In contrast, >35% of the isolates from the OP group were identified as AP-PCR genotype 2, while only 5% of the isolates from the YP group were identified as AP-PCR genotype 2 (Figure 3). In addition, independent of age, >50% of the AP-PCR genotype 1 isolates were distributed within the high proportion group, while none of the other AP-PCR type isolates was detected in this group.
Figure 3.
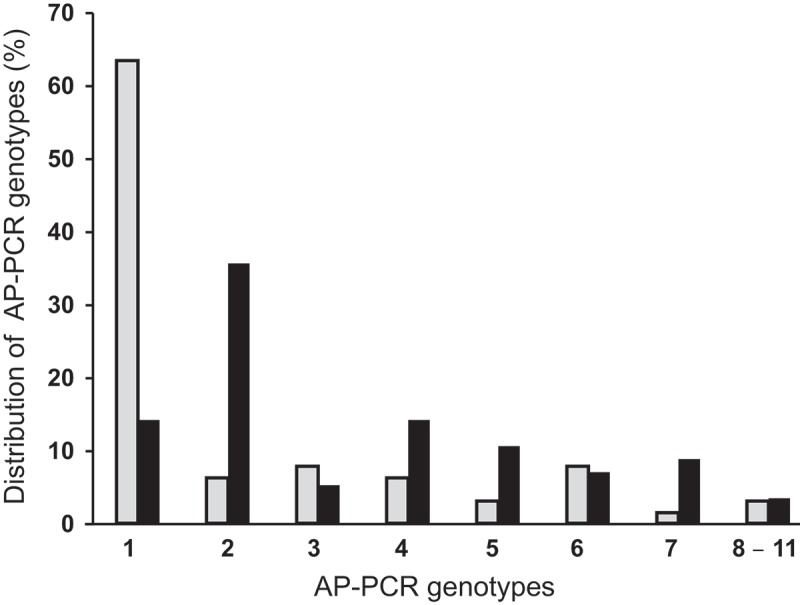
Proportions of serotype b isolates of A. actinomycetemcomitans from patients among YP (gray columns; n = 63) and OP (black columns; n = 56) when distributed in different AP-PCR genotypes, 1–11. AP-PCR genotype 1 was significantly more prevalent among YP than it was among OP (p < 0.001).
Discussion
A high number of reports on the prevalence and proportions of periodontitis-associated bacterial species in infected periodontal pockets are available [8,33,34]. However, the diversity of methods used in these and other studies complicates the comparison of results and conclusions [35–37]. Being aware of these problems, the present study shows, based on cultivation, that the detection frequency of A. actinomycetemcomitans was two times higher among YP than among OP living in Sweden. Furthermore, the characteristics of the A. actinomycetemcomitans isolates differed by the age of the patients.
In line with the ecological plaque hypothesis, this study also investigated the proportions of of A. actinomycetemcomitans in samples from periodontal pockets [24]. It is suggested that the capacity of A. actinomycetemcomitans to multiply more efficiently in periodontal pockets among YP could be due to enviromental factors. In deep pockets, which is one characteristic for periodontitis among OP, A. actinomycetemcomitans may be outcompeted by strictly anaerobic bacterial species such as Fusobacterium nucleatum, Prevotella intermedia, Treponema denticola, Porphyromonas gingivalis, and so on [38–41].
Six serotypes of A. actinomycetemcomitans (a–f) were detected among both YP and OP. However, serotype b, which is more virulent than the other serotypes [10,12], was more prevalent among YP. Serotype b seems more likely to survive in both younger and older patients in an environment that otherwise does not promote the existence of this bacterium. In other studies, the diversity among different serotypes in the binding to other bacterial species and to host molecules has been studied [42].
In the present study, 17 JP2 genotype carriers (i.e. 1.2% out of 1,357 patients) were detected. The majority of the JP2 genotype carriers were Caucasians, which is not in line with earlier reports that JP2 genotype carriers almost exclusively are found in individuals of African origin [18]. Almost 90% of the JP2 genotype carriers belonged to the YP group. Also, in other studies, the detection rate of the JP2 genotype among OP is low [19].
Spreading of the JP2 genotype between family members is reported [18,20]. Also, in this study, a family-related prevalence was detected. Three isolates (3, 4, and 5) were collected from a family comprising a mother and her two daughters, while two isolates (7 and 8) were collected from two brothers. The leukotoxin promoter region of one isolate (14) is missing 640 bp instead of 530 bp [21]. Taken together, it is surprising that a relatively high number of JP2 genotype carriers were identified among Swedish periodontitis patients. It is tempting to speculate that the carriage of the JP2 genotype is when young patients with bone loss visit the dentist. This suggests that it might be valuable to characterize A. actinomycetemcomitans at the subspecies level in isolates from these patients. Risk for a family-related spreading of the JP2 genotype should also be taken in consideration. Apparently, more studies are needed to identify carriers of the JP2 genotype in non-African populations, since it seems they have been underestimated.
When this study monitored the genetic diversity within serotype b isolates of A. actinomycetemcomitans with the AP-PCR technique, it was found that the majority of the isolates from the YP were distributed in the AP-PCR 1 genotype group, while the prevalence of isolates from OP were four times lower in this AP-PCR group. Since AP PCR genotype 1 is reported to be more leukotoxic than other AP-PCR genotypes, it is suggested that serotype b isolated from YP are more virulent than those that colonize OP [12]. This hypothesis is further supported by the fact that the JP2 genotype belongs to AP-PCR type 1. In addition, non-JP2 genotype isolates with an AP-PCR pattern that was identical with the AP-PCR pattern of the JP2 genotype were more prevalent among patients with localized aggressive periodontitis than among adult periodontitis patients [43].
Since younger individuals are at an enhanced risk of developing the aggressive form of periodontitis, it could be speculated that early colonization with A. actinomycetemcomitans of AP-PCR genotype 1 is a risk factor for developing the disease later in life. Further investigation of this genotype showed that the presence of an intact cagE gene identified as a marker for AP-PCR genotype 1 and was suggested as a target for early identification of risk individuals [23].
For diagnosis and treatment purposes, proportions of periodontitis-associated bacterial species in samples from periodontitis patients have been determined for many years [44–48]. Today, the utility of these analyzes is controversial [48]. However, in non-responsive cases, sampling and quantification of periodontitis-associated bacterial species such as A. actinomycetemcomitans may be helpful for the determination of more successful treatment strategies [49–51].
The results of the present study indicate that analysis of subgingival plaque samples from younger individuals with periodontal AL could be beneficial. Due to increased globalization and migration, a higher prevalence of young individuals with periodontal disease may occur in countries with earlier reported low prevalence of aggressive periodontitis.
In summary, the prevalence of A. actinomycetemcomitans was significally higher among YP than among OP. Furthermore, additional individuals of apparently non-African origin, colonized with the JP2 genotype of A. actinomycetem-comitans, have been identified in the present study. In addition, it was shown that a cluster of A. actinomycetemcomitans, with shared genetic characteristics, contained both the JP2 and the non-JP2 genotype. This cluster of the bacterium was significantly more prevalent among YP with periodontitis.
Acknowledgments
The authors thank laboratory technicians Ewa Engbo-Strömqvist and Chrissie Roth for their valuable contribution to the work in the laboratory, including cultivation and characterization of the A. actinomycetemcomitans isolates.
Biographies
Dr Rolf Claesson, PhD, is a microbiologist in Dental School, Umeå, Sweden. He has been studying the oral microflora and its association to oral diseases for more than 35 years. His publication list also contains articles about the function of neutrophils in the oral cavity. Since 1998 he is involved in a research project about the role of the bacterium Aggregatibacter actinomycetemcomitans in periodontitis.
Dr Carola Höglund Åberg, PhD, is a senior consultant in Periodontology at the department of Molecular Periodontology, Medical Faculty, at Umeå University, Sweden. She has studied the microbial profile of young individuals affected with aggressive periodontitis, with specific focus on virulence characteristics of different genotypes of Aggregatibacter actinomycetemcomitans.
Professor Dorte Haubek, PhD, is working at Section for Pediatric Dentistry, Department of Dentistry and Oral Health, Aarhus University, Aarhus, Denmark. Her main research focus is on the bacterium Aggregatibacter actinomycetemcomitans with special reference to the impact of the leukotoxin and the association to the development of periodontitis in adolescents. She has been intensively involved in periodontal research projects in Morocco, Ghana and Kenya.
Dr Anders Johansson, PhD, is a Senior Researcher at the Department of Molecular Periontology, Umeå University Sweden. He is a Principal Investigator of a research group at Umeå University that for several decades had focused on the virulence of Aggregatibacter actinomycetemcomitans and its leukotoxin.
Funding Statement
This study was supported by the County Council of Västerbotten, Sweden.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Pihlstrom BL, Michalowicz BS, Johnson NW.. Periodontal diseases. Lancet. 2005;366(9499):1809–9. DOI: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- [2].Kolenbrander PE, Palmer JRJ, Periasamy S, et al. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8(7):471–480. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20514044 [DOI] [PubMed] [Google Scholar]
- [3].Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–490. Available from: http://www.nature.com/nrmicro/jo/v8/n7/full/nrmicro2337.html [DOI] [PubMed] [Google Scholar]
- [4].Fine DH, Markowitz K, Fairlie K, et al. A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J Clin Microbiol. 2013;51(9):2850–2861. DOI: 10.1128/JCM.00729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Offenbacher S, Divaris K, Barros SP, et al. Genome-wide association study of biologically informed periodontal complex traits offers novel insights into the genetic basis of periodontal disease. Hum Mol Genet. 2016;25(10):2113–2129. DOI: 10.1093/hmg/ddw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Henderson B, Ward JM, Ready D. Aggregatibacter (Actinobacillus) actinomycetemcomitans: a triple A* periodontopathogen? Periodontol 2000. 2010;54:78–105. DOI: 10.1111/j.1600-0757.2009.00331.x. [DOI] [PubMed] [Google Scholar]
- [7].Fine DH, Markowitz K, Furgang D, et al. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: longitudinal cohort study of initially healthy adolescents. J Clin Microbiol. 2007;45:3859–3869. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2168549/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Könönen E, Muller HP. Microbiology of aggressive periodontitis. Periodontol 2000. 2014;65(1):46–78. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24738586 [DOI] [PubMed] [Google Scholar]
- [9].Brigido JA, da Silveira VRS, Rego RO, et al. Serotypes of Aggregatibacter actinomycetemcomitans in relation to periodontal status and geographic origin of individuals – a review of the literature. Med Oral Patol Oral Cir Bucal. 2014;19(2):e184–e191. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24316700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zambon JJ, Slots J, Genco RJ. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect Immun. 1983;41(1):19–27. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC264736/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yang HW, Asikainen S, Dogan B, et al. Relationship of Actinobacillus actinomycetemcomitans serotype b to aggressive periodontitis: frequency in pure cultered isolates. J Periodontol. 2004;75(4):592–599. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15152825 [DOI] [PubMed] [Google Scholar]
- [12].Höglund Åberg C, Haubek D, Kwamin F, et al. Leukotoxic activity of Aggregatibacter actinomycetemcomitans and periodontal attachment loss. Plos One. 2014;9(8):e104095 Available from: http://journals.plos.org/plosone/article?id=10.1371%2Fjournal.pone.0104095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Johansson A. Aggregatibacter actinomycetemcomitans leukotoxin: A powerful tool with capacity to cause imbalance in the host inflammatory response. Toxins. 2011;3:242–259. Available from: http://www.mdpi.com/2072-6651/3/3/242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Spitznagel JRJ, Kraig E. Kolodrubetz. Regulation of leukotoxin in leukotoxic and nonleukotoxic strains of Actinobacillus actinomycetemcomitans. Infect Immun. 1991;59(4):1394–1401. Available from: http://iai.asm.org/content/59/4/1394.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Brogan JM, Lally ET, Poulsen K, et al. Regulation of Actinobacillus actinomycetemcomitans leukotoxin expression: analysis of the promoter regions of the leukotoxic and minimally leukotoxic strains. Infect Immun. 1994;62(2):501–508. Available from: http://iai.asm.org/content/62/2/501.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Haubek D, Ennibi OK, Poulsen K, et al. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Marocco: a prospective longitudinal cohort study. Lancet. 2008;371:237–242. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18207019 [DOI] [PubMed] [Google Scholar]
- [17].Höglund Åberg C, Kwamin F, Claesson R, et al. Progression of attachment loss is strongly associated with the presence of the JP2 genotype of Aggregatibacter actinomycetemcomitans: a prospective cohort study of a young adolescent population. J Clin Periodontol. 2014;41(3):232–241. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24304011 [DOI] [PubMed] [Google Scholar]
- [18].Haubek D, Poulsen K, Kilian M. Microevolution and patterns of dissemination of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect Immun. 2007;75(6):3080–3088. Available from: http://iai.asm.org/content/75/6/3080.full [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Guthmiller JM, Lally ET, Korostoff J. Beyond the specific plaque hypothesis: are highly leukotoxic strains of Actinobacillus actinomycetemcomitans a paradigm for periodontal pathogenesis? Crit Rev Oral Biol Med. 2001;12(2):116–124. Available from: http://cro.sagepub.com/content/12/2/116 [DOI] [PubMed] [Google Scholar]
- [20].Claesson R, Lagervall M, Höglund Åberg C, et al. Detection of the highly leucotoxic JP2 clone of Aggreatibacter actinomycetemcomitans in members of a Caucasian family living in Sweden. J Clin Periodontol. 2010;38(2):115–121. DOI: 10.1111/j.1600-051X.2010.01643.x/abstract. [DOI] [PubMed] [Google Scholar]
- [21].Claesson R, Gudmundson J, Aberg CH, et al. Detection of a 640-bp deletion in the Aggregatibacter actinomycetemcomitans leukoxin promoter region in isolates from an adolescent of Ethiopian origin. J Oral Microbiol. 2015;7:26974 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4400299/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Laiko L, Kuula H, Dogan B, et al. Actinobacillus actinomyetemcomitans proportion of subgingival bacterial flora in relation to its clonal type. Eur J Oral Sci. 2002;110:212–217. DOI: 10.1034/j.1600-0447.2002.201238.x/pdf. [DOI] [PubMed] [Google Scholar]
- [23].Johansson A, Claesson R, Höglund Åberg C, et al. The cagE gene sequence as a diagnostic markewr to identify JP2 and non-JP2 highly leukotoxic Aggregatibacter actinomycetemcomitans serotype b strains. J Periodontal Res. 2017:1–10. https://www.ncbi.nlm.nih.gov/pubmed/28397250 [DOI] [PubMed] [Google Scholar]
- [24].Rosier BT, Jager MD, Zaura E, et al. Historical and contemporary hypotheses on the development of oral diseases: are we there yet? Front Cell Infect Microbiol. 2014;4(92):1–11. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25077073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Loesche WJ, Grossman N. Periodontal disease as a specific, albeit chronic, infection: diagnosis and treatment. Clin Microbiol Rev. 2001;14:727–752. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11585783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Möller ÅJR. Microbiological examination of root canals and periapical tissues of human teeth. Scand Dent J. 1966;74(5–6). Available from: http://www.ncbi.nlm.nih.gov/pubmed/5335186. [PubMed] [Google Scholar]
- [27].Johansson E, Claesson R, van Dijken JWV. Antibacterial effect of ozone on cariogenic bacterial species. J Dent. 2009;37(6):449–453. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19342147 [DOI] [PubMed] [Google Scholar]
- [28].Höglund Åberg C, Sjödin B, Laiko L, et al. Presence of Aggregatibacter actinomycetemcomitans in young individuals: a 16-year clinical and microbiological follow-up study. J Clin Periodotol. 2009;36(10):815–822. DOI: 10.1111/j.1600-051X.2009.01457.x/abstract. [DOI] [PubMed] [Google Scholar]
- [29].Suzuki N, Nakano Y, Yoshida Y, et al. Identification of Actinobacillus actinomycetemcomitans serotypes by multiplex PCR. J Clin Microbiol. 2001;39(5):2002–2005. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC88070/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kaplan JB, Perry MB, MacLean LL, et al. Structural and genetic analyses of O polysaccharide from Actinobacillus actinomycetemcomitans serotype f. Infect Immunol. 2001;69(9):5375–5384. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11500407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Poulsen K, Ennibi OK, Haubek D. Improved PCR for detection of the highly leukotoxic JP2 clone of Actinobacíllus actinomycetemcomitans in subgingival plaque samples. J Clin Microbiol. 2003;41(10):4829–4832. Available from: http://jcm.asm.org/content/41/10/4829.full [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dogan B, Saarela MH, Jousimies-Somer H, et al. Actinobacillus actinomycetemcomitans serotype e-biotypes, genetic diversity and distribution in relation to periodontal status. Oral Microbiol Immunol. 1999;14(2):98–103. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10219168 [DOI] [PubMed] [Google Scholar]
- [33].Haubek D, Johansson A. Pathogenicity of the highly leukotoxic JP2 clone of Aggregatibacter actinomycetemcomitans and its geographic dissemination and role in aggressive periodontitis. J Oral Microbiol. 2014;6 Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4139931/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rylev M, Kilian M. Prevalence and distribution of principal periodontal pathogens worldwide. J Clin Periodontol. 2008;35(8):346–361. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18724862 [DOI] [PubMed] [Google Scholar]
- [35].van Winkelhof AJ, de Groot P, Abbas F, et al. Quantitative aspects of the subgingival distribution of Actinobacillus actinomycetemcomitans in patients with localized juvenile periodontitis. J Clin Periodontol. 1994;21:199–202. DOI: 10.1111/j.1600-051X.1994.tb00304.x/pdf. [DOI] [PubMed] [Google Scholar]
- [36].Jervoe-Storm PM, AIAhdab H, Koltzscher M, et al. Quantification of periodontal pathogens by paper point sampling from coronal and apical aspect of periodontal lesions by real-time PCR. Clin Oral Invest. 2010;14(5):533–541. Available from: http://link.springer.com/article/10.1007/s00784-009-0333-x [DOI] [PubMed] [Google Scholar]
- [37].Göhler A, Hetzer A, Holtfreter B, et al. Quantitative molecular detection of putative periodontal pathogens in clinically healthy and periodontally diseased subjects. Plos One. 2014;9(7):e99244 Available from: http://journals.plos.org/plosone/article?id=10.1371%2Fjournal.pone.0099244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nishihara T, Koseki T. Microbial etiology of periodontitis. Periodontol 2000. 2004;36:14–26. DOI: 10.1111/j.1600-0757.2004.03671.x. [DOI] [PubMed] [Google Scholar]
- [39].Takasaki K, Fujise O, Miura M, et al. Porpyromonas gingivalis displays a competive advantage over Aggregatibacter actinomycetemcomitans in co-cultured biofilm. J Periodontal Res. 2013;48(3):286–292. DOI: 10.1111/jre.12006/abstract. [DOI] [PubMed] [Google Scholar]
- [40].Johansson A, Hänström L, Kalfas S. Inhibition of Actinobacillus actinomycetemcomitans leukotoxocity by bacteria from the subgingival flora. Oral Microbiol Immunol. 2000;15:218–225. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11154406 [DOI] [PubMed] [Google Scholar]
- [41].Haraguchi A, Miura M, Fujise O, et al. Porphyromonas gingivalis gingipain is involved in the detachment and aggregation of Aggregatibacter actinomycetemcomitans biofilm. Mol Oral Microbiol. 2014;29(3):131–143. DOI: 10.1111/omi.12051/abstract. [DOI] [PubMed] [Google Scholar]
- [42].Rosen G, Nisimov I, Helcer M, et al. Actinbacillus actinomycetemcomitans serotype b lipopolysaccharide mediates coaggregation with Fusobacterium nucleatum. Infect Immun. 2003;71(6):3652–3656. Available from: http://wolfson.huji.ac.il/purification/PDF/Publications/Rosen2003.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Paju S, Carlson P, Jousimies-Somer H, et al. Heterogeneity of Actinobacillus actinomycetemcomitans strains and relationships between serotype, genotype, antimicrobial susceptibility. J Clin Microbiol. 2000;38:79–84. Available from: http://jcm.asm.org/content/38/1/79.full [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zambon JJ, Haraszthy VI. The laboratory diagnosis of periodontal infections. Periodontol 2000. 1995;7:69–82. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9567931 [DOI] [PubMed] [Google Scholar]
- [45].Saddi-Ortega L, Carvalho MAR, Cisalpino PS, et al. Actinobacillus actinomycetemcomitans genetic heterogeneity: amplification of JP2 like ltx promoter pattern correlated with specific arbitrarily primed polymerase reaction (AP-PCR) genotypes from human but not marmoset Brazilian isolates. Can J Microbiol. 2002;48(7):602–610.PMID: 12224559 [DOI] [PubMed] [Google Scholar]
- [46].Asikainen S, Chen C, Saarela M, et al. Clonal specificity of Actinobacillus actinomycetemcomitans in destructive periodontal disease. Clin Infect Dis. 1997;25(2):S227–S229. Available from: http://www.jstor.org/stable/4481258 [DOI] [PubMed] [Google Scholar]
- [47].Ezzo PJ, Cutler CW. Microorganisms as risk indicators for periodontal disease. Periodontol 2000. 2003;32:24–35. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12756031 [DOI] [PubMed] [Google Scholar]
- [48].Sanz M, Lau L, Herrera D, et al. Methods of detection of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis in periodontal microbiology, with special emphasis on advanced molecular techniques: a review. J Clin Periodontol. 2004;31(12):1034–1037. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15560803 [DOI] [PubMed] [Google Scholar]
- [49].Faveri M, Figueiredo LC, Duarte PM, et al. Microbiological profile of untreated subjects with localized aggressive periodontitis. J Clin Periodontol. 2009;36(9):739–749. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19637996 [DOI] [PubMed] [Google Scholar]
- [50].D’Ercole S, Catamo G, Piccolomini R. Diagnosis in periodontology: A further aid trough microbiological tests. Crit Rev Microbiol. 2008;34:33–41. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18259979 [DOI] [PubMed] [Google Scholar]
- [51].Shaddox LM, Walker C. Microbial testing in periodontitis: value, limitations and future directions. Periodontol 2000. 2009;50:25–38. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19388951 [DOI] [PubMed] [Google Scholar]


