Abstract
Genus Iotatorquevirus consists of 2 species, Torque teno sus virus 1a and Torque teno sus virus 1b, which are ubiquitous in swine populations, and are widely reported in association with porcine circovirus associated disease (PCVAD). To evaluate the relationship with PCVAD, 100 formalin-fixed paraffin-embedded tissue samples were used to detect both Iotatorquevirus species by nested PCR and sequencing. Sixty-eight PCVAD cases were selected as well as 32 porcine circovirus type 2 (PCV2) non-affected cases. Overall, 33 of the 100 cases were positive for Torque teno sus virus 1a and 8 of 100 were positive for Torque teno sus virus 1b. Only 24 of 68 (35%) PCVAD cases were positive for Torque teno sus virus 1a; 39% (9/23) of post-weaning multisystemic wasting syndrome, and 33% (15/45) of PCV2-associated reproductive failure cases. Among PCV2 non-affected cases, 28% were positive for Torque teno sus virus 1a and 6% were positive for Torque teno sus virus 1b. Torque teno sus virus 1b was not detected in PCV2-associated reproductive failure cases. Regardless of the PCV2-status, a lower frequency of both Iotatorquevirus species was found than depicted in other reports and there was no statistical relationship with PCVAD (χ2 < 0.01). Given the worldwide genomic variability of Iotatorquevirus species, it is feasible that species prevalent in Mexico share a lower nucleotide sequence identity, leading to different pathogenic potential.
Résumé
Le genre Iotatorquevirus consiste en deux espèces, le virus Torque teno sus 1a et le virus Torque teno sus 1b, qui sont ubiquitaires dans la population porcine, et couramment rapportés en association avec la maladie associée au circovirus porcin (MACVP). Afin d’évaluer la relation avec MACVP, 100 échantillons de tissus fixés dans la formaline et enrobés de paraffine ont été utilisés pour détecter les deux espèces de Iotorquevirus par réaction d’amplification en chaîne par la polymérase nichée et séquençage. Soixante-huit cas de MACVP ont été sélectionnés ainsi que 32 cas non-affectés d’infection par le circovirus porcin de type (CVP2). Globalement, 33 des 100 cas étaient positifs pour le virus Torque teno sus 1a et 8 des 100 étaient positifs pour le virus Torque tenos sus 1b. Seulement 24 des 68 (35 %) cas de MACVP étaient positifs pour le virus Torque tenos sus 1a; 39 % (9/23) du syndrome de dépérissement post-sevrage, et 33 % (15/45) des cas de problèmes reproducteurs associés au CVP2. Parmi les cas non-affectés de CVP2, 28 % étaient positifs pour le virus Torque teno sus 1a et 6 % étaient positifs pour le virus Torque tenos sus 1b. Le virus Torque tenos sus 1b n’a pas été détecté dans les cas de problèmes reproducteurs associés au CVP2. Indépendamment du statu vis-à-vis le CVP2, une fréquence plus basse des deux espèces d’Iotatorquevirus fut trouvée comparativement à ce qui est décrit dans d’autres études et il n’y avait pas de relation statistiquement significative avec MACVP (χ2 < 0,01). Étant donné la variabilité génomique mondiale des espèces d’Iotatorquevirus il est possible que les espèces prévalentes au Mexique partagent une plus faible identité de séquences nucléotidiques, entrainant ainsi un potentiel pathogène différent.
(Traduit par Docteur Serge Messier)
Introduction
Torque teno virus (TTV) is a circular single-stranded negative sense DNA virus that enclosed 4 open reading frames (ORF); ORF1, ORF2, ORF 1/1, and ORF 2/2 (former ORF3), and a GC-rich region within an un-translated region (UTR), which was first discovered in human samples with post-transfusion hepatitis of unknown etiology in 1997 (1). Currently, based on the International Committee on Taxonomy of Viruses (2), all human and animal TTV belong to Anelloviridae family. Genus Iotatorquevirus comprises of 2 species, TTSuV1a (former TTSuV1) and TTSuV1b (former TTSuV2). The phylogenetic relationship of incomplete sequences has proposed 4 biotypes (a, b, c, and d) of TTSuV1a, and 2 biotypes (a and b) of TTSuV1b (3,4). Genus Kappatorquevirus, however, includes only one species: Torque teno sus virus κ2 (2). High prevalence of co-infection between TTSuV1a and TTSuV1b has been documented worldwide. Iotatorquevirus species are ubiquitous in domestic and wild pigs, and have been identified in Europe (Hungary, Italy, France, and Spain), Asia (China, Korea, Japan, and Thailand), and North America (Canada and USA).
Transmission among pigs is horizontal and mainly via fecal-oral, but transmission through other routes may be important (3). It is unknown whether TTSuV1a and TTSuV1b infection promotes a specific disease as a primary agent or in co-infection with other pathogens (5). However, it has been suggested that in co-infections with other viruses, TTSuV1a might promote increased disease severity or virulence and TTSuV1b might be associated with reproductive failure (1). In this scenario, several studies have suggested an involvement of both species with porcine circovirus associated disease (PCVAD) since cases of post-weaning multisystemic wasting syndrome (PMWS) have shown a high prevalence of Iotatorquevirus species (6,7). Moreover, clinical manifestations and characteristic lesions of porcine dermatitis and nephropathy syndrome (PDNS) have been reproduced by TTSuV1a inoculation in gnotobiotic pigs (8). In fact, TTSuV1a has been proposed as the additional factor (X-factor) for the development of PCVAD (9). Porcine circovirus associated disease is economically important to the swine industry since it has an impact on production and reproduction parameters. Although all clinical presentations of PCVAD [PMWS, PDNS, porcine circovirus 2-associated reproductive failure (PCV2-RF), and granulomatous enteritis] in Mexico have been confirmed by in situ hybridization (10), the prevalence of Iotatorquevirus species or their possible association with PCVAD has not been recorded. The aim of the present work was to identify TTSuV1a and/or TTSuV1b from well-documented cases of PCVAD in order to assess their potential relationship with the occurrence of PCVAD in an unvaccinated population.
Materials and methods
Case selection
Archived formalin-fixed paraffin-embedded porcine tissues (lymph node, spleen, tonsils, and fetal hearts) from 2001 to 2009 were selected from 100 swine cases. Sixty-eight cases were from non-vaccinated swine with confirmed PCVAD on the basis of clinical signs, characteristic microscopic lesions, and in situ hybridization (11). The cases were subdivided as follows: 23 PMWS-affected tissues (lymph nodes and spleen), depicting severe lymphoid depletion and granulomatous inflammation, and a diffuse pattern of PCV2-positive in situ hybridization (ISH) and 45 cases of PCV2-RF, consisting of fetal hearts with non-suppurative myocarditis as well as a random ISH pattern positive for PCV2. The PCVAD-positive cases were submitted from 13 states of the Mexican Republic with a high-density swine population (Figure 1). Additionally, 32 PCV2 non-affected cases were evaluated and consisted of 12 tissues (lymph node and tonsil) negative for PCV2 by ISH from age-matched, clinically normal, non-vaccinated pigs from a PCV2 non-affected farm according to clinical criteria (12) and 20 hearts from PCVAD non-infected aborted fetuses submitted for diagnosis. Each tissue was tested individually.
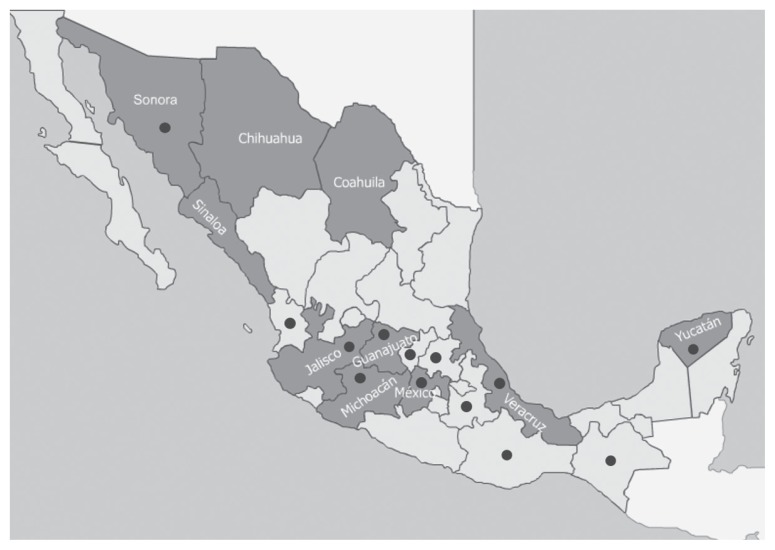
Figure 1.
Geographical distribution of porcine population in the Mexican Republic. ■ Mexican states with higher swine production (27). ● States from which samples were submitted.
Primer design
Due to genomic variability among available sequences, degenerate primers that target ORF1 of TTSuV1a and TTSuV1b were designed using computer software [Primer3 imput program (v.3.0.0; Institute for Biomedical Research, Boston, Massachusetts, USA) (13) and Bioedit software program (v7.2.5; Ibis Bioscience, Carlsbad, California, USA) (14)]. Ten TTSuV1a sequences from different countries available in the NCBI database were used to design the primers (GenBank accession numbers: HM633249, HM633253, HM633258, AY823990, HM633257, AB076001, GU188045, GU456383, GU456384, GQ120664). For TTSuV1b, 12 sequences available in the NCBI database from different countries were used to design the primers (GenBank accession numbers: HM633230, JX173484, HQ204188, GU376737, KC461227, JQ782385, HM633218, GU456386, GU188046, GU570207, AY823991, NC014092). First-round reverse primer contains only 1 degenerate base. Primers were synthesized commercially (IDT Integrated DNA Technologies, Coralville, Iowa, USA).
Nested polymerase chain reaction (PCR)
DNA extraction from all tissues was done separately using commercial kits according to the manufacturer’s instructions (QIAamp DNAFFPE Tissue kit; Qiagen, Germany). Briefly, DNA was eluted in a volume of 200 μL molecular grade water, and stored at −20°C. The lowest limit of detection was determined by serial dilution (1:2) and was of 12.5 ng/μL. Nested 50-μL polymerase chain reaction (PCR) were done using the sets of degenerate primers (Table I) in a thermocycler (Eppendorf, Hamburg, Germany) containing 2.5 UTaq DNA polymerase (GoTaq Flexi DNA polymerase; Promega Corporation, Madison, Wisconsin, USA) PCR buffer 1×, magnesium chloride 2.25 mM (TTSuV1a)/1.5 mM (TTSuV1b), 0.2 mM of each deoxynucleotide (dNTP), 100 pmol of each primer, and 20 ng of template. The following thermal cycle was as follows: the initial activation step at 94°C for 3 min followed by 40 cycles of 1 min at 94°C, 1 min at 56°C (TTSuV1a) or 53°C (TTSuV1b) and 1 min at 72°C, finally last extension step of 10 min at 72°C. The PCR products were electrophoretically separated in a 1.5% agarose gel stained with ethidium bromide. The gel was visualized under ultraviolet light (Apollo Instrumentation, Claremont, California, USA) and photodocumented (Doc-It System; UVP BioImaging Systems, Cambridge, UK).
Table I.
Sequences of the primers utilized for nested polymerase chain reaction (PCR)
| Species | Sequence | Primer | Length |
|---|---|---|---|
| TTSuV1a | 5′-AACTGGCAGGACCACCTATG-3′ | Forward | 481 bp |
| 5′-AGTGTBACHTCHCCACTYC-3′ | Reverse | ||
| 5′-AAAGAGACGCTATGGCTGGA-3′ | Forward nested | 255 bp | |
| 5′-TGYTTTTCWGTGTCCCAYTGC-3′ | Reverse nested | ||
| TTSuV1b | 5′-ATGCCTTACAGACGCTATC-3′ | Forward | 605 bp |
| 5′-TGTGATGTTAATTTTGGTGGA-3′ | Reverse | ||
| 5′-AAGCTCCGGTCATACAATG-3′ | Forward nested | 211 bp | |
| 5′-GCTGTCCATATATTTCTCCAG-3′ | Reverse nested |
Sequences of the primers utilized the detection to the TTSuV1a and TTSuV1b for nested PCR. Bold letters indicate degenerate base. All primers were synthesized by another source (Integrated DNA Technologies, Coralville, Iowa, USA).
Sequencing
Two amplified products from each species were randomly selected and purified from agarose gel using a commercial kit (Min Elute Gel Extraction kit; Qiagen) following the manufacturer’s instructions. The sequencing of the purified PCR products was done using high fidelity, processing, and specificity enzyme kits (Taq Platinum Polymerase; High Fidelity, Carlsbad, California, USA). Internal primers were used to sequence partial sequences of ORF1, using a sequencer (Model 3100; Applied Biosystems, Foster, California, USA).
Data analysis
Nucleotide sequences were edited, aligned, and analyzed using computer software (Bioedit software v7.2.0) (13). Phylogenetic analysis were done using molecular evolutionary genetics analysis version 6 (MEGA6) (15). The maximum likelihood tree was computed using Tamura-Neg Gamma distance. The test of phylogeny was carried out through bootstrap method with 1000 number of replication and the gaps/missing data treated with pairwise deletion. Two by four contingency frames were made based on PCR results to evaluate the relationship of TTSuV1a and TTSuV1b with PCVAD using a Chi-squared test at a trust interval of 99% (P < 0.01).
Results
The PCR protocols using specific degenerate primers to amplify TTSuV1a and TTSuV1b were optimized to obtain of 255 bp (Figure 2A), and 211 bp (Figure 2B), respectively, from PCVAD cases. The sequences of 2 nested products of each virus species proved to be specific (GenBank accession numbers: KU891810 and KU891811 TTSuV1a, KU891808 TTSuV1b and KU891809 TTSuVκ2). At alignment, the amplified regions of each species were highly conserved among available sequences. The topography of the phylogenetic tree of TTSuV1a (Figure 3) revealed that the amplified sequences belong to the species 1a, conforming a well-defined cluster (Mexican, German, and Chinese sequences) standing supported with bootstrap values of 89. Conversely, the phylogenetic tree of TTSuV1b (Figure 4) showed that one Mexican sequence clustered with American and Chinese sequences, but another sequence was located in a different clade, revealing a major phylogenetic relationship with TTSuVκ2 Brazilian sequences (GenBank accession numbers: AY823991, NC014092).
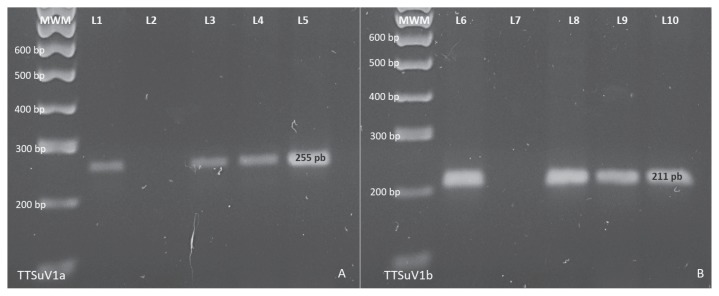
Figure 2.
Iotatorquevirus species nested polymerase chain reaction (PCR) from porcine circovirus associated disease (PCVAD) positive cases. A — TTSuV1a nested PCR from FR-PCV2 positive cases. One hundred base pairs molecular weight marker (MWM). Lanes 1 positive control, lane 2 negative control, line 3, 4, and 5 showing 255 bp amplified products. B — TTSuV1b nested PCR from post weaning multisystemic wasting syndrome (PMWS) positive cases. One hundred base pairs MWM. Lanes 5 positive control, lane 7 negative control, line 8, 9 and 10 display 211 bp amplified products, 2% agarose gel.
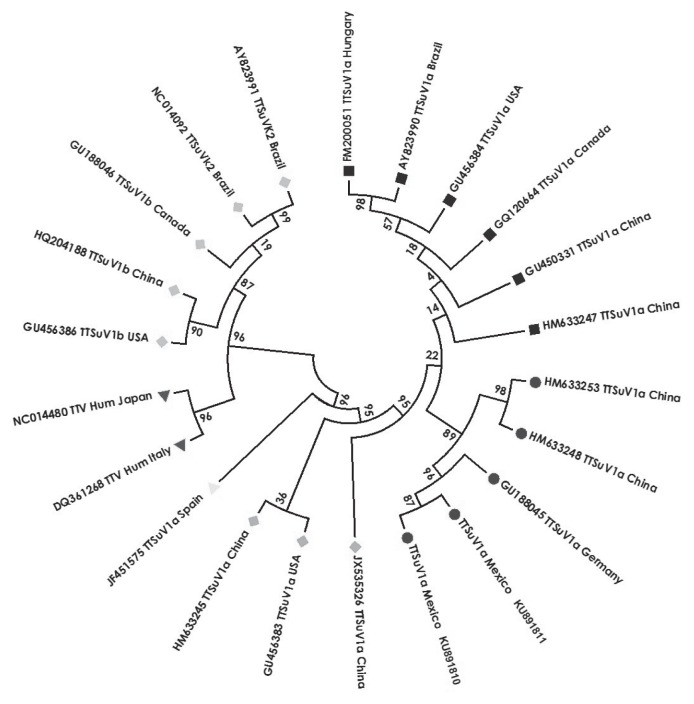
Figure 3.
Phylogenetic tree of the TTSuV1a showing the nucleotide sequences of different identified Torque teno virus with GenBank accession number, and country of origin. The sequences were distributed in 3 TTSuV1a sequence groups (■ Canadian, Chinese, American, Brazilian, and Hungarian; ● Mexican, German, and Chinese;
 Chinese and American;
Chinese and American;
 Spanish). TTSuV1b sequences (
Spanish). TTSuV1b sequences (
 Canadian, Chinese, American, and Brazilian), and TTV human sequences (▼ Italian and Japanese) are included. The Mexican sequences (● KU891810 and KU891811) belong to the 2 TTSuV1a positive cases described in this work. The length of the branch represents the genetic distance among the sequences.
Canadian, Chinese, American, and Brazilian), and TTV human sequences (▼ Italian and Japanese) are included. The Mexican sequences (● KU891810 and KU891811) belong to the 2 TTSuV1a positive cases described in this work. The length of the branch represents the genetic distance among the sequences.
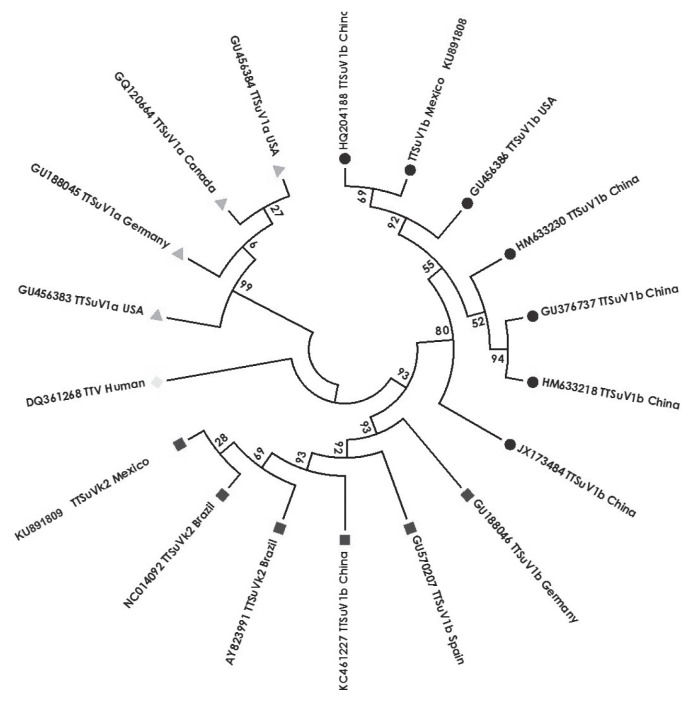
Figure 4.
Phylogenetic tree of the TTSuV1b showing the nucleotide sequences of different identified Torque teno virus with GenBank accession number, and country of origin. The sequences were distributed in 2 TTSuV1b sequence groups (● Chinese, American, and Mexican; ■ German, Spanish, Brazilian, Chinese, and Mexican). TTSuV1a sequences (
 American, German, and Canadian) and TTV human sequences (
American, German, and Canadian) and TTV human sequences (
 Italian) are included. The Mexican sequences (● KU891808 and ■ KU891809) belong to the 2 TTSuV1b positive cases described in this work. The length of the branch represents the genetic distance among the sequences.
Italian) are included. The Mexican sequences (● KU891808 and ■ KU891809) belong to the 2 TTSuV1b positive cases described in this work. The length of the branch represents the genetic distance among the sequences.
In the current retrospective study, 100 cases were used (68 PCVAD-affected, and 32 PCVAD-non affected). Overall, 33% (33/100) were positive to TTSuV1a (TTSuV1a+) and 8% (8/100) were positive to TTSuV1b (TTSuV1b+). From all the PCVAD cases, 35% (24/68) were TTSuV1a+ and 9% (6/68) were TTSuV1b+, whereas 28% (9/32) and 6% (2/32) of the non-affected cases were positive to TTSuV1a and TTSuV1b, respectively (Table II). The frequency of PMWS-affected cases compared to age-matched clinically normal piglets is shown in Table III. Thirty-one percent (11/35) of total cases were TTSuV1a+ and 22.8% (8/35) were TTSuV1b+. Thirty-nine percent (9/23) of PMWS-affected cases were positive for TTSuV1a, 26% (6/23) were TTSuV1b+. Of the clinically normal pigs evaluated, 17% (2/12) were TTSuV1a+ and 17% (2/12) were TTSuV1b+. Co-infection was found only in PMWS cases, 3% of the total cases, and 9% (3/35) of PMWS cases. Concerning cases of reproductive failure in sows (Table IV), 34% (22/65) of cases and 33% (15/45) of PCV2-RF cases were TTSuV1a+. Similarly, 35% (7/20) of the reproductive failure cases not associated with PCV2 were positive for TTSuV1a+. No case of reproductive failure was positive for TTSuV1b. There was no statistical relationship between the manifestation of PCVAD and the presence of TTSuV1a or TTSuV1b by Ji2 test.
Table II.
Overall nested polymerase chain reaction (PCR) results for open reading frame (ORF1) region of TTSuV1a and TTSuV1b (n = 100)
| TTSuV1a+ TTSuV1b+ |
TTSuV1a+ TTSuV1b− |
TTSuV1a− TTSuV1b+ |
TTSuV1a− TTSuV1b− |
Total | |
|---|---|---|---|---|---|
| PCVAD affected | 3 | 21 | 3 | 41 | 68 |
| PCVAD non-affected | 0 | 9 | 2 | 21 | 32 |
| Totala | 3 | 30 | 5 | 62 | 100 |
Not statistically different between porcine circovirus-associated disease (PCVAD)-affected and PCVAD non-affected at P-value 0.01 (X2 test).
Table III.
Post-weaning multisystemic wasting syndrome (PMWS) nested polymerase chain reaction (PCR) results for open reading frame (ORF1) region of TTSuV1a and TTSuV1b
| TTSuV1a+ TTSuV1b+ |
TTSuV1a+ TTSuV1b− |
TTSuV1a− TTSuV1b+ |
TTSuV1a− TTSuV1b− |
Total | |
|---|---|---|---|---|---|
| PMWS-affected | 3b | 6 | 3 | 11 | 23 |
| Age-matched clinically normal piglets | 0 | 2 | 2 | 8 | 12 |
| Totala | 3 | 8 | 5 | 19 | 35 |
Not statistically different between PMWS-affected and age matched healthy piglets at P-value 0.01 (X2 test).
Co-infection only in PMWS-affected.
Table IV.
Reproductive failure nested polymerase chain reaction (PCR) results for open reading frame (ORF1) region of TTSuV1a and TTSuV1b
| TTSuV1a+ TTSuV1b+ |
TTSuV1a+ TTSuV1b− |
TTSuV1a− TTSuV1b+ |
TTSuV1a− TTSuV1b− |
Total | |
|---|---|---|---|---|---|
| PCV2-RF | 0 | 15 | 0 | 30 | 45 |
| non PCV2-RF | 0 | 7 | 0 | 13 | 20 |
| Totala | 0 | 22 | 0b | 43 | 65 |
Not statistically different between porcine circovirus type 2-associated reproductive failure (PCV2-RF) and non-PCV2-RF at P-value 0.01 (X2 test).
Not TTSuV1b positive cases.
Discussion
Several reports have suggested that co-infection of Iotatorquevirus species with other viruses might increase severity of disease as a result of synergy (1,5,16). Consequently, both species have been the target of research for the study of multifactorial diseases, such as porcine respiratory disease complex (17–20) and PCVAD (6–8,21–23).
In natural cases of PMWS, a high frequency of co-infection between TTSuV1a and PCV2 has been described (6). In gnotobiotic pig models, TTSuV1a was proposed to act as an aggravating factor of PMWS (7) and characteristic PDNS lesions were reproduced in PCV2-negative pigs after inoculation with TTSuV1a and porcine reproductive and respiratory syndrome virus (8). In PMWS-affected pigs, higher prevalence and increase viral load have been found with TTSuV1b than with TTSuV1a (6,22). In addition, a possible association of TTSuV1b with reproductive failure in sows has been proposed (1,24). Taken altogether, current information not only reveals high worldwide prevalence of Iotatorquevirus species but also a close association of its occurrence in cases of PCVAD. However, a relationship of both TTSuV1a and TTSuV1b with development of PCVAD is still unclear.
In the present study, the degenerate nested PCR proved to amplify TTSuV1a and TTSuV1b ORF1 specific sequences. The nested PCR results revealed that TTVSuV1a and TTSuV1b are widely distributed in Mexican states with a high cluster of pig population, as it is described for PCV2 (25). Also, both species were amplified from cases of PCVAD. In the current scenario, only 35% of total cases were positive to TTSuV1a. Similar findings were obtained regardless of presentation, 39% (9/23) of PMWS cases and 33% (15/45) of PCV2-RF cases (Table II). The global frequencies of the present work are far lower than reported in other countries, such as Spain (90%), Korea (85%), and China (80%), but comparable to frequencies in Thailand and the United States, which are reported to be 40% and 33%, respectively (26). In the same case series, frequencies in Canada were found to be highly variable (46% to 100%); it was suggested that differences in pig density might influence TTSuV1a prevalence (26). However, we observed a low prevalence of TTSuV1a from Mexican states with the highest proportion of swine farms (27). With regard to TTSuV1b, the gathered data of the present work showed more disparity since European countries have reported higher frequencies (6,21,22,28).
Detection of TTSuV1a and/or TTSuV1b has been strongly associated with cases of PMWS, particularly in Europe. Among PMWS-affected pigs, TTSuV1a seroprevalence of 66% to 76% has been reported in Spain (6,22,23,28). Likewise, TTSuV1a frequencies of 77% and 71.4% were reported in Sweden (21) and Slovakia (29), respectively. Whereas TTSuV1a detection was 41% and 58% in Great Britain from fresh tissues and sera, respectively (30). Frequency of TTSuV1b among PMWS-affected cases, however, showed even more discrepancy since European countries, such as Spain, have reported frequencies of 91% (6,28) and 100% (22) that are consistent with the Sweden prevalence of 94% (21). Moreover, TTSuV1b occurrence of 71% and 64.3% were found in Great Britain (30) and Slovakia (29), respectively.
Altogether, PMWS-affected findings of the present work are not consistent with prevalence in Europe, but are comparable to TTSuV1a prevalence of 48%, 40%, and 30% reported in Brazil (31), Cuba (16), and Japan (18), respectively. However, the latter 2 studies were performed on emaciated pigs without further laboratory confirmation of PCVAD status in affected tissues by ISH or immunohistochemistry. An explanation might be linked to geographic relationship within Asia and North America, but TTSuV1b prevalence of 94.7% found in Brazil (30) from PCV2-affected pigs is not in agreement with that hypothesis. Prevalence rates of 37.5% and 31% reported in Cuba (16) and Japan (18), respectively, are still closer to our findings.
Differences in Iotatorquevirus species prevalence rates among studies might be related to target tissue. For instance, lower frequencies were found (41% for TTSuV1a and 79% for TTSuV1b) using pools of fresh lung, liver, kidney, spleen, and lymph node, but higher prevalence of both species (77% for TTSuV1a and 94% for TTSuV1b) was obtained from fresh lymph nodes (21). Such differences are most likely associated with PCV2-target tissues because lymph nodes and spleen are regarded as the main target of PMWS-affected pigs, displaying higher PCV2 loads (32). Therefore, testing fresh lymph node samples alone might increase the likelihood of detecting TTSuV species (6,22). In the current study, results were considerably lower compared to most reports despite the fact that severely and diffusely PCV2-affected lymphoid tissues were used. Consequently, the prevalence of TTSuV1a and TTSuV1b from PMWS-affected pigs appears to be low in Mexico.
Prevalence of Iotatorquevirus species in cases of PMWS is reported as higher than that of clinically normal pigs (1,21). Results indicate that the prevalence is noticeably lower with PCVAD, however, though the same trend was noted, it is not statistically significant. Co-infection of both species in PMWS cases was much lower than that reported in Spain (76%) (22), but similar to that reported in Japan (10%) (18). However, in the current work, co-infection of age-matched clinically normal pigs was not observed (1,22,24).
Immune suppression caused by PCV2 infection has been suggested as a predisposing factor for the finding of TTSuV1b in PMWS cases because an increased viral load of TTSuV1b was seen in PMWS-affected pigs compared to the viral load of clinically normal pigs. Thus, clinically normal pigs might restrain infection with both Iotatorquevirus species and PCV2 (28). Such statements are in agreement with the data presented herein since co-infection, though in low proportion, was only found in PMWS-affected cases.
The role of Iotatorquevirus in reproductive failure has been proposed. To the authors’ knowledge, there are no reports of TTSuV1a nor TTSuV1b in cases of PCV2-RF. A higher seroprevalence of TTSuV1a (60% to 75%), compared to TTSuV1b (30% to 34%), has been detected in healthy sows (24,33). Litters from clinically normal sows infected with TTSuV1a or TTSuV1b showed that 43% and 19% of piglets were TTSuV1a+ and TTSuV1b+, respectively (34). Likewise, 50% of stillbirths were TTSuV1a+ but 7% of stillbirths were TTSuV1b+, suggesting that both species may cause in utero infection (33). Viremia in sows is a prerequisite for vertical transmission, with the heart being a frequently affected tissue (17). Histopathology of fetal hearts from cases of reproductive failure in sows showed both non suppurative myocarditis and PCV2-specific ISH (35) in 39% of PCV2-RF and 47% of transplacental transmission. These tissues were included in the present work, revealing a low frequency of TTSuV1a and an absence of TTSuV1b with no significant statistical relation between the occurrence of PCV2-RF and the presence of TTSuV1a.
Using tissue pools from aborted fetuses, the frequency rates of TTSuV1a and TTSuV1b were 17% and 30%, respectively. Heart tissue was part of this tissue pool, but the precise site of infection could not be identified (36). Fetal hearts were one of the individual tissues evaluated in the current study, but other additional tissues that could potentially harbor Iotatorquevirus species were not evaluated, thus Iotatorquevirus species could not be definitively ruled out in cases of PCV2-RF. Nevertheless, since the heart is the main target organ for vertical transmission, preliminary findings suggest that participation of TTSuV1b regarding PCV2-RF is unlikely.
The current retrospective work is noteworthy because a non-vaccinated porcine population was evaluated from a time period prior to the start of through and worldwide immunization programs. Therefore, it may elucidate distinct viral dynamics as influenced by changing immune status and add to the understanding of the evolving nature of viruses. In addition, a TTSuVκ2 sequence was amplified based on a new taxonomy (2). Thus, further classification of TTSuV1b is essential to separate it from TTSuVκ2 species.
Taken together, the data reported herein are in agreement with a lack of relationship between Iotatorquevirus species and occurrence of PCVAD. Currently, a broad phylogenetic study is being done to ascertain prevailing swine Anelloviridae species in Mexico as well as its genomic variability that might account for a distinctive pathogenic potential.
Acknowledgments
The authors thank Karina Enriquez-Ramirez MSc. for her excellent technical contribution to PCV2-in situ hybridization. The authors are grateful to the CONACYT scholarship program for its commitment to support graduate students. This work was financially supported by a grant from DGAPA-UNAM (PAPIIT IN203309).
References
- 1.Gallei A, Pesch S, Esking WS, Keller C, Ohlinger VF. Porcine Torque teno virus: Determination of viral genomic loads by genogroup-specifics multiplex rt-PCR, detection of frequent multiple infections with genogroups 1 or 2, and establishment of viral full-length sequences. Vet Microbiol. 2010;143:202–212. doi: 10.1016/j.vetmic.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Davison AJ. ICTV: International Committee on Taxonomy of Viruses. Virus taxonomy. 2015. [Last accessed May 26, 2017]. [Website on Internet]. Available from: http://www.ictvonline.org/virusTaxonomy.asp.
- 3.Cortey M, Macera L, Segalés J, Kekarainen T. Genetic variability and phylogeny of Torque teno sus virus 1 (TTSuV1) and 2 (TTSuV2) based on complete genomes. Vet Microbiol. 2011;148:125–131. doi: 10.1016/j.vetmic.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 4.Cadar D, Kiss T, Ádám D, Cságola A, Novosel D, Tuboly T. Phylogeny, spatio-temporal phylodynamics and evolutionary scenario of Torque teno sus virus 1 (TTSuV1) and 2 (TTSuV2) in wild boars: Fast dispersal and high genetic diversity. Vet Microbiol. 2013;166:200–213. doi: 10.1016/j.vetmic.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Huang YW, Harrall KK, Dryman BA, et al. Expression of the putative ORF1 capsid protein of Torque teno sus virus 2 (TTSuV2) and development of western blot and ELISA serodiagnostic assay: Correlation between TTSuV2 viral load and IgG antibody level in pigs. Virus Res. 2011;158:79–88. doi: 10.1016/j.virusres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Kekarainen T, Sibila M, Segalés J. Prevalence of swine Torque teno virus in post-weaning multisystemic wasting syndrome (PMWS)-affected and no-PMWS-affected pigs in Spain. J Gen Virol. 2006;87:833–837. doi: 10.1099/vir.0.81586-0. [DOI] [PubMed] [Google Scholar]
- 7.Ellis JA, Allan G, Krakowka S. Effect of coinfection with genogroup 1 porcine torque teno virus on porcine circovirus type 2-associated post weaning multisystemic wasting syndrome in gnotobiotic pigs. Am J Vet Res. 2008;69:1608–1614. doi: 10.2460/ajvr.69.12.1608. [DOI] [PubMed] [Google Scholar]
- 8.Krakowka S, Ellis JA. Evaluation of induction of porcine dermatitis and nephropathy syndrome in gnotobiotic pigs with negative results for porcine circovirus type 2. Am J Vet Res. 2008;69:1615–1622. doi: 10.2460/ajvr.69.12.1615. [DOI] [PubMed] [Google Scholar]
- 9.Segalés J, Martínez-Guinó L, Cortey M, et al. Retrospective study on swine Torque teno virus genogroups 1 and 2 infection from 1985 to 2005 in Spain. Vet Microbiol. 2009;134:199–207. doi: 10.1016/j.vetmic.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 10.García-Camacho LA, Enriquez-Ramírez K, Araiza-Nava D, Rangel-Rodríguez IC, Quintero-Ramírez V, García-Reyna PB. Porcine circovirus 2-associated syndromes in Mexican farms as detected by in situ hybridization. Com 57th and 41st Annual Meet Am Coll Vet Pathol and Am S Vet Clin Pathol Vet Pathol. 2006;43:819. [Google Scholar]
- 11.Chae C. A review of porcine circovirus 2-associated syndromes and diseases. Vet J. 2005;169:326–336. doi: 10.1016/j.tvjl.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 12.McIntosh K, Harding J, Parker S, Krakowka S, Allan G, Ellis J. Quantitative polymerase chain reaction for Porcine circovirus-2 in swine feces in a Porcine circovirus disease-affected commercial herd and a nonaffected commercial herd. Can Vet J. 2008;49:1189–1194. [PMC free article] [PubMed] [Google Scholar]
- 13.Untergrasser A, Cutcutache I, Koressaar T, et al. Primer3 — New capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 15.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez LJ, Díaz de Arce H, Frías MT, Perera CL, Ganges L, Nuñez JI. Molecular detection of torque teno sus virus in lymphoid tissues in concomitant infections with other porcine viral pathogens. Res Vet Sci. 2011;91:e154–e157. doi: 10.1016/j.rvsc.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Pensaert B, Sanchez RE, Ladekjaer-Mikkelsen AS, Allan GM, Nauwynck HJ. Viremia and effect of fetal infection with porcine viruses with special reference to porcine circovirus 2 infection. Vet Microbiol. 2004;98:175–183. doi: 10.1016/j.vetmic.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Taira O, Ogawa H, Nagao A, Tuchiya K, Nunoya T, Ueda S. Prevalence of swine Torque teno virus genogrups 1 and 2 in Japanese swine with suspected post-weaning multisystemic wasting syndrome and porcine respiratory disease complex. Vet Microbiol. 2009;139:347–350. doi: 10.1016/j.vetmic.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Lee S, Shin J, Kim C, Lyoo YS. Comparison of Torque teno sus Virus (TTSuV) viral load in Porcine circovirus type 2 vaccinated and non-vaccinated pig herds. Res Vet Sci. 2011;93:1039–1041. doi: 10.1016/j.rvsc.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Rammohan L, Xue L, Wang C, Chittick W, Ganesan S, Ramamoorthy S. Increased prevalence of torque teno viruses in porcine respiratory disease complex affected pigs. Vet Microbiol. 2012;157:61–68. doi: 10.1016/j.vetmic.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Blömstrom A, Belák S, Fossum C, Fuxler L, Wallgren P, Berg M. Studies of porcine circovirus type 2, porcine boca-like virus and torque teno virus indicate the presence of multiple viral infections in post weaning multisystemic wasting syndrome pigs. Virus Res. 2010;152:59–64. doi: 10.1016/j.virusres.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Aramouni M, Segalés J, Sibila M, Martin-Valls GE, Nieto D, Kekarainen T. Torque teno sus virus 1 and 2 viral loads in post weaning multisystemic wasting syndrome (PMWS) and porcine dermatitis and nephropathy syndrome (PDNS) affected pigs. Vet Microbiol. 2011;153:377–381. doi: 10.1016/j.vetmic.2011.05.046. [DOI] [PubMed] [Google Scholar]
- 23.Aramouni M, Kekarainen T, Ganges L, Tarradas J, Segalés J. Increased viral load and prevalence of Torque teno sus virus 2 (TTSuV2) in pigs experimentally infected with classical swine fever virus (CSFV) Virus Res. 2013;172:81–84. doi: 10.1016/j.virusres.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 24.Sibila M, Martínez-Guinó L, Huerta E, et al. Torque teno virus (TTV) infection in sows and suckling piglets. Vet Microbiol. 2009;137:354–358. doi: 10.1016/j.vetmic.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Quintero-Ramirez V, Romero Y, Enriquez K, García-Camacho LA. Reproductive failure associated to PCV2 and its distribution in the Mexican Republic. Proc 21st IPVS Congress; 2010; p. 470. [Google Scholar]
- 26.McKweon NE, Fenaux M, Halbur PG, Meng XJ. Molecular characterization of porcine TT virus, an orphan virus, in pigs from six different countries. Vet Microbiol. 2004;104:113–117. doi: 10.1016/j.vetmic.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 27.México exportó 86 mil 294 toneladas de carne de cerdo en 2013. Secretariat of Agriculture, Livestock, Rural Development, Fisheries and Food; [Last accessed on April 25, 2017]. [Website on the Internet]. Available from: http://www.sagarpa.gob.mx/saladeprensa/2012/Paginas/2014B401.aspx#. [Google Scholar]
- 28.Nieto D, Aramouni M, Grau-Roma L, Segalés J, Kekarainen T. Dynamics of Torque teno sus virus 1 (TTSuV1) and 2 (TTSuV2) DNA loads in serum of healthy and post weaning multisystemic wasting syndrome (PMWS) affected pigs. Vet Microbiol. 2011;152:284–290. doi: 10.1016/j.vetmic.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 29.Vlasakova M, Leskova V, Sliz I, Jackova A, Vilcek S. The presence of six potentially pathogenic viruses in pigs suffering from post-weaning multisystemic wasting syndrome. BMC Vet Res. 2014;10:221. doi: 10.1186/s12917-014-0221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMenamy MJ, McKillen J, McNair I, et al. Detection of a porcine boca-like virus in combination with porcine circovirus type 2 genotypes and torque teno sus virus in pigs from post weaning multisystemic wasting syndrome (PMWS)-affected and non-PMWS-affected farms in archival samples from Great Britain. Vet Microbiol. 2013;164:293–298. doi: 10.1016/j.vetmic.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Teixeira TF, Dezen S, Cibulski SP, et al. Torque teno sus virus (TTSuV) in tissues of pigs and its relation with the occurrence of postweaning multisystemic wasting syndrome. Vir Gen. 2013;47:276–281. doi: 10.1007/s11262-013-0940-0. [DOI] [PubMed] [Google Scholar]
- 32.Rosell C, Segalés J, Plana-Durán J, et al. Pathological, immunohistochemical, and in-situ hybridization studies of natural cases of postweaning multisystemic wasting syndrome (PMWS) in pigs. J Comp Pathol. 1999;120:59–78. doi: 10.1053/jcpa.1998.0258. [DOI] [PubMed] [Google Scholar]
- 33.Martínez-Guinó L, Kekarainen T, Segalés J. Evidence of Torque teno virus (TTV) vertical transmission in swine. Theriogenology. 2009;71:1390–1395. doi: 10.1016/j.theriogenology.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 34.Sibila M, Martínez-Guinó L, Huerta E, et al. Swine torque teno virus (TTV) infection and excretion dynamics in conventional pig farms. Vet Microbiol. 2009;139:213–218. doi: 10.1016/j.vetmic.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 35.Enríquez K. MSc disseration. Cuautitlán Izcalli, Estado de México: National University of Mexico; 2012. Evaluation of participation of Porcine Circovirus type 2 (PCV2) in reproductive failure; pp. 38–40. [Google Scholar]
- 36.Martínez-Guinó L, Kekarainen T, Maldonado J, Aramouni M, Llorens A, Segalés J. Torque teno sus virus (TTV) detection in aborted and slaughterhouse collected fetuses. Theriogenology. 2010;74:277–281. doi: 10.1016/j.theriogenology.2010.02.011. [DOI] [PubMed] [Google Scholar]